Summary
Background and objectives
Atypical hemolytic uremic syndrome is a disease associated with mutations in the genes encoding the complement regulators factors H and I. In addition, factor H autoantibodies have been reported in ∼10% of patients with atypical hemolytic uremic syndrome. This study searched for the presence of factor I autoantibodies in atypical hemolytic uremic syndrome.
Design, setting, participants, & measurements
This study screened 175 atypical hemolytic uremic syndrome patients for factor I autoantibodies using ELISA with confirmatory Western blotting. Functional studies using purified immunoglobulin from one patient were subsequently undertaken.
Results
Factor I autoantibodies were detected in three patients. In one patient with a high titer of autoantibody, the titer was tracked over time and was found to have no association with disease activity. This study found evidence of an immune complex of antibody and factor I in this patient, but purified IgG, isolated from current serum samples, had only a minor effect on fluid phase and cell surface complement regulation. Genetic analysis of the three patients with factor I autoantibodies revealed that they had two copies of the genes encoding factor H–related proteins 1 and 3 and therefore, did not have a deletion commonly associated with factor H autoantibodies in atypical hemolytic uremic syndrome. Two patients, however, had functionally significant mutations in complement factor H.
Conclusions
These findings reinforce the concept of multiple concurrent risk factors being associated with atypical hemolytic uremic syndrome but question whether autoantibodies per se predispose to atypical hemolytic uremic syndrome.
Introduction
Atypical hemolytic uremic syndrome (aHUS) is a disease characterized by overactivation of the alternative complement pathway (1). Mutations in the genes encoding complement regulatory proteins complement factor H (CFH) (2–5), complement factor I (CFI) (6–10), and membrane cofactor protein (CD46) (6,11–14) and complement components C3 (C3) (15) and complement factor B (CFB) (16) are associated with aHUS.
As well as inherited defects in complement regulation, acquired defects in the form of autoantibodies to CFH have been described (17–20). These autoantibodies mainly bind to the C-terminal end of CFH, where aHUS-associated mutations cluster (21). This region of the molecule binds to C3b and glycosaminoglycans and is responsible for cell surface complement regulation (22). CFH autoantibodies have been shown to impair cell surface complement regulation, thus mimicking the action of the CFH mutations seen in aHUS (17–20,23).
CFI is a serine protease that cleaves C3b and C4b in the presence of its cofactor proteins, CFH (24), C4 binding protein (25), CD46 (26), and complement receptor 1 (27). By inactivating C3b and C4b through limited proteolytic cleavage and thereby preventing the formation of the C3 and C5 convertases, CFI inhibits the alternative and classic complement pathways. CFI consists of a light chain (which carries the catalytic site) and a heavy chain (of unclear function) linked by a disulphide bond.
Mutations in CFI have been reported in 2–12% of aHUS patients (6–10). Although they are distributed throughout the molecule, they do cluster in the serine protease domain (21). Most aHUS-associated CFI mutations result in decreased secretion, resulting in a quantitative defect in complement regulation. Functional analysis of CFI mutants that are secreted normally has revealed a loss of alternative and classic pathway cofactor activity, both in the fluid phase and on cell surfaces (7,28,29).
Here, we describe the presence of CFI autoantibodies in the Newcastle aHUS cohort, investigate their functional impact, and show that these autoantibodies occur in the presence of additional genetic risk factors.
Materials and Methods
Subjects
Paired serum and DNA samples were available from 175 patients with aHUS and 100 healthy blood donors (blood donor controls). The study was approved by the Northern and Yorkshire Multi-Center Research Ethics Committee, and informed consent was obtained in accordance with the Declaration of Helsinki.
Genetic Screening
In individuals with CFI autoantibodies, mutation screening of CFH, CD46, CFI, CFB, C3, and thrombomodulin (THBD) was or had previously been undertaken by direct fluorescent sequencing as described (2,8,12,15,30,31). Variants discovered in these genes were assessed in DNA samples from 300 normal control individuals within the Wellcome Trust Patient Control Consortium (32,33). Genotyping of the following single nucleotide polymorphisms was undertaken by direct sequencing: CD46 −652A>G (rs2796267), CD46 −366A>G (rs2796268), CD46 c.4070T>C (rs7144), CFH −331C>T (rs3753394), CFH c.2016A>G p.Gln672Gln (rs3753396), and CFH c.2808G>T p.Glu936Asp (rs1065489).
CFHR1 and -3 copy number was measured by multiplex ligation-dependent probe amplification with a kit from MRC Holland (SALSA MLPA kit P236-A1 ARMD). CFHR4 copy number was measured by multiplex PCR assay as described (20). Screening for CFH autoantibodies was performed as previously described (20,34).
ELISA
The anti-CFI ELISA was carried out essentially as previously described for factor H (34), except that 5 µg/ml CFI (purified from pooled serum samples) (35) was substituted for CFH herein and a standard curve was generated using a polyclonal goat anti-CFI (Comptech) followed by rabbit anti-goat horseradish peroxidase (HRP) (Stratech Scientific). The OD450 value for the 1/5000 dilution of goat anti-human CFI was given an arbitrary value of 100,000 relative units (RU). Alternatively, protein A/G column was used to isolated patient and control Ig from sera following manufacturer’s instructions (Pierce, United Kingdom), and the presence of CFI in the samples was detected using 1 µg/ml Medical Research Council of the United Kingdom (MRC) OX21 (gift from Bob Sim, Oxford, United Kingdom) by standard sandwich ELISA of samples.
Western Blotting
Purified CFI (35 µg/ml) was diluted in solubilizing buffer, and 20 ml was loaded onto a 10% SDS-PAGE preparative gel and transferred to nitrocellulose, which was then cut into 0.5- to 1-cm-wide strips. After blocking in 5% nonfat milk/PBS, strips were then incubated with individual sera samples (1/25 to 1/100 as appropriate) overnight at 4°C. After extensive washing in PBS/Tween 0.02%, bound autoantibody was detected using goat anti-human IgG-HRP (Stratech Scientific). Alternatively, for CFI immune complex detection, pre- or postcolumn sera (equivalent to 1/20 dilution of fresh serum) or purified Ig (using protein A/G column; Pierce; Thermo Scientific) was concentrated (using 30-kD cutoff spin columns; Sartorius Stedim Biotech) and adjusted to 1 mg/ml after quantification by bicinchoninic acid assay (Pierce; Thermo Scientific) was loaded on SDS-PAGE and blotted. MRC OX21 was used to identify the presence of CFI. Blots were developed using an enhanced chemiluminescence substrate according to the manufacturer’s specifications (Pierce; Thermo Scientific).
Complement Assays
C3 and C4 levels were measured by rate nephelometry (Beckman Array 360). CFH and CFI levels were measured by radioimmunodiffusion (Binding Site). Cell surface expression of CD46 was measured by flow cytometry as previously described (12,36).
Alternative Pathway Assays
Cell-bound complement activity was carried out essentially as previously described with minor modifications (37). Briefly, rabbit red blood cells (TCS Biologic) were washed several times in PBS and subsequently transferred to alternative pathway buffer (APB; 3.12 mM Barbital, 0.9 mM Na Barbital, 145 mM NaCl2, 7.83 mM MgCl2, 0.25 mM CaCl2, 10 mM EGTA, and 0.1% w/v gelatin, final pH 7.2). Cells were resuspended at 0.1% v/v, and 100 µl were plated out on round-bottomed 96-well plates containing 100 µl triplicate serial dilutions of normal human serum or patient sera in APB. Wells were supplemented with purified patient or normal control Ig (100 µg/well), CFI (0.7 µg/well), or CFH (5 µg/well) before adding rabbit red blood cells. Plates were incubated at 37°C for 30 minutes before red cells were pelleted at 500 g for 5 min. Absorbance of supernatant was measured at OD410.
Fluid Phase C3b Breakdown
Fluid phase complement activity was established as previously described (38). Briefly, purified C3b (4 µg), CFH (0.5 µg), and CFI (titration from 1 µg; obtained from Comptech, TX) were mixed with APB. Patient and control Ig (25 µg) in APB were mixed with the C3b- and CFH-containing solution before addition of CFI. Samples were then taken to 37°C for 3 min before being heated to 95°C for 5 min; 10% SDS-PAGE gels were stained with Coomassie blue or alternatively subjected to Western blotting as appropriate. C3b breakdown was visualized using a sheep anti-C3 at 1/500 (gift from B. P. Morgan, Cardiff, United Kingdom) followed by 1/2000 donkey anti-sheep HRP (Stratech Scientific). Western blots were visualized for 30 seconds. Image analysis was carried out on scanned gels and autorad films as follows. A set grid was used to compare pixel intensity (grayscale gradient) for each band within a lane. Results were standardized for loading based on the β-chain of C3.
Results
CFI Autoantibodies
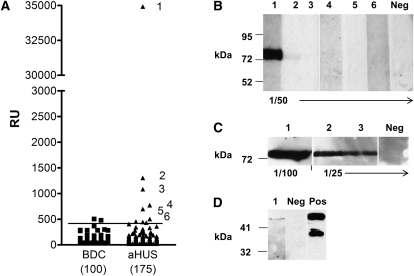
In both the blood donor controls (BDCs) and aHUS patients, CFI autoantibody titer (in RU) was not normally distributed (Figure 1A) (determined by the Kolmogorov–Smirnov test). The median (range; mean) antibody titer in BDC and aHUS patients was 64 RU (32–504; 97) and 56 RU (22–34,921; 310), respectively. Compared with the BDC group, aHUS patients had increased levels of CFI autoantibody (P<0.02). We used the 0.975 fractile of the BDC group to determine autoantibody positivity as recommended by the International Federation of Clinical Chemistry for data with non-normal distribution (39). This use equated to 423 RU, and six patients were above this threshold (Figure 1A). However, our experience with anti-CFH autoantibodies has suggested that it is prudent to confirm presence of autoantibodies using a second technique (34). Thus, Western blotting was used to confirm the presence of autoantibodies in samples with a titer ≥423. Only the three patients with the highest titer of CFI autoantibodies were confirmed to have CFI autoantibodies after Western blotting analysis. Detection of autoantibodies in patient 1’s serum required a shorter film exposure (Figure 1B) and was readily detectable when less serum was used than for patients 2 and 3 (Figure 1C), consistent with the ELISA results. Additional Western analysis indicated the CFI autoantibody detected in patient 1 bound to the heavy chain of CFI (Figure 1D). Using specific HRP-conjugated secondaries (mouse monoclonal antibodies: MH17–15, -22, -32, -42; Invitrogen, United Kingdom) in our ELISA, P1 and P3 autoantibodies were established as being predominately IgG1 subclass, whereas P2 autoantibodies were IgG3 subclass (data not shown). Thus, of the 175 aHUS patients that we screened, 3 patients were confirmed to possess significant levels of CFI autoantibodies.
Figure 1.
Complement factor I (CFI) autoantibodies are found in atypical hemolytic uremic syndrome (aHUS) patients. (A) Antibodies against purified CFI in aHUS patients were detected using ELISA as described in Materials and Methods; 100 normal healthy blood donor controls (BDCs) and 175 aHUS patients were screened, and nonspecific background signal was subtracted. A standard curve was generated from a polyclonal control antibody and given an arbitrary titer of 100,000 relative unit (RUs) from a 1/5000 dilution. The horizontal bar represents the threshold for positivity based on the 97.5 percentile of the control cohort (423 RU). Purified CFI was run out on 10% SDS-PAGE and transferred to nitrocellulose. Strips of nitrocellulose were then incubated with sera collected from subjects. Patients are listed one to six, and the dilution of serum used is indicated below blots. Enhanced chemiluminescence Western blotting substrate was used to visualize bound antibody. Autorad film was exposed for 20 seconds (B) and 10 minutes (C) before developing. A known CFI autoantibody negative (−ve) was used as a control. (D) shows a Western blot of a preparative gel with reduced purified CFI loaded. Sera from patient 1 (lane 1) and a goat anti-CFI as positive control (lane PC) were incubated with the nitrocellulose, and a 30-second exposure is shown. Molecular mass markers are shown, and autorad pictures are of the individual strips of a single blot reassembled/aligned before exposure. Irrelevant intervening strips have been cropped out where appropriate. Data shown is representative of several blots.
Clinical Details: Genotyping and Background Analysis of the Three Patients with CFI Autoantibodies
A summary of the clinical details of the three patients with CFI autoantibodies is shown in Table 1, with a full summary available in Supplemental Material. Briefly, all required renal replacement therapy at initial presentation and plasma therapy was instituted. However, only patient 3 recovered renal function. In the remaining two patients, a total of three renal transplants were undertaken in the absence of any specific therapy to remove autoantibodies. Patient 1 showed recurrent aHUS in the allograft. Table 1 also shows the values for serum levels of C3, C4, CFH, and CFI, the results of screening for CFH autoantibodies, and the measurement of CD46 expression on the original samples. In all individuals, systemic alternative pathway complement activation was shown with low levels of C3, but in only one individual was the C4 level low. Mutation screening showed that patient 1 had a heterozygous mutation in CFH (c.3468dupA) and a sequence variant in CFI (c.1657 C>T; p.Pro553Ser) that was present in normal controls at a frequency of 5/574 chromosomes (Table 2). Patient 2 had a heterozygous mutation in CFH (c.2018G>A; p.Cys673Tyr). No mutations were detected in genes previously associated with aHUS in patient 3, and all three patients had two copies of CFHR1, -3, and -4. Genotyping for CFH and CD46 susceptibility factors revealed that both patients 1 and 2 are heterozygous for the at-risk haplotype CFHTGTGGT (H3) haplotype (20), whereas patient 2 is homozygous and patient 1 is heterozygous for the at-risk CD46GGAAC haplotype (40) (Table 3).
Table 1.
Clinical details of patients with factor I autoantibodies
| Patient ID | 1 | 2 | 3 |
|---|---|---|---|
| Age at presentation | 26 yr | 5 yr | 13 mo |
| Sex | Female | Female | Female |
| Precipitant | Postpartum | Unknown | C. difficile and rotavirus diarrhea |
| Family history | Yes | No | No |
| Outcome | ESRF | ESRF | Recovered renal function |
| Renal transplant | Yes | Yes | N/A |
| One lost to recurrent aHUS at 1 mo | No recurrence at 65 mo | N/A | |
| One lost to transplant glomerulopathy at 50 mo | N/A | ||
| C3 (0.68–1.38 g/L) | 0.66 | 0.61 | 0.56 |
| C4 (0.18–0.60 g/L) | 0.32 | 0.22 | 0.08 |
| Factor H (0.35–0.59 g/L) | 0.34 | 0.52 | 0.58 |
| Factor H auto-Ab | Negative | Negative | Negative |
| Factor I (38–58 mg/L) | 89 | 48 | 54 |
| MCP expression | Normal | Normal | Normal |
Normal range in parentheses. aHUS, atypical hemolytic uremic syndrome; ESRF, end stage renal failure; MCP, membrane cofactor protein.
Table 2.
Mutation screening of the genes encoding complement factor H, membrane cofactor protein, complement factor I, complement factor B, complement components C3, and thrombomodulin and measurement of the copy number of the genes encoding complement factor H–related proteins 1, 3, and 4
| Patient ID | CFH | CD46 | CFI | CFB | C3 | THBD | CFHR1 Copy Number | CFHR3 Copy Number | CFHR4 Copy Number |
|---|---|---|---|---|---|---|---|---|---|
| 1 | c.3468dupA | nmd | c.1657 C>T; p.Pro553Ser | nmd | nmd | nmd | 2 | 2 | 2 |
| 2 | c.2018G>A; p.Cys673Tyr | nmd | nmd | nmd | nmd | nmd | 2 | 2 | 2 |
| 3 | nmd | nmd | nmd | nmd | nmd | nmd | 2 | 2 | 2 |
CFH, complement factor H; CD46, membrane cofactor protein; CFI, complement factor I; CFB, complement factor B; C3, complement component C3; THBD, thrombomodulin; CFHR1, 3, and 4; complement factor H–related proteins 1, 3, and 4; nmd, no mutation detected.
Table 3.
CD46 and CFH susceptibility factors
| Patient ID | CD46 −652A>G (rs2796267) | CD46 −366A>G (rs2796268) | CD46 c.4047T>C (rs7144) | CFH −332C>T (rs3753394) | CFH c.2016A>G Q672Q (rs3753396) | CFH c.2808G>T E936D (rs1065489) | CFH c.1204 C>T H402Y (rs1061170) |
|---|---|---|---|---|---|---|---|
| 1 | AG | AG | TC | TT | GG | GT | TT |
| 2 | GG | GG | CC | CT | GG | TT | TT |
| 3 | AG | AA | TT | CC | AG | GT | CT |
Time Course of Ig Class Switching and Titer
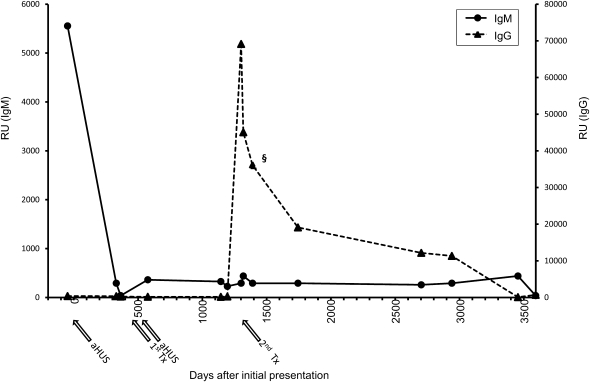
Patient 1 showed a high titer of IgM CFI antibodies at the time of initial presentation with aHUS (Figure 2), but by the time of the first renal transplant (2 years after presenting with aHUS), the titer of IgM CFI autoantibodies was at background levels, where they have remained. Notably, the first renal transplant was lost to recurrent aHUS, and at that time, there were no detectable IgM or IgG CFI autoantibodies. By 42 months after the initial presentation and 24 months after the post-transplant recurrent episode of aHUS, IgG CFI autoantibodies are present in high titers. This finding coincides with the second renal transplant, which was not associated with recurrent aHUS. The sample used in our cohort analysis was 6 months after the peak IgG response (Figure 2), and levels had dropped to 50% maximal detected response by this time. The titer of the IgG CFI autoantibodies then declined slowly and currently rests at the ELISA positive cutoff.
Figure 2.
Patient 1 autoantibody titer changes markedly over time and does not associate with disease. Antibodies against purified CFI in archived samples from patient 1 were detected using ELISA as described in Materials and Methods. Both IgG and IgM autoantibodies were assayed. Nonspecific background signal was subtracted. OD results were transformed as previously described. Results shown are the mean of three separate ELISAs. The chronology of the two transplants and two episodes of aHUS are shown with arrows underneath the x axis. The sample assayed in Figure 1A is denoted (§).
CFI and CFI Autoantibodies Form a Circulating Immune Complex
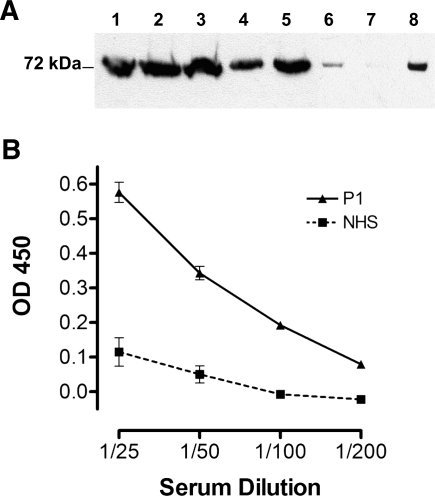
Despite the apparent lack of correlation between CFI autoantibody titer and the clinical course of the disease in patient 1, we wished to establish if the presence of CFI autoantibodies might be disease-modifying. We hypothesized that immune complexes of CFI and CFI autoantibodies could both lead to the generation of additional proinflammatory stimuli (41) and elude detection by standard ELISA. Western blot analysis of a column-purified IgG sample from freshly obtained patient 1 serum (i.e., anti-CFI at 650 RU) showed that CFI had remained associated with IgG (Figure 3A). The post-IgG affinity column sample from patient 1 had less CFI than the normal control sample (despite similar precolumn levels established by anti-CFI sandwich ELISA), suggesting that it was bound out of the serum with IgG and then eluted from the IgG during the wash. In a second approach, we determined that Ig was associated with captured CFI extracted from archived serum samples from patient 1 (Figure 3B). Therefore, we consistently detect immune complexes of IgG autoantibody and CFI in patient 1, which may contribute to disease.
Figure 3.
Complement factor I (CFI) and autoantibodies are found in immune complexes. (A) Both P1 and control serum, confirmed to have equivalent CFI levels using sandwich ELISA, were loaded onto a protein A/G affinity column to purify IgG. Pre- and postcolumn samples and purified Ig were run out on 10% SDS-PAGE and transferred to nitrocellulose. The presence of CFI in these samples was determined using anti-CFI (Medical Research Council of the United Kingdom OX21; Bob Sim, Oxford, United Kingdom) and goat anti-mouse IgG horseradish peroxidase (HRP). Enhanced chemiluminescence Western blotting substrate was used to visualize bound antibody. Column 1, pure CFI (1 µg). Column 2, P1 precolumn serum. Column 3, control precolumn sample. Column 4, P1 postcolumn sample. Column 5, control postcolumn sample. Column 6, 25 µg purified control Ig. Column 7, 1 µg purified P1 Ig. Column 8, 25 µg purified P1 Ig. Molecular mass markers are shown, and the data are representative of three independent experiments. (B) Serum from P1 or normal controls (normal human serum) were incubated on anti-CFI (MRC OX21) -coated ELISA plates and doubly diluted from an initial 1/25 load. Goat anti-human IgG HRP was then used to detect CFI-associated IgG. Data shown is mean ± SD of combined data from three independent serum collections for patient 1 or controls. Data shown is representative of two experiments.
Purified Patient Ig Interferes with CFI Function in a Fluid Phase C3b Breakdown Assay
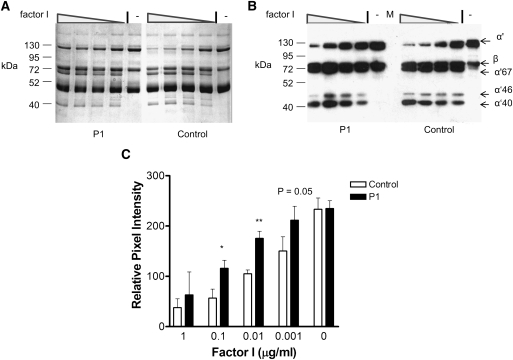
Detection of immune complexes suggests that CFI autoantibodies readily associate with CFI in the fluid phase. Therefore, we next assessed whether purified total Ig (containing CFI autoantibodies) isolated from patient 1 could alter CFI function in a fluid phase C3b breakdown assay. Purified C3b, CFH, and CFI were incubated with control or patient total Ig. The presence of Ig from patient 1 slowed the breakdown of C3b in the fluid phase (Figure 4). A 50% reduction in C3b breakdown over the 3-minute time period was found when limiting concentrations of CFI were mixed with patient Ig before the addition of the C3b to the reaction mixture (Figure 4C).
Figure 4.
Fluid phase breakdown of complement components C3b is modified after addition of purified P1 IgG. Purified C3b (4 µg), complement factor H (CFH; 0.5 µg), and CFI (4×1/10 double dilution from 1 µg, left to right on gels, and no CFI; designated by −) were incubated with patient or control Ig (25 µg) at 37°C for 3 minutes before the addition of 2× reducing SDS-PAGE sample solution and incubation at 95°C for 5 minutes. (A) Samples (10 µl) were loaded on 10% SDS-PAGE gel for staining with Coomassie blue. (B) Alternatively, 10 µl 1/20 dilution of sample were used for SDS-PAGE destined for Western blotting. C3b breakdown was then visualized using a sheep anti-C3 at 1/500 followed by appropriate secondary. Shown is a 30-second autorad exposure. M indicates the marker lane; sizes are illustrated on the left side, and C3 breakdown fragments are illustrated by labeled arrows on the right. The data shown is representative of four experiments. (C) Image analysis was carried out on scanned autorad films and Coomassie gels using a defined area to compare the pixel intensity (on a grayscale gradient) of bands in each lane. Results shown are the values obtained for the α′-band after being standardized for loading variation using the values obtained for the β-chain of C3. Data shown are the mean plus SD of the four experiments. Mann–Whitney t test was used to establish statistical significance at each concentration. *P<0.05, **P<0.01.
Alternative Pathway Hemolytic Activity Significantly Increased in Patient 1 Largely Because of Lower CFH Levels
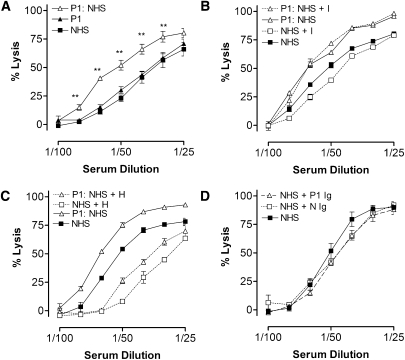
From the fluid phase assays, there was evidence that CFI autoantibody did interfere with CFI function. However, cell surface complement regulation is critical in aHUS (42). Using standard alternative pathway hemolysis assays (37), freshly isolated patient 1 serum had similar hemolytic activity compared with normal human serum (NHS) (Figure 5A). This finding was counterintuitive considering that the patient serum had both lower CFH levels and CFI autoantibody, albeit currently at low levels. We surmised that the low serum C3 levels in this patient could be undermining this assay. Therefore, we mixed patient and NHS serum 1:1 to replenish C3 levels. In this analysis, the 1:1 mix gave significantly greater lysis than NHS only, suggesting failure to control complement activation in serum from patient 1. Addition of purified CFI (Figure 5B) or CFH (Figure 5C) into this mixed sample indicated that the majority of the defect was caused by the loss of CFH. To test whether the CFI autoantibody had any effect on complement regulation, we supplemented NHS with 100 µg purified patient or control Ig. Addition of patient Ig had no impact on hemolysis compared with control Ig (Figure 5D). These findings were also replicated in the sheep red blood cell alternative pathway assay (43), and addition of purified CFI autoantibody to a standard classic pathway hemolytic assay (37) again showed no clear increase in lysis as a result of interference with factor I function (data not shown).
Figure 5.
Complement factor I (CFI) autoantibodies had minimal/no detectable effect on cell surface complement regulation. (A) Alternative pathway mediate rabbit red blood cell lysis was established as per standard protocols. Patient 1 sera (P1), normal human sera (NHS), and a 1:1 mix of patient 1 and NHS (P1:NHS) are shown. (B) A 1:1 mix of patient 1 and NHS (P1:NHS) or NHS alone was supplemented with purified CFI (0.7 µg/well) before the addition of cells and hemolytic assay. (C) A 1:1 mix of patient 1 and NHS (P1:NHS) or NHS alone was supplemented with purified CFH (5 µg/well) before the addition of cells and hemolytic assay. (D) NHS was supplemented with purified Ig (100 µg/well) from a normal control (N Ig), patient 1 (P1 Ig), or buffer only as indicated. Percentage lysis was calculated from the OD410 readings of released haem compared with 100% lysis of rabbit red blood cells in dH2O. Mean and SD of triplicate analysis are shown. Results are representative of several independent experiments. Mann–Whitney test was used to establish significance. **P<0.01.
Discussion
In this study, we report for the first time the presence of CFI autoantibodies in aHUS patients. There was, however, little evidence to correlate the genesis of IgG isotype CFI autoantibodies with the course of aHUS in the patient with the highest recorded titer of CFI autoantibodies. Furthermore, functional analysis of freshly isolated CFI autoantibodies suggests that, currently, their presence results in only a minor modification of complement regulator capacity of CFI. This finding leads us to question whether these autoantibodies are disease-modifying or an epiphenomenon.
At a frequency of ∼2% (3/175) in the Newcastle aHUS cohort, CFI autoantibodies are less frequent than CFH autoantibodies (5–10%) (17–20). In patient 1, we have for the first time seen evidence of the class switch event in a complement protein autoantibody. Intriguingly, there is a large interval between the initial IgM and IgG response (Figure 2). Review of the patient’s history has provided no clear indication of the trigger for either the initial or subsequent events, and critically, the levels of autoantibody do not seem to be associated with the course of the disease. That we show that CFI autoantibodies exist both free in solution and in an immune complex with CFI suggests the autoantibodies may have low to intermediate affinity to native CFI.
A deletion incorporating CFHR1 and -3 is strongly associated with CFH autoantibodies in aHUS (19,44). Here, we found that all three individuals with CFI autoantibodies have two copies of CFHR1 and -3, suggesting that their absence only plays a specific role in the development CFH autoantibodies. That two of three individuals with CFI autoantibodies also carried mutations in CFH supports the hypothesis that chronic increased complement activation may predispose to the generation of autoantibodies against complement components. In patient 1, there was a heterozygous duplication of a single base pair (c.3468dupA), leading to a frame shift and premature stop codon in CFH. This finding is consistent with the low levels of CFH seen in this patient. Furthermore, patients with CFI deficiency also display low CFH levels. Therefore, the low levels of CFH seen in this patient could result from both immune complex removal of CFH associated with excess C3b (45) as well as the effects of the CFH mutation. Patient 2 has a heterozygous nonsynonymous CFH mutation (c.2018G>A; p.Cys673Tyr) that would be predicted to result in failure of secretion from that allele (3), but CFH levels, presumably produced from other allele, were found to be in the normal range (Table 1). In addition to mutations and autoantibodies, several studies have now identified CFH and CD46 risk haplotypes for aHUS. Intriguingly, both patients 1 and 2 do possess at-risk haplotypes on CFH and CD46 (Table 3) (CFHTGTGGT [H3] haplotype [20] and CD46GGAAC haplotype [40]), whereas patient 3, who had the best clinical outcome, did not have additional genetic risk factors for aHUS, a phenomenon previously noted in aHUS patients with CFI mutations (29).
Thus, as in patients with CFH autoantibodies, individuals with CFI autoantibodies may have additional genetic risk factors predisposing to aHUS. Furthermore, despite having a functionally significant mutation in CFH, this latent predisposition to disease did not manifest in patient 1 until the age of 26 years when she developed the disease in association with pregnancy (46). Likewise, patient 3 only developed aHUS after a diarrheal illness (not shiga toxin-associated). Thus, these three patients also show that, in addition to inherited and acquired susceptibility factors, a trigger is needed for the disease to be manifest. Recent analysis of cohorts of aHUS patients with complement mutations has identified upper respiratory tract infections (6), viruses (36), pregnancy (6,47), drugs (6), and non-Escherichia coli diarrheal illnesses (48) as potential triggers.
We have found CFI autoantibodies in ∼2% of the Newcastle aHUS cohort. These autoantibodies were associated with other susceptibility factors and support the theory that multiple hits are necessary in most patients before aHUS presents clinically (49). Importantly, CFI autoantibodies do not seem to track with disease in the patient with highest antibody titer, raising the possibility that these autoantibodies could be a marker of disease rather than a direct factor in disease development.
Disclosures
T.H.J.G. is a scientific advisor for Alexion Pharmaceuticals and has received grant support from the same company.
Supplementary Material
Acknowledgments
This work was supported by the Medical Research Council (G0701325), Kidney Research UK, the Northern Counties Kidney Research Fund, the Mason Medical Research Foundation, and the Academy of Medical Sciences. D.K. is a Wellcome Intermediate Clinical Fellow. We acknowledge use of DNA from the UK Blood Services Collection of Common Controls funded by Wellcome Trust Grant 076113/C/04/Z, Juvenile Diabetes Research Foundation Grant WT061858, and the National Institutes of Health Research of England. The collection was established as part of the Wellcome Trust Patient Control Consortium.
Footnotes
D.K. and I.Y.P. contributed equally to this work.
Published online ahead of print. Publication date available at www.cjasn.org.
This article contains supplemental material online at http://cjasn.asnjournals.org/lookup/suppl/doi:10.2215/CJN.05750611/-/DCSupplemental.
References
- 1.Kavanagh D, Richards A, Atkinson J: Complement regulatory genes and hemolytic uremic syndromes. Annu Rev Med 59: 293–309, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH: Factor H mutations in hemolytic uremic syndrome cluster in exons 18-20, a domain important for host cell recognition. Am J Hum Genet 68: 485–490, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dragon-Durey MA, Frémeaux-Bacchi V, Loirat C, Blouin J, Niaudet P, Deschenes G, Coppo P, Herman Fridman W, Weiss L: Heterozygous and homozygous factor h deficiencies associated with hemolytic uremic syndrome or membranoproliferative glomerulonephritis: Report and genetic analysis of 16 cases. J Am Soc Nephrol 15: 787–795, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Caprioli J, Bettinaglio P, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M; Itaslian Registry of Familial and Recurrent HUS/TTP: The molecular basis of familial hemolytic uremic syndrome: Mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J Am Soc Nephrol 12: 297–307, 2001 [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Caballero D, González-Rubio C, Gallardo ME, Vera M, López-Trascasa M, Rodríguez de Córdoba S, Sánchez-Corral P: Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet 68: 478–484, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caprioli J, Noris M, Brioschi S, Pianetti G, Castelletti F, Bettinaglio P, Mele C, Bresin E, Cassis L, Gamba S, Porrati F, Bucchioni S, Monteferrante G, Fang CJ, Liszewski MK, Kavanagh D, Atkinson JP, Remuzzi G; International Registry of Recurrent and Familial HUS/TTP: Genetics of HUS: The impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood 108: 1267–1279, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kavanagh D, Richards A, Noris M, Hauhart R, Liszewski MK, Karpman D, Goodship JA, Fremeaux-Bacchi V, Remuzzi G, Goodship TH, Atkinson JP: Characterization of mutations in complement factor I (CFI) associated with hemolytic uremic syndrome. Mol Immunol 45: 95–105, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Kavanagh D, Kemp EJ, Mayland E, Winney RJ, Duffield JS, Warwick G, Richards A, Ward R, Goodship JA, Goodship TH: Mutations in complement factor I predispose to development of atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 2150–2155, 2005 [DOI] [PubMed] [Google Scholar]
- 9.Sellier-Leclerc AL, Fremeaux-Bacchi V, Dragon-Durey MA, Macher MA, Niaudet P, Guest G, Boudailliez B, Bouissou F, Deschenes G, Gie S, Tsimaratos M, Fischbach M, Morin D, Nivet H, Alberti C, Loirat C; French Society of Pediatric Nephrology: Differential impact of complement mutations on clinical characteristics in atypical hemolytic uremic syndrome. J Am Soc Nephrol 18: 2392–2400, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Fremeaux-Bacchi V, Dragon-Durey MA, Blouin J, Vigneau C, Kuypers D, Boudailliez B, Loirat C, Rondeau E, Fridman WH: Complement factor I: A susceptibility gene for atypical haemolytic uraemic syndrome. J Med Genet 41: e84, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fremeaux-Bacchi V, Moulton EA, Kavanagh D, Dragon-Durey MA, Blouin J, Caudy A, Arzouk N, Cleper R, Francois M, Guest G, Pourrat J, Seligman R, Fridman WH, Loirat C, Atkinson JP: Genetic and functional analyses of membrane cofactor protein (CD46) mutations in atypical hemolytic uremic syndrome. J Am Soc Nephrol 17: 2017–2025, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Richards A, Kemp EJ, Liszewski MK, Goodship JA, Lampe AK, Decorte R, Müslümanoğlu MH, Kavukcu S, Filler G, Pirson Y, Wen LS, Atkinson JP, Goodship TH: Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc Natl Acad Sci USA 100: 12966–12971, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards A, Kathryn Liszewski M, Kavanagh D, Fang CJ, Moulton E, Fremeaux-Bacchi V, Remuzzi G, Noris M, Goodship TH, Atkinson JP: Implications of the initial mutations in membrane cofactor protein (MCP; CD46) leading to atypical hemolytic uremic syndrome. Mol Immunol 44: 111–122, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Sullivan M, Erlic Z, Hoffmann MM, Arbeiter K, Patzer L, Budde K, Hoppe B, Zeier M, Lhotta K, Rybicki LA, Bock A, Berisha G, Neumann HP: Epidemiological approach to identifying genetic predispositions for atypical hemolytic uremic syndrome. Ann Hum Genet 74: 17–26, 2010 [DOI] [PubMed] [Google Scholar]
- 15.Frémeaux-Bacchi V, Miller EC, Liszewski MK, Strain L, Blouin J, Brown AL, Moghal N, Kaplan BS, Weiss RA, Lhotta K, Kapur G, Mattoo T, Nivet H, Wong W, Gie S, Hurault de Ligny B, Fischbach M, Gupta R, Hauhart R, Meunier V, Loirat C, Dragon-Durey MA, Fridman WH, Janssen BJ, Goodship TH, Atkinson JP: Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood 112: 4948–4952, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, Carreras L, Arranz EA, Garrido CA, López-Trascasa M, Sánchez-Corral P, Morgan BP, Rodríguez de Córdoba S: Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc Natl Acad Sci USA 104: 240–245, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragon-Durey MA, Loirat C, Cloarec S, Macher MA, Blouin J, Nivet H, Weiss L, Fridman WH, Frémeaux-Bacchi V: Anti-factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 555–563, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Józsi M, Strobel S, Dahse HM, Liu WS, Hoyer PF, Oppermann M, Skerka C, Zipfel PF: Anti factor H autoantibodies block C-terminal recognition function of factor H in hemolytic uremic syndrome. Blood 110: 1516–1518, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Józsi M, Licht C, Strobel S, Zipfel SL, Richter H, Heinen S, Zipfel PF, Skerka C: Factor H autoantibodies in atypical hemolytic uremic syndrome correlate with CFHR1/CFHR3 deficiency. Blood 111: 1512–1514, 2008 [DOI] [PubMed] [Google Scholar]
- 20.Moore I, Strain L, Pappworth I, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Staniforth SJ, Holmes LV, Ward R, Morgan L, Goodship TH, Marchbank KJ: Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood 115: 379–387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavanagh D, Goodship T: Genetics and complement in atypical HUS. Pediatr Nephrol 25: 2431–2442, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt CQ, Herbert AP, Kavanagh D, Gandy C, Fenton CJ, Blaum BS, Lyon M, Uhrín D, Barlow PN: A new map of glycosaminoglycan and C3b binding sites on factor H. J Immunol 181: 2610–2619, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Ferreira VP, Herbert AP, Cortés C, McKee KA, Blaum BS, Esswein ST, Uhrín D, Barlow PN, Pangburn MK, Kavanagh D: The binding of factor H to a complex of physiological polyanions and C3b on cells is impaired in atypical hemolytic uremic syndrome. J Immunol 182: 7009–7018, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whaley K, Ruddy S: Modulation of C3b hemolytic activity by a plasma protein distinct from C3b inactivator. Science 193: 1011–1013, 1976 [DOI] [PubMed] [Google Scholar]
- 25.Nagasawa S, Stroud RM: Mechanism of action of the C3b inactivator: Requirement for a high molecular weight cofactor (C3b-C4bINA cofactor) and production of a new C3b derivative (C3b’). Immunochemistry 14: 749–756, 1977 [DOI] [PubMed] [Google Scholar]
- 26.Liszewski MK, Post TW, Atkinson JP: Membrane cofactor protein (MCP or CD46): Newest member of the regulators of complement activation gene cluster. Annu Rev Immunol 9: 431–455, 1991 [DOI] [PubMed] [Google Scholar]
- 27.Medof ME, Iida K, Mold C, Nussenzweig V: Unique role of the complement receptor CR1 in the degradation of C3b associated with immune complexes. J Exp Med 156: 1739–1754, 1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nilsson SC, Kalchishkova N, Trouw LA, Fremeaux-Bacchi V, Villoutreix BO, Blom AM: Mutations in complement factor I as found in atypical hemolytic uremic syndrome lead to either altered secretion or altered function of factor I. Eur J Immunol 40: 172–185, 2010 [DOI] [PubMed] [Google Scholar]
- 29.Bienaime F, Dragon-Durey MA, Regnier CH, Nilsson SC, Kwan WH, Blouin J, Jablonski M, Renault N, Rameix-Welti MA, Loirat C, Sautés-Fridman C, Villoutreix BO, Blom AM, Fremeaux-Bacchi V: Mutations in components of complement influence the outcome of Factor I-associated atypical hemolytic uremic syndrome. Kidney Int 77: 339–349, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Kavanagh D, Burgess R, Spitzer D, Richards A, Diaz-Torres ML, Goodship JA, Hourcade DE, Atkinson JP, Goodship TH: The decay accelerating factor mutation I197V found in hemolytic uraemic syndrome does not impair complement regulation. Mol Immunol 44: 3162–3167, 2007 [DOI] [PubMed] [Google Scholar]
- 31.Kavanagh D, Goodship TH: Membrane cofactor protein and factor I: Mutations and transplantation. Semin Thromb Hemost 32: 155–159, 2006 [DOI] [PubMed] [Google Scholar]
- 32.Wellcome Trust Case Control Consortiumperson-group>: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447: 661–678, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, Todd JA, Donnelly P, Barrett JC, Davison D, Easton D, Evans DM, Leung HT, Marchini JL, Morris AP, Spencer CC, Tobin MD, Attwood AP, Boorman JP, Cant B, Everson U, Hussey JM, Jolley JD, Knight AS, Koch K, Meech E, Nutland S, Prowse CV, Stevens HE, Taylor NC, Walters GR, Walker NM, Watkins NA, Winzer T, Jones RW, McArdle WL, Ring SM, Strachan DP, Pembrey M, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Hamshere ML, Holmans PA, Jones IR, Kirov G, Moskivina V, Nikolov I, O’Donovan MC, Owen MJ, Collier DA, Elkin A, Farmer A, Williamson R, McGuffin P, Young AH, Ferrier IN, Ball SG, Balmforth AJ, Barrett JH, Bishop TD, Iles MM, Maqbool A, Yuldasheva N, Hall AS, Braund PS, Dixon RJ, Mangino M, Stevens S, Thompson JR, Bredin F, Tremelling M, Parkes M, Drummond H, Lees CW, Nimmo ER, Satsangi J, Fisher SA, Forbes A, Lewis CM, Onnie CM, Prescott NJ, Sanderson J, Matthew CG, Barbour J, Mohiuddin MK, Todhunter CE, Mansfield JC, Ahmad T, Cummings FR, Jewell DP, Webster J, Brown MJ, Lathrop MG, Connell J, Dominiczak A, Marcano CA, Burke B, Dobson R, Gungadoo J, Lee KL, Munroe PB, Newhouse SJ, Onipinla A, Wallace C, Xue M, Caulfield M, Farrall M, Barton A, Bruce IN, Donovan H, Eyre S, Gilbert PD, Hilder SL, Hinks AM, John SL, Potter C, Silman AJ, Symmons DP, Thomson W, Worthington J, Dunger DB, Widmer B, Frayling TM, Freathy RM, Lango H, Perry JR, Shields BM, Weedon MN, Hattersley AT, Hitman GA, Walker M, Elliott KS, Groves CJ, Lindgren CM, Rayner NW, Timpson NJ, Zeggini E, Newport M, Sirugo G, Lyons E, Vannberg F, Hill AV, Bradbury LA, Farrar C, Pointon JJ, Wordsworth P, Brown MA, Franklyn JA, Heward JM, Simmonds MJ, Gough SC, Seal S, Stratton MR, Rahman N, Ban M, Goris A, Sawcer SJ, Compston A, Conway D, Jallow M, Newport M, Sirugo G, Rockett KA, Bumpstead SJ, Chaney A, Downes K, Ghori MJ, Gwilliam R, Hunt SE, Inouye M, Keniry A, King E, McGinnis R, Potter S, Ravindrarajah R, Whittaker P, Widden C, Withers D, Cardin NJ, Davison D, Ferreira T, Pereira-Gale J, Hallgrimsdo’ttir IB, Howie BN, Su Z, Teo YY, Vukcevic D, Bentley D, Brown MA, Compston A, Farrall M, Hall AS, Hattersley AT, Hill AV, Parkes M, Pembrey M, Stratton MR, Mitchell SL, Newby PR, Brand OJ, Carr-Smith J, Pearce SH, McGinnis R, Keniry A, Deloukas P, Reveille JD, Zhou X, Sims AM, Dowling A, Taylor J, Doan T, Davis JC, Savage L, Ward MM, Learch TL, Weisman MH, Brown M; Wellcome Trust Case Control Consortium Australo-Anglo-American Spondylitis Consortium (TASC) Biologics in RA Genetics and Genomics Study Syndicate (BRAGGS) Steering Committee Breast Cancer Susceptibility Collaboration (UK): Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 39: 1329–1337, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhillon B, Wright AF, Tufail A, Pappworth I, Hayward C, Moore I, Strain L, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Armbrecht AM, Laude A, Deary IJ, Staniforth SJ, Holmes LV, Goodship TH, Marchbank KJ: Complement factor h autoantibodies and age-related macular degeneration. Invest Ophthalmol Vis Sci 51: 5858–5863, 2010 [DOI] [PubMed] [Google Scholar]
- 35.Roversi P, Johnson S, Caesar JJ, McLean F, Leath KJ, Tsiftsoglou SA, Morgan BP, Harris CL, Sim RB, Lea SM: Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc Natl Acad Sci USA 108: 12839–12844, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bento D, Mapril J, Rocha C, Marchbank KJ, Kavanagh D, Barge D, Strain L, Goodship TH, Meneses-Oliveira C: Triggering of atypical hemolytic uremic syndrome by influenza A (H1N1). Ren Fail 32: 753–756, 2010 [DOI] [PubMed] [Google Scholar]
- 37.Morgan BP: Measurement of Complement Hemolytic Activity, Generation of Complement-Depleted Sera, Production of Hemolytic Intermediates. In: Complement Methods and Protocols, edited by Morgan BP, Totowa, NJ, Humana Press, 2000, pp 61–72 [DOI] [PubMed] [Google Scholar]
- 38.Sahu A, Isaacs SN, Soulika AM, Lambris JD: Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J Immunol 160: 5596–5604, 1998 [PubMed] [Google Scholar]
- 39.Solberg HE: International Federation of Clinical Chemistry. Scientific committee, Clinical Section. Expert Panel on Theory of Reference Values and International Committee for Standardization in Haematology Standing Committee on Reference Values. Approved recommendation (1986) on the theory of reference values. Part 1. The concept of reference values. Clin Chim Acta 165: 111–118, 1987 [DOI] [PubMed] [Google Scholar]
- 40.Esparza-Gordillo J, Goicoechea de Jorge E, Buil A, Carreras Berges L, López-Trascasa M, Sánchez-Corral P, Rodríguez de Córdoba S: Predisposition to atypical hemolytic uremic syndrome involves the concurrence of different susceptibility alleles in the regulators of complement activation gene cluster in 1q32. Hum Mol Genet 14: 703–712, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Holers VM: The spectrum of complement alternative pathway-mediated diseases. Immunol Rev 223: 300–316, 2008 [DOI] [PubMed] [Google Scholar]
- 42.Roumenina LT, Loirat C, Dragon-Durey MA, Halbwachs-Mecarelli L, Sautes-Fridman C, Fremeaux-Bacchi V: Alternative complement pathway assessment in patients with atypical HUS. J Immunol Methods 365: 8–26, 2011 [DOI] [PubMed] [Google Scholar]
- 43.Sánchez-Corral P, González-Rubio C, Rodríguez de Córdoba S, López-Trascasa M: Functional analysis in serum from atypical Hemolytic Uremic Syndrome patients reveals impaired protection of host cells associated with mutations in factor H. Mol Immunol 41: 81–84, 2004 [DOI] [PubMed] [Google Scholar]
- 44.Zipfel PF, Edey M, Heinen S, Józsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, Goodship JA, Licht C, Goodship TH, Skerka C: Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet 3: e41, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naked GM, Florido MP, Ferreira de Paula P, Vinet AM, Inostroza JS, Isaac L: Deficiency of human complement factor I associated with lowered factor H. Clin Immunol 96: 162–167, 2000 [DOI] [PubMed] [Google Scholar]
- 46.George JN: The association of pregnancy with thrombotic thrombocytopenic purpura-hemolytic uremic syndrome. Curr Opin Hematol 10: 339–344, 2003 [DOI] [PubMed] [Google Scholar]
- 47.Goodship TH, Kavanagh D: Pulling the trigger in atypical hemolytic uremic syndrome: The role of pregnancy. J Am Soc Nephrol 21: 731–732, 2010 [DOI] [PubMed] [Google Scholar]
- 48.Fakhouri F, Roumenina L, Provot F, Sallée M, Caillard S, Couzi L, Essig M, Ribes D, Dragon-Durey MA, Bridoux F, Rondeau E, Frémeaux-Bacchi V: Pregnancy-associated hemolytic uremic syndrome revisited in the era of complement gene mutations. J Am Soc Nephrol 21: 859–867, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodríguez de Córdoba S: aHUS: A disorder with many risk factors. Blood 115: 158–160, 2010 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.