In the DNA damage checkpoint, the sensor kinase Mec1 must be activated by Ddc1 or Dpb11. However, Ddc1 and Dpb11 are dispensable for the related replication checkpoint. Instead, colocalization of Mec1 and the replisome component Mrc1 is the minimal signal required to activate the replication checkpoint and allow survival of replication stress.
Abstract
When DNA is damaged or DNA replication goes awry, cells activate checkpoints to allow time for damage to be repaired and replication to complete. In Saccharomyces cerevisiae, the DNA damage checkpoint, which responds to lesions such as double-strand breaks, is activated when the lesion promotes the association of the sensor kinase Mec1 and its targeting subunit Ddc2 with its activators Ddc1 (a member of the 9-1-1 complex) and Dpb11. It has been more difficult to determine what role these Mec1 activators play in the replication checkpoint, which recognizes stalled replication forks, since Dpb11 has a separate role in DNA replication itself. Therefore we constructed an in vivo replication-checkpoint mimic that recapitulates Mec1-dependent phosphorylation of the effector kinase Rad53, a crucial step in checkpoint activation. In the endogenous replication checkpoint, Mec1 phosphorylation of Rad53 requires Mrc1, a replisome component. The replication-checkpoint mimic requires colocalization of Mrc1-LacI and Ddc2-LacI and is independent of both Ddc1 and Dpb11. We show that these activators are also dispensable for Mec1 activity and cell survival in the endogenous replication checkpoint but that Ddc1 is absolutely required in the absence of Mrc1. We propose that colocalization of Mrc1 and Mec1 is the minimal signal required to activate the replication checkpoint.
INTRODUCTION
To avoid passing on damaged DNA, cells activate checkpoints under conditions that threaten the genome. The better studied of the DNA integrity checkpoints, the DNA damage checkpoint, is activated when initial processing of a wide variety of DNA lesions reveals stretches of single-stranded DNA (Garvik et al., 1995; Lee et al., 1998). In Saccharomyces cerevisiae, the mechanism by which this DNA structure activates checkpoint signaling has been well delineated: RPA-coated single-stranded DNA recruits the sensor kinase Mec1 through its binding partner, Ddc2 (Rouse and Jackson, 2002; Zou and Elledge, 2003; Ball et al., 2005), and the junction between RPA-coated single-stranded and double-stranded DNA independently (Edwards et al., 1999; Melo et al., 2001) recruits the 9-1-1 clamp (Majka and Burgers, 2003; Majka et al., 2006a; Zou et al., 2003; Ellison and Stillman, 2003). Ddc1, a subunit of 9-1-1, then increases the kinase activity of Mec1 both directly and by recruiting Dpb11, another Mec1 activator (Majka et al., 2006b; Mordes et al., 2008; Navadgi-Patil and Burgers, 2008, 2009). Mec1 phosphorylates histone H2A, creating a mark referred to as gamma-H2A, which is the correlate of the mark made on the metazoan H2A variant H2AX (Downs et al., 2000, 2004). Along with a constitutive methylation on H3K79, gamma-H2A promotes the recruitment of Rad9, the checkpoint mediator (Huyen et al., 2004; Nakamura et al., 2004; Giannattasio et al., 2005). Alternatively, Rad9 can be recruited by a 9-1-1–Dpb11 complex (Saka et al., 1997; Furuya et al., 2004; Puddu et al., 2008; Pfander and Diffley, 2011). Rad9 phosphorylation by Mec1 promotes Rad9's association with the checkpoint effector kinase Rad53, which binds these Rad9 phosphorylations through its FHA domains (Emili, 1998; Sun et al., 1998; Vialard et al., 1998; Schwartz et al., 2002). This is thought to position Rad53 for Mec1 phosphorylation (Sweeney et al., 2005), leading to Rad53 autophosphorylation and activation (Gilbert et al., 2001; Usui et al., 2009). Activated Rad53 can then diffuse away from the site of damage and phosphorylate downstream effectors of the checkpoint. Tel1, a sensor kinase related to Mec1, can perform some of the same activities as Mec1. Tel1 phosphorylates and activates Rad53 using the mediator Rad9 (P. Garber and David P. Toczyski, unpublished results). However, Tel1 does not require activation by Ddc1 or Dpb11 (Giannattasio et al., 2002; reviewed in Mordes and Cortez, 2008).
The DNA replication checkpoint uses much of the same machinery as the DNA damage checkpoint. However, it responds to stalled replication forks during S phase (reviewed in Tourrière and Pasero, 2007). Both canonical Mec1 activators Ddc1 and Dpb11 may play a role in the replication checkpoint (Wang and Elledge, 2002), although it is not clear whether they are absolutely required (Navadgi-Patil and Burgers, 2009; Puddu et al., 2011). Mec1 is recruited to stalled replication forks, probably through an interaction with RPA-coated single-stranded DNA (Katou et al., 2003; Osborn and Elledge, 2003). A significant difference between the replication checkpoint and the DNA damage checkpoint is that a different mediator protein is used. Rad9 does not appear to participate in the replication checkpoint; instead, Mrc1 acts as the checkpoint mediator (Alcasabas et al., 2001). Mrc1 is part of the replication machinery and travels with replication forks during every S phase (Osborn and Elledge, 2003) and therefore does not need to be specifically recruited to stalled replication forks. On fork stalling, Mec1 phosphorylates Mrc1, which promotes the recruitment and activation of Rad53 (Osborn and Elledge, 2003). As in the DNA damage checkpoint, active Rad53 leads to arrest of the cell cycle at mitosis, destruction of the ribonucleotide reductase inhibitor Sml1, and transcriptional regulation (reviewed in Tourrière and Pasero, 2007). However, these activities of Rad53 are less important for cell survival after acute, as opposed to chronic, replication stress. Rad53 also stabilizes stalled replication forks so that they can restart efficiently after replication stress is over, in part by blocking the activity of Exo1 (Segurado and Diffley, 2008). This fork stabilization is the checkpoint function essential for cell survival after acute replication stress (Desany et al., 1998; Tercero et al., 2003).
In vitro experiments using purified proteins demonstrate that Dpb11 and Ddc1 activate Mec1 directly (Majka et al., 2006b; Mordes et al., 2008; Navadgi-Patil and Burgers, 2008). In addition, artificial colocalization of Ddc1 and Mec1 on chromatin promotes Mec1 activity in vivo (Bonilla et al., 2008). This colocalization was achieved through a system in which an array of lac operator repeats (LacO) was integrated into the genome and Ddc1 and the Mec1 binding partner Ddc2 were fused to lac repressor (LacI). Ddc2-LacI was used instead of directly tethering Mec1 to LacI because C-terminal Mec1 fusions are not functional. Colocalization of Ddc1 and Ddc2 LacI fusions promoted phosphorylation of Rad9 and Rad53 and cell cycle arrest.
We used a similar approach to investigate Mec1 activation in the replication checkpoint. We fused Ddc2 and Mrc1 to LacI and showed that this replication-checkpoint mimic can promote phosphorylation of Rad53. The Mec1 activator Dpb11 has an essential role in the initiation of DNA replication, confounding attempts to examine its checkpoint signaling role in isolation. Because the replication-checkpoint mimic enacts checkpoint signaling in the absence of DNA replication, it provides an ideal setting in which to examine Dpb11's role in Mec1 activation. We show that Mec1 activity in the replication-checkpoint mimic does not depend on Dpb11 or Ddc1. Furthermore, Mec1 can act through Mrc1 to phosphorylate Rad53 in the endogenous replication checkpoint, even in a ddc1 dpb11-1 strain, and this activity is sufficient to maintain viability after acute replication stress. Therefore we propose that, whereas Ddc1 and Dpb11 aid in replication-checkpoint activation, colocalization of Mec1 and Mrc1 at stalled replication forks promotes Rad53 activation sufficient to stabilize the replisome during transient replication stress.
RESULTS
Development of a replication-checkpoint mimic
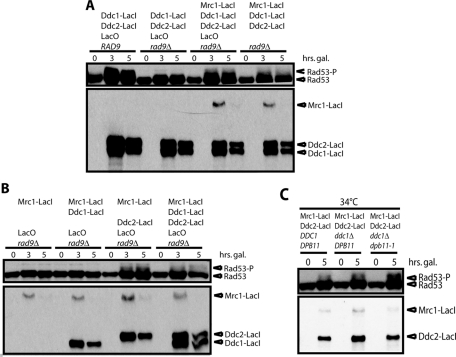
Colocalization of Mec1 and the 9-1-1 complex through the induction of Ddc2–green fluorescent protein (GFP)–LacI and Ddc1-GFP-LacI promotes phosphorylation of Rad53 in the absence of DNA damage. This is dependent on Rad9 (Bonilla et al., 2008; Figure 1A), since Mrc1 is not recruited to the LacO array. To generate a mimic of the replication checkpoint, we fused Mrc1 to GFP-LacI. Mrc1-GFP-LacI (hereafter referred to as Mrc1-LacI) can substitute for Rad9 and allow Rad53 phosphorylation in a strain lacking Rad9 (Figure 1A). Of importance, this replication-checkpoint mimic signaling was assayed in nocodazole-arrested cells, so it is independent of DNA replication and of S phase.
FIGURE 1:
Colocalization of Mec1-Ddc2 and Mrc1 promotes Rad53 phosphorylation independent of Ddc1 and Dpb11. (A, B) Strains with or without a LacO array and with the indicated combination of Mrc1-LacI, Ddc2-LacI, and Ddc1-LacI under the control of Gal promoter were grown in raffinose and arrested in nocodazole for 3 h. Galactose was pulsed for 1 h before transcription was inhibited with dextrose, and then samples were collected at the indicated time points from galactose addition and blotted for Rad53 and the LacI fusion proteins. (C) As in A and B, except that strains were grown at 23°C, arrested with nocodazole, and either kept at 23°C or shifted to 34°C at 1 h before galactose induction.
Recruitment of Rad9 is mediated by histone modifications. Therefore, even though Ddc1-LacI and Ddc2-LacI are able to associate through dimerization of LacI (and possibly GFP), this is not sufficient for Rad9 activation, and a LacO array is required. In contrast, Rad53 phosphorylation mediated by Mrc1-LacI should not require chromatin. Therefore we determined whether the LacO array was required for activation of the replication-checkpoint mimic. While activation of this replication-checkpoint mimic was enhanced by integration of an array of LacO, Rad53 was partially phosphorylated in a strain without a LacO array (Figure 1A).
Next we tested whether all three LacI fusions were required for checkpoint activation (Figure 1B). As expected, neither Mrc1-LacI alone nor Mrc1-LacI/Ddc1-LacI promoted Rad53 phosphorylation. However, colocalization of just Mrc1-LacI and Ddc2-LacI was sufficient to promote Rad53 phosphorylation without Ddc1-LacI.
Mec1 activators in the replication-checkpoint mimic
Having shown that the LacI fusion of the Mec1-activating 9-1-1 component Ddc1 was not required for the replication-checkpoint mimic, we tested whether Ddc1 or the other known Mec1 activator, Dpb11, was required at all. We could not delete DPB11 because it is essential for DNA replication. In vitro studies have shown that the Mec1-activating domain of Dpb11 lies at the C-terminus, between amino acids 572 and 764 (Mordes et al., 2008; Navadgi-Patil and Burgers, 2008). The protein encoded by dpb11-1 is truncated after amino acid 582. Although dpb11-1 has been reported to have checkpoint defects (Araki et al., 1995; Wang and Elledge, 2002; Puddu et al., 2011), it is formally possible that the 11 amino acids between 572 and 582 could partially activate Mec1, especially since similar-sized domains of Ddc1 have been shown to activate Mec1 (Navadgi-Patil and Burgers, 2009). Therefore we tested the activity of the replication-checkpoint mimic in the dpb11-1 ddc1∆ mutant at 34°C, a nonpermissive temperature for dpb11-1 (Supplemental Figure S1). Rad53 is phosphorylated as strongly in the ddc1∆ dpb11-1 strain as in a DDC1 DPB11 strain (Figure 1C). Thus we conclude that neither Ddc1 nor Dpb11 is required for activity of the replication-checkpoint mimic.
Optimization and further characterization of the replication-checkpoint mimic
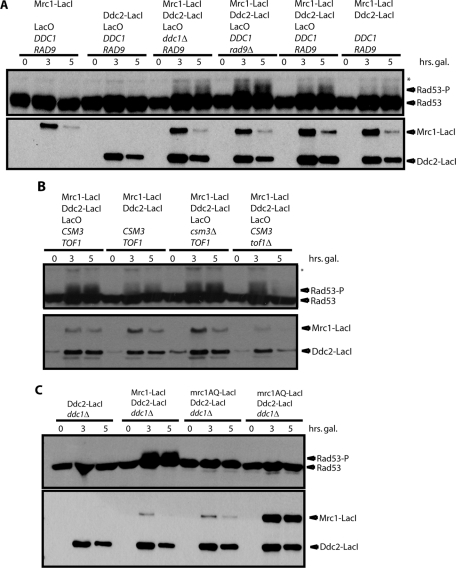
As shown in Figure 1B, the Ddc2-LacI/Mrc1-LacI system phosphorylated Rad53 less efficiently than the original Ddc1-LacI/Ddc2-LacI DNA-damage-checkpoint mimic. We hypothesized that this resulted from low expression of Mrc1-LacI relative to Ddc2-LacI (Figure 1, A and B). Therefore we expressed Mrc1-LacI from a stronger promoter (Gal instead of GalS), such that its levels are almost as high as Ddc2-LacI. This resulted in more robust Rad53 phosphorylation (unpublished data and Figure 2A).
FIGURE 2:
The replication-checkpoint mimic faithfully reproduces qualities of the replication checkpoint. (A) As in Figure 1, but Mrc1-LacI expression was increased so that it was similar to that of Ddc2-LacI. (B) The replication-checkpoint mimic was examined, as in A, in cells lacking the Mrc1 binding partners Csm3 or Tof1. (C) A ddc1∆ strain containing Ddc2-LacI and LacO was transformed with no additional fusion protein, Mrc1-LacI, mrc1AQ-LacI, or high levels of mrc1AQ-LacI and assayed as in A.
In this optimized replication-checkpoint mimic, again, neither Mrc1-LacI nor Ddc2-LacI alone is sufficient to activate Rad53. Deletion of RAD9 or DDC1 in the mimic strain did not have a strong impact on Rad53 phosphorylation (Figure 2A). It is likely that Ddc1 cannot be recruited to the LacO array, since there is no junction between doubled-stranded and single-stranded DNA, and therefore it is not surprising that the status of the 9-1-1 complex is not important. Rad9 is not phosphorylated in response to stalled replication forks in an MRC1 wild-type strain (Alcasabas et al., 2001), and so this feature of the replication-checkpoint mimic matches the endogenous checkpoint. However, it is unclear why Rad9 cannot be recruited to either a natural stalled replication fork or the LacO array in our system; we investigate this question further in Figure 4. In this optimized system, as in Figure 1A, some Rad53 phosphorylation was seen in the absence of the LacO array.
FIGURE 4:
Rad53 phosphorylation in the absence of 9-1-1 requires Mrc1. (A) Cells of the indicated genotype were treated with HU for 2.5 h at 30°C. All strains carry the sml1-1 mutation, which suppresses lethality of mec1∆. Rad53 phosphorylation was visualized by SDS–PAGE and Western blot. Cdc28 served as a loading control. (B) After HU treatment as described in A for the indicated time, HU was washed out and cells were plated on rich medium to measure viability. (C) Cells of the indicated genotype were treated with HU for 2.8 h at 30°C and processed as in A. (D) Cells of the indicated genotype were treated with MMS for 4 h at 22.5°C, and phosphorylation of FLAG-tagged Rad9 and Rad53-HA was visualized by SDS–PAGE and Western blotting.
Most of the Mrc1 in cells is associated with the proteins Csm3 and Tof1. Both Csm3 and Tof1 are required for normal localization of Mrc1 to replication forks (Katou et al., 2003; Bando et al., 2009), and tof1∆ cells have been reported to be unable to activate the replication checkpoint (Foss, 2001). However, csm3∆ and tof1∆ cells activated the replication-checkpoint mimic as efficiently as wild-type cells (Figure 2B), suggesting that these proteins play no direct role in the replication checkpoint and that the checkpoint defects observed when they are mutated are the result of mislocalization of Mrc1.
In the endogenous replication checkpoint, phosphorylation of Mrc1 by Mec1 is required to recruit Rad53 and promote its phosphorylation. Therefore the mrc1AQ mutant protein, in which all potential Mec1 phosphorylation sites are removed, cannot promote Rad53 phosphorylation (Osborn and Elledge, 2003). In agreement with this, mrc1AQ-LacI could not promote Rad53 phosphorylation in the replication-checkpoint mimic (Figure 2C). The mrc1AQ-LacI protein could be nonspecifically hypomorphic, for example, by being partially unfolded. Therefore we screened for integrants expressing higher levels of mrc1AQ-LacI and showed that these also failed to phosphorylate Rad53 (Figure 2C, fourth strain).
Mec1 activity during replication stress
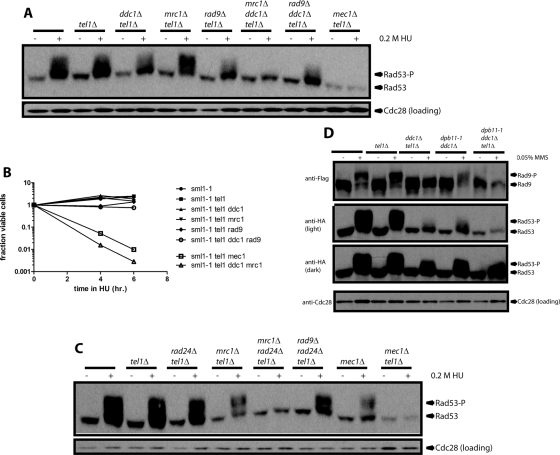
Because Mec1 phosphorylation of Rad53 in the replication-checkpoint mimic did not depend on known Mec1 activators, we tested whether these activators were required during replication stress induced by treatment with the ribonucleotide reductase inhibitor hydroxyurea (HU) for 4 h at 23°C. To make sure that we observed Mec1 activity only, we deleted the MEC1 orthologue TEL1. Rad53 phosphorylation in response to HU treatment was reduced when DDC1 was deleted or DPB11 was mutated, but it was only slightly more reduced when these two mutations were combined (Figure 3A). Consistent with what we observed in the replication-checkpoint mimic, the ddc1∆ dpb11-1 tel1∆ strain still displayed significant Rad53 phosphorylation upon HU treatment while, as expected, a mec1∆ tel1∆ mutant did not phosphorylate Rad53 (Figure 3A). Levels of Rad53 phosphorylation in these mutants observed by gel shift (Figure 3A) were recapitulated when the same samples were tested for Rad53 kinase activity by in situ assay (Supplemental Figure S2). This is consistent with both the replication checkpoint and the minimal endogenous checkpoint relying exclusively on Mec1 and Mrc1, although unknown proteins could be required in both cases, since these experiments are performed in vivo.
FIGURE 3:
Rad53 can be phosphorylated in response to replication stress in the absence of 9-1-1 and Dpb11. (A) Strains with the indicated genotype were grown asynchronously and then treated with 0.2 M HU for 4 h at 22.5°C. All strains carry the sml1-1 mutation, which suppresses lethality of a mec1∆. Rad53 phosphorylation was visualized by SDS–PAGE and Western blot. Cdc28 serves as a loading control. (B) After HU treatment as described in A for the indicated time, HU was washed out and cells plated on rich medium to determine viability. The mean of three independent experiments is plotted; error bars reflect SE. (Error bars that cannot be seen are thinner than the line.)
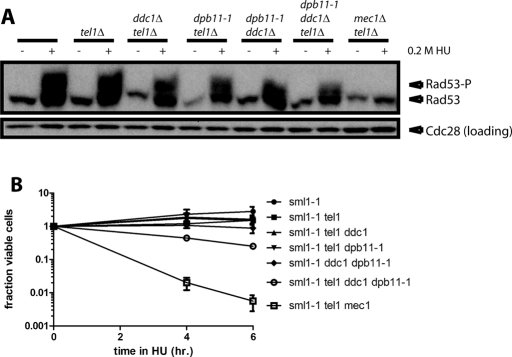
To test the physiological relevance of the levels of Rad53 phosphorylation we observed in these mutants, we treated them with HU for 4 and 6 h at 23°C and then washed out the drug and plated cells on rich medium to test viability (Figure 3B). Wild-type cells and all single and double mutants of ddc1∆, tel1∆, and dpb11-1 retained >75% viability after HU treatment. Similarly, the ddc1∆ dpb11-1 tel1∆ triple mutant retained ∼50% viability, as compared with an almost complete loss of viability in mec1∆ tel1∆ strains, which lack all checkpoint signaling.
Previous studies showed that Mec1-dependent Rad53 phosphorylation requires DDC1 in cells arrested in G1 with alpha factor or in mitosis with nocodazole (Paciotti et al., 1998; Navadgi-Patil and Burgers, 2009). Therefore we tested whether Ddc1-independent phosphorylation of Rad53 by Mec1 depended on Mrc1, which is active only in S phase. An mrc1∆ ddc1∆ tel1∆ strain could not phosphorylate Rad53, suggesting that 9-1-1 is required for Rad9-mediated Rad53 phosphorylation but not Mrc1-mediated Rad53 phosphorylation (Figure 4A) or activity (Supplemental Figure S3). Consistently, this triple mutant cannot survive a 2.5-h treatment with HU at 30°C (Figure 4B). As expected, inactivation of 9-1-1 by deletion of the clamp loader RAD24 gives the same result as deletion of DDC1 (Figure 4C), consistent with an earlier observation in a TEL1 strain (Bjergbaek et aI., 2005). The slightly increased Rad53 phosphorylation observed when MRC1 is deleted in a tel1∆ strain is consistent with a report that mrc1∆ strains phosphorylate Rad53 even in the absence of replication-stressing agents such as HU, likely because Mrc1 has a checkpoint-independent role in promoting replisome stability (Alcasabas et al., 2001).
The requirement for Mrc1 in the absence of 9-1-1 suggested that the other checkpoint mediator, Rad9, cannot function for some reason at stalled replication forks or in the absence of 9-1-1. One possibility is that 9-1-1 is physically required for recruitment or function of Rad9. When Rad9 is recruited to the site of DNA damage, it is phosphorylated by Mec1. Then it mediates Rad53 autophosphorylation and is phosphorylated by Rad53 in the process. Therefore we tested whether Rad9 could be phosphorylated in various mutants (Figure 4D). For these experiments, we used methyl methanesulfonate (MMS) instead of HU, since Rad9 is not phosphorylated upon HU treatment in MRC1 strains. It is not surprising that both Rad53 and Rad9 are robustly phosphorylated in wild-type or tel1∆. However, in MMS, Rad53 phosphorylation is even more significantly reduced in all ddc1∆ strains than it was in HU (Figure 4D). This is unsurprising since checkpoint signaling in HU depends most on Mrc1 (and thus is independent of Ddc1), whereas signaling in MMS is more dependent on Rad9 (Alcasabas et al., 2001; Komata et al., 2009). The levels of Rad53 phosphorylation (Figure 4D) and activity (Supplemental Figure S4) are comparable in the ddc1∆tel1∆ and ddc1∆ dpb11-1 strains, but levels of Rad9 phosphorylation vary significantly. Rad9 exhibits no phospho shift in ddc1∆tel1∆ and a striking shift in ddc1∆ dpb11-1. We know that Rad9 activity is not absolutely dependent on Tel1, since the mrc1∆ tel1∆ strain can still phosphorylate Rad53 (Figure 4A). Therefore we conclude that phosphorylation of Rad9 requires either Tel1 or Mec1 and Ddc1, reinforcing the idea that the activation of Mec1 by 9-1-1 is critical for Rad9- but not Mrc1-mediated activity.
DISCUSSION
Here we report Mec1-dependent Rad53 phosphorylation in the absence of the canonical Mec1 activators Ddc1 and Dpb11, both in response to replication stress and in an artificial in vivo system. The essential role of Dpb11 in replication limits the ability to examine its role in the replication checkpoint. We eliminated this issue by using a replication-checkpoint mimic, which allows us to assay Mrc1-dependent checkpoint activation outside of S phase. Others reported that Rad53 phosphorylation in a ddc1∆ dpb11-1 strain is restricted to S phase (Navadgi-Patil and Burgers, 2009; Puddu et al., 2011). To show that this represented Mec1 activity in the absence of its activators, we demonstrated Rad53 phosphorylation in a ddc1∆ dpb11-1 tel1∆ strain. Our results also suggest the mechanism by which activator-independent Mec1 activity is restricted to S phase: in the absence of Ddc1, phosphorylation of Rad53 is absolutely dependent on Mrc1, the S-phase–specific checkpoint mediator that is a constitutive part of the DNA replisome (Figure 4). Furthermore, forced colocalization of Mec1 and Mrc1 is sufficient to phosphorylate Rad53 in metaphase-arrested cells undergoing neither DNA replication nor DNA damage (Figure 1). We propose that a high local concentration of Mrc1, both at a stalled replication fork and in the replication-checkpoint mimic, allows unactivated Mec1 to phosphorylate Mrc1, which leads to Rad53 recruitment and a high local concentration of Rad53, allowing unactivated Mec1 to phosphorylate Rad53. Alternatively, an undiscovered, replication-specific Mec1 activator could operate at stalled replication forks. Of importance, the Rad53 phosphorylation we observe is sufficient to promote survival of acute replication stress (Figure 3B). Here we place our results in the context of the extensive literature showing replication checkpoint defects in dpb11-1 and ddc1∆ dpb11-1 strains, outline the molecular events implied by our results and model, and suggest likely cellular consequences of the Mec1-dependent, activator-independent Rad53 phosphorylation we observe.
We propose that the replication-checkpoint phenotypes associated with dpb11-1 and ddc1∆ dpb11-1 strains can be understood as a function of the dual role of Dpb11 in promoting DNA replication initiation and checkpoint signaling or of the difference between levels of Rad53 activation required for growth under chronic replication stress and levels sufficient for survival of acute replication stress. Dpb11 was first implicated in checkpoint signaling because dpb11-1 cells, which are incompetent for DNA replication at the restrictive temperature, fail to arrest in mitosis when exposed to replication stress, even at the permissive temperature (Araki et al., 1995). At the restrictive temperature, dpb11-1 cells could not phosphorylate Rad53 in response to replication stress (Wang and Elledge, 1999). Later Mordes et al. (2008) and Navadgi-Patil and Burgers (2008) showed that the product of dpb11-1 is a truncation that lacks almost all of the domain required for Mec1 activation. The phenotypes of dpb11-1 certainly reflect some genuine defect in Rad53 phosphorylation in dpb11-1 cells. However, impairment of replication initiation can nonspecifically prevent activity of the replication checkpoint by reducing the number of replication forks, even if the mutated protein does not participate directly in checkpoint signaling (Shimada et al., 2002). dpb11-1 cells have defective replication initiation at the permissive and restrictive temperatures (Kamimura et al., 1998). Therefore the replication-checkpoint phenotypes of dpb11-1 probably reflect a combination of defective replication initiation and defective checkpoint signaling. We avoided this conflation by using the replication-checkpoint mimic, which does not require the replication function of Dpb11, to show that neither Dpb11 nor Ddc1 is required for Mec1 phosphorylation of Rad53.
The phenotypes of ddc1∆ dpb11-1 double mutants likely also reflect the high level of Rad53 phosphorylation required to grow in the presence of chronic replication stress. It was proposed that Ddc1 and Dpb11 act independently to promote Mec1 activation in response to replication stress, because either single mutant can grow well in the presence of hydroxyurea but the double mutant cannot (Wang and Elledge, 2002). Furthermore, Ddc1 and Dpb11 can activate Mec1 independently of each other (Navadgi-Patil and Burgers, 2009). Puddu et al. (2011) showed that Rad53 phosphorylation in a ddc1∆ dpb11-1 strain is insufficient to allow growth under chronic replication stress, but this level of Rad53 phosphorylation can promote cell survival after acute replication stress. Because they did not observe Rad53 phosphorylation in a ddc1∆ dpb11-1 tel1∆ triple mutant, they attributed Rad53 phosphorylation in the ddc1∆ dpb11-1 strain to Tel1. We do see Rad53 phosphorylation in the ddc1∆ dpb11-1 tel1∆ strain. Moreover, our ddc1∆ dpb11-1 tel1∆ cells can survive acute replication stress two orders of magnitude better than mec1∆ tel1∆ cells, indicating that Mec1 must have significant function remaining in the absence of activation by Ddc1 and Dpb11.
The unique structure of a stalled replication fork, as opposed to a processed double-strand break or other lesion, may explain why checkpoint signaling can be activated in the ddc1∆ dpb11-1 tel1∆ strain by HU treatment during S phase but not outside of S phase. Mec1 is recruited to stalled forks, whereas Mrc1 is already present (Osborn and Elledge, 2003; Katou et al., 2003). If colocalization of Mec1 and Mrc1 is the only requirement for the reduced but significant level of checkpoint activation we observe in the ddc1∆ dpb11-1 tel1∆ strain, then Mec1 recruitment to a stalled fork is the only molecular event required to activate the checkpoint. In vitro results suggest that this model is possible, since Mrc1 purified from bacteria can promote Mec1 phosphorylation of Rad53 even in the absence of Mec1 activators (Chen and Zhou, 2009). Moreover, some Rad53 phosphorylation is seen in the replication-checkpoint mimic in the absence of a LacO array, suggesting that the Ddc2-LacI/Mrc1-LacI heterodimer is sufficient to promote Rad53 phosphorylation (Figures 1A and 2A). Alternatively, if a replication-specific activator of Mec1 exists and is required for all Mec1 phosphorylation of Rad53, our replication-checkpoint mimic results suggest that 1) it must be recruited by either Mrc1 or Mec1-Ddc2, 2) it does not require that Mec1-Ddc2 be bound to chromatin, and 3) it does not depend on 9-1-1, Dpb11, Tof1, or Csm3.
The ability of Mrc1, but not the other checkpoint mediator, Rad9, to promote 9-1-1–independent Mec1 phosphorylation of Rad53 is probably explained in one of two ways: Mrc1 could be a better Mec1 substrate, either intrinsically or because it is present at a high local concentration at stalled forks, or Rad9 might not be recruited to stalled replication forks in the absence of 9-1-1 and Tel1. Indeed, we found that Rad9 could not be phosphorylated in response to MMS unless either 9-1-1 or Tel1 was intact (Figure 4C). Consistent with this, a recent article shows that the 9-1-1–Dpb11 complex is one pathway by which Rad9 can be recruited to Mec1 (Pfander and Diffley, 2011).
DNA integrity checkpoints protect the genome in four ways: by arresting the cell cycle until damage can be repaired or replication can complete, changing transcription to promote DNA repair and cell-cycle arrest, inhibiting late-origin firing, and acting at stalled replication forks to stabilize them. This last activity is the only one required for survival of acute replication stress, and it may occur locally at the stalled fork that activated the checkpoint. Puddu et al. (2011) showed that some of the other checkpoint readouts may be impaired in ddc1∆ dpb11-1 mutants, but survival of acute replication stress is not impaired. We showed significant survival in the ddc1∆ dpb11-1 tel1∆ strain. Perhaps the Mec1 phosphorylation of Rad53 that we observe in this strain provides, in wild-type cells, a way to stabilize transiently stalled forks without engaging the entire checkpoint machinery.
MATERIALS AND METHODS
Yeast strains
For complete genotypes, see Table 1. All strains are from the W303-1a background (rad5 mutation uncorrected). LacI fusions were integrated at the marker locus, and Rad53 was tagged at its endogenous locus. For strains containing LacO arrays, array length was measured by Southern blot. Genomic DNA was digested with BglII and probed with the XbaI fragment of pAFS52, which contains the LacO repeat. All Mrc1-LacI strains within the same experiment have the same array length, and the length is noted in the genotype. Rad9 was tagged with a PCR fragment containing appropriate homology, the 3xFLAG tag, and the gene for hygromycin B resistance. MEC1, TEL1, and RAD24 were disrupted with the TRP1 sequence. In replication-checkpoint mimic strains, DDC1 was deleted with a URA3 cassette that was subsequently looped out, and RAD9 was deleted by transformation with a PCR fragment containing appropriate homology and the hygromycin B resistance gene. In other TBY strains, DDC1, MRC1, and RAD9 were deleted by transformation with a PCR fragment containing appropriate homology and genes for resistance to G418, nourseothricin, and hygromycin B, respectively. In PGY strains, RAD9 was disrupted with HIS3, and MRC1 was disrupted with g418R.
TABLE 1:
Yeast strains.
| Figure location | Strain name | Genotype | Source |
|---|---|---|---|
| 1A, strain 1 | CBY88 | Mat a ade2-1 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 GalS-Ddc1-LacI::URA3 ddc1∆ | Bonilla et al. (2008) |
| 1A, strain 2 | CBY90 | Mat a ade2-1 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 GalS-Ddc1-LacI::URA3 ddc1∆ rad9∆::hygR | Bonilla et al. (2008) |
| 1A, strain 3; 1B, strain 4 | TBY66 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO180::TRP1 GalS-Ddc1-LacI::URA3 ddc1∆ rad9∆::hygR | This study |
| 1A, strain 4 | TBY63 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 trp1 GalS-Ddc1-LacI::URA3 ddc1∆ rad9∆::hygR | This study |
| 1B, strain 1 | TBY60 | Mat a GalS-Mrc1-LacI::ADE2 his3-11,15 Rad53-HA::LEU2 LacO180::TRP1 ura3-1 ddc1∆ rad9∆::hygR | This study |
| 1B, strain 2 | TBY61 | Mat a GalS-Mrc1-LacI::ADE2 his3-11,15 Rad53-HA::LEU2 LacO180::TRP1 GalS-Ddc1-LacI::URA3 ddc1∆ rad9∆::hygR | This study |
| 1B, strain 3 | TBY65 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO180::TRP1 ura3-1 ddc1∆ rad9∆::hygR | This study |
| 1C, strains 1 and 4 | TBY79 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO180::TRP1 ura3-1 | This study |
| 1C, strains 2 and 5 | TBY80 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO180::TRP1 ura3-1 ddc1∆ | This study |
| 1C, strains 3 and 6 | TBY81 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO180::TRP1 ura3-1 ddc1∆ dpb11-1 | This study |
| 2A, strain 1 | TBY217 | Mat a Gal-Mrc1-LacI::ADE2 his3-11,15 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 | This study |
| 2A, strain 2; 2C, strain 2 | TBY206 | Mat a ade2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 | This study |
| 2A, strain 3 | TBY34 | Mat a Gal-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 rad9∆::hygR | This study |
| 2A, strain 4 | TBY214 | Mat a Gal-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 ddc1∆ | This study |
| 2A, strain 5 | TBY205 | Mat a Gal-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 | This study |
| 2B, strain 1 | |||
| 2A, strain 6 | TBY207 | Mat a Gal-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 trp1-1 ura3-1 | This study |
| 2B, strain 2 | |||
| 2B, strain 3 | TBY36 | Mat a Gal-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 csm3∆::g418R | This study |
| 2B, strain 4 | TBY38 | Mat a Gal-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 tof1∆::g418R | This study |
| 2C, strain 1 | TBY236 | Mat a GalS-Mrc1-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 | This study |
| 2C, strain 3 | TBY238 | Mat a GalS-Mrc1AQ-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 | This study |
| 2C, strain 4 | TBY239 | Mat a GalS-Mrc1AQ-LacI::ADE2 Gal-Ddc2-LacI::HIS3 Rad53-HA::LEU2 LacO256::TRP1 ura3-1 | This study |
| 3, A and B, strain 1 | PGY1824 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 ura3-1 sml1-1 | Peter Garber, Toczyski lab |
| 4, A and B, strain 1 | |||
| 4C, strain 1 | |||
| 3, A and B, strain 2 | PGY2525 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1∆::URA3 sml1-1 | Peter Garber, Toczyski lab |
| 4, A and B, strain 2 | |||
| 4C, strain 2 | |||
| 3, A and B, strain 3 | TBY326 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 ddc1∆::g418R | This study |
| 4, A and B, strain 3 | |||
| 3, A and B, strain 4 | TBY233 | Mat alpha ade2-1 leu2-3,112 his4 RAD53-HA::TRP1 tel1::URA3 sml1-1 dpb11-1 | This study |
| 3, A and B, strain 5 | TBY380 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 ura3-11,15 sml1-1 ddc1∆::g418R dpb11-1 | This study |
| 3, A and B, strain 6 | TBY327 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 ddc1∆::g418R dpb11-1 | This study |
| 3, A and B, strain 7 | TBY143 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 mec1::TRP1 tel1::URA3 sml1-1 | This study |
| 4, A and B, strain 8 | |||
| 4C, strain 8 | |||
| 4, A and 4B, strain 4 | TBY51 | Mat a ade2-1 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 mrc1∆::natR | This study |
| 4, A and B, strain 5 | TBY49 | Mat a ade2-1 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 rad9∆::hygR | This study |
| 4, A and B, strain 6 | TBY50 | Mat a ade2-1 leu2-3,112 RAD53-HA::TRP1 tel1::URA3 sml1-1 ddc1∆::g418R mrc1∆::natR | This study |
| 4, A and B, strain 7 | TBY371 | Mat a ade2-1 leu2-3,112 RAD53-HA::TRP1 tel1::URA3 sml1-1 ddc1∆::g418R rad9∆::hygR | This study |
| 4C, strain 4 | PGY2383 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 mrc1::g418R | Peter Garber, Toczyski lab |
| 4C, strain 5 | PGY2387 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 rad24::TRP1 tel1::URA3 sml1-1 mrc1::g418R | Peter Garber, Toczyski lab |
| 4C, strain 6 | PGY2215 | Mat a ade2-1 rad9::HIS3 leu2-3,112 lys2 RAD53-HA::TRP1 rad24::TRP1 tel1::URA3 sml1-1 | Peter Garber, Toczyski lab |
| 4D, strain 1 | TBY409 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 ura3-1 sml1-1 RAD9-3xFLAG::hygR | This study |
| 4D, strain 2 | TBY410 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 RAD9-3xFLAG::hygR | This study |
| 4D, strain 3 | TBY411 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 RAD9-3xFLAG::hygR ddc1∆::g418R | This study |
| 4D, strain 4 | TBY412 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 sml1-1 RAD9-3xFLAG::hygR ddc1∆::g418R dpb11-1 | This study |
| 4D, strain 5 | TBY413 | Mat a ade2-1 his3-11,15 leu2-3,112 lys2 RAD53-HA::TRP1 tel1::URA3 sml1-1 RAD9-3xFLAG::hygR ddc1∆::g418R dpb11-1 | This study |
| S1, strain 1 | ADR21 | Mat a ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | Adam Rudner, A. Murray lab |
| S1, strain 2 | Mat a ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 dpb11-1 | Gift of H. Araki |
Activation of replication-checkpoint mimic
All experiments were performed at 30°C, except that shown in Figure 1C, which is described later. Cells were grown to late log phase in YM1 (Hartwell, 1967) + 2% dextrose, collected, and resuspended in YM1 + 2% raffinose and grown for 2 h. Nocodazole was added to 10 μg/ml, and cells were arrested for 3 h. An amount corresponding to 0.75 OD600 was collected and flash-frozen on dry ice for the “0-h” time point. Then 2% galactose was added to promote transcription of the LacI fusions, and cells were grown for a further hour, at which point 2% dextrose was added to shut off transcription. Further time points were collected at 3 and 5 h after galactose addition.
For Figure 1C, the protocol was as described, except that the initial parts of the experiment were performed at 23°C. After 3 h of arrest in nocodazole, cells were moved to 34°C for an additional hour before addition of galactose.
Treatment with hydroxyurea and methyl methanesulfonate
Experiments for Figures 3 and 4D were performed at 22.5°C, and experiments for Figure 4, A–C, were performed at 30°C. To cells in early log phase were added 0.2 M HU (H8627; Sigma-Aldrich, St. Louis, MO), 0.05% MMS (156890050; Acros Organics, Geel, Belgium), or medium alone. For Western blotting, cells were incubated in medium alone for 2.5 h (Figures 3 and 4D) or 1.5 h (Figure 4, A and C) and in HU or MMS for 4 h (Figures 3 and 4D), 2.5 h (Figure 4A), or 2.8 h (Figure 4C). Then pellets equivalent to an OD600 of 0.75 were collected and flash-frozen on dry ice.
For viability experiments, the same volume of cells for a given strain was plated before HU treatment and after 4 and 6 h of HU at 22.5°C (Figure 3B) or 2.5 h of HU at 30°C (Figure 4B). Colonies were counted, and the number at 4 and 6 h was divided by the number at 0 h to give the fraction of viable cells.
Western blotting and antibodies
Cell pellets were lysed in 20% trichloroacetic acid with glass beads, and protein was precipitated and resuspended in SDS sample buffer. SDS–PAGE gels to be blotted for Rad53-HA or Rad9-3xFLAG were Tris-HCl 6% acrylamide (37.5:1). All other gels were Criterion Tris-HCl 4–20% gradients (345-0034; Bio-Rad, Hercules, CA). Rad53-HA was detected with 1:1000 mouse monoclonal anti–hemagglutinin 16B12 (MMS-101P; Covance, Berkeley, CA) for Figures 1C, 2, B and C, 3, and 4, or 1:2000 rabbit anti-Rad53 (DAB001, kind gift of D. Durocher, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, ON, Canada) for Figures 1, A and B, and 2A. Rad9-3xFLAG was detected with 1:1000 mouse monoclonal anti–FLAG M2 (F3165; Sigma-Aldrich), X-GFP-LacI fusions with 1:1000 mouse monoclonal anti–GFP JL-8 (632380; Clontech, Mountain View, CA), and Cdc28 with 1:1000 goat anti–Cdc28 yC-20 (sc-6709; Santa Cruz Biotechnology, Santa Cruz, CA).
Supplementary Material
Acknowledgments
We thank H. Araki (Division of Microbial Genetics, National Institute of Genetics, Shizuoka, Japan) for the dpb11-1 mutant, D. Durocher for the anti-Rad53 antibody, and Nevan Krogan, Joachim Li, and members of the Toczyski lab for helpful conversations. This work was funded by National Institutes of Health Grant GMO59691 to D.P.T.
Abbreviations used:
- HU
hydroxyurea
- MMS
methyl methanesulfonate
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-10-0852) on February 1, 2012.
REFERENCES
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat Cell Biol. 2001;11:958–965. doi: 10.1038/ncb1101-958. [DOI] [PubMed] [Google Scholar]
- Araki H, Leem SH, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc Natl Acad Sci USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball HL, Myers JS, Cortez D. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol Biol Cell. 2005;16:2372–2381. doi: 10.1091/mbc.E04-11-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, Shirahige K. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem. 2009;284:34355–34365. doi: 10.1074/jbc.M109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla CY, Melo JA, Toczyski DP. Colocalization of sensors is sufficient to activate the DNA damage checkpoint in the absence of damage. Mol Cell. 2008;30:267–276. doi: 10.1016/j.molcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SH, Zhou H. Reconstitution of Rad53 activation by Mec1 through adaptor protein Mrc1. J Biol Chem. 2009;284:18593–18604. doi: 10.1074/jbc.M109.018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Côté J. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- Edwards RJ, Bentley NJ, Carr AM. A Rad3-Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. doi: 10.1038/15623. [DOI] [PubMed] [Google Scholar]
- Ellison V, Stillman B. Biochemical characterization of DNA damage checkpoint complexes: clamp loader and clamp complexes with specificity for 5′ recessed DNA. PLoS Biol. 2003;1:E33. doi: 10.1371/journal.pbio.0000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- Foss EJ. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics. 2001;157:567–577. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Poitelea M, Guo L, Caspari T, Carr AM. Chk1 activation requires Rad9 S/TQ-site phosphorylation to promote association with C-terminal BRCT domains of Rad4TOPBP1. Genes Dev. 2004;18:1154–1164. doi: 10.1101/gad.291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Sommariva E, Vercillo R, Lippi-Boncambi F, Liberi G, Foiani M, Plevani P, Muzi-Falconi M. A dominant-negative MEC3 mutant uncovers new functions for the Rad17 complex and Tel1. Proc Natl Acad Sci USA. 2002;99:12997–13002. doi: 10.1073/pnas.202463999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- Hartwell LH. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93 doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA, Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- Komata M, Bando M, Araki H, Shirahige K. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Mol Cell Biol. 2009;18:5008–5019. doi: 10.1128/MCB.01934-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Majka J, Binz SK, Wold MS, Burgers PM. Replication protein A directs loading of the DNA damage checkpoint clamp to 5′-DNA junctions. J Biol Chem. 2006a;281:27855–27861. doi: 10.1074/jbc.M605176200. [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM. Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc Natl Acad Sci USA. 2003;100:2249–2254. doi: 10.1073/pnas.0437148100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM. The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell. 2006b;24:891–901. doi: 10.1016/j.molcel.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JA, Cohen J, Toczyski DP. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 2001;15:2809–2821. doi: 10.1101/gad.903501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Cortez D. Activation of ATR and related PIKKs. Cell Cycle. 2008;7:2809–2812. doi: 10.4161/cc.7.18.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordes DA, Nam EA, Cortez D. Dpb11 activates the Mec1-Ddc2 complex. Proc Natl Acad Sci USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TM, Du LL, Redon C, Russell P. Histone H2A phosphorylation controlsCrb2 recruitment at DNA breaks, maintains checkpoint arrest, and influences DNA repair in fission yeast. Mol Cell Biol. 2004;14:6215–6230. doi: 10.1128/MCB.24.14.6215-6230.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J Biol Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navadgi-Patil VM, Burgers PM. The unstructured C-terminal tail of the 9-1-1 clamp subunit Ddc1 activates Mec1/ATR via two distinct mechanisms. Mol Cell. 2009;36:743–753. doi: 10.1016/j.molcel.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–1767. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciotti V, Lucchini G, Plevani P, Longhese MP. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Diffley JFX. Dpb11 coordinates Mec1 kinase activation with cell cycle-regulated Rad9 recruitment. EMBO J. 2011;30:4897–4907. doi: 10.1038/emboj.2011.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Granata M, Di Nola L, Balestrini A, Piergiovanni G, Lazzaro F, Giannattasio M, Plevani P, Muzi-Falconi M. Phosphorylation of the budding yeast 9-1-1 complex is required for Dpb11 function in the full activation of the UV-induced DNA damage checkpoint. Mol Cell Biol. 2008;28:4782–4793. doi: 10.1128/MCB.00330-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M. Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase ε. PLoS Genet. 2011;7:e1002022. doi: 10.1371/journal.pgen.1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP. Lcd1p recruits Mec1p to DNA lesions in vitro and in vivo. Mol Cell. 2002;9(4):857–869. doi: 10.1016/s1097-2765(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz MF, Duong JK, Sun Z, Morrow JS, Pradhan D, Stern DF. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol Cell. 2002;9:1055–1065. doi: 10.1016/s1097-2765(02)00532-4. [DOI] [PubMed] [Google Scholar]
- Segurado M, Diffley JF. Separate roles for the DNA damage checkpoint protein kinases in stabilizing DNA replication forks. Genes Dev. 2008;22:1816–1827. doi: 10.1101/gad.477208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Pasero P, Gasser SM. ORC and the intra-S-phase checkpoint: a threshold regulates Rad53p activation in S phase. Genes Dev. 2002;16:3236–3252. doi: 10.1101/gad.239802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Hsiao J, Fay DS, Stern DF. Rad53FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D. Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol. 2005;15:1364–1375. doi: 10.1016/j.cub.2005.06.063. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–1336. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- Tourrière H, Pasero P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amst) 2007;6:900–913. doi: 10.1016/j.dnarep.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Usui T, Foster SS, Petrini JH. Maintenance of the DNA-damage checkpoint requires DNA-damage-induced mediator protein oligomerization. Mol Cell. 2009;33:147–159. doi: 10.1016/j.molcel.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vialard JE, Gilbert CS, Green CM, Lowndes NF. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Elledge SJ. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Elledge SJ. Genetic and physical interactions between DPB11 and DDC1 in the yeast DNA damage response pathway. Genetics. 2002;160:1295–1304. doi: 10.1093/genetics/160.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA. 2003;100:13827–13832. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.