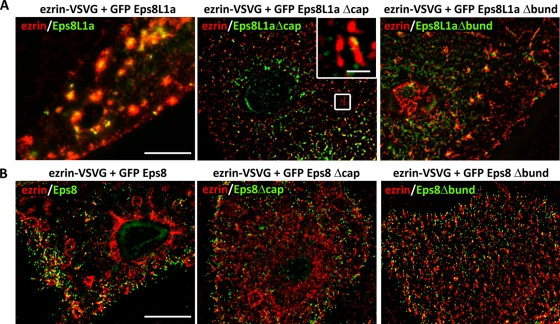
The membrane cytoskeleton linker ezrin differentially regulates the activity of Eps8 and Eps8L1a in microvillar actin-F assembly. Eps8L1a displays F-actin capping activity, therefore controlling microvillus length, whereas, as previously shown, Eps8 displays bundling activity.
Abstract
The mechanisms that regulate actin filament polymerization resulting in the morphogenesis of the brush border microvilli in epithelial cells remain unknown. Eps8, the prototype of a family of proteins capable of capping and bundling actin filaments, has been shown to bundle the microvillar actin filaments. We report that Eps8L1a, a member of the Eps8 family and a novel ezrin-interacting partner, controls microvillus length through its capping activity. Depletion of Eps8L1a leads to the formation of long microvilli, whereas its overexpression has the opposite effect. We demonstrate that ezrin differentially modulates the actin-capping and -bundling activities of Eps8 and Eps8L1a during microvillus assembly. Coexpression of ezrin with Eps8 promotes the formation of membrane ruffles and tufts of microvilli, whereas expression of ezrin and Eps8L1a induces the clustering of actin-containing structures at the cell surface. These distinct morphological changes are neither observed when a mutant of ezrin defective in its binding to Eps8/Eps8L1a is coexpressed with Eps8 or Eps8L1a nor observed when ezrin is expressed with mutants of Eps8 or Eps8L1a defective in the actin-bundling or -capping activities, respectively. Our data show a synergistic effect of ezrin and Eps8 proteins in the assembly and organization of actin microvillar filaments.
INTRODUCTION
Intestinal and renal absorptive epithelial cells display at their luminal surface a brush border formed of densely packed microvilli that are uniform in length and diameter. Microvilli are membrane protrusions each supported by a bundle of actin filaments that are linked laterally to the membrane and anchored to the tip of the microvilli. A specific repertoire of actin-binding proteins that participate in the assembly and dynamics of these highly organized structures has been characterized (Revenu et al., 2004). It includes proteins responsible for the organization of actin filaments into bundles and those that tether actin filaments to the membrane. However, the factors that trigger and control actin polymerization resulting in the assembly of microvilli with uniform length are not known.
The membrane–cytoskeleton linker ezrin is an essential component in the morphogenesis of the apical domain of epithelial cells (Crepaldi et al., 1997; Saotome et al., 2004; Tamura et al., 2005). Ezrin−/− mice show defects in the terminal web and the brush border of intestinal microvilli that are irregular in length and shape (Saotome et al., 2004). Ezrin belongs to the band 4.1 superfamily, whose members share a common domain named the FERM domain (four-point one, ezrin/radixin/moesin). Ezrin, as well as the closely related proteins radixin and moesin, provides a regulated linkage between the plasma membrane and the actin cytoskeleton. These proteins are composed of an ∼300–amino acid FERM domain in the amino-terminus, followed by a region rich in α-helices, and a carboxy-terminal domain that contains an F-actin–binding site. The binding of ezrin to the membrane and to the actin cytoskeleton is strictly regulated. The protein exists in the cytoplasm in an inactive conformation due to an intramolecular interaction between the FERM domain and the ∼80 carboxy-terminal amino acids (also called N-/C-ERM association domains; Gary and Bretscher, 1995). The relief of this intramolecular association is triggered by the sequential binding of the FERM domain to the phosphoinositides and the phosphorylation of a conserved threonine residue present in the F-actin–binding site (T567 in ezrin; Fiévet et al., 2004). On activation, the FERM domain of ezrin can bind to the cytoplasmic tail of transmembrane proteins and/or to the scaffolding proteins of the Na+–H+ exchanger regulatory factor (NHERF) family (Fehon et al., 2010), whereas the C-terminal domain interacts with actin filaments (Turunen et al., 1994). Through its interaction with its numerous partners, ezrin has the ability to organize multiprotein complexes, therefore participating in the organization and function of specific domains of the plasma membrane (Arpin et al., 2011).
How ezrin contributes to microvillar morphogenesis is not understood. Ezrin recruitment to the apical surface of epithelial cells, with its concomitant phosphorylation at the regulatory threonine 567, represents an essential step in microvillar biogenesis (Yonemura et al., 1999; Gautreau et al., 2000; ten Klooster et al., 2009). In this activated state, ezrin presumably interacts with components that participate in processes essential for brush border assembly. Thus ezrin interaction with ERM-binding phosphoprotein-50 (EBP50, also known as NHERF1) is required for microvillar assembly and maintenance (Hanono et al., 2006; Garbett et al., 2010; Lalonde et al., 2010). Indeed, expression of an EBP50 construct lacking the ezrin binding site in cells in which endogenous EBP50 is knocked down does not restore normal microvilli, whereas expression of wild-type EBP50 does (Hanono et al., 2006; Garbett et al., 2010).
Ezrin is the target of receptor and nonreceptor tyrosine kinases (Krieg and Hunter, 1992; Fazioli et al., 1993b; Crepaldi et al., 1997; Heiska and Carpen, 2005). Its phosphorylation at specific tyrosine residues triggers the formation of multimolecular complexes, and therefore we searched for partners of phosphorylated ezrin. We identified epidermal growth factor receptor substrate 8–like (Eps8L) proteins, which are members of the Eps8 family, as partners of phosphorylated ezrin. In mammals, the Eps8 family comprises four genes coding for Eps8 and three Eps8-like proteins (L1–L3; Fazioli et al., 1993a; Tocchetti et al., 2003; Offenhauser et al., 2004). In addition, at least three Eps8L1 variants (L1a–c) that arise from alternative splicing have been identified. Eps8 and Eps8L1a display the same modular organization. They contain a phosphotyrosine-binding (PTB) domain at the N-terminus and an SH3 domain, followed by a C-terminus, actin binding domain or effector domain (Figure 1A). Eps8 transduces signals elicited by growth factor, leading to actin cytoskeleton remodeling (Scita et al., 1999). The Eps8 effector domain has been shown to exert both actin filament barbed-end capping and bundling activities (Croce et al., 2004; Disanza et al., 2004, 2006; Hertzog et al., 2010). Genetic analyses indicated that the actin bundling but not the capping activity of Eps8 is essential for the proper organization of intestinal microvilli (Croce et al., 2004; Tocchetti et al., 2010). A capping activity has also been described in vitro for Eps8-like proteins (Disanza et al., 2004). Of interest, the capping/bundling activities are masked in the full-length protein, and it has been proposed that protein–protein interaction might relieve this autoinhibition (Scita et al., 2001; Disanza et al., 2006; Manor et al., 2011). For example, Abi1 has been shown to modulate Eps8-dependent actin dynamics (Disanza et al., 2004; Roffers-Agarwal et al., 2005).
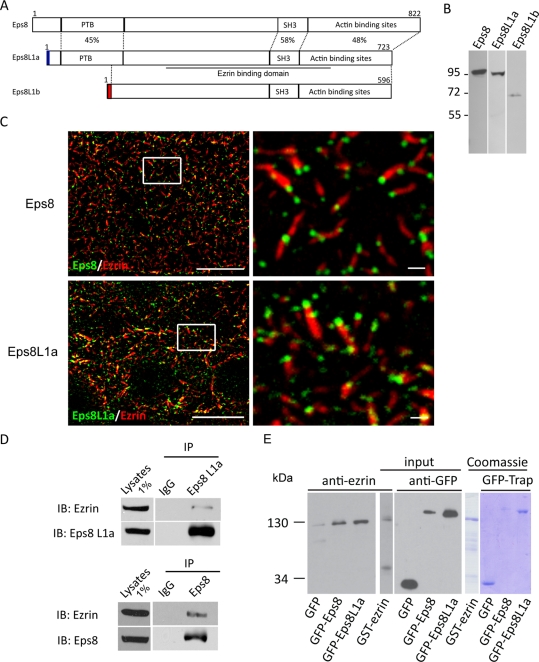
FIGURE 1:
Eps8 and Eps8L1a are components of the brush border microvilli. (A) Schematic representation of the structure of Eps8 and Eps8L1a and b isoforms. The percentage of identity of Eps8 domains with that of Eps8L1a and b is indicated. The position of the peptides used to generate antibodies against Eps8L1a and b is represented by the blue and red boxes, respectively. The region of Eps8L1a and b found to interact with ezrin in the two-hybrid screen is underlined. (B) Western blot was performed on LLC-PK1 cell extracts using anti-Eps8, anti-Eps8L1a, and anti-Eps8L1b antibodies. (C) Localization of Eps8 proteins in LLC-PK1 cells. Double immunofluorescence was performed with an anti-ezrin antibody (red) and either anti-Eps8 (top) or Eps8L1a (bottom) antibodies (green). Images were taken with a three-dimensional wide-field optical sectioning microscope. The images on the left correspond to a maximum-intensity Z projection, performed after deconvolution, of 10 slices starting from the top of the cells. Bar, 10 μm. Microvilli in the rectangles are shown at high magnification on the right. Bar, 1 μm. (D) Immunoprecipitation was performed with control immunoglobulin G (IgG) or anti-Eps8L1a (top) and anti-Eps8 (bottom) antibodies, respectively, from LLC-PK1 cell lysates. Thirty percent of the immunoprecipitates were loaded on the gel. The Western blot performed with an anti-ezrin antibody indicates that ezrin coimmunoprecipitates with Eps8 and Eps8L1a. (E) In vitro interaction of GFP-Eps8 or GFP-Eps8L1a with ezrin. GFP, GFP-Eps8, or GFP- Eps8L1a bound to GFP-Trap_A beads were incubated with purified GST-ezrin. Left, the immunoblot performed with the anti-ezrin antibody shows that ezrin interacts with Eps8 and Eps8L1a but not with GFP alone. One-fifth of the beads were loaded on the gel. Middle, 1/300 and 1/25 of purified GST-ezrin and GFP proteins used for the interaction were loaded on the gel and analyzed with anti-ezrin and anti-GFP antibodies, respectively. Right, Coomassie staining of purified proteins.
Whereas Eps8 functions have been studied in detail in vitro and in vivo, no studies have been performed on Eps8-like proteins. Here, we investigate the role of ezrin/Eps8 protein interaction in microvillus morphogenesis. We show that ezrin positively regulates the activities of these proteins through its interaction with their SH3 domain. Of importance, we report that Eps8 and Eps8L1a exert distinct activities—actin bundling and capping, respectively—in the organization of the brush border microvilli.
RESULTS
Ezrin interacts with Eps8 and Eps8-like proteins
To identify proteins that preferentially bind to ezrin phosphorylated by Src family kinases, we performed a modified yeast two-hybrid screen using yeast strains transformed or not with the Lyn kinase (Naba et al., 2008). A truncated form of ezrin deleted of the last 52 amino acids was used as bait since most of the partner binding sites are masked in full-length ezrin. In this screen, a fragment corresponding to an amino acid sequence of Eps8L1 was found to interact with phosphorylated ezrin. The sequence isolated in the two-hybrid screen is common to Eps8L1a (amino acids 258–607) and Eps8L1b (amino acids 131–480) but not to Eps8L1c. Eps8L1a and b share the same primary sequence except in their N-terminal domains. Eps8L1a displays a 126–amino acid extension that comprises a PTB domain absent in Eps8L1b. The sequence of the first 16 N-terminal amino acids of Eps8L1b diverges from the corresponding sequence in Eps8L1a (Figure 1A).
To further analyze the expression and localization of these proteins, we generated antibodies specific for Eps8L1a and Eps8L1b. These antibodies were tested in LLC-PK1 cells, a nontransformed porcine cell line derived from the kidney proximal tubule. These cells display brush border microvilli at their apical surface that have in common with intestinal microvilli the same highly organized arrays of actin filaments and share most of the actin-binding proteins. All three proteins—Eps8 and Eps8L1a and b—are expressed in LLC-PK1 cells. The anti-Eps8L1a and anti-Eps8L1b antibodies each recognized a single band of the expected molecular weight (Figure 1B). The specificity of our anti-Eps8L1a antibody was further confirmed by immunofluorescence performed with cells depleted for this protein (see later discussion of Figure 7). Because the localization of endogenous Eps8L1 proteins had not been reported, we performed immunofluorescence on LLC-PK1 cells with our specific antibodies, as well as with the anti-Eps8 antibody. As observed previously in intestinal cells from Caenorhabditis elegans (Croce et al., 2004), Eps8 is concentrated at the tip of the microvilli (Figure 1C). Of interest, Eps8L1a is also present at the tip of the microvilli (Figure 1C). Unlike Eps8 proteins, ezrin is distributed along the microvilli but seems to be excluded from the tip. A colocalization of ezrin with Eps8 proteins could only be observed in a restricted area—at the interface between the tip and the microvillus. In LLC-PK1 cells, Eps8L1b is absent from the microvilli but present on vesicular structures (data not shown). Because we were interested in microvillus morphogenesis, we concentrated our studies on Eps8L1a and Eps8.
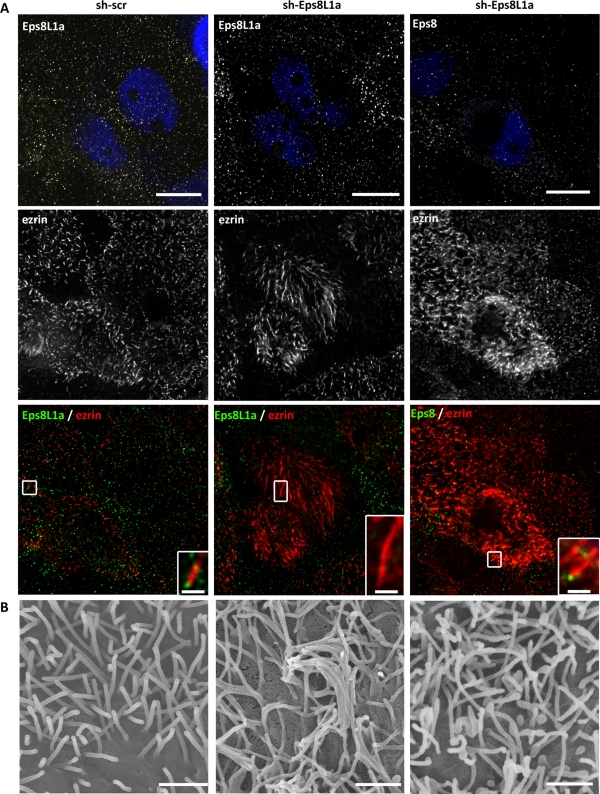
FIGURE 7:
Depletion of Eps8L1a induces the formation of long microvilli in LLC-PK1 cells. Cells transfected with plasmids coding the reporter gene GFP and shRNA, nontargeting (scr; left) or targeting Eps8L1a (middle and right) were analyzed by immunofluorescence. Transfected cells were monitored through the expression of GFP (artificially rendered in blue). Immunofluorescence was performed with anti-ezrin (red) and anti-Eps8L1a and anti-Eps8 (green) antibodies. Images correspond to a maximum-intensity Z-projection performed after deconvolution. Eps8 is still localized at the tip of the microvilli (right). Bars, 10 μm; inset, 1 μm. (B) CL-SEM shows microvilli of cells expressing scramble (left) or Eps8L1a shRNA. Bar, 1 μm. Both immunofluorescence and CL-SEM indicate that depletion of Eps8L1a leads to the formation of long microvilli.
To confirm the interaction of ezrin with Eps8L1a, we performed an immunoprecipitation with the anti-Eps8L1a antibody, and we observed a coimmunoprecipitation of endogenous ezrin (Figure 1D, top). The Eps8L1 ezrin-binding domain identified in the two-hybrid screen comprises the SH3 domain, which is well conserved in Eps8 (58% identity). We thus tested whether ezrin interacts with Eps8 as well, and we found that endogenous ezrin interacted with Eps8 (Figure 1D, bottom). We then determined whether green fluorescent proteins GFP-Eps8/GFP-Eps8L1a purified from mammalian cells using the GFP-Trap_A assay interact with recombinant ezrin (Figure 1E). Glutathione S-transferase (GST)–ezrin was found to interact with GFP-Eps8 and GFP-Eps8L1a but not with GFP alone, suggesting that the association between ezrin and Eps8/Eps8L1a is direct (Figure 1E).
Mapping the interacting domains reveals a distinct mode of association between ezrin and Eps8 or Eps8L1a
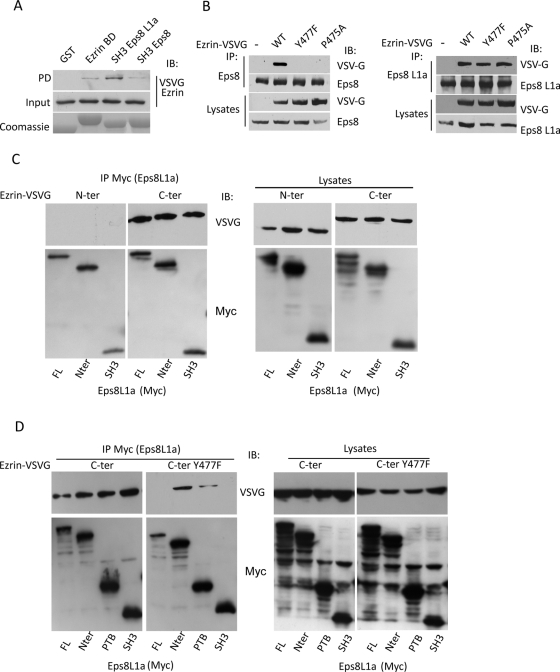
We sought to dissect the mechanisms of interaction between ezrin and Eps8 and Eps8L1a. To do so, we performed pull-down experiments with the ezrin-binding domain of Eps8L1a or the SH3 domain of either Eps8L1a or Eps8 fused to GST and lysates of cells expressing vesicular stomatitis virus glycoprotein (VSVG)-tagged ezrin. As shown in Figure 2A, ezrin interacted with the ezrin-binding domain identified in the two-hybrid screen and more precisely with the SH3 domains of Eps8L1a. The ezrin/Eps8 interaction was also mediated by Eps8 SH3 domain. The SH3 domain of Eps8 has been shown to interact preferentially with the consensus motif PXXDY or PPPY/W (Mongiovi et al., 1999). Because ezrin contains a PPPVY477 motif, we determined whether this motif was involved in the binding of ezrin to Eps8 and Eps8L1a. LLC-PK1 cells stably expressing either wild-type ezrin or the mutants of ezrin Y477F or P475A were used to test the capacity of these different forms to interact with endogenous Eps8 and Eps8L1a. As shown in Figure 2B, Eps8 interaction with ezrin mutants Y477F or P475A was completely abolished, indicating that the PPPVY477motif in ezrin is indeed the binding site for Eps8. On the contrary, Eps8L1a could still interact with ezrin P475A or Y477F, suggesting that either the PPPVY477 motif in ezrin was not involved in the binding to Eps8L1a or that there was a second binding site in ezrin that could mediate its interaction with Eps8L1a. To test these hypotheses, we coexpressed in 293T cells either the amino- or carboxy-terminal domains of ezrin with Myc-tagged, full-length Eps8L1a or its various domains and performed an immunoprecipitation with an anti-Myc antibody. No interaction of the amino-terminal domain of ezrin was observed with Eps8L1a full length or any of its domains (Figure 2C). The carboxy-terminal domain of ezrin was found to interact with both the amino-terminal domain of Eps8L1a and the SH3 domain (Figure 2C). Further mapping indicated that the PTB domain of Eps8L1a interacts with ezrin carboxy-terminal domain (Figure 2D). Moreover, we demonstrated that Eps8L1a SH3 domain interacted with the PPPVY477 motif in ezrin, as the introduction of the Y477F mutation in the carboxy-terminal domain of ezrin inhibited the interaction with the SH3 domain of Eps8L1a (Figure 2D). Of interest, the second interaction site mediated by the PTB domain of Eps8L1a was not affected by the introduction of the Y477F mutation in ezrin carboxy-terminal domain. Taken together these data indicated that Eps8 and ezrin interact through the consensus PPPVY477 motif of ezrin and the SH3 domain of Eps8. In contrast, two binding sites mediate the interaction between Eps8L1a and ezrin: one binding site involved the SH3 domain of Eps8L1a and the PPPVY477 motif in ezrin, and the other one the Eps8L1a amino-terminal domain and the ezrin carboxy-terminal domain.
FIGURE 2:
Mapping of interaction sites between ezrin and Eps8 proteins. (A) Immobilized GST or GST fused to ezrin-binding domain on Eps8L1a (BD), the SH3 domain of Eps8L1a, and the SH3 domain of Eps8 were incubated with lysates of LLC-PK1 cells expressing ezrin-VSVG. Western blot was performed with an anti-VSVG antibody. Bottom, the Coomassie staining of GST-fused proteins. (B) Left, immunoprecipitation performed with an anti-Eps8 antibody on lysates of LLC-PK1 cells stably transfected with the empty vector (–) or with cDNA coding ezrin wild type (WT), ezrinY477F, or P475A tagged with VSVG. Immunoblots were performed with an anti-VSVG antibody to detect ezrin. Eps8 interacts only with wild-type ezrin. Right, the immunoprecipitation was performed as before, but with an anti-Eps8L1a antibody. Eps8L1a interacts with wild-type ezrin, as well as with ezrin Y477F and ezrin P475A. (C) 293T cells were cotransfected with cDNA coding the VSVG-tagged carboxy-terminal (C-ter) or amino-terminal (N-ter) domains of ezrin and Myc-tagged full-length Eps8L1a (FL) or its various domains. Left, immunoprecipitation performed with an anti-Myc antibody, followed by an immunoblot on immunoprecipitated proteins with anti-Myc and anti-VSVG antibodies. Right, cell lysates analyzed with anti-VSVG and anti- Myc antibodies. (D) 293T cells were cotransfected with cDNA coding the VSVG tagged carboxy-terminal domain of ezrin carrying or not the mutation Y477F and Myc-tagged full-length Eps8L1a or its various domains. Left, immunoprecipitation performed with an anti-myc antibody, followed by Western blots with anti-VSVG and anti-Myc antibodies. Right, cell lysates analyzed with anti-VSVG and anti-Myc antibodies. The introduction of the Y477F mutation in the carboxy-terminal domain of ezrin abolishes the interaction of Eps8L1a SH3 domain with ezrin.
Ezrin and Eps8 proteins cooperate in the control of microvillus formation
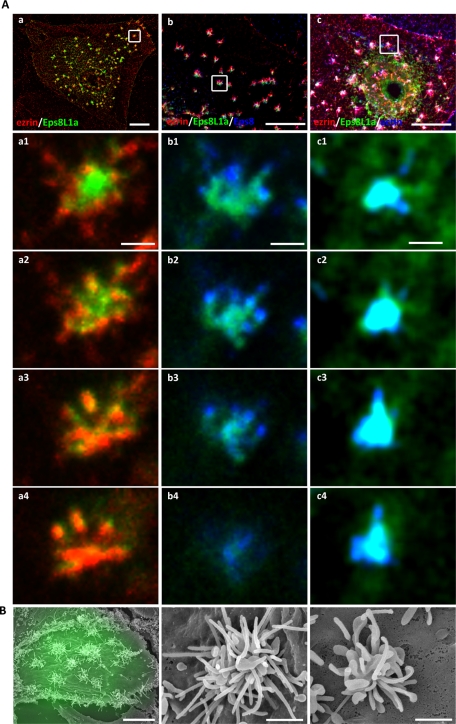
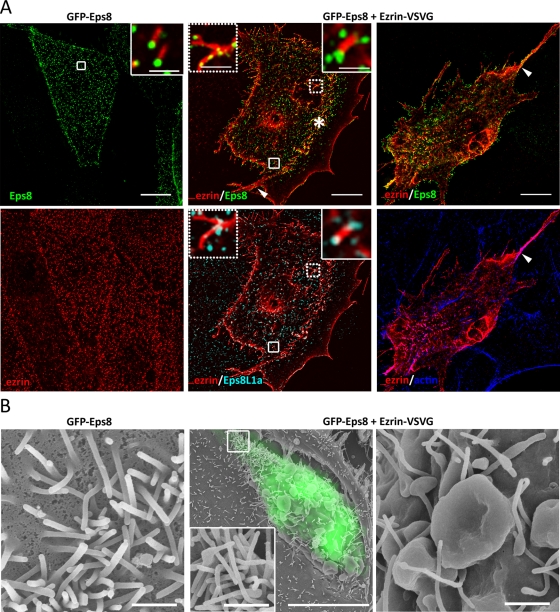
Eps8 displays multiple functions in actin filament organization presumably through its interaction with distinct actin-containing regulatory complexes (Croce et al., 2004; Disanza et al., 2004; Roffers-Agarwal et al., 2005). However, such a requirement for interacting proteins in Eps8L1a activities has not been established. To test the role of ezrin and Eps8/Eps8L1a interaction in the formation of the microvilli, we overexpressed either Eps8L1a or Eps8 alone or together with ezrin in LLC-PK1 cells. Cells were analyzed by immunofluorescence and correlative light scanning electron microscopy (CL-SEM). For CL-SEM, in the first instance we performed immunofluorescence to detect cells coexpressing ezrin-VSVG and GFP-Eps8 or GFP-Eps8L1a. Because morphological changes were observed only when both ezrin and either Eps8 or Eps8L1a were coexpressed, we subsequently monitored transfected cells only through the expression of GFP-Eps8 or GFP-Eps8L1a to avoid damaging cell membrane by permeabilization. The overexpression of ezrin-VSVG or -GFP tagged alone had no striking effect on microvillar morphology (Figure 3, A and B, left) as compared with untransfected cells or cells expressing GFP alone (Figure 3, bottom). The expression of Eps8L1a led to an important reduction in the microvillus length as observed by immunofluorescence and CL-SEM (Figure 3, A and B, right). About 70% of cells expressing Eps8L1a displayed shorter microvilli as assessed by immunofluorescence, suggesting that Eps8L1a prevents microvillus elongation. In addition, the width of the microvilli measured from SEM images was increased by ∼10% in cells expressing Eps8L1a as compared with cells expressing GFP (112.4 ± 2.000 nm vs. 104.0 ± 1.499 nm, respectively). It is striking that coexpression of ezrin and GFP-Eps8L1a induced morphological changes at the apex of the cells characterized by the clustering of actin-containing structures in which ezrin, Eps8L1a, and Eps8 were present (Figure 4, A and B). This clustering was observed in nearly 100% of cells coexpressing both proteins. Close examination of the clusters by SEM indicated that they comprise enlarged membrane extensions. Microvilli emerged from some but not from all of these enlarged structures. Moreover, the number of microvilli that form in these structures varied from one cell to another, likely due to different levels of Eps8L1a expression (Figure 4B). A strong staining of Eps8L1a and actin was observed by immunofluorescence at the base of the clusters, presumably in the enlarged structures observed by SEM (Figure 4A, a1–a4 and c1–c4). Although ezrin and Eps8 colocalized with Eps8L1a, both proteins were more abundant in the microvilli that originate from the enlarged structures (Figure 4A, a1–a4 and b1–b4). A faint staining of Eps8L1a was also observed along the microvilli.
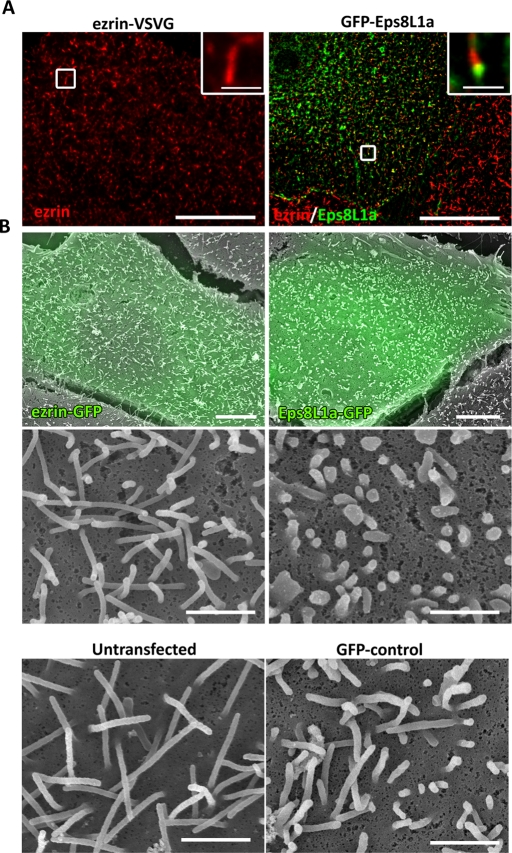
FIGURE 3:
Expression of Eps8L1a blocks microvillus elongation. (A) Immunofluorescence was performed on LLC-PK1 cells expressing ezrin-VSVG (left) or GFP-Eps8L1a (right). Ezrin (in red) was detected with an anti-VSVG antibody. Images were taken as in Figure 1B and correspond to a maximum-intensity Z-projection performed after deconvolution. Insets show the localization of ezrin along the microvilli (left) and Eps8L1a at the tip (right). Bar, 10 μm; insets, 1 μm. (B) CL-SEM was performed on cells expressing ezrin-GFP or GFP-Eps8L1a. Transfected cells (in green) were tracked by fluorescence and subsequently analyzed by scanning electron microscopy. The size of the microvilli is considerably reduced in cells expressing Eps8L1a as compared with that of cells expressing ezrin-GFP or of untransfected cells (bottom). Bars, first row,10 μm; others, 1 μm.
FIGURE 4:
Coexpression of ezrin and Eps8L1a induces microvillar clustering at the apical surface of LLC-PK1 cells. LLC-PK1 cells were cotransfected with plasmids coding for ezrin-VSVG and GFP-Eps8L1a. (A) Immunofluorescence was performed with an anti-VSVG antibody to detect ezrin (a, b, c, a1–a4; red) and an anti-Eps8 antibody (b, b1–b4; blue). Actin was detected with phalloidin (c, c1–c4; blue). Images correspond to a maximum-intensity Z-projection performed after deconvolution. Bottom, four successive slices of the clusters in the insets from a–c are shown from the bottom (a1, b1, c1) to the top of the structure (a4, b4, c4). Eps8L1a is mainly localized in the enlarged membrane structures (see scanning electron microscopy) and ezrin in the microvilli that originate from them. Bars, a–c, 10 μm; a1–c4, 1 μm. (B) CL-SEM. Low (left) and high (middle and right) magnification of the surface of LLC-PK1 cells expressing ezrin-VSVG and GFP-Eps8L1a. Clusters from two different cells are shown at high magnification. Transfected cells are monitored by fluorescence through GFP fluorescence. Bars, left, 10 μm; middle and right, 1 μm.
Distinct morphological effects were induced when we overexpressed Eps8 with ezrin. In cells overexpressing Eps8 alone, the microvilli appear similar to that of cells expressing GFP alone as observed by CL-SEM (Figures 3B and 5B). The major phenotype observed in most cells when ezrin was coexpressed with Eps8 was the formation of circular dorsal ruffles and membrane waves on the dorsal surface of the cells (Figure 5, A and B). Moreover, arc-shaped ridges covered with abundant microvilli were observed at the edge of the cells (∼30% of cells; Figure 5A, asterisk), as well as bundles of actin filaments (∼40% of cells; Figure 5A, arrowhead). In 60% of cells, tufts of microvilli were detected mostly at the cell periphery when ezrin was coexpressed with Eps8. Both Eps8L1a and Eps8 remained localized at the tip of the microvilli in cells coexpressing ezrin and Eps8 and gave a punctate staining in the membrane ruffles (Figure 5A, middle, insets).
FIGURE 5:
Coexpression of ezrin and Eps8 induces the formation of membrane ruffles and tufts of microvilli at the apical surface of LLC-PK1 cells. (A) Immunofluorescence of cells expressing GFP-Eps8 alone or GFP-Eps8 and ezrin-VSVG. Images correspond to a maximum-intensity Z-projection performed after deconvolution. Dorsal circular ruffles, arc-shaped ridges covered with abundant microvilli (asterisk, middle), and actin bundles (arrow, right) are observed in cells expressing both proteins. Insets (right of the middle) show the localization of Eps8 and Eps8L1a at the tip of the microvilli. A punctate staining of Eps8 and Eps8L1a is observed in membrane ruffles (left insets, middle). Bars, 10 μm; inset, 1 μm. (B) CL-SEM. Left, microvilli of cells overexpressing GFP-Eps8. Middle and right, low and high magnification of the surface of cells expressing ezrin and GFP-Eps8. Extensive dorsal ruffles are observed. The inset shows the tufts of microvilli observed at the edge of the cells. Bars, left, 1 μm; middle, 10 μm (inset, 1 μm); right, 1 μm.
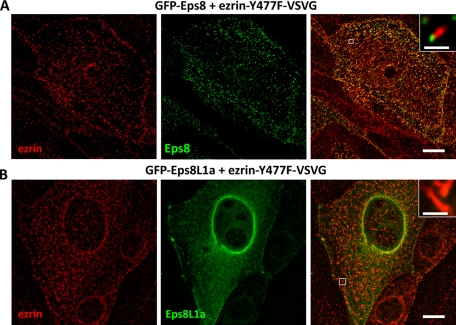
To confirm that the ezrin/Eps8 proteins interaction was responsible for the observed morphological changes, we coexpressed ezrin Y477F with Eps8L1a or Eps8 since both proteins interact with ezrin via the PPPVY477 motif. The morphological changes detected when wild-type ezrin was coexpressed with either Eps8 or Eps8L1a were rarely observed with ezrin Y477F. In both cases, transfected cells displayed microvilli at their apical surface. However, whereas Eps8 was still present at the tip of the microvilli, Eps8L1a was not and remained diffuse in the cytoplasm. Taken together, these experiments indicated that ezrin binding to Eps8/Eps8L1a promotes their activities, resulting in the morphological changes described (Figure 6, A and B). Moreover, the interaction of Eps8L1a with the PPPVY477 motif in ezrin is critical for its activities and localization.
FIGURE 6:
Ezrin Y477F fails to promote Eps8 protein activities. Immunofluorescence was performed on LLC-PK1 cells coexpressing ezrin Y477F and (A) GFP-Eps8 or (B) GFP-Eps8L1a with an anti-VSVG antibody to detect ezrin (red). No morphological changes are observed in cells expressing both proteins. (A) Eps8 remains localized at the tip of the microvilli (inset, right). (B) Eps8L1a is diffuse in the cytoplasm and absent from the tip of the microvilli (inset, right). Bars, 10 μm; inset, 1 μm.
Eps8 and Eps8L1a play distinct roles in microvillar assembly
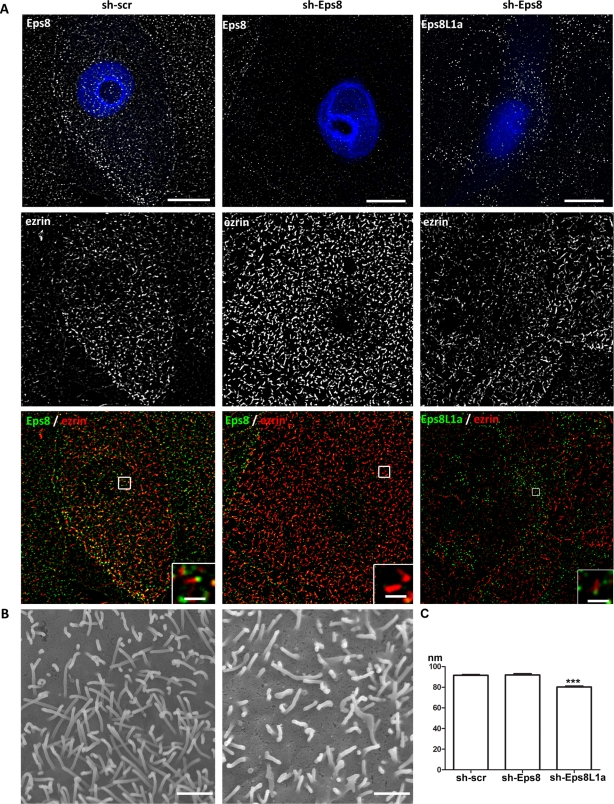
The foregoing coexpression experiments suggested that Eps8 and Eps8L1a play distinct roles in the organization of the microvillar actin cytoskeleton. To confirm these observations, we inhibited the expression of either Eps8L1a or Eps8. The depletion of Eps8/Eps8L1a was performed with two independent short hairpin RNAs (shRNAs) in all experiments and gave identical results. Cells transfected with the plasmids encoding the shRNA were monitored through the expression of GFP. Efficient depletion of Eps8L1a or Eps8 was achieved, as shown by an almost complete disappearance of Eps8L1a and Eps8 staining (Figures 7 and 8). Cells depleted for Eps8L1a exhibited very long microvilli as compared with cells expressing a nontargeting shRNA (scrambled sequence [scr]; Figure 7A). This observation was confirmed by CL-SEM, which showed that the cells were covered with very long and bent microvilli that were thinner than the microvilli observed at the surface of cells producing scrambled shRNA (Figures 7B and 8C). No changes in the localization of either ezrin or Eps8 were observed in these microvilli, with ezrin being distributed along the microvilli and Eps8 at their tip (Figure 7A). On the contrary, Eps8 depletion did not reveal striking effects on microvilli structure as observed by immunofluorescence (Figure 8A). However, CL-SEM indicated that the microvilli were more irregular in shape and length as than the microvilli of cells transfected with the scramble shRNA (Figure 8B). In absence of Eps8, Eps8L1a was still localized at the tip of the microvilli (Figure 8A, right). Finally, we sought to analyze the phenotype of cells after depletion of both Eps8 and Eps8L1a. Cells were transfected with plasmids coding shRNA targeting Eps8L1a/Eps8 and the reporter genes mCherry and GFP, respectively. Cells expressing both shRNAs were rounded, suggesting a defect in cell adhesion (Supplemental Figure). Furthermore, their surface displayed abundant membrane ruffles and heterogeneous microvilli. We observed in several cells very long microvilli often localized at the center of the cells, whereas abundant shorter microvilli were seen at the junction between the cells. It is worth mentioning that, in LLC-PK1 cells, Eps8 but not Eps8L1a is present at the cell–cell junctions. These observations might be an indication that the functional requirements for Eps8 and Eps8L1a are not the same in different regions of the cells.
FIGURE 8:
LLC-PK1 cells depleted for Eps8 display irregular microvilli at their apical surface. (A) Immunofluorescence and (B) CL-SEM were performed on cells expressing shRNA scr (left) or targeting Eps8 (middle and right). Transfected cells were detected through the expression of GFP (artificially rendered in blue). Eps8L1a localizes to the tip of the microvilli (right). Bars, 10 μm; inset, 1 μm. (B) CL-SEM indicates that cells depleted for Eps8 display shorter and more irregular microvilli. Bar, 1 μm. (C) Graph showing the width of the microvilli determined from CL-SEM images of cells transfected with Scr, Eps8, or Eps8L1a psiRNA. The values in the histogram are means ± SE. Statistical significance was determined by one-way analysis of variance with a Dunnett multiple comparison test. The width (nm) of the microvilli in cells depleted of Eps8L1a is significantly thinner than in cells transfected with nontargeting or Eps8-targeting shRNA, with p < 0.005.
The capping activity of Eps8L1a and the bundling activity of Eps8 are involved in microvillar actin filament assembly
Both the depletion of Eps8L1a and the coexpression of ezrin and Eps8L1a strongly suggested that Eps8L1a displayed an actin-capping activity in microvillar actin cytoskeleton assembly. Moreover, the formation of tufts of microvilli when ezrin was coexpressed with Eps8 suggested that Eps8 actin bundling activity plays a role in the assembly of microvilli as previously described in animal models (Croce et al., 2004; Tocchetti et al., 2010). Recently the domains of Eps8 involved in actin filament-capping and -bundling activities have been mapped and validated by mutagenesis (Hertzog et al., 2010). We introduced the mutations that abrogate capping or bundling activities in human Eps8 (V690D/L694D; Eps8Δcap) and (L758A/K760A; Eps8Δbund). So far only the capping activity of Eps8L1a has been demonstrated in vitro (Disanza et al., 2004). We observed that the motifs identified as critical for the capping and bundling activities of Eps8 are conserved in Eps8L1a. Therefore we introduced mutations in Eps8L1a (V601D/L605D; Eps8L1aΔcap) and (L667A/K669A; Eps8L1a Δbund). To determine which of these activities were involved in the observed phenotypes, we coexpressed ezrin with Eps8 or Eps8L1a in which the capping or the bundling activity was suppressed. Coexpression of ezrin with Eps8L1aΔcap prevented the clustered phenotype observed upon expression of wild-type Eps8L1a, and the surface of the cells was covered with microvilli (Figure 9A). It is striking that these clustered structures were still observed when ezrin was coexpressed with Eps8L1aΔBund, although they were less abundant (Figure 9A). Of interest Eps8L1aΔcap was not localized to the tip of the microvilli but rather at the microvilli side. This suggests that Eps8L1a interacts, through its capping site, with the actin filament barbed ends (Figure 9, A and B).
FIGURE 9:
Actin-capping and -bundling activities of Eps8L1a and Eps8, respectively, in microvillus assembly. (A) LLC-PK1 cells were transfected with plasmids coding ezrin and GFP-Eps8L1a (left), ezrin and Eps8L1a mutated in its capping site (Eps8L1a∆cap, middle), or ezrin and Eps8L1a mutated in its actin-bundling site (Eps8L1a∆bund, right). Clusters are observed in cells expressing ezrin with either Eps8L1 wild type or Eps8L1aΔbund but not with Eps8L1aΔcap. Inset in the middle shows that Eps8L1a∆cap is localized at the microvilli side and rarely at their tip as observed with Eps8L1a. Bars, 10 μm; inset, 1 μm. (B) LLC-PK1 cells were transfected with plasmids coding ezrin and GFP-Eps8 (left), ezrin and GFP-Eps8 mutated in its capping site (middle, Eps8Δcap), or ezrin and Eps8 mutated in its actin-bundling site (right, Eps8Δbund). Circular dorsal ruffles are observed on the surface of the cells coexpressing ezrin and Eps8 or Eps8Δcap but not in cells expressing ezrin and Eps8Δbund. Bars, 10 μm; inset, 1 μm.
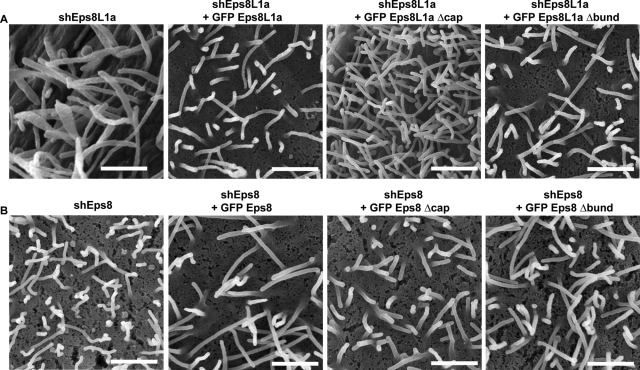
Consistent with the formation of long microvilli following depletion of Eps8L1a or short microvilli upon expression of Eps8L1a, these experiments suggested that the capping activity of Eps8L1a was involved in the control of the microvillus length. To further confirm that Eps8L1a controls the microvillus length through its capping activity, we sought to rescue the defects observed in LLC-PK1 cells after depletion of Eps8L1a by expressing wild-type Eps8L1a, Eps8L1aΔcap, or Eps8L1aΔbund (Figure 10A). Whereas microvilli with normal length were observed by CL-SEM and immunofluorescence (data not shown) in cells depleted for endogenous Eps8L1a and expressing shRNA-resistant, wild-type human Eps8L1a or Eps8L1aΔbund, long microvilli were still observed in the cells expressing Eps8L1aΔcap. Quantification of these phenotypes by immunofluorescence indicated that ∼85% of the cells depleted of Eps8L1a display long microvilli. This number dropped to ∼20% in depleted cells expressing wild-type Eps8L1a, whereas 60% of cells expressing Eps8L1aΔcap had long microvilli. Together these data converge toward a role for Eps8L1a's capping activity in the control of microvilli morphogenesis.
FIGURE 10:
The defects caused by depletion of either Eps8 or Eps8L1a can be reverted by expressing wild-type Eps8 or Eps8L1a, respectively, but not their mutant forms. (A) LLC-PK1 cells expressing shRNA targeting Eps8L1a were transfected with human GFP-Eps8L1a wild type, GFP-Eps8L1aΔcap, or GFP-Eps8L1aΔbund. Long microvilli are observed in cells depleted for Eps8L1a with or without expression of Eps8L1aΔcap, whereas shorter microvilli are observed in cells expressing the shRNA targeting Eps8L1a and Eps8L1a wild type or Eps8L1aΔbund. (B) LLC-PK1 cells expressing shRNA targeting Eps8 were transfected with human GFP-Eps8 wild type, GFP-Eps8Δcap, or GFP-Eps8Δbund. Cells expressing shRNA targeting Eps8 or cells expressing shRNA targeting Eps8 and GFP-Eps8Δbund display irregular, bent microvilli. In contrast, cells expressing shRNA targeting Eps8 and either wild-type GFP-Eps8 or GFP-Eps8 Δcap show stiff microvilli. Bar, 10 μm.
We next examined which of the Eps8 actin filament-capping or -bundling activity was responsible for the morphological changes observed in cells expressing ezrin and Eps8. Coexpression of ezrin and Eps8Δcap resulted in the formation of circular ruffles in nearly 100% of cells, but their number per cell was reduced, as was the presence of tufts of microvilli (∼20% of cells) or arc-shaped ridges (∼10% of cells) when compared with cells expressing wild type Eps8 and ezrin. Eps8Δcap was no longer present in membrane ruffles but was still observed at the tip of the microvilli. In contrast, coexpression of ezrin with Eps8Δbund prevented the formation of these structures, and the cells were covered with microvilli (Figure 9B). We next examined by CL-SEM the surface of Eps8-depleted cells expressing a shRNA-resistant form of human wild-type Eps8, Eps8Δcap, or Eps8Δbund. Cells expressing wild-type Eps8 or Eps8Δcap displayed straight microvilli, whereas cells expressing Eps8Δbund were irregular and bent (Figure 10B). Taken together, our data indicate that both Eps8 and Eps8L1a cooperate with ezrin to regulate the actin organization within microvilli through the bundling and capping activities of Eps8 and Eps8L1a, respectively.
DISCUSSION
Of finger-like protrusions observed in metazoans, the microvilli present at the luminal surface of absorptive cells represent one of the most highly ordered actin-based structures, yet the mechanisms controlling their assembly is the least well understood. This is partly due to the fact that microvillus assembly at the apical surface of epithelial cells is a slow process that requires well-differentiated cells, which precludes a dynamic analysis. In particular, the mechanisms involved in microvillar actin filament growth and in the control of their uniform length are not known. In this study, we uncovered a role for ezrin, Eps8, and Eps8L1a in the control of microvillus morphogenesis. First, we observed distinct functions for Eps8 and Eps8L1a in microvilli organization. We reported for the first time a role for Eps8L1a in the control of microvillar length due to its capping activity. Second, we showed that the interaction of ezrin with Eps8 or Eps8L1a promotes at least in part their activities.
A long-standing question concerns the regulatory mechanism by which the length of the microvillus actin filaments is maintained constant. Because Eps8 displays an actin-capping activity, it was a potential candidate for regulating microvillus length (Croce et al., 2004; Disanza et al., 2004). Genetic analyses of Eps8 function in the intestine of two evolutionary distant organisms, C. elegans and mouse, led to the observation that Eps8 is required for the correct organization of intestinal microvilli. However, it was proposed that the bundling rather than the capping activity of Eps8 was involved in the regulation of the microvilli organization (Croce et al., 2004; Tocchetti et al., 2010). Our observations are in agreement with these conclusions since expression of Eps8 in our model leads to the formation of tufts of microvilli. Of importance, we report here that the capping activity necessary for the control of microvillar length is exerted by Eps8L1a. Three lines of evidence support this conclusion. First, Eps8L1a localizes at the tip of the microvilli, where actin monomers are added to the barbed end of actin filaments. Second, depletion of Eps8L1a results in the formation of very long microvilli, indicating that actin polymerization proceeds without control. Conversely, overexpression of Eps8L1a results in very short microvilli. Third, Eps8L1a mutated in a site that is identical in amino acid sequence to the actin-capping site in Eps8 does not rescue the phenotype observed in absence of endogenous Eps8L1a, namely the formation of long microvilli, whereas wild-type Eps8L1a does.
The coexpression of ezrin and Eps8/Eps8L1a indicates that ezrin tailors Eps8 protein functions. This is illustrated by the distinct morphological changes—formation of microvilli clusters or circular dorsal ruffles and tufts of microvilli—when ezrin is coexpressed with Eps8L1a or Eps8, respectively. It was previously shown that Eps8 is required for the formation of circular dorsal ruffles in response to receptor tyrosine kinase growth factor stimulation (Scita et al., 1999; Innocenti et al., 2003; Offenhauser et al., 2004; Goicoechea et al., 2006). Here we report that coexpression of ezrin and Eps8 induced these circular dorsal ruffles in the absence of growth factor stimulation. This implies that ezrin, together with Eps8, can trigger the assembly of these structures by recruiting the components involved in actin cytoskeleton remodeling. The exchange factor for the GTPases RhoG/Rac, PLEKHG6, might be one of these components, since we previously showed that its recruitment to the apical surface of epithelial cells by ezrin triggers the formation of membrane ruffles (D'Angelo et al., 2007).
The distinct morphological changes triggered by the expression of ezrin with Eps8 or Eps8L1a could be correlated with the actin-capping and -bundling activities of Eps8L1a and Eps8, respectively. Eps8 possesses intrinsically these dual functions. Eps8L1a likely displays a bundling activity in addition to the capping activity since it contains a motif identical to that characterized in Eps8 (Hertzog et al., 2010). How is the switch between the two activities bundling or capping regulated? Several hypotheses can be envisioned to explain our data. The mode of interaction between ezrin and Eps8 proteins may differentially regulate their activities toward actin. Both Eps8 and Eps8L1a interact with the PPPVY477 motif in ezrin through their SH3 domains, and this interaction is critical for Eps8 and Eps8L1a activities. However, the interaction between Eps8L1a and ezrin is also mediated through an additional binding site. Therefore ezrin may trigger distinct conformational changes in Eps8 proteins that expose either their actin-capping or -bundling sites. Furthermore, the phosphorylation of ezrin and/or Eps8 proteins might regulate their binding to each other since both are the targets of receptor and nonreceptor tyrosine kinases (Bretscher, 1989; Fazioli et al., 1993a; Heiska and Carpen, 2005; Menna et al., 2009). Ezrin phosphorylation at tyrosine 477 by Src kinases (Heiska and Carpen, 2005) may differentially regulate the binding to Eps8 proteins since Eps8L1a but not Eps8 was found in a phospho-dependent screen performed with yeast transformed with Src family kinase. Further supporting this observation, we found that Eps8L1a but not Eps8 binds preferentially ezrin phosphorylated by Src kinases in a coimmunoprecipitation assay (data not shown). However, because tyrosine 477 is part of the PPPVY477 motif involved in the binding to the SH3 domains of Eps8 proteins, we were not able to firmly determine whether its phosphorylation regulates the binding since mutation of any amino acid in the motif abolishes the interaction between ezrin and Eps8 proteins. Finally, the distinct mode of interaction might, in turn, promote the formation of distinct molecular complexes that regulate different steps of microvillus actin filament assembly. It is worth mentioning, however, that the rescue with Eps8L1aΔbund in cells depleted of Eps8L1a or with Eps8Δcap in cells depleted of Eps8 is not as efficient as with wild-type Eps8L1a or Eps8, respectively. This suggests that the two activities—bundling and capping—are not completely separable.
Our studies suggest that ezrin and Eps8 proteins interact at an early step of microvilli assembly. Indeed, the coexpression of ezrin and Eps8L1a leads to the formation of clustered actin structures in which both Eps8 proteins are present. Which factors contribute to this clustering is not clear, but overexpression of ezrin and Eps8L1a may promote the clustering of PIP2 in specific membrane domains since binding to PIP2 represents the first step in the activation mechanism of ezrin. The requirement for ezrin/Eps8L1a interaction in the formation of these structures is confirmed by our observation that they are neither observed when ezrin or Eps8L1a is expressed alone nor when ezrin Y477F is coexpressed with Eps8L1a. Moreover, the formation of these structures suggests that in restricted domains of the apical membrane ezrin together with Eps8L1a recruits the machinery allowing the assembly of the microvilli. One component of this machinery might be the scaffolding protein EBP50, as its interaction with ezrin has been shown to be critical for microvilli assembly (Garbett et al., 2010). Furthermore, microvillus clustering has been observed at the surface of JEG cells expressing mutant forms of EBP50 (Garbett et al., 2010).
The proteins recruited at the membrane by ezrin and Eps8L1a would be part of a complex reminiscent of the “tip complex” observed in filopodia or in microvilli (Mooseker et al., 1982; Svitkina et al., 2003). In this complex, Eps8 proteins would participate in the initiation and elongation of microvillar actin filaments through their capping and bundling activities. As for filopodia, formins are believed to play an important role in the elongation of microvillar actin bundles (Chhabra and Higgs, 2007). Formins associate with the barbed ends of filaments, protecting them from excess of capping proteins. It is therefore possible that Eps8L1a competes with formins for the elongation of actin filaments. This hypothesis is consistent with our observation that coexpression of Eps8L1a and ezrin leads to the formation of enlarged structure that likely results from an excess of Eps8L1a capping activity, which prevents microvilli elongation. The presence or not of microvilli emerging from these enlarged structures likely depends on the relative amount of Eps8L1a capping activity. Our results indicate that Eps8 is also required at an early step of microvillus assembly, presumably through its bundling activity. Of interest, the bundling activity of VASP has been shown to be necessary for the elongation of actin filaments driven by Diaphanous-related formin dDia2 and to provide the force to push the membrane outward (Schirenbeck et al., 2006). Therefore Eps8 would bundle actin filaments near their barbed ends, and microvillar actin filaments would be subsequently stabilized by other bundling proteins, including villin, fimbrin, and espin (Revenu et al., 2004).
The robust biochemical and biological interactions between ezrin and Eps8 or Eps8L1a that we uncovered seem in contradiction with the almost exclusive distribution of these proteins in the microvilli. Only a restricted colocalization of these proteins is observed at the interface between the distal end of the microvilli and their core. These observations further support our hypothesis that the interaction of ezrin may occur at an early step of microvillus assembly. In this scenario, ezrin would recruit Eps8 proteins and participate in their activation in the tip complex. Whereas Eps8 proteins would exert their capping and bundling activities at the barbed ends, ezrin would remain associated with the actin filament bundles as they elongate.
In conclusion, we identified a new component of the brush border microvilli, Eps8L1a, which controls microvillus length through its capping activity. Our data emphasize the remarkably fine regulation of Eps8 protein activities, which display in the same subcellular compartment complementary, nonredundant functions. Whether ezrin interacts with and regulates the activities of the other Eps8-related proteins such as Eps8L2 and Eps8L3 remains to be investigated. However, the versatile interaction of ezrin with Eps8 proteins that we uncovered underlines the key role of ezrin in organizing molecular complexes essential for the morphogenesis of the apical domain of epithelial cells and provides new insights into the mechanisms of microvillus formation.
MATERIALS AND METHODS
Antibodies
The following primary antibodies were used: affinity-purified, rabbit polyclonal anti-VSVG and anti-ezrin antibodies (Algrain et al., 1993); monoclonal anti-ezrin antibody clone 4G11B5, a kind gift from G. Choquet-Kastylevsky (New Markers Department, BioMérieux, France); monoclonal anti-VSVG (clone P5D4; Kreis, 1986), polyclonal anti-Myc antibody generated in the laboratory; mouse monoclonal anti-Myc (clone 9E10); mouse monoclonal anti–α-tubulin (Sigma-Aldrich, St. Louis, MO); mouse monoclonal anti-Eps8 (BD Biosciences, Franklin Lakes, NJ); rabbit polyclonal antibodies anti-Eps8L1a and anti-Eps8L1b were raised by Biogenes (Berlin, Germany) against the peptide CAPKPSAKSIYEQRKR corresponding to amino acids 10–24 in Eps8L1a and against the peptide CMNRTWPRRIWGSSQ corresponding to amino acids 1–14 in Eps8L1b and affinity purified against their respective peptides. Secondary antibodies were as follows: horseradish peroxidase– and Cy3-, Texas Red–, and Cy5-conjugated goat anti-rabbit and anti-mouse (Jackson ImmunoResearch Laboratories, West Grove, PA); Alexa 488–conjugated goat anti-rabbit and anti-mouse (Molecular Probes, Invitrogen, Carlsbad, CA). Phalloidin was purchased from Invitrogen Life Technologies (Paisley, United Kingdom).
cDNA constructs
VSVG-tagged ezrin constructs in pcB6 and GST-fused ezrin constructs in pGEX-2T were previously described (Algrain et al., 1993; Andreoli et al., 1994). pEGFP-ezrin was a kind gift from Richard Lamb (University of Alberta, Canada). The single-point mutants ezrin Y477F and ezrin P475A were obtained using the QuikChange Mutagenesis Kit (Stratagene, La Jolla, CA). The cDNAs encoding human Eps8 (IMAGE 4820933) and Eps8L1a (NITE clone AK075098) were cloned into pCS2-Myc or pCS2-GFP vectors (pCS2 vectors were a gift from Alex Gautreau, Laboratoire d'Enzymologie et Biochimie Structurales, Gif-sur-Yvette, France). cDNAs encoding Eps8L1a ezrin-binding domain (amino acids [aa] 258–607) and SH3 domain (aa 480–534) were obtained by PCR amplification and cloned into the pGEX-CS2 vector to produce GST-fusion proteins. pGEX-CS2 encoding the SH3 domain of Eps8 (aa 528–596) was a kind gift from Alexis Gautreau. cDNAs encoding Eps8L1a N-ter (aa 1–476), SH3 (aa 480–534), and PTB (aa 1–176) domains were obtained by PCR amplification and cloned into the pCS2-Myc vector. Mutants in the capping and bundling activities of Eps8 proteins (Hertzog et al., 2010) were obtained using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent Technologies, La Jolla, CA).
Cell culture and transfection
LLC-PK1 cells (CCL 101; American Type Culture Collection, Manassas, VA) were grown in DMEM (Invitrogen) containing 10% fetal bovine serum. LLC-PK1 cells were transiently transfected by electroporation. The stable LLC-PK1 cell lines expressing ezrin WT, ezrinY477F, and ezrin P475A were generated as previously described (Naba et al., 2008). 293T cells were transiently transfected by the calcium phosphate method and lysed 24 h after transfection.
Eps8, Eps8L1a, and ezrin knockdown by RNA interference
For depletion of the proteins, oligonucleotides were inserted into the psiRNA-h7SK-GFP-zeo vector to express shRNA and the reporter GFP (Invitrogen). For some experiments we used the same plasmid in which we replaced the reporter GFP by mCherry. The following oligonucleotides derived from porcine expressed sequence tags and genomic sequences were used to target Eps8 and Eps8L1a:
shEps8, #0: 5′ACCTCGGCAAATGTAACCCGTCAGAATCAAGAGTTCTGACGGGTTACATTTGCCTT3′.
shEps8, #2: 5′ACCTCGAAAGCCGCATTAGAGGATAATCAAGAGTTATCCTCTAATGCGGCTTTCTT3′.
shEps8L1a, #4: 5′ACCTCGAGGAAGCGATATACCACTGATCAAGAGTCAGTGGTATATCGCTTCCTCTT3′.
shEps8L1a, #11: 5′ACCTCAGATGCTGCTGTGCGTGTCTTTCAAGAGAAGACACGCACAGCAGCATCTTT3′.
Plasmids encoding scrambled shRNA were previously described (Naba et al., 2008). LLC-PK1 cells were transfected by electroporation with the plasmids, and cells were analyzed 2 d after transfection. Expression of the reporter GFP or mCherry allows monitoring of transfected cells by fluorescence microscopy. For the rescue experiments, cells were transfected at the same time with plasmids encoding shRNA targeting porcine Eps8L1a and myc-tagged human Eps8L1a wild type or mutated. For CL-SEM analysis, cells were transfected as described with plasmids encoding shRNA expressing mCherry and GFP-Eps8 or GFP-Eps8L1a.
Protein interactions
For GST pull-down experiments, cells were lysed in RIPA buffer A containing 50 mM 4- (2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES) (pH 7.5), 150 mM NaCl, 10 mM ethylene glycol-bis(2-aminoethylether)-N,N,N′,N′-tetraacetic acid), 1.5 mM MgCl2, 10% glycerol, 0.1% SDS, 0.5% sodium deoxycholate, and protease inhibitor cocktail (Sigma-Aldrich). Clarified supernatants were incubated for 2 h at 4°C with immobilized GST fusion proteins on glutathione–Sepharose 4B beads (GE Healthcare, Chalfont St Giles, United Kingdom). For immunoprecipitation, cells were lysed in RIPA lysis buffer B (50 mM HEPES, pH 7.5, 150 mM NaCl, 10 mM EDTA, 0.1% SDS, 0.5% sodium deoxycholate, 1% NP40, and protease inhibitor cocktail). Clarified supernatants were incubated for 2 h at 4°C with antibody and protein G–Sepharose beads (Perbio, Aalst, Belgium). For the interaction of GST-ezrin with GFP-Eps8 or GFP-Eps8L1a, GFP or GFP-fused proteins were purified from lysates of 293T cells transfected with the corresponding plasmids. Cells were lysed with RIPA buffer A containing 300 mM NaCl, and the supernatant was incubated with 10 μl of GFP-Trap_A beads (Chromotek, Planegg-Martinsried, Germany). After washing with RIPA buffer containing 300 mM NaCl, beads were incubated with purified GST-ezrin for 15 h. After six washes with RIPA buffer containing 300 mM NaCl, the bound material was analyzed by SDS–PAGE.
Immunofluorescence
Cells were fixed with paraformaldehyde (4%) and processed for immunofluorescence. Images were taken with a Nikon 90i upright microscope equipped with PIFOC objective stepper and a 100×/1.4 numerical aperture Plan Apochromat objective (Nikon, Champigny sur Marne, France). Slices were acquired along the z-axis every 0.2 μm. Deconvolution was done by the MetaMorph module (Roper Scientific, Sarasota, FL) using the Meinel algorithm. Image processing and analyses were performed with ImageJ (National Institutes of Health, Bethesda, MD; Abramoff et al., 2004).
Correlative light scanning electron microscopy
Cells plated on 35 mm Grid-500 dishes (Ibidi, Martinsried, Germany) were fixed in 4% formaldehyde. Cells transfected with plasmids expressing functional shRNAs (psiRNAs) were monitored through the expression of GFP. Cells coexpressing ezrin-VSVG and either Eps8-GFP or Eps8L1a-GFP were processed for immunofluorescence with a VSVG antibody. Images over a 2 cm2 area were taken by light and fluorescence microscopy. Cells were then fixed for 30 min in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 7.2, and postfixed with 1% tannic acid and 2% osmium tetroxide (both from Electron Microscopy Sciences, Hatfield, PA). Samples were dehydrated in a graded ethanol series and critical point dried with CO2 in a Leica CPD-030 apparatus (Leica, Wetzlar, Germany). Specimens were coated with 5 nm of gold/palladium in a Gatan Ion Beam Coater 681 (Gatan, Pleasanton, CA). Fluorescent cells were tracked based on their position relative to letters/numbers on the grid and observed in a JEOL JSM-6700f field emission scanning electron microscope (JEOL, Tokyo, Japan). Image processing and analyses were performed with ImageJ.
Supplementary Material
Acknowledgments
We thank Maud Hertzog (Institut Curie), Danijela Vignjevic (Institut Curie), Sergio Marco (Institut Curie), and Alexis Gautreau (Laboratoire d'Enzymologie et Biochimie Structurales, Gif-sur-Yvette) for helpful discussions, insights, and reagents. We acknowledge Vincent Fraisier for image deconvolution and advice, the Nikon Imaging Center at the Institut Curie-Centre National de la Recherche Scientifique, and Fabrice Cordelières and the Plateforme d'Imagerie Cellulaire et Tissulaire (Institut Curie). Scanning electron microscopy was performed at the Plateforme de Microscopie Ultrastructurale/Imagopole (Institut Pasteur, Paris, France). We thank the Hybrigenics staff for yeast two-hybrid experiments. This work was supported by grants from the Association pour la Recherche sur le Cancer (3267, 4823) and the Agence Nationale de la Recherche (05BLAN033001) and by a GenHomme Network Grant (02490-6088) to Hybrigenics and the Institut Curie. I.Z. was a recipient of fellowships from the Agence Nationale de la Recherche (05BLAN033001) and the Fondation pour la Recherche Médicale. A.N. was a recipient of fellowships from the Ministère de l'Education Nationale, de la Recherche et de la Technologie and from the Association pour la Recherche sur le Cancer. M.C. is a recipient of fellowships from the Institut Curie and the Fondation pour la Recherche Médicale.
Abbreviations used:
- CL-SEM
correlative light scanning electron microscopy
- Eps8
EGF receptor pathway substrate 8
- ERM
ezrin, radixin, moesin
- FERM
protein 4.1, ezrin, radixin, moesin
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- shRNAs
short hairpin RNAs
- VSVG
vesicular stomatitis virus glycoprotein G
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0588) on January 19, 2012.
REFERENCES
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Algrain M, Arpin M, Louvard D. Wizardry at the cell cortex. Cur Biol. 1993;3:451–454. doi: 10.1016/0960-9822(93)90354-q. [DOI] [PubMed] [Google Scholar]
- Andreoli C, Martin M, Leborgne R, Reggio H, Mangeat P. Ezrin has properties to self-associate at the plasma membrane. J Cell Sci. 1994;107:2509–2521. doi: 10.1242/jcs.107.9.2509. [DOI] [PubMed] [Google Scholar]
- Arpin M, Chirivino D, Naba A, Zwaenepoel I. Emerging role for ERM proteins in cell adhesion and migration. Cell Adhes Migration. 2011;5:199–206. doi: 10.4161/cam.5.2.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN. The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- Crepaldi T, Gautreau A, Comoglio PM, Louvard D, Arpin M. Ezrin is an effector of hepatocyte growth factor-mediated migration and morphogenesis in epithelial cells. J Cell Biol. 1997;138:423–434. doi: 10.1083/jcb.138.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce A, Cassata G, Disanza A, Gagliani MC, Tacchetti C, Malabarba MG, Carlier M-F, Scita G, Baumeister R, Di Fiore PP. A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol. 2004;6:1175–1179. doi: 10.1038/ncb1198. [DOI] [PubMed] [Google Scholar]
- D'Angelo R, Aresta S, Blangy A, Del Maestro L, Louvard D, Arpin M. Interaction of ezrin with the novel guanine nucleotide exchange factor PLEKHG6 promotes RhoG-dependent apical cytoskeleton rearrangements in epithelial cells. Mol Biol Cell. 2007;18:4780–4793. doi: 10.1091/mbc.E06-12-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disanza A, Carlier M-F, Stradal TEB, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G. Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol. 2004;6:1180–1188. doi: 10.1038/ncb1199. [DOI] [PubMed] [Google Scholar]
- Disanza A, et al. Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol. 2006;12:1337–1347. doi: 10.1038/ncb1502. [DOI] [PubMed] [Google Scholar]
- Fazioli F, Minichiello L, Matoska V, Castagnino P, Miki T, Wong WT, Di Fiore PP. Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 1993a;12:3799–3808. doi: 10.1002/j.1460-2075.1993.tb06058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazioli F, Wong WT, Ullrich SJ, Sakaguchi K, Appella E, Di FP. The ezrin-like family of tyrosine kinase substrates: receptor-specific pattern of tyrosine phosphorylation and relationship to malignant transformation. Oncogene. 1993b;8:1335–1345. [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiévet BT, Gautreau A, Roy C, Del Maestro L, Mangeat P, Louvard D, Arpin M. Phosphoinositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J Cell Biol. 2004;164:653–659. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Lalonde DP, Bretscher A. The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J Cell Biol. 2010;191:397–413. doi: 10.1083/jcb.201004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary R, Bretscher A. Ezrin self-association involves binding of an N-terminal domain to a normally masked C-terminal domain that includes the F-actin binding site. Mol Biol Cell. 1995;6:1061–1075. doi: 10.1091/mbc.6.8.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautreau A, Louvard D, Arpin M. Morphogenic effects of ezrin require a phosphorylation-induced transition from oligomers to monomers at the plasma membrane. J Cell Biol. 2000;150:193–203. doi: 10.1083/jcb.150.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea S, Arneman D, Disanza A, Garcia-Mata R, Scita G, Otey CA. Palladin binds to Eps8 and enhances the formation of dorsal ruffles and podosomes in vascular smooth muscle cells. J Cell Sci. 2006;119:3316–3324. doi: 10.1242/jcs.03076. [DOI] [PubMed] [Google Scholar]
- Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiska L, Carpen O. Src phosphorylates ezrin at tyrosine 477 and induces a phosphospecific association between ezrin and a kelch-repeat protein family member. J Biol Chem. 2005;280:10244–10252. doi: 10.1074/jbc.M411353200. [DOI] [PubMed] [Google Scholar]
- Hertzog M, et al. Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PloS Biol. 2010;8:e1000387. doi: 10.1371/journal.pbio.1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti M, Frittoli E, Ponzanelli I, Falck JR, Brachmann SM, Di Fiore PP, Scita G. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis TE. Microinjected antibodies against the cytoplasmic domain of vesicular stomatitis virus glycoprotein block its transport to the cell surface. EMBO J. 1986;5:931–941. doi: 10.1002/j.1460-2075.1986.tb04306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg J, Hunter T. Identification of the two major epidermal growth factor-induced tyrosine phosphorylation sites in the microvillar core protein ezrin. J Biol Chem. 1992;267:19258–19265. [PubMed] [Google Scholar]
- Lalonde DP, Garbett D, Bretscher A. A regulated complex of the scaffolding proteins PDZK1 and EBP50 with ezrin contribute to microvillar organization. Mol Biol Cell. 2010;21:1519–1529. doi: 10.1091/mbc.E10-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Disanza A, Grati M, Andrade L, Lin H, Di Fiore PP, Scita G, Kachar B. Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr Biol. 2011;21:167–172. doi: 10.1016/j.cub.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menna E, et al. Eps8 regulates axonal filopodia in hippocampal neurons in response to brain-derived neurotrophic factor (BDNF) PLoS Biol. 2009;7:e1000387. doi: 10.1371/journal.pbio.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongiovi AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Di Fiore PP. A novel peptide-SH3 interaction. EMBO J. 1999;18:5300–5309. doi: 10.1093/emboj/18.19.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, Pollard TD, Wharton KA. Nucleated polymerization of actin from the membrane-associated ends of microvillar filaments in the intestinal bush border. J Cell Biol. 1982;95:223–233. doi: 10.1083/jcb.95.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A, Reverdy C, Louvard D, Arpin M. Spatial recruitment and activation of the Fes kinase by ezrin promotes HGF-induced cell scattering. EMBO J. 2008;27:38–50. doi: 10.1038/sj.emboj.7601943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offenhauser N, Borgonovo A, Disanza A, Romano P, Ponzanelli I, Iannolo G, Di Fiore PP, Scita G. The eps8 family of proteins links growth factor stimulation to actin reorganization generating functional redundancy in the Ras/Rac pathway. Mol Biol Cell. 2004;15:91–98. doi: 10.1091/mbc.E03-06-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C, Athman R, Robine S, Louvard D. The co-workers of actin filaments: from cell structures to signals. Nat Rev Mol Cell Biol. 2004;5:635–646. doi: 10.1038/nrm1437. [DOI] [PubMed] [Google Scholar]
- Roffers-Agarwal J, Xanthos JB, Miller JR. Regulation of actin cytoskeleton architecture by Eps8 and Abi1. BMC Cell Biol. 2005;6:36–50. doi: 10.1186/1471-2121-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome I, Curto M, McClatchey AI. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev Cell. 2004;6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J. The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA. 2006;103:7694–7699. doi: 10.1073/pnas.0511243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G, Nordstrom J, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegård M, Betsholtz C, Di Fiore PP. EPS8 and E3B1 transduce signals from Ras to Rac. Nature. 1999;401:290–293. doi: 10.1038/45822. [DOI] [PubMed] [Google Scholar]
- Scita G, Tenca P, Areces LB, Tocchetti A, Frittoli E, Giardina G, Ponzanelli I, Sini P, Innocenti M, Di Fiore PP. An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J Cell Biol. 2001;154:1031–1044. doi: 10.1083/jcb.200103146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S-I, Vasiliev JM, Borisy GG. Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol. 2003;160:409–421. doi: 10.1083/jcb.200210174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura A, Kikuchi S, Hata M, Katsuno T, Matsui T, Hayashi H, Suzuki Y, Noda T, Tsukita S, Tsukita S. Achlorhydria by ezrin knockdown: defects in the formation/expansion of apical canaliculi in gastric parietal cells. J Cell Biol. 2005;169:21–28. doi: 10.1083/jcb.200410083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Klooster JP, et al. Mst4 and ezrin induce brush borders downstream of the Lkb1/Strad/Mo25 polarization complex. Dev Cell. 2009;16:551–562. doi: 10.1016/j.devcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Tocchetti A, Confalonieri S, Scita G, Di Fiore PP, Betsholtz C. In silico analysis of the EPS8 gene family: genomic organization, expression profile, and protein structure. Genomics. 2003;81:234–244. doi: 10.1016/s0888-7543(03)00002-8. [DOI] [PubMed] [Google Scholar]
- Tocchetti A, et al. Loss of the actin remodeler Eps8 causes intestinal defects and improved metabolic status in mice. Plos One. 2010;5:e9468. doi: 10.1371/journal.pone.0009468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen O, Wahlström T, Vaheri A. Ezrin has a COOH-terminal actin-binding site that is conserved in the ezrin protein family. J Cell Biol. 1994;126:1445–1453. doi: 10.1083/jcb.126.6.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Tsukita S, Tsukita S. Direct involvement of ezrin/radixin/moesin (ERM)- binding membrane proteins in the organization of microvilli in collaboration with activated proteins. J Cell Biol. 1999;145:1497–1509. doi: 10.1083/jcb.145.7.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.