A localization atlas is provided for 66 of 90 mammalian GFP-tagged deubiquitinases (DUBs). USP21 is the only DUB in the panel that localizes to both microtubules and the centrosome. Functional data suggest a key role for USP21 in the choreography of microtubule reorganization.
Abstract
Ubiquitination is a reversible modification that influences a broad range of physiological processes. There are approximately 90 deubiquitinases (DUBs) encoded in the human genome, of which 79 are predicted to have catalytic activity. We tagged 66 DUBs with green fluorescent protein and systematically surveyed their subcellular distribution, identifying enzymes specific to the nucleus, plasma membrane, and secretory and endocytic pathways. USP21 is unique in showing clear association with both centrosomes and microtubules. Using an in vitro assay, we show that microtubule binding is direct and identify a novel microtubule-binding motif encompassed within amino acids 59–75 of the N-terminus of USP21. Our functional studies indicate a key role for USP21 in the governance of microtubule- and centrosome-associated physiological processes: Depletion of USP21 in A549 cells compromises the reestablishment of a radial array of microtubules during recovery from cold-induced depolymerization and also reduces the probability of primary cilium formation, whereas USP21 knockdown in PC12 cells inhibits nerve growth factor–induced neurite outgrowth.
INTRODUCTION
Ubiquitination is a reversible posttranslational modification involved in most complex cellular processes, including cell division, DNA repair, membrane trafficking, and cell signaling. Deubiquitination is mediated by specific deubiquitinases (DUBs), of which there are approximately 79 active enzymes that can be predicted from the human genome sequence (Komander et al., 2009; Reyes-Turcu et al., 2009). They can be subdivided into five families: ubiquitin C-terminal hydrolases (UCHs), ubiquitin-specific proteases (USPs), ovarian tumor, Josephins (Machado–Joseph disease), and JAB1/MPN/Mov34 metalloenzymes (JAMM/MPN+). The first four families are cysteine proteases, whereas the JAMM family belongs to the zinc metalloproteases. In general, DUBs may act to rescue proteins from ubiquitin-dependent degradation processes or to reverse ubiquitin signals.
We surveyed the subcellular localization of >60 green fluorescent protein (GFP)–tagged DUBs in HeLa cells and subsequently focused our attention on the unique example of clear centrosome and microtubule association, namely USP21. This little-studied member of the USP family has been suggested to influence cell growth and to display dual specificity for both ubiquitinated and neddylated conjugates (Gong et al., 2000). However, a recent study clearly showed promiscuous activity toward ubiquitin chain types and a high degree of discrimination against Nedd8-conjugated substrates (Ye et al., 2011). A microarray study observed that USP21 levels changed during liver regeneration and proposed the relevant target to be monoubiquitinated histone H2A (Nakagawa et al., 2008). Recently, USP21 was also proposed to be a negative regulator of NFκB signaling (Xu et al., 2010).
Reversible ubiquitination is fundamental to mitosis and cytokinesis, but other links between the ubiquitin/proteasome system and microtubule cell biology are emerging. Ubiquitination sites for α- and β-tubulin have been identified using mass spectrometry (Meierhofer et al., 2008). The DUB CYLD can associate with microtubules through a CAP-Gly domain, regulate their dynamics, and control entry into mitosis (Stegmeier et al., 2007; Gao et al., 2008; Wickstrom et al., 2010), whereas a ubiquitin C-terminal hydrolase, UCHL1, has also been found associated with microtubules in certain cell types (Bheda et al., 2010). The E3 ligase BRCA1 is required for γ-tubulin localization to the centrosome (Sankaran et al., 2007). Following proteasome inhibition, various proteins accrue at the pericentriole, and the cells become impaired in microtubule aster formation (Didier et al., 2008). In postmitotic neurons, a centrosomal Cdc20-APC/Cyclosome ubiquitin signaling pathway drives dendrite formation (Kim et al., 2009).
In this study, we show that USP21 associates with both centrosomes and microtubules. We map the sequence determinants responsible for such localization, demonstrate direct microtubule-binding properties, and identify several microtubule-associated proteins as USP21 binding partners. We go on to conduct functional studies in which we implicate USP21 in microtubule network regeneration following cold depolymerization, in primary cilia formation, and in neurite outgrowth from rat pheochromocytoma PC12 cells.
RESULTS
Analysis of subcellular localization of DUBs
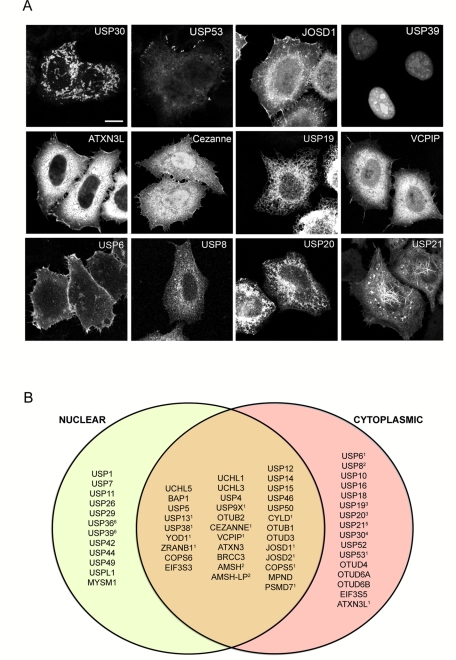
We undertook a systematic analysis of the subcellular localization of GFP-tagged DUBs in HeLa cells (Figure 1 and Supplemental Figure S1 and Table S1). A significant number accumulate in the nucleus (12 exclusively nuclear, nine predominantly nuclear), including some that are enriched in identifiable subnuclear structures such as the nucleolus (USP36; Endo et al., 2009; Figure 1 and Supplemental Figure S1 and Table S1). Many are cytosolic under our conditions of overexpression, but clear association with identifiable structures can be seen in several cases, including endosomes (AMSH/STAMBP, AMSH-LP/STAMBPL, UBPY/USP8; McCullough et al., 2004; Nakamura et al., 2006; Row et al., 2006), mitochondria (USP30; Nakamura and Hirose, 2008), plasma membrane (USP6; Martinu et al., 2004), and endoplasmic reticulum (USP19; Hassink et al., 2009), which we verified through colocalization with other markers. By its very nature, our screening procedure is prone to the occasional example in which the appended GFP tag interferes with the correct localization. Indeed, our N-terminally GFP-tagged CYLD localized to the plasma membrane and cytosol but did not show noticeable microtubule staining, in contrast to the clear microtubule localization of CYLD tagged with GFP at the C-terminus and of endogenous protein reported by others (Supplemental Figure S1; Stegmeier et al., 2007; Gao et al., 2008; Wickstrom et al., 2010).
FIGURE 1:
Subcellular localization of enhanced GFP (EGFP)–tagged DUBs. (A) Selection of DUBs showing distinctive subcellular distributions. All images correspond to confocal sections and are shown at identical magnification. Scale bar, 10 μm. (B) Venn diagram representing the relative distribution of EGFP-tagged DUBs between nuclear and cytoplasmic compartments. Those DUBs also localizing to clearly identifiable organelles or structures are indicated with a number corresponding to the following locations: 1, plasma membrane; 2, endosome; 3, endoplasmic reticulum; 4, mitochondria; 5, microtubules and centrosome; 6, nucleoli. Note that plasma membrane localization may be confined to particular structures, for example, filipodia or lamellipodia. The full data set corresponding to fluorescence images of 61 EGFP-DUB family members is provided in Supplemental Figure S1 and Table S1.
USP21 associates with the microtubular cytoskeletal network
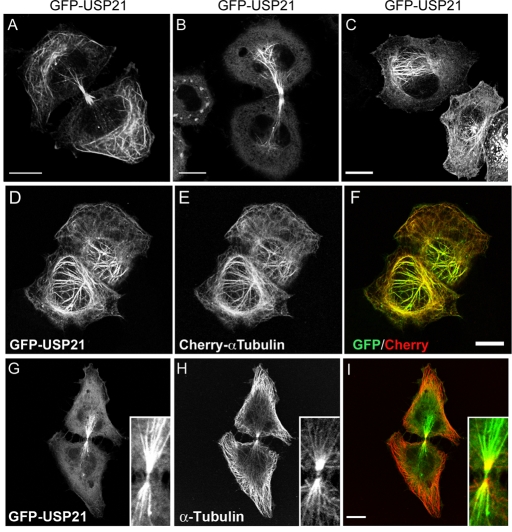
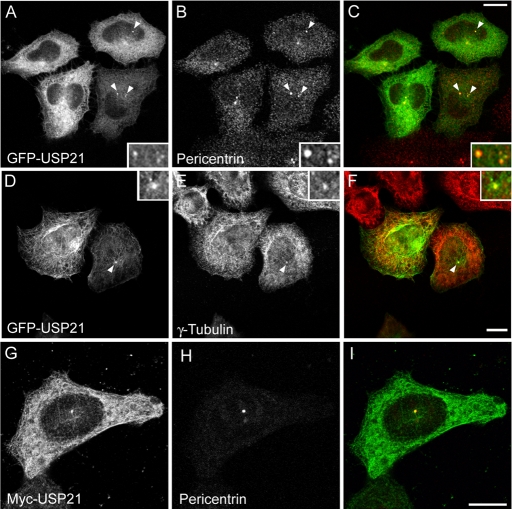
Out of 66 GFP-tagged DUBs, only GFP-USP21 (or Myc-tagged USP21; see Supplemental Figure S2) displayed a distribution that was suggestive of microtubular localization (Figure 2, A–C). In interphase cells, expressing moderate levels of GFP-USP21, fluorescent microtubules were mostly located in close proximity to the nucleus (Figure 2, C–F, and Supplemental Figure S2, D–F). Microtubule-association of GFP-USP21 was also prominent in late telophase and in cells undergoing cytokinesis; in these cells USP21 appears to redistribute to the polar and midbody microtubule network (Figure 2, A, B, and G–I). At low expression levels, GFP-USP21 and Myc-USP21 are predominantly cytosolic, together with a clear accumulation at the centrosome, where they colocalize both with pericentrin and γ-tubulin, suggesting that the microtubule-organizing center/centrosome provides a high-affinity recruitment site for USP21 (Figure 3).
FIGURE 2:
GFP-USP21 associates with microtubular structures. HeLa cells were transfected for 24 h with GFP-USP21. (A–C) Different aspects of GFP-USP21 microtubular staining. (D–F) HeLa cells stably expressing Cherry–α-tubulin transiently transfected with GFP-USP21. (G–I) Colocalization with α-tubulin in a HeLa cell undergoing cytokinesis. Scale bar, 10 μm.
FIGURE 3:
USP21 localizes to centrosomes at low expression levels. HeLa cells were transfected with GFP-USP21 (A–F) or myc-USP21 (G–I) and counterstained with pericentrin (A–C, G–I) or γ-tubulin antibodies (D–E). Insets show threefold enlargement of the pericentriolar region (indicated by arrows). Scale bar, 10 μm.
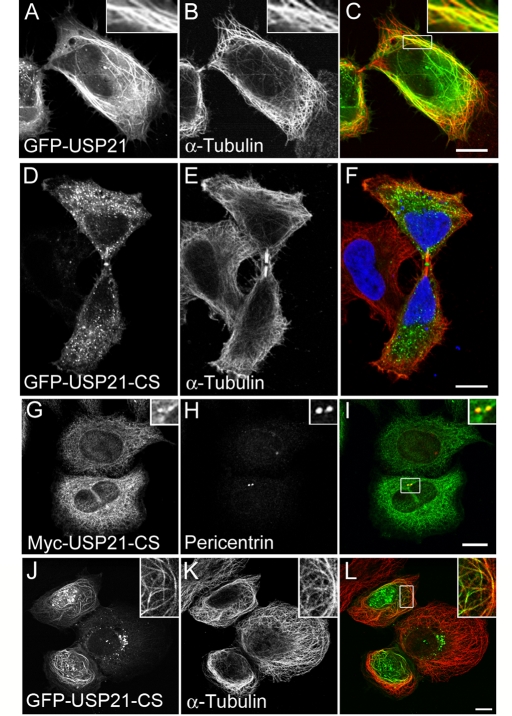
High levels of GFP-USP21 promote microtubule bundling in a manner commonly observed for microtubule-associated proteins (MAPs; Figure 4, A–C; Olson et al., 1995). It is intriguing that a catalytically inactive mutant of USP21 (C221S; Supplemental Figure S3) is not effectively recruited to microtubules in HeLa cells but retains the primary localization to the midbody (Figure 4, D–F) as well as to centrosomes (Figure 4, G–I). We did observe, however, some microtubule localization for this inactive mutant in the osteosarcoma cell line U2OS, albeit at high expression levels only (Figure 4, J–L).
FIGURE 4:
Catalytic activity favors microtubule association but is not required for centrosome or midbody colocalization. HeLa cells were transfected for 24 h with GFP-USP21 wild type or catalytically inactive mutant (CS) tagged with GFP or myc. (A–C) Colocalization with microtubule bundles in cells expressing high levels of GFP-USP21. (D–F) GFP-USP21-CS localizes to the midbody but is absent from cytosolic microtubules. (G–I) myc-USP21-CS colocalization with pericentrin. (J–L) High-level expression of GFP-USP21-CS in U2OS cells promotes its localization to microtubules. Scale bar, 10 μm.
Mapping the requirements for USP21 association with centrosomes and microtubules
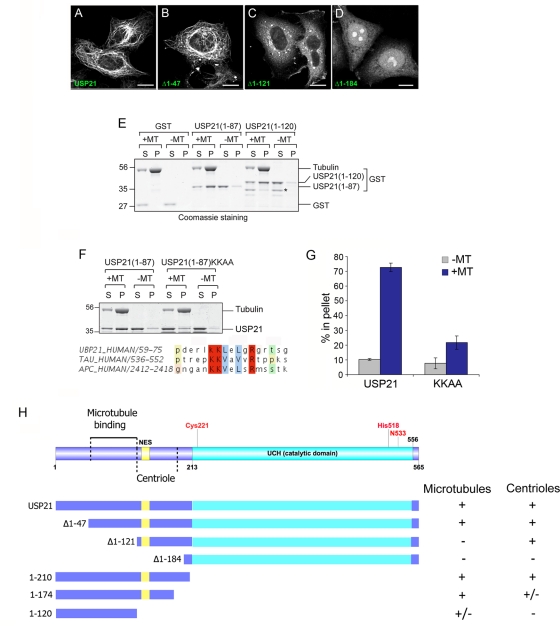
Whereas the C-terminus of USP21 contains its catalytic domain, the amino terminus (1–212) is intrinsically disordered and contains multiple proline-rich sequences that are commonly involved in protein–protein interactions. We therefore constructed a set of N-terminal and C-terminal deletion mutants for analysis of USP21 associations in HeLa and U2OS cells (Figure 5 and Supplemental Figure S4). Deletion of the first 184 amino acids (USP21∆1–184) abolishes microtubule and centrosomal localization. This construct partially localizes to the nucleus with enrichment in nucleoli, consistent with the loss of a CRM1-dependent nuclear export sequence (NES; amino acids 134–147). USP21∆1–121 can still localize to centrosomes but does not extend along microtubules or bundle them (Figure 5 and Supplemental Figure S4). USP21∆1–47 behaves as full length with respect to microtubule bundling, centrosome, and microtubule association. Our C-terminal deletion constructs appeared largely cytosolic in HeLa cells but retained microtubular localization in U2OS cells to different degrees (Supplemental Figure S5). Removal of the catalytic domain confirmed that the first 210 amino acids are sufficient to target GFP to microtubules. Further truncation to amino acid 174 did not affect microtubule association, whereas truncation to the first 120 amino acids markedly reduced microtubular localization.
FIGURE 5:
Mapping the requirements for USP21 localization and binding to microtubules. (A–D) HeLa cells were transfected for 24 h with full-length GFP-USP21 wild type or the indicated N-terminal truncation mutant. Scale bar, 10 μm. Note that this localization was also confirmed in U2OS cells. (E) In vitro spin-down assay for direct binding to purified microtubules (MTs). Binding of both USP21 N-terminal fragments (1–87 and 1–120) to microtubules is evidenced by redistribution from supernatant (S) to pellet fraction (P). Asterisk indicates a shorter truncation, which has lost microtubule binding. (F) Alignment of USP21 residues 59–75 with motifs in tau and APC proteins: mutation of matching lysine residues diminishes in vitro binding to microtubules. (G) Quantitation of data presented in F. Bars indicate range of two distinct experiments. (H) Summary of constructs used and properties reported in this figure and Supplemental Figures S4 and S5.
We next tested for direct binding to microtubules using an in vitro assay that depends on redistribution of purified protein in the presence of microtubules to a pellet fraction following high-speed ultracentrifugation (Figure 5E). Full-length protein was not amenable to this analysis owing to its tendency to pellet independently of microtubules under the conditions of this assay. We therefore took a fragment-based approach. Both glutathione S-transferase (GST)–USP21(1–120) and GST-USP21(1–87) showed substantial microtubule association. It is serendipitous that our GST-USP21(1–120) preparation contains a lower–molecular weight degradation product, fractionally smaller than the first 87 amino acids, which has lost the ability to bind microtubules. Analysis of this sequence revealed high homology with motifs previously identified in tau and adenomatous polyposis coli (APC) proteins. The tau motif is also involved in microtubule binding (Goode et al., 1997), whereas the APC motif is part of a larger region known to bind microtubules (Deka et al., 1998). Mutation of matching lysine residues within this motif abrogates microtubule binding of the 1–87 N-terminal fragment in vitro (Figure 5, F and G). Of interest, this motif is adjacent to the major sites of phosphorylation that we were able to detect within USP21 using mass spectrometry (S93, S111, S113, and S115; Supplemental Table S2). Although this sequence is essential for in vitro binding of amino acids 1–87, expression of the full-length protein bearing these mutations in HeLa and U2OS cells nevertheless retained a microtubular disposition indistinguishable from that of wild-type USP21, suggesting that multiple binding modes must exist, reflecting a common feature of microtubule-associated proteins (Supplemental Figure S5). Mutation of the microtubule-binding motif in the context of the (1–210) N-terminal fragment reduces microtubule association, whereas mutation of the shorter (1–120) fragment largely abolished the residual level of microtubule association observed with this construct (Supplemental Figure S5).
Candidate interacting partners
In a recent comprehensive proteomic survey of DUB interaction partners, Sowa et al. (2009) found that all four microtubule-affinity regulating kinases (MARKs) coimmunoprecipitate with hemagglutinin (HA)-tagged USP21, stably expressed in HEK293 cells. We confirmed this interaction by Western blotting following coimmunoprecipitation of GFP-USP21 and HA-tagged MARK1 and 2 and mapped this interaction to the same region that was required for microtubule localization in intact cells (Supplemental Figure S6). Our mass spectroscopic analysis of GFP-USP21 interactors in HeLa and HEK293T cells also suggested association with endogenous MARK2 and MARK3, as well as with MAP4, GEF-H1 (ARHGEF2), and CKAP5, which are all annotated as microtubule associated (Supplemental Table S3). Despite multiple attempts to generate antipeptide and anti–fusion protein antibodies against USP21 in three different species, we failed to obtain a high-affinity antibody capable of detecting endogenous levels of USP21 by immunofluorescence. We were, however, able to determine the efficacy of three out of four small interfering RNA (siRNA) oligonucleotides targeting USP21 by RT-PCR and by Western blotting, using our affinity-purified anti-USP21 antibody on concentrated RIPA buffer cell lysates (Supplemental Figure S7, A and B). However, significant knockdown of USP21 did not result in changes to the protein expression levels of any of these USP21 interacting proteins, nor did we observe an upshift in protein mass that might be indicative of enhanced ubiquitination (Supplemental Figure S7, C and D).
USP21 regulates microtubule regrowth and primary cilium formation in A549 cells
Knockdown of USP21 had no effect on the levels of γ-tubulin, a centrosomal component known to influence microtubule aster formation, or on a range of proteins known to be associated with interphase microtubule dynamics (Didier et al., 2008; Supplemental Figure S7C). We also assessed a potential role for USP21 in mitosis but did not find any significant alteration in the distribution of cells within the cell cycle (as judged by fluorescence-activated cell sorting analysis), in accordance with published data (Garnett et al., 2009), although a modest effect on cell growth/viability could be detected after prolonged knockdown (Supplemental Figure S8, A and B).
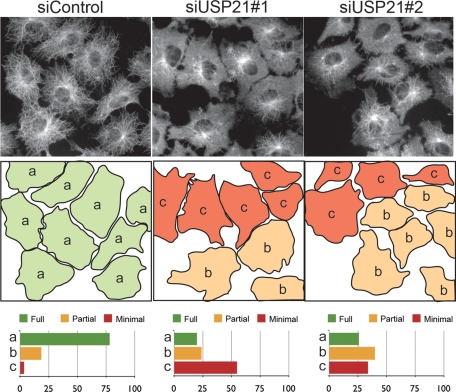
A549 lung adenocarcinoma cells treated with siRNA oligos targeting USP21 display an extensive microtubule network comparable to that of control cells. Given its preferential centrosomal localization at low expression levels, we wondered whether USP21 might play a role in nucleating new microtubule growth radiating from the centrosome. We used a well-established assay measuring regrowth of microtubules after cold-induced depolymerization to assess the effect of depleting USP21 by siRNA (Figure 6). Within 2.5 min, control cells reestablish a radial array of new microtubules extending to the periphery of the cell. Following USP21 knockdown with either of two independent oligonucleotides, this process is markedly impaired. In those cells that do establish a microtubule network it is usually less ordered, only rarely displaying a clear radial pattern emanating from the centrosome and extending all the way to the cell periphery.
FIGURE 6:
USP21 depletion impairs the reestablishment of a microtubule network after cold depolymerization. A549 lung cells were transfected with a nontargeting siRNA oligonucleotide (siControl) or one of two individual siRNA oligonucleotides (siUSP21#1, siUSP21#2) specifically targeting USP21 for 120 h prior to experimentation. After cold-induced microtubule depolymerization, cells were incubated for 2.5 min at 37°C, followed by methanol fixation and staining with α-tubulin antibodies. Cells were classified into three categories of microtubule regrowth status: (a) green, advanced (full) regrowth; (b) orange, partially defective regrowth; and (c) red, severely defective (minimal) regrowth. Shown are the results of a representative experiment in which 287 (siControl), 323 (siUSP21#1), and 385 (siUSP21#2) cells were counted, respectively.
One particular aspect of centrosome function in resting cells is the establishment and maintenance of the primary cilium—a sensory organelle emanating from a modified centriole, the basal body, and consisting of a discrete array of particularly stable microtubules—which is a key site of multiple signal transduction cascades (Goetz and Anderson, 2010). On the basis of our observation that USP21 is required for effective microtubule regrowth from centrosomes, we wondered whether its depletion might also negatively affect the generation of a primary cilium upon serum starvation. Indeed, we observed a significant reduction in the number of A549 cells developing a primary cilium upon serum starvation when USP21 was depleted with two independent siRNA oligos (Figure 7). This reduction was comparable (∼60% in our hands) to that observed with a known regulator of ciliogenesis, VHL (Thoma et al., 2007). We also confirmed these results in telomerase-immortalized retinal pigmented epithelial (hTERT-RPE1) cells, in which a higher percentage of cells respond to serum starvation by forming a primary cilium: 69% in cells treated with nontargeting control oligo, compared with 47% in cells treated with siUSP21 oligo 1, 39% with oligo 2, and 43% with oligo 4. We did not observe any significant changes in primary cilium length in USP21-depleted cell populations.
FIGURE 7:
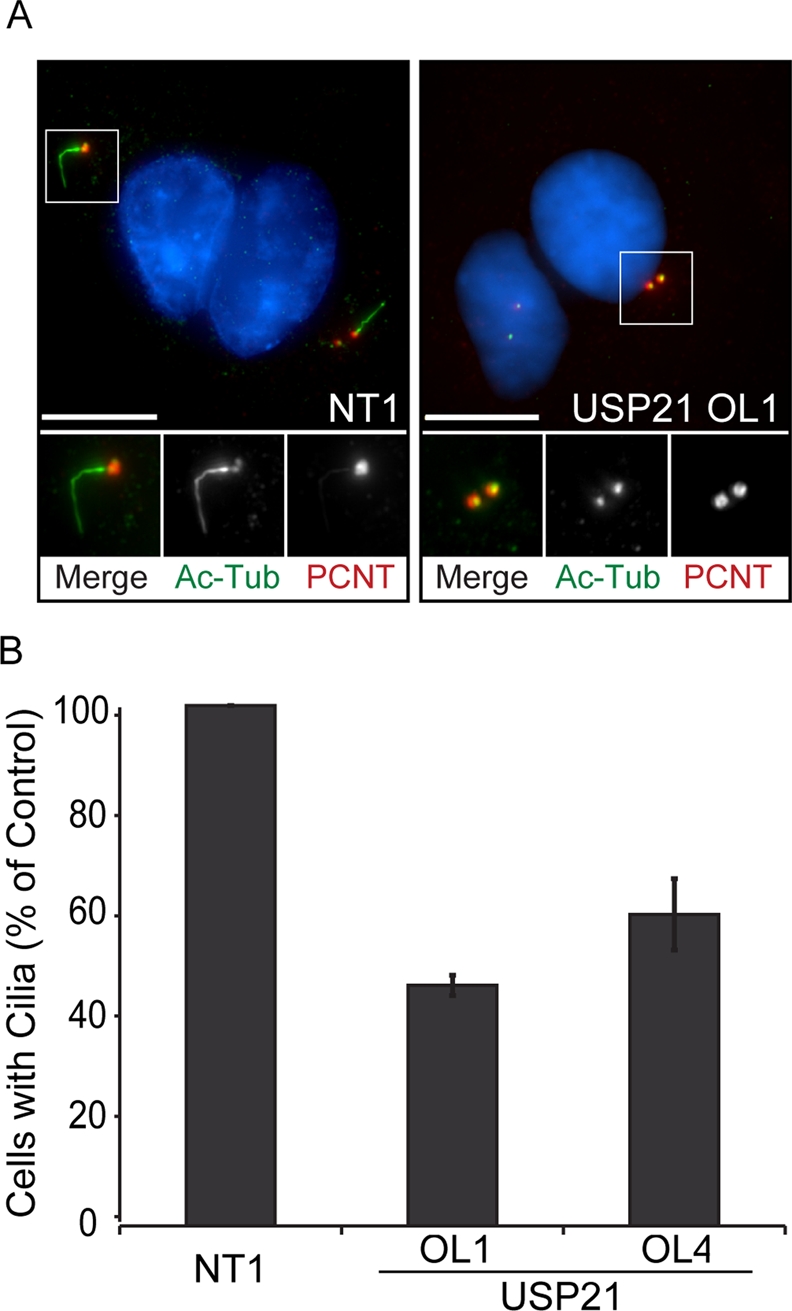
USP21 is required for primary cilium formation. A549 cells were transfected with siRNA specific to USP21 (OL1, OL4) or nontargeting siRNA (NT1) and serum starved for 48 h to induce primary cilium formation. (A) A maximal projection of NT1 and USP21-targeting, siRNA (OL1)-transfected cells is shown after staining of acetylated α-tubulin as a marker for primary cilia (Ac Tub, green) and pericentrin (PCNT, red). DNA was stained with DAPI (blue). Scale bar, 10 μm. (B) The percentage of cells with a primary cilium (∼800 cells counted per condition) is significantly reduced for cells transfected with siRNA specific to USP21 (error bars, SD; n = 3 independent experiments).
Requirement of USP21 for neurite outgrowth in PC12 cells
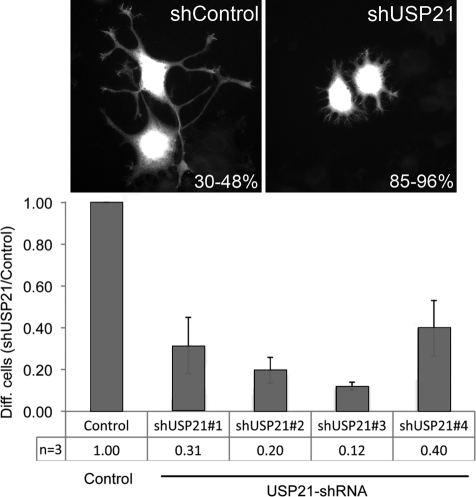
We turned to another physiological process intimately connected with microtubule reorganization, that of neurite outgrowth. Rat pheochromocytoma PC12 cells respond to application of nerve growth factor (NGF) by the outgrowth of branching neurites, which become visible after 2–3 d (Black et al., 1986). We transfected cells with HUSH plasmids expressing short hairpin RNA (shRNA) targeting USP21 together with red fluorescent protein (RFP) translated from the same transcript using an internal ribosomal entry site. After 3 d in the presence of NGF, neurite length was measured relative to the cell body (Figure 8). We observed a significant decrease for all four shRNA targeting plasmids in the number of cells with neurites, defined as extensions larger than the maximum chord length across the cell body.
FIGURE 8:
USP21 depletion interferes with NGF-induced neurite outgrowth in PC12 cells. PC12 cells were transfected with pRFP-C-RS vector expressing either nontargeting shRNA (NC15, Control) or HuSH-29 shRNA targeting rat USP21 (shUSP21#1–4). Transfected cells coexpress RFP. Neurite outgrowth was induced by application of 100 ng/ml NGF 48 h after transfection, and 3 d later cells were fixed and counterstained with Alexa Fluor 488–labeled phalloidin to visualize nontransfected cells. Cells expressing RFP were scored as differentiated if one or more neurites had reached a length larger than the diameter of the cell body (30–48% of control cells, compared with 4–15% in USP21-depleted cells). The data are derived from three independent experiments, in each of which between 130 and 350 cells were scored for each condition. Error bars, ±SD.
DISCUSSION
Systematic analysis of interaction partners (Sowa et al., 2009) or subcellular localization of DUBs can provide clues to cellular processes within which they might function. In this study, we provide the most comprehensive analysis of mammalian DUB localization hitherto undertaken, providing a unique resource for future studies of this important class of enzymes. Our data are complementary to a recent survey of DUB localization within fission yeast (Kouranti et al., 2010). However, in that organism there are only 20 family members, and assignment of mammalian orthologues is not straightforward.
We focused our further attention on USP21, which presented in our screen of GFP-tagged DUBs as the only microtubule- and centrosome-associated protein. Further analysis revealed that the centrosome represents the site of highest concentration at low expression levels, suggesting that this might be the primary recruitment or highest-affinity binding site for USP21. Whereas CYLD and UCHL1 have also been shown to associate with microtubules (Stegmeier et al., 2007; Gao et al., 2008; Bheda et al., 2010), this is a unique example of a centrosome-associated DUB. Because we have been unable to generate an antibody that is capable of detecting endogenous USP21 by immunofluorescence, our localization data are limited to tagged proteins. Whereas these are strongly supported by our in vitro binding and interaction data, as well as by functional evidence discussed later, the localization of endogenous USP21 will require further validation in future studies as and when new tools become available. We were able to show that binding to microtubules is direct and specified in part by a region of the N-terminal domain distinct from the sequence necessary for centrosome binding. Of interest, the USP21 sequence suggested to encompass a microtubule-binding site contains a KKLeLgR motif juxtaposed to a proline-rich region, similar to a motif required for microtubule binding of Tau (KKVaVvR; Goode et al., 1997).
Our identification of USP21 as a DUB associated with centrosomes and microtubules, and functioning in several physiological processes depending on microtubule reorganization ties in well with the mass spectrometric DUB interaction survey data provided by Sowa et al. (2009), which indicated an association with all four members of the microtubule affinity–regulating kinase family (MARK1–4). The DUB USP9X was shown to regulate MARK4 activity through its ubiquitination status (Al-Hakim et al., 2008). We confirmed interactions with MARK1 and MARK2 but were unable to provide any parallel observation in this case using coexpressed MARKs, nor did we observe any changes in steady-state levels of endogenous MARK1 and MARK3 proteins. We also reported three novel interactions with microtubule-associated proteins: MAP4, GEF-H1 (ARHGEF2), and CKAP5. Although these do not appear to be direct targets of USP21, as their stability and ubiquitination status are not altered upon manipulation of USP21 expression, they lend further credence to the association of USP21 with microtubule-dependent processes.
We observed no major influence on cell growth or in distribution of cell population between cell-cycle phases. We therefore pursued a functional role of USP21 in regulating microtubule dynamics of interphase cells. Our data suggest a physiological role for USP21 in the establishment and organization of a new microtubule network following cold-induced depolymerization. A similar observation was reported in HeLa cells following knockdown of CYLD (Gao et al., 2008). Dominant-negative effects that we observe upon overexpression of USP21 (unpublished data) have so far thwarted our attempts to demonstrate a direct requirement for catalytic activity in this assay. USP21 depletion in both resting A549 and hTERT-RPE1 cells also reduces the probability of serum starvation–induced primary cilium formation, further corroborating a requirement for USP21 in centriole (basal body)–dependent microtubule outgrowth in interphase cells. Major microtubule reorganization is also an essential component of NGF-dependent neurite extension but is not required for the transcription-dependent “priming” of this remodeling event (Greene et al., 1982). We propose a role for USP21 in this physiologically important process that is concordant with its governance of microtubule dynamics. Taken together these data indicate a complex and fundamental role for reversible ubiquitination events in the choreography of microtubule regrowth.
Future work will need to address the full range of ubiquitination events necessary for centrosome and microtubule function in nonmitotic cells, as well as the salient targets of USP21, which have so far remained elusive. We anticipate that as a unique centrosomal DUB and a rare microtubule-associated enzyme, USP21 will be one of the major DUBs controlling these processes.
MATERIALS AND METHODS
Cell culture and transfection
A549, HeLa, U2OS, and HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% nonessential amino acids. HeLa cells stably expressing either mCherry–α-tubulin or GFP-EB1 were kindly provided by Anna Akhmanova (Erasmus University, Rotterdam, Netherlands). hTERT-RPE1 cells were provided by Francis Barr (University of Oxford, Oxford, UK) and cultured in DMEM/F12 supplemented with 10% FBS and 1% nonessential amino acids. PC12 cells were provided by Giampietro Schiavo (Cancer Research UK, London Research Institute, London, United Kingdom) and grown in DMEM, 7.5% FBS, 7.5% filtered horse serum, 4 mM glutamine, and 1% penicillin/streptomycin. HeLa, U2OS, and HEK293T cells were transfected with plasmids using GeneJuice (Novagen, EMD4Biosciences, Gibbstown, NJ) and fixed or lysed after 24 h. PC12 cells were transfected with HuSH-29 constructs using Lipofectamine LTX reagent at a 3:1 ratio and Plus reagent (Invitrogen, Carlsbad, CA). For siRNA experiments, A549, HeLa, and hTERT-RPE1 cells were treated with nontargeting (NT1) or target-specific siRNA oligos (Dharmacon On-Target Plus oligos, Thermo Fisher Scientific, Waltham, MA) at 40–50 nM final concentration using Oligofectamine Transfection Reagent (Invitrogen) twice over a period of 120 h.
Plasmids, antibodies and other reagents
The human USP21 open reading frame (ORF; NM_012475.4) was amplified from human brain cDNA library (Clontech, Mountain View, CA) using a two-step PCR with KOD Hot Start DNA Polymerase (Novagen) and fully sequence verified; primers are available upon request. The ORF was inserted into pDONR223 and then shuttled into Gateway-converted pEGFP-C, pCMV-Myc, and pGEX6P expression vectors. A catalytically inactive mutant was constructed by QuikChange mutagenesis introducing a T-to-A mutation at position 661, resulting in Cys-221 to Ser (CS) conversion. Lys-64 and Lys-65 were converted to Ala using QuikChange mutagenesis (KKAA). All mutant ORFs were sequenced and subcloned into relevant vectors. Full-length and C- as well as N-terminal truncation mutants of USP21 (1–87, 1–120, 1–174, 1–210, ∆1–47, ∆1–121, ∆1–184) were amplified, inserted into pCR4-TOPO (Invitrogen), and subcloned into pGEX6P1 and/or pEGFP-C1 vectors. HA-MARK1 and 2 constructs were kindly provided by Dario Alessi (University of Dundee, Dundee, Scotland). Rabbit polyclonal anti-pericentrin (ab4448), β-tubulin (ab6046), and γ-tubulin (ab11317) antibodies were purchased from Abcam (Cambridge, MA). Mouse anti-myc (4A6) was obtained from Millipore (Billerica, MA). Rabbit anti-actin (A2066), Myc (C3956), and ubiquitin (U5379), mouse anti–α-tubulin (T5168 and DMA1), γ-tubulin (T6557), and mouse acetylated α-tubulin, as well as poly-l-lysine, were obtained from Sigma-Aldrich (St. Louis, MO). Secondary donkey anti–mouse and anti–rabbit IRDye (680 and 800 nm) antibodies were purchased from LI-COR Biosciences (Lincoln, NE), and Alexa Fluor 488 and 594–labeled secondary antibodies were from Invitrogen (Molecular Probes, Invitrogen). Sheep anti-GFP was a gift from Ian Prior (University of Liverpool, Liverpool, United Kingdom). rHuNGF was purchased from First Link (Birmingham, United Kingdom)UK. On-Target Plus USP21 siRNA oligos were purchased from Thermo Fisher Scientific. The target sequences are as follow: #1 (J-006071-05), CCACCCACUUUGAGACGUA; #2 (J-006071-06), GAUCCAAGCUACCAUUUGC; #3 (J-006071-07), CGAGAGCCACCTGTTAATA; and #4 (J-006071-08), CUUCGGGACUUCUGUCUGA. HuSH-29 constructs expressing four individual shRNAs targeting rat USP21, together with RFP, were purchased from OriGene (Rockville, MD).
GFP-DUBs
GFP-USP8 and GFP-AMSH have been described elsewhere (McCullough et al., 2004; Row et al., 2006). GFP-A20 and GFP-USP9X were kind gifts of Paul Evans (Imperial College, London, United Kingdom) and Dario Alessi (University of Dundee), respectively. A subset of DUBs was obtained in a Gateway-compatible entry vector from the human ORFeome collection, version 1.1, courtesy of Mark Vidal (Harvard Medical School, Boston, MA). The rest of the DUB collection was assembled by PCR amplifying the ORFs of the reference sequences from 1) human cDNA libraries (liver, brain or testis; Clontech), 2) human IMAGE clones (Mammalian Gene Collection, http://mgc.nci.nih.gov/), or 3) constructs kindly donated by Rohan T. Baker (The Australian National University, Canberra, Australia; USP4), René Bernards (The Netherlands Cancer Institute, Amsterdam, The Netherlands; CYLD), Boudewijn M. T. Burgering (University Medical Center Utrecht, The Netherlands; USP7), Erich E. Wanker (Max Delbrueck Center for Molecular Medicine, Berlin, Germany; USP13), Sirano Dhe-Paganon (University of Toronto, Canada; USP50), and Paul Evans (Imperial College London, UK; Cezanne, ZRANB1). ORFs were cloned into the Gateway-based entry vector pDONR223 and subsequently shuttled by LR reaction into a Gateway-converted pEGFP-C vector. USP30 was cloned by restriction enzyme and ligation-based cloning into pEGFP-N3, resulting in a C-terminal GFP tag. MYSM1 and AMSH-LP were cloned by restriction enzyme and ligation–based cloning into pEGFP-C1. All ORFs were fully sequence verified, and all complete sequences are available on request. A complete description of this library will be published elsewhere.
Protein purification and DUB assays
Rosetta cells (Invitrogen) were transformed with pGEX6p1-USP21 constructs encoding various GST-tagged, USP21-derived proteins and purified with glutathione–Sepharose 4B (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions. For DUB assays, purified GST-USP21 or GST-USP21(C221S) (both at 100 nM) were incubated with K48- or K63-linked tetra-ubiquitin chains (Boston Biochem, Boston, MA; both at 0.4 μM) in DUB assay buffer (50 mM Tris, pH 7.5, 25 mM KCl, 5 mM MgCl2, and 1 mM dithiothreitol) at 37°C for 2 h. Samples were subjected to NuPAGE analysis (Invitrogen), followed by immunoblotting with anti-ubiquitin antibodies.
Microtubule spin-down assay
Microtubule Binding Protein Spin-down Assay Kit (BK029) was purchased from Cytoskeleton (Denver, CO). GST, GST-USP21(1–87), GST-USP21(1–87)KKAA, or GST-USP21(1–120) was incubated with or without in vitro–assembled microtubules at 21°C for 30 min. The reaction mixture was centrifuged at 100,000 × g for 40 min at 25°C. Supernatant and pellet fractions were collected and analyzed by SDS–PAGE and staining with Coomassie blue.
Cell lysis and immunoblotting
Cultured cells were lysed with either Nonidet P-40 (NP-40) buffer (0.5% NP-40, 25 mM Tris, pH 7.5, 100 mM NaCl, and 50 mM NaF), RIPA buffer (1% NP-40, 10 mM Tris, pH 7.5, 150 mM NaCl, 1% sodium deoxycholate, and 0.1% SDS) supplemented with proteinase and phosphatase inhibitors, or “hot lysis buffer” (1% SDS, 50 mM NaF, and 1 mM EDTA at 110°C). In some cases, N-ethylmaleimide was added to a final concentration of 10 mM to inactivate cysteine proteinases. Proteins were resolved by SDS–PAGE and transferred to nitrocellulose. Visualization of Western blots was achieved using an Odyssey infrared scanner (LI-COR Biosciences).
Immunofluorescence microscopy
Cells were processed for immunofluorescence by fixation either with 3% paraformaldehyde in phosphate-buffered saline (PBS) followed by permeabilization with 0.2% Triton in PBS (localization of GFP-tagged DUBs) or with −20°C methanol (microtubule staining). Cells were stained as indicated with primary antibodies for 30–60 min at room temperature in 3% BSA followed by Alexa Fluor 488 or 594–labeled secondary donkey anti-mouse and anti-rabbit antibodies. Confocal images were captured with a Leica confocal SP2 AOBS (HCX PL APO CS, 63×, 1.40 oil objective) for all colocalization experiments and the localization of the GFP-DUB collection. Microtubule regrowth experiments were visualized using a Leica DMIL (Wetzlar, Germany) or a Nikon Ti-Eclipse (Melville, NY) microscope.
Microtubule regrowth assay
A549 cells seeded on coverslips were incubated for 90 min on ice to depolymerize microtubules in ice-cold PBS++ (PBS supplemented with 1 mM CaCl2 and 1 mM MgCl2), and then the buffer was exchanged for prewarmed PBS++ (37°C) and held at this temperature for 2.5 min before fixation with −20°C Methanol (5 min). Cells were then stained with anti–α-tubulin and anti-pericentrin antibodies, followed by secondary labeled antibodies.
Primary cilium assay
A549 cells (3.5 × 105 cells per 6-cm dish) were treated with nontargeting or USP21-targeting siRNA oligos (40 nM) twice over 120 h. At 24 h after the second hit, cells were seeded at 1 × 105 cells per well onto 22 × 22 mm coverslips and serum starved for a further 48 h to induce primary cilium formation. hTERT-RPE1 cells (5 × 105 cells per 6-cm dish with 22 × 22 mm coverslip) were treated with siRNA (100 nM) at 12 h after seeding and serum starved for 48 h. Cells were then washed with PBS++ and incubated in PBS++ on ice for 60 min (hTERT-RPE1) or 90 min (A549) to promote microtubule depolymerization. Cells were fixed with methanol (A549) or 4% paraformaldehyde, followed by Triton permeabilization (hTERT-RPE1), and stained with antibodies against pericentrin and acetylated tubulin to identify primary cilia. Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI), and slides were mounted with p-phenylenediamine. Wide-field microscopy was performed with a Nikon Ti-Eclipse microscope (CFI Plan Apochromat VC, 100× oil objective, N2, numerical aperture 1.4). Per condition, 800 and 100 cells were counted for A549 and hTERT-RPE1 cells, respectively. The percentage of cells developing a primary cilium was ∼20% in A549 and 70% in hTERT-RPE1 cells.
Neurite outgrowth assay
PC12 cells were seeded at 100,000 cells per well of a six-well plate and transfected with 1 μg of rHuSH-29 plasmid DNA expressing either nontargeting (NC15; control) or rat USP21-targeting shRNA (77, 78, 79, 80) together with RFP. Two days after transfection, cells were reseeded onto poly-l-lysine–coated coverslips situated in a six-well plate at 40,000 cells/well. On attachment, cells were incubated in full growth medium supplemented with 100 ng/ml NGF. Cells were fixed after 3 d of NGF treatment with 3% PFA in PBS and counterstained with Alexa Fluor 488–labeled phalloidin (Invitrogen). Coverslips were mounted using Mowiol infused with DAPI, and cells were visualized using an epifluorescence microscope and a 50× objective. Between 130 and 350 RFP-expressing cells were assessed for each condition and scored as differentiated if one or more neurites had reached a length larger than the maximum chord length of the cell body.
Supplementary Material
Acknowledgments
This work was supported by a Cancer Research UK Senior Fellowship award to S.U. and a project grant from the Wellcome Trust. S.D.H. was supported by a Wellcome Trust studentship. We thank the many colleagues listed in Materials and Methods for reagents; Dario Alessi, Franck Perez, and Giampietro Schiavo for helpful discussions; Zehra Akerman, Sara Cadeco, and Rebecca Eccles for technical assistance; and Monika Chojnowska, Ewan Macdonald, Carla Bento, and Matthew Edmonds for help in producing some constructs and preliminary experiments.
Abbreviations used:
- AMSH
associated molecule with the SH3 domain of STAM
- APC
adenomatous polyposis coli
- CKAP5
cytoskeleton-associated protein 5
- CYLD
cylindromatosis (turban tumor syndrome)
- DUB
deubiquitinase
- GEF-H1
guanine nucleotide exchange factor H1
- GFP
green fluorescent protein
- EGFP
enhanced GFP
- GST
glutathione S transferase
- hTERT
telomerase-immortalized retinal pigmented epithelial
- MAP4
microtubule-associated protein 4
- MARK
microtubule-affinity regulating kinase
- NES
nuclear export sequence
- NF-kB
nuclear factor kappa B
- NGF
nerve growth factor
- RFP
red fluorescent protein
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- UCH
ubiquitin C-terminal hydrolase
- USP
ubiquitin-specific protease
- VHL
von Hippel Lindau
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0668) on February 1, 2012.
REFERENCES
- Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR. Control of AMPK-related kinases by USP9X and atypical Lys29/Lys33-linked polyubiquitin chains. Biochem J. 2008;411:249–260. doi: 10.1042/BJ20080067. [DOI] [PubMed] [Google Scholar]
- Bheda A, Gullapalli A, Caplow M, Pagano JS, Shackelford J. Ubiquitin editing enzyme UCH L1 and microtubule dynamics: implication in mitosis. Cell Cycle. 2010;9:980–984. doi: 10.4161/cc.9.5.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MM, Aletta JM, Greene LA. Regulation of microtubule composition and stability during nerve growth factor-promoted neurite outgrowth. J Cell Biol. 1986;103:545–557. doi: 10.1083/jcb.103.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deka J, Kuhlmann J, Muller O. A domain within the tumor suppressor protein APC shows very similar biochemical properties as the microtubule-associated protein tau. Eur J Biochem. 1998;253:591–597. doi: 10.1046/j.1432-1327.1998.2530591.x. [DOI] [PubMed] [Google Scholar]
- Didier C, Merdes A, Gairin JE, Jabrane-Ferrat N. Inhibition of proteasome activity impairs centrosome-dependent microtubule nucleation and organization. Mol Biol Cell. 2008;19:1220–1229. doi: 10.1091/mbc.E06-12-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Matsumoto M, Inada T, Yamamoto A, Nakayama KI, Kitamura N, Komada M. Nucleolar structure and function are regulated by the deubiquitylating enzyme USP36. J Cell Sci. 2009;122:678–686. doi: 10.1242/jcs.044461. [DOI] [PubMed] [Google Scholar]
- Gao J, Huo L, Sun X, Liu M, Li D, Dong JT, Zhou J. The tumor suppressor CYLD regulates microtubule dynamics and plays a role in cell migration. J Biol Chem. 2008;283:8802–8809. doi: 10.1074/jbc.M708470200. [DOI] [PubMed] [Google Scholar]
- Garnett MJ, Mansfeld J, Godwin C, Matsusaka T, Wu J, Russell P, Pines J, Venkitaraman AR. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat Cell Biol. 2009;11:1363–1369. doi: 10.1038/ncb1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Kamitani T, Millas S, Yeh ET. Identification of a novel isopeptidase with dual specificity for ubiquitin- and NEDD8-conjugated proteins. J Biol Chem. 2000;275:14212–14216. doi: 10.1074/jbc.275.19.14212. [DOI] [PubMed] [Google Scholar]
- Goode BL, Denis PE, Panda D, Radeke MJ, Miller HP, Wilson L, Feinstein SC. Functional interactions between the proline-rich and repeat regions of tau enhance microtubule binding and assembly. Mol Biol Cell. 1997;8:353–365. doi: 10.1091/mbc.8.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene LA, Burstein DE, Black MM. The role of transcription-dependent priming in nerve growth factor promoted neurite outgrowth. Dev Biol. 1982;91:305–316. doi: 10.1016/0012-1606(82)90037-9. [DOI] [PubMed] [Google Scholar]
- Hassink GC, Zhao B, Sompallae R, Altun M, Gastaldello S, Zinin NV, Masucci MG, Lindsten K. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009;10:755–761. doi: 10.1038/embor.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim AH, Puram SV, Bilimoria PM, Ikeuchi Y, Keough S, Wong M, Rowitch D, Bonni A. A centrosomal Cdc20-APC pathway controls dendrite morphogenesis in postmitotic neurons. Cell. 2009;136:322–336. doi: 10.1016/j.cell.2008.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, Roberts-Galbraith RH, Gould KL. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 2010;8:e1000471. doi: 10.1371/journal.pbio.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinu L, Masuda-Robens JM, Robertson SE, Santy LC, Casanova JE, Chou MM. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol Cell Biol. 2004;24:9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clague MJ, Urbe S. AMSH is an endosome-associated ubiquitin isopeptidase. J Cell Biol. 2004;166:487–492. doi: 10.1083/jcb.200401141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meierhofer D, Wang X, Huang L, Kaiser P. Quantitative analysis of global ubiquitination in HeLa Cells by mass spectrometry. J Proteome Res. 2008;7:4566–4576. doi: 10.1021/pr800468j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, et al. Deubiquitylation of histone H2A activates transcriptional initiation via trans-histone cross-talk with H3K4 di- and trimethylation. Genes Dev. 2008;22:37–49. doi: 10.1101/gad.1609708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Tanaka N, Kitamura N, Komada M. Clathrin anchors deubiquitinating enzymes, AMSH and AMSH-like protein, on early endosomes. Genes Cells. 2006;11:593–606. doi: 10.1111/j.1365-2443.2006.00963.x. [DOI] [PubMed] [Google Scholar]
- Nakamura N, Hirose S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol Biol Cell. 2008;19:1903–1911. doi: 10.1091/mbc.E07-11-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson KR, McIntosh JR, Olmsted JB. Analysis of MAP 4 function in living cells using green fluorescent protein (GFP) chimeras. J Cell Biol. 1995;130:639–650. doi: 10.1083/jcb.130.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Row PE, Prior IA, McCullough J, Clague MJ, Urbe S. The ubiquitin isopeptidase UBPY regulates endosomal ubiquitin dynamics and is essential for receptor down-regulation. J Biol Chem. 2006;281:12618–12624. doi: 10.1074/jbc.M512615200. [DOI] [PubMed] [Google Scholar]
- Sankaran S, Crone DE, Palazzo RE, Parvin JD. BRCA1 regulates gamma-tubulin binding to centrosomes. Cancer Biol Ther. 2007;6:1853–1857. doi: 10.4161/cbt.6.12.5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Sowa ME, Nalepa G, Gygi SP, Harper JW, Elledge SJ. The tumor suppressor CYLD regulates entry into mitosis. Proc Natl Acad Sci U S A. 2007;104:8869–8874. doi: 10.1073/pnas.0703268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma CR, Frew IJ, Hoerner CR, Montani M, Moch H, Krek W. pVHL and GSK3beta are components of a primary cilium-maintenance signalling network. Nat Cell Biol. 2007;9:588–595. doi: 10.1038/ncb1579. [DOI] [PubMed] [Google Scholar]
- Wickstrom SA, Masoumi KC, Khochbin S, Fassler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. EMBO J. 2010;29:131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor alpha-induced nuclear factor kappaB activation via binding to and deubiquitinating receptor-interacting protein 1. J Biol Chem. 2010;285:969–978. doi: 10.1074/jbc.M109.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.