SR splicing factors are distributed in the speckled pattern in the nucleus. Alternative pre-mRNA splicing is regulated through nuclear distribution of phosphorylated SR splicing factors, which is specifically regulated by the RANBP2 system in mammalian cell lines, as well as in mouse tissues.
Abstract
The mammalian cell nucleus is functionally compartmentalized into various substructures. Nuclear speckles, also known as interchromatin granule clusters, are enriched with SR splicing factors and are implicated in gene expression. Here we report that nuclear speckle formation is developmentally regulated; in certain cases phosphorylated SR proteins are absent from the nucleus and are instead localized at granular structures in the cytoplasm. To investigate how the nuclear architecture is formed, we performed a phenotypic screen of HeLa cells treated with a series of small interfering RNAs. Depletion of Ran-binding protein 2 induced cytoplasmic intermediates of nuclear speckles in G1 phase. Detailed analyses of these structures suggested that a late step in the sequential nuclear entry of mitotic interchromatin granule components was disrupted and that phosphorylated SR proteins were sequestered in an SR protein kinase–dependent manner. As a result, the cells had an imbalanced subcellular distribution of phosphorylated and hypophosphorylated SR proteins, which affected alternative splicing patterns. This study demonstrates that the speckled distribution of phosphorylated pre-mRNA processing factors is regulated by the nucleocytoplasmic transport system in mammalian cells and that it is important for alternative splicing.
INTRODUCTION
Mammalian nuclei are highly organized and compartmentalized into a large number of membrane-free structures that increase the local concentrations of essential molecules and facilitate nuclear events, including transcription, pre-mRNA processing, DNA replication, and DNA repair/recombination (Lamond and Earnshaw, 1998; Lanctot et al., 2007; Sexton et al., 2007; Takizawa et al., 2008; Zhao et al., 2009). Spatial and temporal coordination of chromosomes and the nuclear microenvironment significantly influence gene expression. Although the general principles underlying nuclear body formation are not well determined, several mechanisms have been proposed. One uses scaffold proteins to which additional components are added by ordered assembly. The other is by self-organization in which the components simply associate with each other in a stochastic manner (Misteli, 2001; Kaiser et al., 2008). In addition, RNAs or proteins may serve as seeds for nucleation of a nuclear body, followed by either the stochastic or ordered assembly (Mao et al., 2011). Chromosomes with high gene densities, active gene loci, or coordinately regulated gene loci are clustered at, or adjacent to, nuclear speckles, also called interchromatin granule clusters (IGCs; Shopland et al., 2003; Brown et al., 2008; Hu et al., 2008, 2010; Zhao et al., 2009; Spector and Lamond, 2011). Previous proteomics analyses showed that nuclear speckles contain a variety of proteins involved in gene expression, including pre-mRNA splicing and processing, transcription, subunits of RNA polymerase II (RNAPII), mRNA export, nonsense-mediated mRNA decay, and translation, suggesting that these structures provide assembly, modification, and/or storage sites for particular proteins and that they couple the nuclear processes to facilitate efficient gene expression (Saitoh et al., 2004).
The SR family of splicing factors, of which serine/arginine–rich splicing factor 2 (SRSF2; previously known as SC35) is one of the founding members, forms the major components of nuclear speckles (Fu and Maniatis, 1990; Saitoh et al., 2004). SR proteins are involved in both constitutive and alternative pre-mRNA splicing, and they are characterized by RNA recognition motifs and an arginine/serine–rich (RS) domain. The RS domains are phosphorylated; this affects almost all the steps of pre-mRNA splicing via modulation of their protein–protein and protein–RNA interactions (Caceres et al., 1997; Xiao and Manley, 1997; Misteli, 1999; Lai et al., 2001; Lin et al., 2005). Several kinases, including SR protein kinases 1 and 2 (SRPK1 and SRPK2) and Cdc2-like kinase (CLK/STY), have been identified to phosphorylate SR proteins (Gui et al., 1994; Colwill et al., 1996; Wang et al., 1998). During mitosis, SR proteins become diffusely distributed in the cytosol, but later in mitosis, nuclear speckles are assembled into cytoplasmic structures with unknown function, called mitotic interchromatin granules (MIGs; Ferreira et al., 1994), which disappear around telophase, concomitant with the reconstruction of nuclear speckles just after nuclear envelope/lamina formation (Ferreira et al., 1994; Prasanth et al., 2003). In this process, the MIG constituents are sequentially released from the structure at the transition from mitosis to early G1 and are translocated to the nucleus, along with the other transcription machinery, through the newly formed nuclear pore complex (NPC; Ferreira et al., 1994; Prasanth et al., 2003). Some proteins, including a subset of heteronuclear ribonucleoproteins (hnRNPs) and SR proteins, relocate at an early time point, but the phosphorylated forms of SR proteins and Ser2-phosphorylated RNAPII remain in MIGs until early G1 (Ferreira et al., 1994; Prasanth et al., 2003). Although these observations illustrate an ordered process for the establishment of subnuclear architectures and regulatory mechanisms, the exact contribution of nucleocytoplasmic transport factors to nuclear speckle formation has not been demonstrated.
Nucleocytoplasmic transport of a macromolecule is generally controlled by the asymmetric distribution of RAN, a Ras-like GTPase. The GTP-bound form of RAN predominates in the nucleus due to the chromatin association of the regulator of chromosome condensation 1 (RCC1), which is a Ran guanine nucleotide-exchange factor. Conversely, localization of RAN GTPase activating-protein 1 (RANGAP1) at the cytoplasmic face of the NPC through Ran-binding protein 2 (RANBP2) maintains a high concentration of RAN-GDP in the cytoplasm. Under this RAN-GTP gradient, nuclear transport receptors carry cargo molecules across the nuclear membrane (Lee et al., 2006; Stewart, 2007). Among the importin β family members is transportin 3 (TNPO3), which is required for the nuclear import of the SR proteins (Kataoka et al., 1999; Lai et al., 2001). Association between TNPO3 and the SR proteins is regulated by phosphorylation; appropriate levels of phosphorylation of the SR proteins are critical for their nuclear translocation (Sanford and Bruzik, 2001; Zhong et al., 2009). Ran-independent nuclear transport has also been reported for several proteins, including small nuclear ribonucleoproteins (snRNPs; Wohlwend et al., 2007).
To gain insight into the mechanisms of nuclear architecture formation and its physiological role, we investigated nuclear speckle formation in various mouse tissues and performed a phenotypic screen using HeLa cells treated with a series of small interfering RNAs (siRNAs). We addressed the molecular mechanisms of altered nuclear speckle formation and influence on gene expression.
RESULTS
The subcellular distribution of phosphorylated SR proteins is developmentally regulated
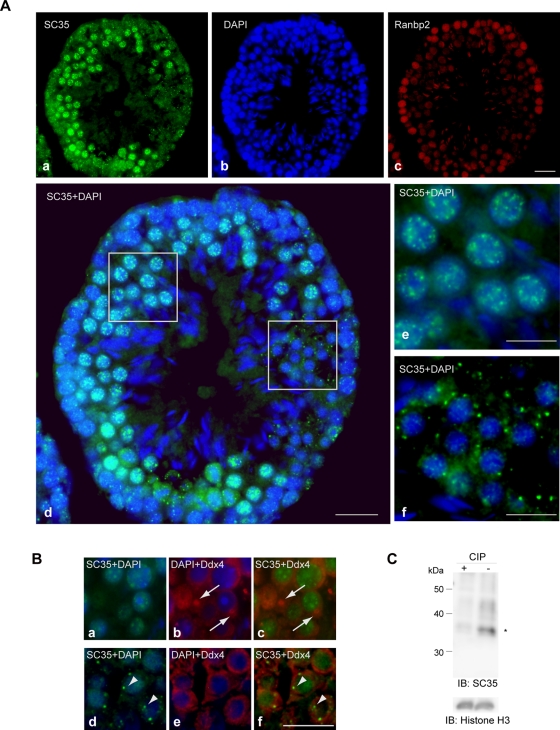
To address nuclear speckle formation in a whole animal, we performed a series of immunostaining experiments on sections derived from various mouse organs, including the testis, stomach, and blood, using the mouse monoclonal antibodies SC35 and 1H4, which are well-known markers of nuclear speckles (Figure 1 and Supplemental Figure S1). Cross-sections of seminiferous tubules in the adult mouse testis showed that nuclear speckle formation was limited to certain cell types, including spermatocytes, that are located inside the tubules (Figure 1A). In agreement with a previous report (Nagai et al., 2011), the nucleoporin Ranbp2 was highly expressed in cells adjacent to the basement membrane, and it became down-regulated toward the inner lumen, where spermatogenesis progresses (Figure 1A, c). In the region where Ranbp2 expression was low and spermatids were enriched, SC35 antigens formed granular structures in the cytoplasm (Figure 1A). In many mammalian germline cells, cytoplasmic RNP-containing structures called germinal granules or nuage exist and are recognized by antibodies against DEAD box polypeptide 4 (Ddx4; Chuma et al., 2009). In cells with germinal granules, nuclear speckles formed normally, indicating that the two are distinct (Figure 1B). As previously reported for cell lines (Fu and Maniatis, 1990), SC35 antibody recognized phosphorylated srsf2 in this tissue (Figure 1C).
FIGURE 1:
Distribution of phosphorylated SR proteins in adult mouse testis. (A) The distribution of phosphorylated SR proteins and expression of Ranbp2 are developmentally regulated. Representative views of cross-sections of seminiferous tubules stained with the mouse monoclonal antibody SC35 (a), DAPI (b), and anti-Ranbp2 (c). (d–f) Merged images of SC35 and DAPI. (e, f) Enlarged pictures of the boxed areas in d. SC35 antigens were present in a speckled pattern in the nucleus of spermatocytes (e), whereas they formed granular structures in the cytoplasm in the spermatids (f). The differentiation stage was determined based on morphology under DAPI staining. (B) An adult mouse testis section was immunostained with SC35 (green) and antibodies against Ddx4 for nuage/germinal granules (red). Nuage/germinal granules (arrows) were not recognized by SC35 (a–c). In addition, cells with SC35-stained granules (arrowheads) did not form nuage/germinal granules (d–f). Bars, 20 μm (A, a–d), 10 μm (A, e–f, and B). (C) Immunoblot analysis of cell lysate prepared from adult mouse testis. Antibody SC35 recognizes mouse srsf2 (asterisk) in this tissue, but it fails when lysate is treated with phosphatase. Histone H3 serves as a loading control.
In mouse stomach, a few surface epithelial cells in the gastric pits also exhibited granular cytoplasmic structures (Supplemental Figure S1B). In megakaryocytes derived from mouse bone marrow, phospho-SR proteins recognized by the antibody 1H4 were located only in the cytoplasm (Supplemental Figure S1C). Other types of blood cells showed normal nuclear speckles (Supplemental Figure S1C). We found that nuclear speckle formation was developmentally regulated in a whole organism and that granular structures in the cytoplasm could be observed in several mouse tissues, predominantly in the testis, where alternative RNA splicing is abundant (Lander et al., 2001; Johnson et al., 2003).
Nuclear speckle morphology changes upon inhibition of nucleocytoplasmic transport factors
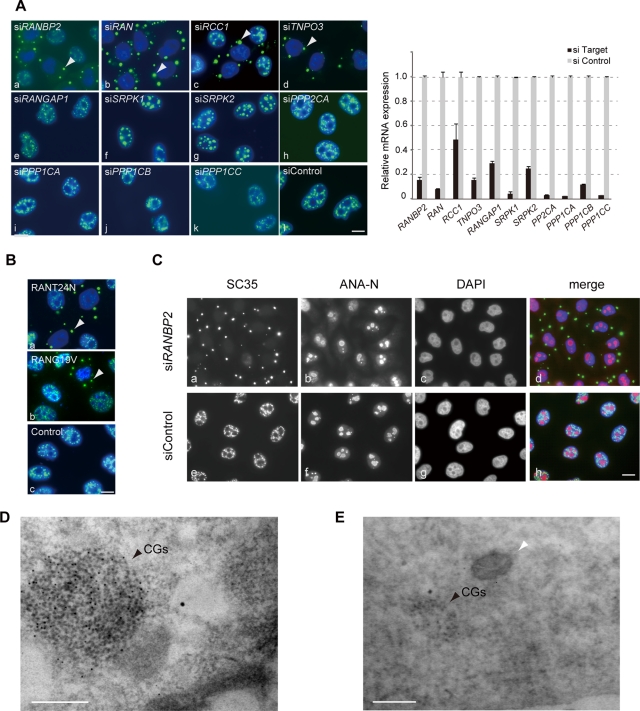
To gain insight into the molecular processes of nuclear speckle formation, we performed phenotypic screening with a series of siRNAs, including those targeting kinases, phosphatases, and other factors in nucleocytoplasmic transport. We treated HeLa cells with siRNAs for 48–72 h and monitored nuclear speckle morphology by immunofluorescence with SC35 (Figure 2A and Supplemental Figure S2A). Striking relocation of the SC35 epitopes was observed when factors involved in RAN nucleocytoplasmic transport system were depleted. The loss of RAN, RCC1, RANBP2, or TNPO3 commonly resulted in the loss of the regular speckled SC35 pattern in nuclei, which instead formed new cytoplasmic structures (Figure 2A, a–d). We termed these structures cytoplasmic granules (CGs). CGs were reminiscent of the MIGs that appear at a specific stage during mitosis (Ferreira et al., 1994) but were substantially larger and were observed in cells in interphase with decondensed chromatin. Depletion of RANGAP1, SRPK1, SRPK2, protein phosphatases (PPP1CA, PPP1CB, PPP1CC, PPP2CA), or small ubiquitin-like modifier protein (SUMO) had little or no effect on the nuclear speckles (Figure 2A, e–k, and Supplemental Figure S2A). We observed the same phenomenon with multiple siRNAs against each target gene and in several cell lines including HCT116 cells (data not shown). To confirm the specific requirement for RAN in nuclear speckle formation, we used dominant-negative mutants of RAN; RANT24N, which is defective in nucleotide binding and directly inhibits RCC1; and RANG19V, which lacks GTPase activity (Dasso et al., 1994; Clarke et al., 1995; Figure 2B). Introduction of either mutant caused marked formation of CGs, supporting the idea that the transport system is required for the proper formation of nuclear speckles (Figure 2B).
FIGURE 2:
Cytoplasmic granules (CGs) appear upon inhibition of the RANBP2-RAN transport system. (A) HeLa cells were transfected with a series of specific siRNAs and immunostained with SC35 (green) and DAPI (blue). Control cells displayed a speckled distribution of SC35 (siControl). In contrast, knockdown of RANBP2, RAN, RCC1, or TNPO3 induced new granular structures, CGs, in the cytoplasm (arrowheads). The relative mRNA levels of the knocked-down genes were examined by quantitative RT-PCR and are shown in the graph. PPP1CA, PPP1CB, PPP1CC, and PPP2CA, protein phosphatases; SRPK1 and SRPK2, SR protein kinases. (B) Exogenous expression of a dominant-negative form of RAN (RANT24N or RANG19V) also produced CGs. Vector (pcDEB) alone had no effect on nuclear speckles. (C) The morphology of the nucleoli and chromatin was visualized with the anti-nucleolar antibody ANA-N and DAPI and was not affected in the RANBP2-knockdown cells that contained CGs. (D) CGs existed as heavily clustered granules in the cytoplasm of the RANBP2-knockdown cells. An ultrathin section of the cell was immunogold labeled with the SC35 antibody by the EDTA regressive method (Spector et al., 1998). (E) CGs are membrane-free cytoplasmic structure. In this electron micrograph, the sample was stained with osmium tetroxide, and membranes were well contrasted, as the mitochondrion (white arrowhead) is observed next to CGs. Bars, 10 μm (A–C) and 200 nm (D, E).
In contrast to the dynamic changes in SC35 localization, RANBP2-knockdown cells showed morphologically intact nuclear architecture, including that of the NPC, nucleolus, and nuclear lamina (Figure 2C and data not shown). This suggests that CG generation is not merely an effect of inhibiting global nuclear import. It has also been reported that, although RANBP2 is the major nuclear pore component, it does not play an essential role in nuclear import, at least in certain cases (Walther et al., 2002; Salina et al., 2003).
We next observed the cytoplasm of RANBP2-knockdown cells under transmission electron microscopy. CGs existed as heavily clustered granules measuring ∼10 nm in diameter, and they were embedded in a fibrogranular environment (Figure 2D). CGs may represent a redistribution of IGCs because they maintained the granular structure found in nuclear speckles and IGCs. Similar to MIGs and other cytoplasmic RNP-containing structures, including germinal granules, stress granules, and processing bodies, CGs are non–membrane-bound structures, as we observed in the electron micrograph, in which membranes were well contrasted (Figure 2E).
We further investigated whether CGs represent any of the previously reported RNP granules, including processing bodies and stress granules (Anderson and Kedersha, 2009). Despite the morphological similarities, CGs did not colocalize with DCP1A, a marker of processing bodies (Supplemental Figure S2B), or with TIA1, a marker for stress granules, even under conditions of cellular stress (Supplemental Figure S2, C and D). RNP granules generally contain RNA molecules, which are required to maintain the morphology, but CGs were resistant to RNase treatment (Supplemental Figure S2E). Collectively, CGs are distinct from the previously described cytoplasmic RNP granules.
CGs appear in G1 phase
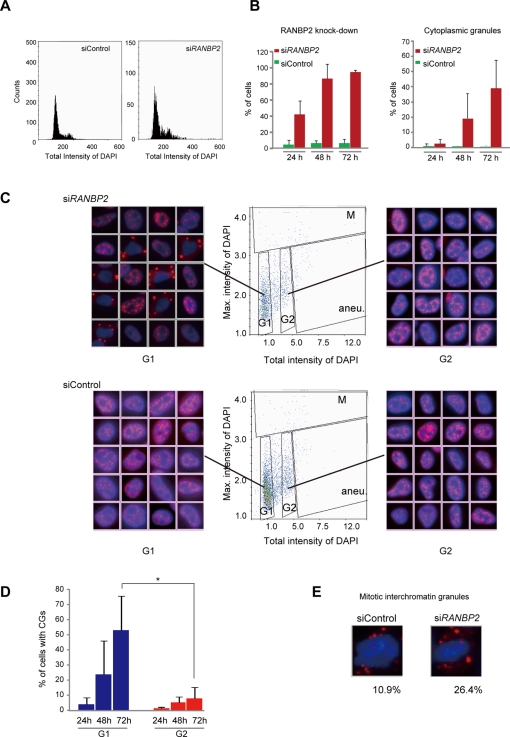
To investigate the mechanisms of CG generation, we analyzed the cell cycle progression of CG-containing cells using imaging cytometry (high-throughput fluorescence microscopy; Figure 3). We grew HeLa cells in multiwell plates, treated them with siRNAs, stained them with anti-RANBP2 and SC35 antibodies plus 4′,6-diamidino-2-phenylindole (DAPI), and then automatically captured images of >1000 cells. Signal intensities in individual cells were measured concurrently with the x, y coordinates of the cell position. Using described methods (Pozarowski et al., 2004; Kawamura et al., 2006), we first checked the cellular DNA content for the cell cycle profile (Figure 3A). Consistent with a previous report (Salina et al., 2003), a small population of cells (mean, 3.88%) became aneuploid upon RANBP2 knockdown, but no major cell cycle arrest or massive cell death was detected under our experimental conditions. We then measured the kinetics of RANBP2 depletion and CG formation over time after RANBP2-siRNA transfection. Automated recognition of SC35 labeling on digital images enabled us to discriminate CGs from normal nuclear speckles (Supplemental Figure S3). We found that RANBP2-negative cells (Figure 3B, left) and cells with CGs (Figure 3B, right) increased over time but with a different time course. The increase in CGs was later than the RANBP2 depletion. In a substantial proportion (∼40%) of cells, RANBP2 was reduced to a nondetectable level within 24 h of introduction of the siRNA, whereas it took 48–72 h for CGs to appear. This suggests that nuclear speckles do not transform into CGs immediately after RANBP2 depletion, but rather that the transition in morphology requires progression through the cell cycle.
FIGURE 3:
CGs appear at the G1 cell cycle stage in the absence of RANBP2. Imaging cytometry analyses. (A) The cell cycle profiles of control and RANBP2-knockdown cells were analyzed based on >1000 DAPI-stained cell images (left). (B) Correlation between RANBP2 knockdown and presence of CGs. HeLa cells were transfected with the indicated siRNAs and were immunostained with anti-RANBP2, SC35, and DAPI. RANBP2-negative cells (left) and CG-containing cells (right) were automatically counted and normalized to the total number of cells (DAPI signals). The values are the means and SDs of three independent experiments of >250 cells each. (C) Scatterplot analysis of cycling cells by imaging cytometry. Based on the total and maximum intensities of the DAPI signals, the cells could be classified into G1, G2, mitosis (M), or aneuploidy (aneu; Pozarowski et al., 2004; Kawamura et al., 2006). Representative immunofluorescence images corresponding to each spot in the respective clusters are shown at the sides. (D) Quantification of cells with CGs in scatterplots. Cells with CGs were increasingly detected with time in the G1 stage and the G2 stage but at a higher frequency in G1. The values are the means and SDs of three independent experiments of >80 cells each. *p < 0.05. (E) Representative images of cells with MIGs. The frequency of the appearance of MIGs among mitotic cells was counted based on the scatterplots in C and is indicated below the images.
To determine when CGs appear in RANBP2-knockdown cells, we analyzed the correlation between speckle morphology and the cell cycle using scattergrams (Figure 3C). An immunofluorescence image corresponding to each spot in the scattergram was relocated according to the cell position, indicating that most cells with CGs were in G1 phase. At 72 h after RANBP2-siRNA transfection, >50% of G1 cells contained CGs, whereas they were present in <10% of G2 cells (Figure 3D). MIGs existed in both RANBP2-knockdown and control cells but more often among mitotic cells with RANBP2 knockdown (Figure 3E). Collectively, our data show that RANBP2-knockdown cells progress through the cell cycle without significant arrest and that the CG-containing cells are in G1 phase, suggesting that CGs initially emerge as MIGs and continue to grow into large granules in the cytoplasm after the cells enter G1 phase.
Impaired nuclear entry of MIG components may induce CG formation
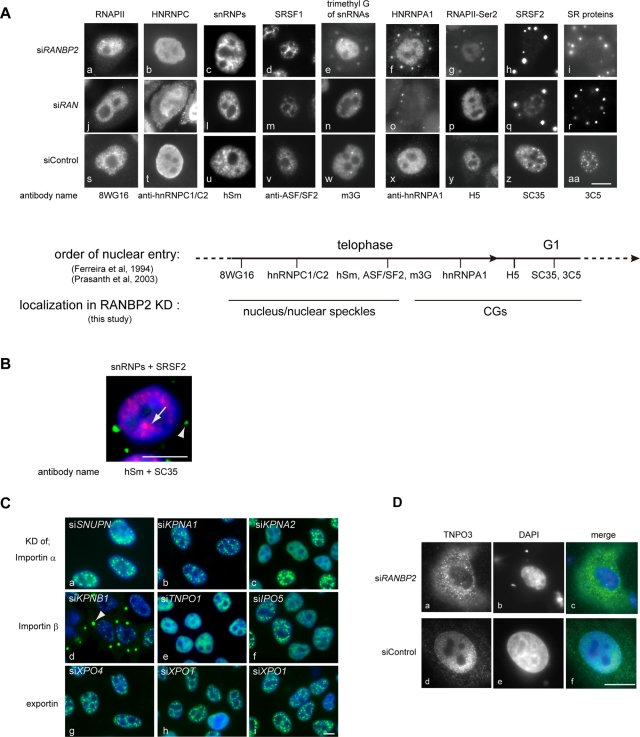
A previous proteomic analysis of nuclear speckles revealed that they contain >100 proteins, including snRNPs, SR proteins, and other RNA-associated proteins (Saitoh et al., 2004). Nuclear speckles also contain a hyperphosphorylated form of RNAPII (Bregman et al., 1995) and nucleus-retained poly(A)+ RNA, including MALAT1 (Huang et al., 1994; Tripathi et al., 2010). In interphase cells, certain speckle proteins selectively shuttle continuously between the nucleus and the cytoplasm, whereas others remain in the nucleus (Pinol-Roma and Dreyfuss, 1992; Caceres et al., 1998). By immunofluorescence and RNA fluorescent in situ hybridization (FISH), we asked whether components of speckles also reside in CGs. We found that not all, but rather a specific set of speckle-associated factors was located in CGs upon depletion of either RANBP2 or RAN (Figure 4 and Supplemental Figure S4). We found that CG localization did not depend on the shuttling properties of the factors (Supplemental Figure S4A). For example, SRSF1 (previously known as ASF/SF2) and HNRNPA1, which are both shuttling proteins, were found in CGs and the nucleus, respectively (Supplemental Figure S4A, a, b, e, and f). Similarly, nonshuttling proteins, including SRSF2 and HNRNPC (hnRNP C1/C2), were found in CGs and the nucleus, respectively, in the absence of RAN or RANBP2 (Supplemental Figure S4A, c, d, g, and h). This indicates that the formation of CGs is not simply a secondary effect of global transport inhibition during interphase. Because cells were treated with siRNAs for 48–72 h, which corresponds to multiple cell cycles, the specific subset of proteins may not be able to enter the re-forming nucleus once the cell goes through nuclear breakdown during mitosis.
FIGURE 4:
Sequential nuclear entry of speckle components at late mitosis was disrupted in RANBP2-depleted cells. (A) CG constituents correlated with those present in MIGs at very late mitosis. Immunofluorescence analyses of RANBP2- or RAN-depleted HeLa cells using various antibodies against speckle-associated components are shown. The subcellular localization of each factor revealed in this study is summarized at the bottom, along with the previously reported sequence of their nuclear import at mitosis (Ferreira et al., 1994; Prasanth et al., 2003). (B) Simultaneous staining of snRNPs (red) and SC35 antigens (green) in RANBP2-depleted cells showed that snRNPs are distributed in a speckled pattern in the nucleus (arrow) without SC35 (arrowhead). (C) HeLa cells were knocked down (KD) for a series of nuclear transport receptor family members and stained with SC35 (green) and DAPI (blue). CGs (arrowhead) were produced only when KPNB1 is depleted. IPO, importin; KPNA and KPNB, karyopherins; SNUPN, snurportin1; TNPO, transportin; XPO, exportin. (D) TNPO3 was concentrated in the cytoplasm and was absent from the nuclear rim in cells depleted of RANBP2. Cells treated with siRANBP2 were transfected with the plasmid SR2/pcDNA3-FLAG-HA, and the localization of TNPO3 was monitored by immunofluorescence using anti-FLAG antibodies. Bars, 10 μm.
Surveying the distributions of the multiple factors highlighted two interesting findings that suggest CG-formation mechanisms. First, the CG constituents were well correlated with those found in MIGs at very late mitosis (Figure 4A). Our immunofluorescence data showed that the late-nucleus-entering factors, including the trimethyl guanosine cap of small nuclear RNAs (m3G antigen), HNRNPA1, and the antigens for H5, SC35, and 3C5, were located in CGs (Figure 4A, e–i and o–r). On the other hand, the early-entering factors, including hypophosphorylated RNAPII (8WG antigen), HNRNPC, snRNPs, and SRSF1, were found in the nucleus (Figure 4A, a–c and j–l). Therefore the absence of the RAN system results in a specific interruption of the sequential nuclear entry of speckle components, and CGs may represent MIG derivatives that are produced by the prolonged sequestration of the late-entering factors. Of note, several speckle components, including snRNPs, SRSF1, and Y14 (Figure 4A, c, d, l, and m, and Supplemental Figure S5A, e), as well as bulk poly(A)+ RNAs (Supplemental Figure S4B, c and e), showed a speckled pattern in the nucleus of RAN- or RANBP2-knockdown cells. Simultaneous visualization confirmed that the CG-containing cells formed partial speckles of snRNPs, early-entering factors in the absence of SC35 antigens (Figures 4B and Supplemental Figure S4B, e). This suggests that a subset of speckle components is transported to the nucleus by RAN-unassisted mechanisms, as described for snRNPs and several other proteins (Fagotto et al., 1998; Nakielny and Dreyfuss, 1998; Truant et al., 1998; Kose et al., 1999; Yokoya et al., 1999; Huber et al., 2002). It is also possible that the effects of RANBP2 knockdown are specific to proteins that enter the nucleus at later time points because they use different transport receptors. To test this, we depleted cells for a series of importin α and β and exportin family members and asked whether these cells still produced CGs (Figure 4C). We found that knockdown of most transport receptors, including SNUPN, which mediates nuclear import of snRNPs, did not cause CG formation (Figure 4C, a). CGs emerged only when cells were depleted of KPNB1 (importin β1), a transport receptor for a wide variety of proteins (Figure 4C, d), or TNPO3 (transportin-SR2), an importin β family protein for SR proteins (Figure 2A, d). Consistent with this observation, the subcellular localization of TNPO3 was dysregulated in RANBP2-knockdown cells, suggesting that RANBP2 is a docking site for TNPO3 and is critical for its regulation (Figure 4D).
CG formation requires phosphorylation
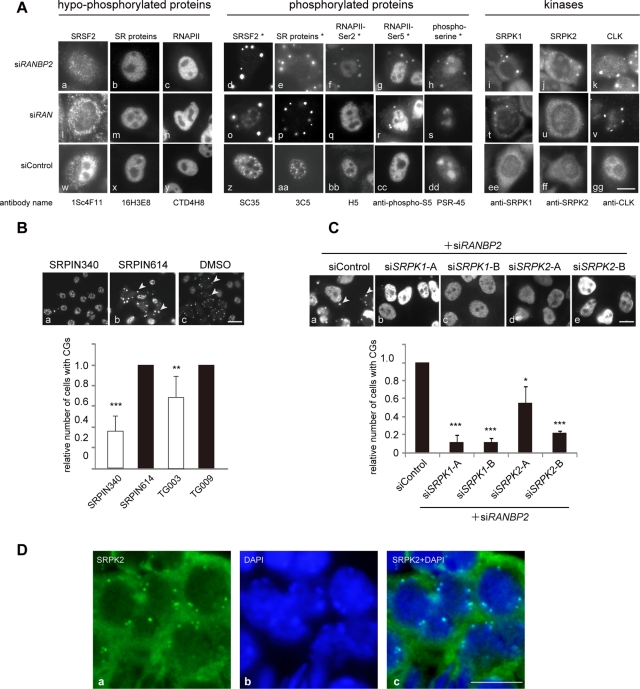
Our second finding from surveying the CG constituents was that CG localization of proteins correlated with their phosphorylation status. CGs were stained with SC35, which recognizes phosphorylated SRSF2 (Fu and Maniatis, 1990; Figure 5A, d and o) but not with the monoclonal antibody 1Sc4F11, which recognizes unphosphorylated SRSF2 (Cavaloc et al., 1999; Figure 5A, a and l). Similarly, CGs were stained with the monoclonal antibodies 3C5, 1H4, and mAb104, which recognize a series of phosphorylated SR proteins (Turner and Franchi, 1987; Roth et al., 1990; Neugebauer and Roth, 1997; Figure 5A, e and p, and Supplemental Figure S5A, a and b), but not with monoclonal antibody 16H3E8, which recognizes a series of dephosphorylated SR proteins (Figure 5A, b and m). In terms of RNAPII, the serine-2– and serine-5–phosphorylated forms (Bregman et al., 1995) were found in CGs (Figure 5A, f, g, q, and r), whereas antibodies CTD4H8 and 8WG16, which predominantly detect the hypophosphorylated form of RNAPII (Thompson et al., 1989), did not detect CGs at all (Figures 5A, c and n, and 4A, a and j). Consistent with these observations, phosphorylated serine, but not threonine, was enriched in CGs (Figure 5A, h and s, and Supplemental Figure S5A, c). Furthermore, the SR protein kinases SRPK1 and 2, as well as CLK, were found in CGs (Figure 5C, i–k). To elucidate the role of protein phosphorylation in CG formation, we treated cells with specific inhibitors of SR protein kinases and asked whether the formation of CGs containing SC35 was affected (Figure 5B). The chemical compound SRPIN340 preferentially inhibits SRPK1 and SRPK2, and TG003 targets CLK family members (Muraki et al., 2004). SRPIN614 and TG009 are structurally analogous to SRPIN340 and TG003, respectively, but do not exhibit any inhibitory effects and were thus used as negative controls. Treatment of RANBP2-knockdown cells with SRPIN340 or TG003 significantly decreased the frequency of CG-containing cells (Figure 5B). Similarly, when SRPKs and RANBP2 were simultaneously depleted, the number of CG-containing cells was dramatically reduced (Figure 5C), confirming that phosphorylation of SR proteins is required for the generation of CGs.
FIGURE 5:
Phosphorylation of SR proteins is involved in CG generation. (A) CG localization of proteins correlated with their phosphorylation status. Cells were stained with antibodies recognizing hypophosphorylated forms (left) and the corresponding phosphorylated forms (middle) of the indicated proteins. Phosphoepitopes are denoted with an asterisk (Thompson et al., 1989; Bregman et al., 1995; Cavaloc et al., 1999). Cells were also stained with antibodies against SR protein kinases (right). (B) RANBP2-knockdown cells were treated with SR protein kinase inhibitors (SRPIN340 and TG003; unfilled bars) or control compounds (SRPIN614 or TG009; filled bars; Kuroyanagi et al., 1998; Muraki et al., 2004) for 20 h and immunostained with SC35. Arrowheads indicate CGs (top). Relative numbers of RANBP2-negative CG-containing cells are shown on the graph. Inhibition of SRPKs led to a reduction in CG generation. The values are the means and SDs of three independent experiments of >250 cells each. **p < 0.01, ***p < 0.001 vs. control compounds. (C) Cells were treated with the indicated combinations of siRNAs and were immunostained with the monoclonal antibody mAb104. Relative numbers of RANBP2-SRPK–negative, CG-containing cells are shown. For knockdown of SRPK1 and SRPK2, two siRNAs with different target sequences were used. Depletion of SRPKs significantly reduced CG generation in RANBP2-knockdown cells. The values are the means and SDs of three independent experiments of >100 cells each. *p< 0.05, ***p < 0.001 vs. controls. (D) Srpk2 was also found in cytoplasmic granular structures in mouse seminiferous tubules. Bars, 10 μm (A–C), 5 μm (D).
Our data showing the colocalization of SR protein kinases SRPK1, SRPK2, and phosphorylated SR proteins in CGs (Figure 5A) and a previous report (Aubol et al., 2003) suggest that sequestration of the processive SR protein kinases in CGs may be responsible for the docking of hyperphosphorylated SR proteins in these structures. Nuclear translocation by TNPO3 depends on appropriate levels of phosphorylation of the SR proteins (Kataoka et al., 1999; Lai et al., 2001; Sanford and Bruzik, 2001; Zhong et al., 2009). It is noteworthy that MIGs in wild-type cells, as well as CGs in mouse seminiferous tubule cells, also accumulate SR protein kinase (Figure 5D and Supplemental Figure S5B), suggesting regulation by phosphorylation in wild-type cell lines, as well as in whole organisms.
RANBP2 is implicated in several molecular processes, including modification with SUMO proteins (Pichler et al., 2002). RANBP2 exerts SUMO E3 ligase activity at the NPC through its internal repeat domain (IR) at the carboxy-terminus (Pichler et al., 2002; Saitoh et al., 2006). To test the role of the SUMOylation activity of RANBP2 in nuclear speckle formation, we asked whether reintroduction of the subdomains of RANBP2 could repress CG generation. We found that a fusion construct containing the IR only moderately reduced the formation of CGs (Supplemental Figure S6). This suggests that the SUMOylation activity of RANBP2 has little effect on CG generation.
In summary, the CG components correlated well with those present in MIGs at very late mitosis. Depletion of RAN or RANBP2 did not interrupt the early stages of the sequential nuclear entry of MIG components, and speckles were partially reconstructed. However, knockdown of RAN or RANBP2 specifically affected the late step of nuclear entry, inducing CGs enriched with phosphorylated components. This suggests a novel regulatory mechanism for nuclear speckle formation involving RANBP2 and phosphorylation.
CG-containing cells have variant alternative splicing patterns
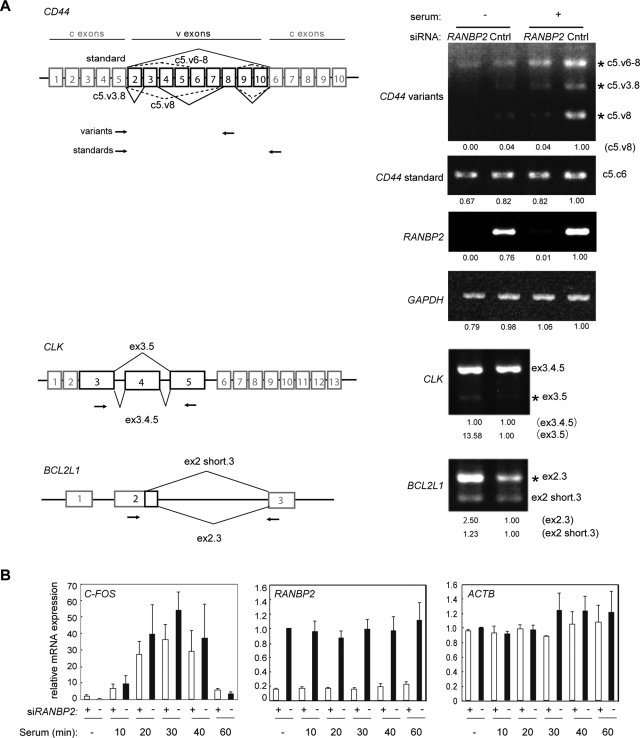
Recent studies suggested that nuclear speckles play a role in the coordinated and enhanced regulation of gene expression (Shopland et al., 2003; Saitoh et al., 2004; Brown et al., 2008; Hu et al., 2008). CGs are enriched in phosphorylated SR proteins and their kinases (Figure 5A), resulting in an imbalanced distribution of the phosphorylated and hypophosphorylated forms of SR proteins, which could affect gene regulation. Moreover, we demonstrated that the recently described nuclear speckle component SON (Sharma et al., 2010) and the long-nucleus-retained regulatory RNA MALAT1 (Tripathi et al., 2010), which are somehow involved in pre-mRNA splicing regulation, were also mislocalized in the absence of RANBP2 (Figures S5A, d, and S4B, a). Therefore we used reverse transcriptase (RT)-PCR to investigate how transcription and/or pre-mRNA splicing is affected in RANBP2-knockdown cells that contain semi-intact nuclear speckles (Figure 6). We first tested the alternative pre-mRNA splicing of human CD44, which contains 10 variable exons residing between the constitutive exons 5 and 6. The most common isoform of CD44, expressed under normal conditions, excludes all the variable exons (Figure 6A). CD44 variants that include different combinations of the variable exons are up-regulated upon serum stimulation signaling (Cheng et al., 2006). Of note, RANBP2 knockdown significantly reduced the levels of the CD44 alternatively spliced variants compared with control cells. On the other hand, the constitutive pre-mRNA splicing of CD44 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was not affected. Similarly, the alternative splicing of CLK and BCL2-like 1 (BCL2L1) was modified in RANBP2-depleted cells. Alternative splicing, but not constitutive transcription or pre-mRNA splicing, was altered in RANBP2-knockdown cells.
FIGURE 6:
RANBP2 knockdown affects pre-mRNA alternative splicing. (A) Alternative splicing patterns reported for the CD44 (Weg-Remers et al., 2001; Cheng et al., 2006), CLK, and BCL2L1 (Boise et al., 1993) transcripts (left). Variable exons that participate in alternative splicing (v) are indicated by black boxes, and constitutive exons (c) are indicated by gray boxes. Primers used for RT-PCR are shown by arrows. For CD44, the upper and lower pairs of primers amplified the variable and standard isoforms, respectively. HeLa cells were treated with siRNA for 48 h, and total RNA was purified to analyze the splicing patterns (right). For CD44, cells were also serum starved for 24 h prior to stimulation with 20% serum for 4 h. The quantitative band intensity relative to the control is indicated at the bottom. Alternatively spliced products affected by RANBP2 knockdown are marked with asterisks. RANBP2 and GAPDH were used to confirm the knockdown efficiency of RANBP2 and unaffected transcription, respectively. (B) Quantitative RT-PCR of the C-FOS gene under serum induction. The expression of C-FOS, an archetypal immediate early gene, was unaffected by RANBP2 knockdown. RANBP2 and ACTB served as controls for the knockdown efficiency and constant transcription, respectively. The values are the means and SDs of three independent experiments.
Transcription of the proto-oncogene C-FOS is markedly upregulated upon serum induction through a signaling cascade including the Rho family GTPase RHOA and phosphatidylinositol 3-kinase. Quantitative RT-PCR showed that the kinetics of this gene activation was almost identical in control and RANBP2-knockdown cells (Figure 6B), implying that transcriptional induction and serum-responsive signal transduction were intact in the RANBP2-knockdown cells. In conclusion, we found that alternative splicing is affected by the physical segregation of phosphorylated and hypophosphorylated SR proteins that is induced by RANBP2 knockdown. This suggests that the speckled distribution of phosphorylated SR proteins in the nucleus is important for regulation of alternative splicing of pre-mRNA.
DISCUSSION
The spatial organization of the eukaryotic nucleus reflects its gene expression profile, and the distribution pattern of the nuclear substructures relative to genes may govern genome function. In the present study, we demonstrated that the nucleoporin protein RANBP2 plays a specific role in nuclear speckle formation. Its loss resulted in the absence of SC35-positive nuclear speckles and, instead, the generation of CGs, novel granular structures in the cytoplasm of interphase cells. Phosphorylated forms of a subset of SR proteins and RNAPII, together with SRPKs, accumulated in CGs, and SRPKs were required for CG formation. CGs occurred mostly in G1-phase cells, suggesting that CGs are the remnants of MIGs, which are induced by dysfunction of the nucleocytoplasmic transport of phosphorylated SR proteins. More important, CG-containing cells were capable of constitutive transcription and pre-mRNA splicing, but their alternative splicing patterns were altered. This suggests a specific function of the speckled distribution of phosphorylated SR proteins in determining alternative pre-mRNA splicing patterns. Indeed, the distribution of SR proteins was developmentally regulated, and CGs existed in the mouse testis with reduced levels of Ranbp2 and high alternative splicing activity (Lander et al., 2001; Johnson et al., 2003).
Alternative pre-mRNA splicing provides one of the most powerful ways of gene expression regulation by selecting a combination of exons, leading to the generation of many distinct protein isoforms from a common primary transcript. Genome-wide studies indicated that alternative splicing is fundamental in vivo and that ∼95% of human multiexon genes are alternatively spliced (Pan et al., 2008; Wang et al., 2008). Many pre-mRNA splicing factors, including SR proteins, are phosphorylated; phosphorylation plays a crucial role in both constitutive and alternative splicing. It has been suggested that phosphorylation modulates protein–protein interactions in the spliceosome, which results in switches in splice-site recognition. Alternative splice-site choice is also mediated by changing the ratio of various splicing factors in the nucleus (van der Houven van Oordt et al., 2000; Zhong et al., 2009). In our study, we displaced a selected set of nuclear speckle components from the nucleus by depletion of RANBP2 and anchored them in CGs. Typically, phosphorylated SR proteins were sequestered in CGs in the cytoplasm, whereas hypophosphorylated SRs remained in the nucleus (Figures 4A and 5A, respectively). Therefore it is very likely that we altered the ratios of phosphorylated and hypophosphorylated splicing factors in the nucleus; this could be the major cause of the observed modification of splice-site selection (Figure 6). Constitutive pre-mRNA splicing may be less sensitive to the altered stoichiometry of splicing factors; this was maintained probably due to the snRNPs, hypophosphorylated SR proteins, and other proteins that remained in the nucleus in the absence of RANBP2 (Figures 4, A and B and 5A). We demonstrated cytoplasmic sequestration of phosphorylated SR proteins in several mouse tissues, and the distribution of phosphorylated SR proteins was developmentally regulated (Figure 1 and Supplemental Figure S1). In seminiferous tubules, we observed two distinct cytoplasmic structures: germinal granules and CGs (Figure 1). Because they each appeared during a specific stage of spermatogenesis, sequestration or storage of a set of RNA-binding proteins as granules in the cytoplasm may be a fundamental mechanism for RNA regulation during cell differentiation. The detailed molecular networks involved in nuclear speckle dynamics during developmental processes remain to be investigated.
In cell lines, nuclear speckles change their morphology under conditions such as transcription and pre-mRNA splicing inhibition, heat shock, and osmotic stress (Misteli and Spector, 1997; Zhong et al., 2009). These changes are largely regulated by phosphorylation of the SR proteins, and some are led by altered subcellular partitioning of SR protein kinases (Misteli and Spector, 1997; Zhong et al., 2009). Several of our observations suggest that CG generation is also under the control of phosphorylation. First, SR proteins and RNAPII were phosphorylated in CGs, whereas nonphosphorylated forms of these proteins were located in the nucleus. Second, SRPKs were sequestered in CGs in RANBP2-knockdown cells. Third, the frequency of CG generation was reduced when SRPKs were inhibited. In this context, it is intriguing that SRPKs also localized transiently in MIGs in wild-type cells (Supplemental Figure S5B). In normal cells, the MIG components are sequentially released from the structure and are translocated to the nucleus at the transition from mitosis to early G1 (Ferreira et al., 1994; Prasanth et al., 2003). Hyperphosphorylated SR proteins in MIGs in late mitosis may be dephosphorylated to an intermediate state, which becomes a good cargo substrate for TNPO3 to enter the nucleus using the RAN–RANBP2 system. Knockdown of either RAN or RANBP2 may disconnect the flow of these consecutive events at a specific step, resulting in clusters of hyperphosphorylated SR proteins in CGs. Our analysis of novel cytoplasmic structures, CGs, in a RANBP2-knockdown cell line revealed the regulatory mechanisms for reconstruction of the nuclear architecture after mitosis.
This study is the first demonstration that alternative splicing is governed by nucleocytoplasmic transport mechanisms via correct nuclear substructure formation. On the basis of our observation of CGs in human cell lines and mouse tissues, we propose that the regulatory mechanisms for alternative RNA splicing through an imbalanced distribution of posttranslationally modified splicing factors may be critical in cell differentiation during development.
MATERIALS AND METHODS
Cell culture
HeLa cells were maintained in DMEM and Ham's F-12 medium (Sigma-Aldrich, St. Louis, MO) containing 10% heat-inactivated fetal bovine serum (FBS), penicillin, and streptomycin. For serum stimulation, cells were first cultured for serum starvation in medium containing 0.5% FBS for 24 h and then stimulated with 20% FBS for the indicated period. For treatment of cells with SR protein kinase inhibitors, SRPIN340, SRPIN614, TG003, or TG009 (Kuroyanagi et al., 1998; Muraki et al., 2004) was added to the medium at a final concentration of 50 μM in dimethyl sulfoxide.
Transfection and RNA interference
Cells were transfected with plasmid DNAs using FuGENE 6 and FuGENE HD (Roche, Indianapolis, IN). For siRNA experiments, cells were transfected with siRNA duplex oligonucleotides (Japan Bioservice, Tokyo, Japan, and Nippon EGT, Tokyo, Japan) using RNAiMAX (Invitrogen, Carlsbad, CA). Knockdown cells were analyzed at 24, 48, or 72 h after transfection. The results were confirmed using multiple siRNA duplex oligonucleotides against different sequences. siRNA against firefly luciferase GL3 was used as a control, as described previously (Saitoh et al., 2006). For treatment with SR protein kinase inhibitors (Kuroyanagi et al., 1998; Muraki et al., 2004), cells were first transfected with RANBP2 siRNA, and 28 h later the inhibitors were added at 50 μM and incubated for another 20 h before immunofluorescence analyses.
Immunofluorescence and immuno-RNA FISH
Immunofluorescence of cell lines, including HeLa cells, was performed as described previously (Saitoh et al., 2006). Briefly, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature, washed with PBS, blocked with 0.5% bovine serum albumin in PBS, incubated with specific primary antibodies for 1 h at room temperature, and then incubated with appropriate secondary antibodies for 1 h. Cells were counterstained with DAPI (1 μg/ml) before mounting. For detection of SRSF1 and snRNPs, cells were extracted with 0.2% Triton X-100 in PBS for 5 min on ice prior to fixation. For detection of mAb104 and 3C5, cells were fixed in cold methanol for 3 min at –20°C. For in situ RNase assays, cells were preextracted with 0.2% Triton X-100 in CSK buffer (10 mM 1,4-piperazinediethanesulfonic acid (pH 6.8), 300 mM sucrose, 3 mM MgCl2, 100 mM NaCl) for 5 min on ice, then treated with RNase solution (1/25 diluted RNase cocktail [Applied Biosystems/Ambion, Austin, TX] containing a mixture of 20 U/ml RNase A and 800 U/ml RNase T1 with a protease inhibitor cocktail containing 4-(2-aminoethyl)- benzene-sulfonyl fluoride, chymostatin, leupeptin, pepstatin A, antipain, and aprotinin) for 60 min at 25°C, followed by fixation and immunostaining. For detection of poly(A)+ RNA and MALAT1, FISH was performed as described previously (Huang et al., 1994) prior to immunofluorescence. Briefly, cells were preextracted in CSK buffer containing 0.25% Triton X-100, 1 mM ethylene glycol tetraacetic acid, and 5 mM vanadyl-ribonucleoside complex for 5 min on ice and then fixed with 4% paraformaldehyde in PBS at room temperature for 10 min. The cells were permeabilized in PBS containing 0.5% Triton X-100 and 5 mM vanadyl-ribonucleoside complex for 5 min on ice. Hybridization was carried out with oligo(dT)50 end-labeled with digoxigenin (Roche) or nick-translated MALAT1 cDNA (a gift from K. V. Prasanth, University of Illinois, Urbana–Champaign, IL) in a moist chamber for 16 h. The cells were washed three times in 2× saline–sodium citrate (SSC) buffer containing 50% formamide (pH 7.2) at 37°C for 5 min, then three times in 2× SSC only, followed by incubation with secondary antibodies. Immunofluorescence of mouse tissues was performed essentially as described previously (Watanabe et al., 2009). Briefly, mouse tissues were fixed in 4% paraformaldehyde and embedded in paraffin. Histological sections were sliced at 4 μm and deparaffinized, and antigens were retrieved by autoclaving at 121°C for 5 min in 10 mM sodium citrate buffer (pH 6.0). The slides were immersed in 0.5% BlockAce (Dainippon Sumitomo Pharma, Osaka, Japan) in PBS for 30 min, incubated with primary antibodies for 1 h at room temperature, followed by incubation with secondary antibodies for another 1 h. For immunostaining of a mixed population of blood cells, bone marrow cells were isolated from adult mice and cultured in suspension with medium containing 50 ng/ml thrombopoietin (Chemicon, Temecula, CA) for 4 d. Cells were collected on slides with a cytospin, dried for 30 min at 37°C, fixed in 4% paraformaldehyde in PBS, and processed as for the immunofluorescence of cell lines as described.
Immunoblot analysis
Testes were obtained from adult mice, washed in PBS, and homogenized in extraction buffer (10 mM Tris-HCl [pH 7.5], 500 mM EDTA, 2 mM MgCl2, 2 mM 2-mercaptoethanol, 0.5% NP-40) supplemented with a protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan). Protein concentration was measured by a Pierce 660-nm Protein Assay Kit (Thermo Scientific, Waltham, MA). A 10-μg amount of cell lysate was incubated with 30 U of calf intestinal alkaline phosphatase (Takara Bio, Otsu, Japan) for 30 min at 37°C in the presence of a protease inhibitor cocktail (Nacalai Tesque). Lysate was applied to SDS–PAGE, followed by immunoblot as previously described (Ichimura et al., 2005).
Light microscopy and image analysis
Images were obtained with a microscope (IX-71; Olympus, Tokyo, Japan) equipped with a 60×, numerical aperture 1.0, Plan Apo objective lens, a cooled charged-coupled device camera (Hamamatsu, Hamamatsu, Japan), and image acquisition software (Lumina Vision Version 2.4; Mitani, Fukui, Japan). For imaging cytometry analyses, a CELAVIEW RS100 (Olympus) was used to capture images automatically; images were quantitatively analyzed using CELAVIEW analysis software (Olympus). Specialized software was designed for automated recognition of nuclear speckles, as described for Supplemental Figure S3.
Electron microscopy
Samples were prepared for immuno–electron microscopy as described (Spector et al., 1998). Briefly, cells transfected with siRNAs for 48 h were fixed with 4% paraformaldehyde plus 0.5% glutaraldehyde for 1 h at room temperature. They were then dehydrated with a methanol series and embedded in acrylic resin (Lowicryl K4M). Polymerization was conducted at –20°C within an UV light chamber. Sections were mounted on Formvar-coated gold grids. They were then floated on Tris-buffered saline with 0.2% Tween-20 for 30 min and immediately floated on the first antibody SC35 (Sigma-Aldrich) for 1 h at room temperature within a humid chamber. After rinsing with distilled water, the grids were floated on a solution of monoclonal secondary antibody coupled to 10-nm colloidal gold particles (1:100 dilution) for 1 h in the same conditions as for the first antibody. The grids were contrasted for 2 min with 3% uranyl acetate and for 1 min with 0.3% lead citrate. For electron micrographs of osmium-fixation, cells were prepared as described in standard protocols (Jiménez-García and Segura-Valdez, 2004). Briefly, cells were fixed with 2.5% glutaraldehyde for 1 h at room temperature. They were then postfixed with 1% osmium tetroxide for 1 h and dehydrated with a series of graded ethanol and propylene oxide. Embedding was performed with epoxy resin at 60°C for 16 h. Thin sections were placed on copper grids covered with Formvar. Contrast was conducted with 5% uranyl acetate and 0.5% lead citrate. Grids were observed with a transmission electron microscope (JEOL 1010, JEOL, Peabody, MA) working at 80 kV. Images were obtained with a charge-coupled device camera coupled to the microscope.
Plasmid construction
The cDNA plasmids pcDEBRanT24N, pcDEBRanG19V, pcDEB, and TKS-BP2 were gifts from T. Nishimoto (Kyushu University, Fukuoka, Japan). The cDNA for TNPO3 (SR2/pcDNA3-FLAG-HA) was cloned into pcDNA3-FLAG-HA by amplifying the insert of the plasmid pGST-TRN-SR2 (a gift from T. Woan-Yuh, Academia Sinica, Taiwan). A series of constructs to express hemagglutinin (HA)-NUP88 fused to each region of RANBP2 (Region 1–4 or IR; Supplemental Figure S6) was created as follows. First, a DNA fragment of NUP88 was excised from NUP88-pOTB7 (a gift from S. Yoshimura, Kyoto University, Kyoto, Japan), and then it was inserted into pcDNA3-HA to create pcDNA3-HA-NUP88. To generate pcDNA3-HA-NUP88-Region1–4, each region was amplified by PCR from the template cDNA plasmid TKS-BP2, digested with a restriction enzyme (NheI for Region 1, XbaI for Regions 2–4), and inserted into pcDNA3-HA-NUP88. To generate pcDNA3-HA-NUP88-IR, the IR region (amino acids 2553–2936) was excised from pEGFPC3BP2IR (a gift from H. Saitoh, Kumamoto University, Kumamoto, Japan) with EcoRI and BamHI and inserted into pcDNA3-HA, and the NheI–XbaI fragment containing the IR was further excised and reinserted into the XbaI site of pcDNA3-HA-NUP88.
RT-PCR
Total RNA was isolated using Isogen (Nippon Gene, Tokyo, Japan). For cDNA synthesis, 1 μg of total RNA was reverse transcribed with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative PCR of the target cDNAs was performed using Power SYBR Green PCR Master Mix (Applied Biosystems). Each experiment was performed at least three times. The fold relative enrichment was quantified, with normalization to β-actin (ACTB) or GAPDH.
Supplementary Material
Acknowledgments
We thank Yaeko Murase (Olympus Corporation) and Yuko Nakatsu, Yuko Hino, and Kayo Kinoshita (Kumamoto University) for technical assistance. We thank David L. Spector (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) for valuable discussions and for providing reagents. We thank Tokio Tani, Hisato Saitoh, Kazuaki Tokunaga, Satoshi Tanaka, Yasuka Yamaguchi, Hiroshi Sakamoto, and Minetaro Ogawa (Kumamoto University) for providing reagents and for technical advice. We also thank Takeharu Nishimoto (Kyushu University), Kannanganattu V. Prasanth (University of Illinois), Paula A. Bubulya (Wright State University, Dayton, OH), James Stevenin (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France), Jens Lykke-Andersen (University of Colorado, Boulder, CO), and Woan-Yuh Tarn (Academia Sinica, Taiwan) for kindly providing reagents. We thank Noriko Yasuhara (Osaka University, Osaka, Japan), Kazuyuki Ohbo (Yokohama City University, Kanagawa, Japan), and Nobuyoshi Akimitsu (University of Tokyo, Tokyo, Japan) for helpful discussions and Edith Heard (Institut Curie, Paris, France) for critical reading of the manuscript. This work was partly supported by a Grant-in-Aid for the Global Center of Excellence “Cell Fate Regulation Research and Education Unit,” Kumamoto University, and a Grant-in-Aid for Scientific Research on Priority Areas from the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to N.S. and M.N.).
Abbreviations used:
- FISH
fluorescence in situ hybridization
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase
- IGC
interchromatin granule cluster
- MIG
mitotic interchromatin granule
- NPC
nuclear pore complex
- RANBP2
Ran-binding protein 2
- RCC1
regulator of chromosome condensation
- RNP
ribonucleoprotein
- siRNA
small interfering RNA
- SUMO
small ubiquitin-like modifiers
- TNPO3
transportin 3
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-09-0783) on January 19, 2012.
REFERENCES
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci USA. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boise LH, Gonzalez-Garcia M, Postema CE, Ding L, Lindsten T, Turka LA, Mao X, Nunez G, Thompson CB. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12:55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaloc Y, Bourgeois CF, Kister L, Stevenin J. The splicing factors 9G8 and SRp20 transactivate splicing through different and specific enhancers. RNA. 1999;5:468–483. doi: 10.1017/s1355838299981967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Yaffe MB, Sharp PA. A positive feedback loop couples Ras activation and CD44 alternative splicing. Genes Dev. 2006;20:1715–1720. doi: 10.1101/gad.1430906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Klebe C, Wittinghofer A, Karsenti E. Regulation of Cdc2/cyclin B activation by Ran, a Ras-related GTPase. J Cell Sci. 1995;108((Pt 3)):1217–1225. doi: 10.1242/jcs.108.3.1217. [DOI] [PubMed] [Google Scholar]
- Colwill K, Feng LL, Yeakley JM, Gish GD, Caceres JF, Pawson T, Fu XD. SRPK1 and Clk/Sty protein kinases show distinct substrate specificities for serine/arginine-rich splicing factors. J Biol Chem. 1996;271:24569–24575. doi: 10.1074/jbc.271.40.24569. [DOI] [PubMed] [Google Scholar]
- Dasso M, Seki T, Azuma Y, Ohba T, Nishimoto T. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 1994;13:5732–5744. doi: 10.1002/j.1460-2075.1994.tb06911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Curr Biol. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gui JF, Tronchere H, Chandler SD, Fu XD. Purification and characterization of a kinase specific for the serine- and arginine-rich pre-mRNA splicing factors. Proc Natl Acad Sci USA. 1994;91:10824–10828. doi: 10.1073/pnas.91.23.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci USA. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Plutz M, Belmont AS. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–719. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol. 2002;156:467–479. doi: 10.1083/jcb.200108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura T, Watanabe S, Sakamoto Y, Aoto T, Fujita N, Nakao M. Transcriptional repression and heterochromatin formation by MBD1 and MCAF/AM family proteins. J Biol Chem. 2005;280:13928–13935. doi: 10.1074/jbc.M413654200. [DOI] [PubMed] [Google Scholar]
- Jiménez-García LF, Segura-Valdez ML. Visualizing Nuclear Structure In Situ by Atomic Force Microscopy. Totowa, NJ: Humana Press; 2004. [DOI] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Morita N, Domiki C, Fujikawa-Yamamoto K, Hashimoto M, Iwabuchi K, Suzuki K. Induction of centrosome amplification in p53 siRNA-treated human fibroblast cells by radiation exposure. Cancer Sci. 2006;97:252–258. doi: 10.1111/j.1349-7006.2006.00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. Beta-subunit of nuclear pore-targeting complex (importin-beta) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–3952. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi N, Onogi H, Wakabayashi T, Hagiwara M. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem Biophys Res Commun. 1998;242:357–364. doi: 10.1006/bbrc.1997.7913. [DOI] [PubMed] [Google Scholar]
- Lai MC, Lin RI, Tarn WY. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc Natl Acad Sci USA. 2001;98:10154–10159. doi: 10.1073/pnas.181354098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM. Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell. 2006;126:543–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Xiao R, Sun P, Xu X, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–425. doi: 10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. RNA splicing: what has phosphorylation got to do with it? Curr Biol. 1999;9:R198–R200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Spector DL. Protein phosphorylation and the nuclear organization of pre-mRNA splicing. Trends Cell Biol. 1997;7:135–138. doi: 10.1016/S0962-8924(96)20043-1. [DOI] [PubMed] [Google Scholar]
- Muraki M, et al. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J Biol Chem. 2004;279:24246–24254. doi: 10.1074/jbc.M314298200. [DOI] [PubMed] [Google Scholar]
- Nagai M, Moriyama T, Mehmood R, Tokuhiro K, Ikawa M, Okabe M, Tanaka H, Yoneda Y. Mice lacking Ran binding protein 1 are viable and show male infertility. FEBS Lett. 2011;585:791–796. doi: 10.1016/j.febslet.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Import and export of the nuclear protein import receptor transportin by a mechanism independent of GTP hydrolysis. Curr Biol. 1998;8:89–95. doi: 10.1016/s0960-9822(98)70039-9. [DOI] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- Pozarowski P, Huang X, Gong RW, Priebe W, Darzynkiewicz Z. Simple, semiautomatic assay of cytostatic and cytotoxic effects of antitumor drugs by laser scanning cytometry: effects of the bis-intercalator WP631 on growth and cell cycle of T-24 cells. Cytometry A. 2004;57:113–119. doi: 10.1002/cyto.a.10121. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Sacco-Bubulya PA, Prasanth SG, Spector DL. Sequential entry of components of the gene expression machinery into daughter nuclei. Mol Biol Cell. 2003;14:1043–1057. doi: 10.1091/mbc.E02-10-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh N, Uchimura Y, Tachibana T, Sugahara S, Saitoh H, Nakao M. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp Cell Res. 2006;312:1418–1430. doi: 10.1016/j.yexcr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Salina D, Enarson P, Rattner JB, Burke B. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol. 2003;162:991–1001. doi: 10.1083/jcb.200304080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Bruzik JP. Regulation of SR protein localization during development. Proc Natl Acad Sci USA. 2001;98:10184–10189. doi: 10.1073/pnas.181340498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA. Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell. 2010;21:650–663. doi: 10.1091/mbc.E09-02-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Byron M, McNeil J, Lawrence JB. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Goldman RD, Leinwand LA. Cells: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- Spector DL, Lamond AI. Nuclear speckles. Cold Spring Harb Perspect Biol. 2011;3(2) doi: 10.1101/cshperspect.a000646. pii: a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truant R, Fridell RA, Benson ER, Herold A, Cullen BR. Nucleocytoplasmic shuttling by protein nuclear import factors. Eur J Cell Biol. 1998;77:269–275. doi: 10.1016/s0171-9335(98)80085-x. [DOI] [PubMed] [Google Scholar]
- Turner BM, Franchi L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J Cell Sci. 1987;87:269–282. doi: 10.1242/jcs.87.2.269. [DOI] [PubMed] [Google Scholar]
- van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149:307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Ueda Y, Akaboshi S, Hino Y, Sekita Y, Nakao M. HMGA2 maintains oncogenic RAS-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am J Pathol. 2009;174:854–868. doi: 10.2353/ajpath.2009.080523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weg-Remers S, Ponta H, Herrlich P, Konig H. Regulation of alternative pre-mRNA splicing by the ERK MAP-kinase pathway. EMBO J. 2001;20:4194–4203. doi: 10.1093/emboj/20.15.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlwend D, Strasser A, Dickmanns A, Ficner R. Structural basis for RanGTP independent entry of spliceosomal U snRNPs into the nucleus. J Mol Biol. 2007;374:1129–1138. doi: 10.1016/j.jmb.2007.09.065. [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- Yokoya F, Imamoto N, Tachibana T, Yoneda Y. β-Catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.