Twenty-four Caenorhabditis elegans genes are involved in GPI-anchor synthesis. Based on the isolation of a deletion allele of the PIGA gene mediating the first step of GPI-anchor synthesis, GPI-anchor synthesis in somatic gonads and/or in germline is shown to be indispensable for the normal development of oocytes and eggs.
Abstract
Glycosylphosphatidylinositol (GPI)-anchor attachment is one of the most common posttranslational protein modifications. Using the nematode Caenorhabditis elegans, we determined that GPI-anchored proteins are present in germline cells and distal tip cells, which are essential for the maintenance of the germline stem cell niche. We identified 24 C. elegans genes involved in GPI-anchor synthesis. Inhibition of various steps of GPI-anchor synthesis by RNA interference or gene knockout resulted in abnormal development of oocytes and early embryos, and both lethal and sterile phenotypes were observed. The piga-1 gene (orthologue of human PIGA) codes for the catalytic subunit of the phosphatidylinositol N-acetylglucosaminyltransferase complex, which catalyzes the first step of GPI-anchor synthesis. We isolated piga-1–knockout worms and found that GPI-anchor synthesis is indispensable for the maintenance of mitotic germline cell number. The knockout worms displayed 100% lethality, with decreased mitotic germline cells and abnormal eggshell formation. Using cell-specific rescue of the null allele, we showed that expression of piga-1 in somatic gonads and/or in germline is sufficient for normal embryonic development and the maintenance of the germline mitotic cells. These results clearly demonstrate that GPI-anchor synthesis is indispensable for germline formation and for normal development of oocytes and eggs.
INTRODUCTION
Glycosylphosphatidylinositol (GPI) anchors are used by organisms from archaebacteria to humans. Pathogens such as Trypanosoma cruzi use GPI-anchor variants to escape from host immune surveillance, and the enzymes involved in their synthesis are potential drug targets for the treatment and prevention of disease (Ikezawa, 2002; Tsukahara et al., 2003). Paroxysmal nocturnal hemoglobinuria (PNH) is a human acquired genetic disease caused by “somatic” mutation of the X chromosome–linked phosphatidylinositol glycan-class A (PIGA) gene in hematopoietic progenitor cells (Nishimura et al., 1999). The PIGA gene codes for the major catalytic subunit (subunit A) of the glycosylphosphatidylinositol N-acetylglucosaminyltransferase complex (GPI-GnT; EC 2.4.1.198), which catalyzes the first step of GPI-anchor synthesis. Mutations in this gene would thus inhibit all GPI-anchor synthesis. As a result of the lack of GPI-anchored proteins, the complement system attacks the affected red blood cells, causing hemolysis and hematuria. No heritable form of PNH has been reported, and among the more than 29 genes involved in GPI-anchor synthesis, the only heritable mutations discovered in humans are in the PIGM promoter region (Almeida et al., 2006, 2009) and in the PIGV gene, which is deficient in patients with hyperphosphatasia mental retardation syndrome (Mabry syndrome; Krawitz et al., 2010). One explanation for the rarity of heritable GPI-anchor diseases is that the GPI modification is not essential, and mutations in these genes result in no adverse phenotypes. The other possibility is that the GPI-anchor modification is so indispensable that most mutations in these genes are lethal, so that organisms with such mutations cannot complete gametogenesis and/or embryonic stages. It is unusual to obtain high chimerism from Pig-a (murine PIGA orthologue)–disrupted embryonic stem cells, and no Pig-a–knockout mice have been generated, with the exception of a conditional Cre-loxP knockout (Kawagoe et al., 1996; Rosti et al., 1997; Keller et al., 1999; Nozaki et al., 1999). We previously reported that Ca2+-dependent cell–cell adhesion is mediated by GPI-anchored proteins in Xenopus laevis blastula cells. These proteins have partial amino acid sequences identical to that of cortical granule lectin, which is involved in the inhibition of polyspermy during fertilization (Nomura et al., 1998). On the basis of these results, we planned to study the roles of GPI-anchor synthesis in germline formation and early embryogenesis. Because it is not feasible to observe gametogenesis and early embryogenesis in ovo in mammals, no information on the roles of GPI anchors in these fundamental processes has yet been reported. Owing to the transparency of its body, we instead used Caenorhabditis elegans to study germline cell formation and early embryogenesis. The presence of GPI-anchored proteins in C. elegans was first predicted by sequence analysis (Eisenhaber et al., 2003), and various GPI-anchor protein–predicting algorithms have been developed and used to predict which proteins will be modified by GPI anchors. For instance, 159 worm proteins are now predicted to be “highly probable” GPI-anchored proteins by the FragAnchor GPI predictor, which is based on the tandem use of a neural network predictor and a hidden Markov model predictor (Poisson et al., 2007). By biochemical analysis followed by transfection of protein-coding genes into mammalian cells, several worm proteins (e.g., PHG-1 [ Agostoni et al., 2002] and WRK-1; see later discussion) have been identified to be GPI anchored. Recently Rao et al. (2011) studied proteins in the membrane raft and showed that two proteins are sensitive to phosphatidylinositol-specific phospholipase C (PI-PLC) treatment, which is frequently used to identify GPI-anchored proteins biochemically. Yet, to our knowledge, no systematic study including histological examination of the proteins has been conducted on GPI-anchored proteins in C. elegans. In this study, we examined whether GPI-anchored proteins are present in the C. elegans germline. We found GPI-anchored proteins in oocytes, sperm, and other somatic cells of the nematode with fluorescence microscopy and biochemical analysis. We carried out RNA interference (RNAi) screening of the worm orthologues of human genes involved in GPI-anchor synthesis and found that inhibition of GPI-anchor synthesis results in germline abnormalities. With piga-1–knockout worms devoid of GPI-anchor synthesis, we confirmed the essential roles of GPI-anchor synthesis in germline and eggshell formation.
RESULTS
Identification of GPI-anchored proteins in C. elegans
To determine whether GPI-anchors are present in C. elegans, we initially performed fluorochrome (Alexa 488)–labeled inactivated aerolysin (FLAER) staining. FLAER is an Aeromonas hydrophila toxin that binds specifically to GPI-anchored proteins (Brodsky et al., 2000; Hong et al., 2002). Cell membranes of oocytes and sperm in the dissected gonad were brightly stained by FLAER (Figure 1A), as were distal tip cells (DTCs) and their processes (Figure 1B). Similar bright fluorescence was detected in cell membranes of muscles and neurons in adult hermaphrodites (unpublished data). These results strongly suggest that GPI anchors are present in germline cells and somatic cells of the nematode. Next we fractionated total proteins from wild-type N2 worms to identify GPI-anchored proteins. Potential GPI-anchored proteins were isolated by phase partition with the detergent Triton X-114 (Hooper, 1992; Nomura et al., 1998). In this method, membrane proteins and GPI-anchored proteins are partitioned into the detergent-rich phase, whereas soluble proteins remain in the aqueous phase. The proteins in the detergent-rich phase were treated with PI-PLC to specifically cleave GPI anchors, and the Triton X-114 phase partitioning was repeated. The proteins that moved to the aqueous phase after PI-PLC treatment were analyzed by SDS–PAGE (Figure 1C). Twelve protein bands were cut out of the gel, and the protein sequences were identified using liquid chromatography–electrospray tandem mass spectrometry (LC/MS/MS). Forty-two proteins were identified, and their sequences were scanned for possible GPI-anchor modification signal sequences using the Big-PI (Eisenhaber et al., 2003), GPI-SOM (Fankhauser and Maser, 2005), FragAnchor (Poisson et al., 2007), and PredGPI (Pierleoni et al., 2008) programs. Twenty-two proteins were predicted to be GPI anchored by at least two different programs (Supplemental Table S1).
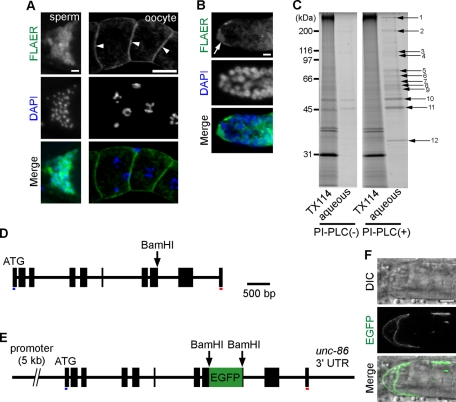
FIGURE 1:
GPI-anchored proteins in wild-type C. elegans germline. (A) Dissected gonadal arm stained with Alexa 488–labeled proaerolysin (FLAER; green). Oocytes (arrowheads) and sperm were strongly labeled by FLAER. Bars, 5 μm (left column) and 20 μm (right column). (B) DTC expressing GPI-anchored proteins. Strong FLAER staining is observed in DTC (arrow) with its cytonemes. Bar, 5 μm. (C) GPI-anchored proteins concentrated by Triton X-114 (TX114) phase partitioning, followed by PI-PLC treatment were analyzed by SDS–PAGE. SDS–PAGE of the detergent-rich phase (TX114) and aqueous phase are shown before (PI-PLC (–)) and after PI-PLC (PI-PLC (+)) treatment. Numbered arrows indicate possible GPI-anchored proteins. (D) Structure of the wrk-1 (F41D9.3) gene isoform b (wrk-1b). Black boxes indicate exons. (E) Schematic of an egfp-tagged wrk-1b reporter gene construct. The coding sequence of EGFP was inserted into the BamHI restriction site in exon 8 of the wrk-1b gene. Blue and red underlines indicate the predicted coding sequences of the N-terminal signal sequence and the C-terminal GPI-anchor modification signal, respectively. (F) A typical GPI-anchored protein, WRK-1, in DTCs. EGFP-tagged WRK-1 proteins are visible on the DTC surface. Bar, 5 μm.
One of the GPI-anchor candidate proteins identified was WRK-1. WRK-1 is an orthologue of the Drosophila Wrapper protein, which belongs to the immunoglobulin superfamily and is GPI anchored (Boulin et al., 2006). The behavior of the WRK-1 protein was examined in vivo by transgenic analysis with enhanced green fluorescent protein (EGFP)–tagged WRK-1 protein (Figure 1, D–F). The tagged protein was detected in the cell membranes of DTCs and muscle cells, and disruption of GPI-anchor synthesis resulted in abnormal accumulation of the protein inside DTCs (see Figure 4D later in the paper). These results demonstrate that WRK-1 is GPI anchored and that the Triton X-114 phase partitioning reliably separated GPI-anchored proteins. Thus C. elegans has GPI-anchored proteins that are widely expressed in oocytes, sperm, DTCs, muscle, neurons, and other cell types.
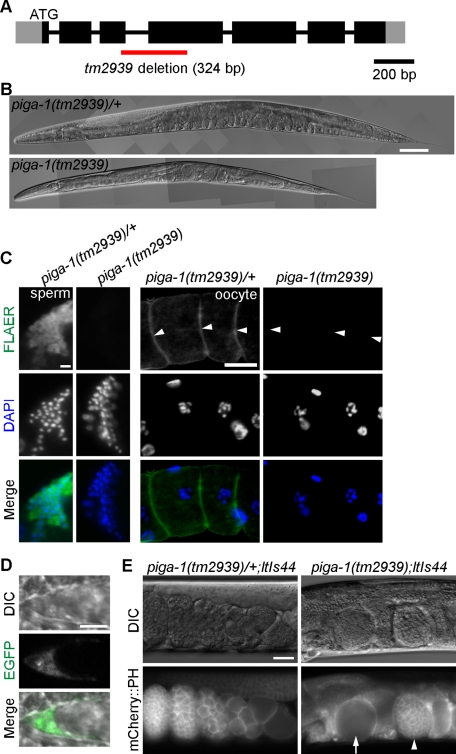
FIGURE 4:
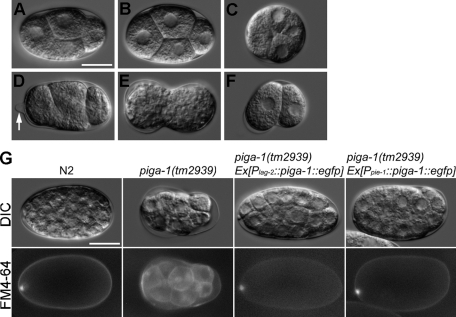
Knockout of piga-1 results in a germline-lethal phenotype. (A) Structure of the piga-1 gene. Black and gray boxes indicate exons and untranslated regions, respectively. The red line indicates the region deleted in the tm2939 allele. (B) Although piga-1(tm2939)/+ worms showed no apparent phenotype, piga-1(tm2939) worms showed reduced body length and various abnormalities. Bar, 100 μm. (C) GPI-anchored proteins were absent in piga-1(tm2939) homozygous worms. No FLAER staining was observed in dissected gonads of the piga-1(tm2939) worms, whereas bright staining was observed in gonads of the piga-1(tm2939)/+ worms. Arrowheads indicate oocyte membranes. Bars, 5 μm (left column) and 20 μm (right column). (D) The GPI-anchored protein WRK-1 was mislocalized in piga-1–knockout worms. EGFP-tagged WRK-1 was not present on the surface of DTCs but instead remained inside the cells. Bar, 5 μm. (E) Abundant abnormal eggs in the uteri of piga-1(tm2939);ltIs44 worms. Nondividing eggs (arrow) and dividing eggs at the multicellular stage (arrowhead) were present, but all were nonviable. Bar, 20 μm.
Knockdown of genes involved in C. elegans GPI-anchor synthesis
After confirming the presence of GPI-anchored proteins in the nematode, we next asked whether genes involved in GPI-anchor synthesis are conserved between nematodes and humans. More than 29 different genes are involved in human GPI-anchor synthesis (Kinoshita et al., 2007). Through a PSI-BLAST search using human gene sequences, we confirmed the presence of 24 orthologous genes in the C. elegans genome, although no orthologues for PIGH, PIGY, PIGL, PIGF, or DPM2 were identified (Figure 2 and Table 1). Knockdown phenotypes of these genes were analyzed by feeding RNAi. Suppression of GPI-anchor synthesis resulted in abnormalities in oogenesis, fertilization, and embryogenesis (Figure 3 and Table 1). Single RNAi against the following orthologous genes resulted in the sterile progeny phenotype, in which no oocytes or eggs were present in adult F1 hermaphrodites (C. elegans sequence names in parentheses): PIGP (Y48E1B.2), PIGV (T09B4.1), PIGO (C27A12.9), PIGK (T05E11.6), PIGU (T22C1.3), GAA1 (F33D11.9), MPDU1 (F38E1.9), and DPM3 (F28D1.11).
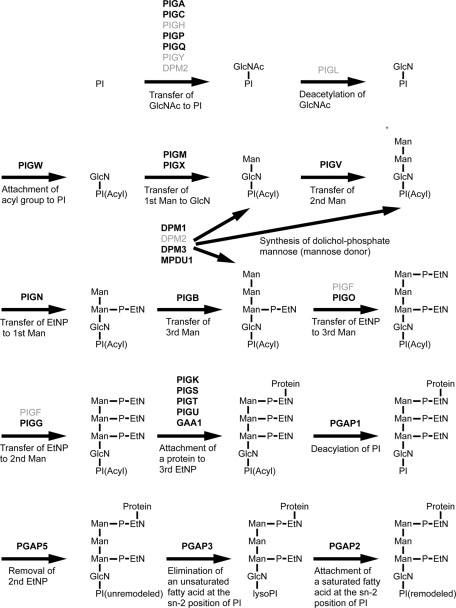
FIGURE 2:
GPI-anchor synthesis in the mammalian cells. The orthologous genes found in the C. elegans genome are shown in bold black text (Table 1). GlcN, glucosamine; GlcNAc, N-acetylglucosamine; Man, mannose; P-EtN, ethanolamine phosphate; PI, phosphatidylinositol.
TABLE 1:
Genes involved in GPI-anchor synthesis in C. elegans.
| Human gene | C. elegans orthologue | Identity (%) | RNAi phenotype | Human gene | C. elegans orthologue | Identity (%) | RNAi phenotype |
|---|---|---|---|---|---|---|---|
| PIGA | piga-1 (D2085.6) | 52 | WT | PIGG | F28C6.4 | 40 | WT |
| PIGC | T20D3.8 | 32 | WT | PIGK | T05E11.6 | 52 | Stpa (15%, n = 510) |
| PIGH | Not found | — | — | PIGS | T14G10.7 | 39 | WT |
| PIGP | Y48E1B.2 | 45 | Stpb (32%, n = 576) | PIGT | F17C11.7 | 26 | WT |
| PIGQ | F01G4.5 | 34 | WT | PIGU | T22C1.3 | 26 | Stpa (14%, n = 542) |
| PIGY | Not found | — | — | GAA1 | F33D11.9 | 25 | Stpa (14%, n = 614) |
| PIGL | Not found | — | — | PGAP1 | T19B10.8 | 29 | WT |
| PIGW | Y110A2AL.12 | 31 | WT | PGAP5 | B0511.13 | 35 | WT |
| PIGM | B0491.1 | 42 | WT | PGAP3 | R01B10.4 | 31 | WT |
| PIGX | F49E7.2 | 42 | WT | PGAP2 | T04A8.12 | 40 | WT |
| PIGV | T09B4.1 | 24 | Stpa (18%, n = 562) | Genes also involved in N-/O-glycosylation: | |||
| PIGN | Y54E10BR.1 | 34 | WT | DPM1 | dpm-1 (Y66H1A.2) | 65 | Sck (100%, n = 408) Lva (92%, n = 408) |
| PIGB | T27F7.4 | 36 | WT | DPM2 | Not found | — | — |
| PIGF | Not found | — | — | DPM3 | dpm-3 (F28D1.11) | 45 | Emb (40%, n = 698) Stp (10%, n = 698) |
| PIGO | C27A12.9 | 41 | Stpa (15%, n = 519) | MPDU1 | F38E1.9 | 40 | Stp (13%, n = 580) |
Identities indicate the percentage of amino acid sequence identities. Emb, embryonic lethal phenotype; Lva, larval arrest phenotype; Sck, sick phenotype; Stp, sterile progeny phenotype; WT, wild-type.
aSterility with the fertilization defect (Figure 3E).
bSterility with the abnormal oocyte maturation (Figure 3, A, C, and D).
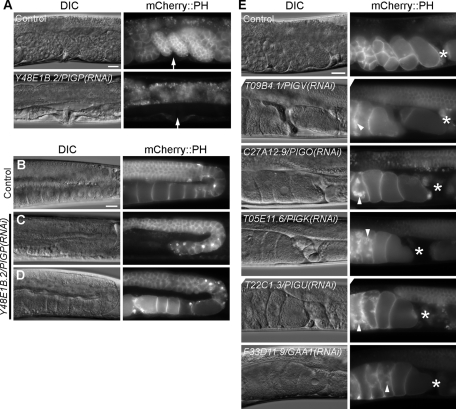
FIGURE 3:
RNAi-mediated knockdown of genes involved in GPI-anchor synthesis results in severe germline phenotypes. (A) In typical Y48E1B.2/PIGP RNAi-treated worms, no eggs were found in the uteri. Arrows indicate the position of the vulva. (B–D) Abnormalities of germline cells in Y48E1B.2/PIGP RNAi-treated worms with no eggs (A). Wild-type gonad with normal oocytes in control RNAi-treated worms (B). Gonad without matured oocytes (C) and with irregularly shaped oocytes (D) in Y48E1B.2/PIGP RNAi-treated worms. (E) Knockdown of T09B4.1/PIGV, C27A12.9/PIGO and the GPI transamidase complex (T05E11.6/PIGK, T22C1.3/PIGU, and F33D11.9/GAA1) resulted in abnormal oocytes just past the spermatheca. Dislocated membrane materials labeled with mCherry::PH were observed around the nuclei and in the cytosol of abnormal oocytes in the uteri (arrowheads). Asterisks indicate the spermatheca. Bars, 20 μm.
To visualize membrane abnormalities in germline cells, we used the OD70 strain (Ppie-1::mCherry::PH (PLC delta1)) for RNAi analysis. PIGP is one of the subunits of the GPI-GnT complex that catalyzes the first step of GPI-anchor synthesis, in which PIGA is the catalytic subunit. PIGP (Y48E1B.2) RNAi yielded F1 adult animals without fertilized or unfertilized eggs (Figure 3A; 32%, n = 576). In the affected F1 animals, oocyte formation was also defective, and the gonads contained no matured oocytes (Figure 3C; 71%, n = 24). In some animals, although oocytes were formed, they were irregular in shape (Figure 3D; 29%, n = 24). In the single-RNAi knockdowns of the PIGV, PIGO, and GPI transamidase orthologues (subunit genes PIGK, PIGU, and GAA1), oocytes just past the spermatheca showed no indication of cleavage, and abnormally dislocated mCherry membrane materials were detected in the cytosol and around the nuclei of oocytes (Figure 3E). Severe defects were observed in eggshell formation. In oocytes just past the spermatheca, no eggshells were visible, and fertilization seemed to be defective. PIGV and PIGO transfer the second mannose and third mannose, respectively, to the glycosylphosphatidylinositol during GPI precursor assembly. The GPI transamidase (PIGK, PIGU, and GAA1) mediates GPI anchoring in the endoplasmic reticulum by replacing a protein's C-terminal GPI attachment signal peptide with a preassembled GPI. Thus RNAi inhibition of the first step of GPI-anchor synthesis (PIGP), of the attachment of mannoses to GPI (PIGV and PIGO), and of the attachment of the GPI-anchor to proteins (PIGK, PIGU, and GAA1) resulted in severe defects in oogenesis. No morphological, growth, or movement abnormalities were observed in the F1 worms. These results indicate that GPI-anchor synthesis is essential for normal development of germline cells, oocytes, and eggs in C. elegans.
Isolation and analysis of piga-1–knockout allele tm2939
Among the 24 orthologous genes involved in GPI-anchor synthesis, DPM1, DPM3, and MPDU1 (SL15) are involved in the synthesis of the sugar donor dolichol phosphate-mannose, which is required in the synthesis of N-linked/O-linked oligosaccharides and for the synthesis of GPI-anchors (Figure 2). Thus the RNAi phenotypes of each gene, including larval arrest, scrawny larvae, and germline defects, could have resulted from N-/O-glycosylation inhibition as well as GPI-anchor inhibition (see Table 1 and Supplemental Figure S1). To distinguish phenotypes solely derived from GPI-anchor synthesis from N-/O-glycosylation defects, it is important to choose genes that are not involved in N-/O-glycosylation for further study. For this reason, we chose the C. elegans gene piga-1 (orthologue of human PIGA), which is conserved among eukaryotes. In humans, the GPI-GnT enzyme complex consists of seven subunits: PIGA, PIGC, PIGH, PIGP, PIGQ, PIGY, and DPM2. There is only one PIGA orthologue (piga-1: D2085.6) in the worm genome (on chromosome II), whose predicted amino acid sequence and catalytic site are well conserved among various species (human PIGA and worm PIGA-1 are 52.3% identical and 76.7% similar; Supplemental Figure S2). Because we observed no abnormalities in embryos treated with piga-1 RNAi, we isolated a deletion allele of the piga-1 gene by screening the trimethylpsoralen (TMP)/UV–induced deletion library with appropriate primers. The deletion allele (tm2939) lacked 324 base pairs of the piga-1 sequence, including exons 3 and 4 (Figure 4A). The deletion introduced a frameshift mutation and should produce a null allele. Homozygous embryos (piga-1(tm2939)) born from heterozygotes (piga-1(tm2939/+)) grew to the adult stage, but the worms moved slowly and had a scrawny phenotype (Figure 4B). The body length of the piga-1(tm2939) adult hermaphrodites was reduced. Homozygous adult worms laid no eggs, and many lethal eggs were observed in the uterus. The lethal eggs included cleavage-arrested embryos at various stages, from “before-first-cleavage” embryos to multicellular embryos (Figure 4E). The lethality of the F1 eggs was 100% (n > 100). No FLAER staining was observed in the gonads of piga-1(tm2939) worms, strongly indicating that GPI-anchor synthesis was totally inhibited (Figure 4C). To confirm that the synthesis of GPI-anchored proteins is inhibited in piga-1–knockout worms, we generated a transgenic tm2939 line expressing EGFP-tagged WRK-1 by transgene microinjection. On the piga-1–null background, membrane sorting of WRK-1 in the DTCs was inhibited, and WRK-1 was not present on the cell surface but was instead retained inside the cell (compare Figure 4D with Figure 1F). Thus GPI-anchor attachment was inhibited in the piga-1–deficient worms.
To gain insights into the nature of the lethality of the affected eggs, we examined whether the eggs from the piga-1–null allele showed changes in osmotic sensitivity (Kaitna et al., 2002). In two-thirds of the eggs dissected from piga-1–null animals, the shape of the eggshell was abnormal (n = 144), suggesting defects in the eggshells (Figure 5, C–F). The integrity of the eggshell barrier with respect to small molecules was examined with the lipophilic dye FM4-64 (Sato et al., 2008), which stains membrane structures. The eggshell of a wild-type embryo is composed of a vitelline layer, a chitinous middle layer, and a lipid-rich inner layer (Rappleye et al., 1999). FM4-64 cannot normally pass through the mature eggshell, preventing staining of the cells within the eggshell. Conversely, embryos with a defective eggshell inner layer do stain with FM4-64 (Rappleye et al., 1999; Johnston et al., 2006). When we stained dissected eggs with FM4-64 dye, the wild-type eggshells were not permeable to the dye, and only eggshells and polar bodies were stained (Figure 5G and Table 3). In 25% of the piga-1(tm2939) embryos, the eggshells were permeable, and the dye stained blastomere membranes. This indicates that loss of GPI-anchor synthesis results in changes in eggshell permeability. Under the standard osmolarity-supported condition (340 mOsm egg buffer; Strange et al., 2007), we still observed abnormalities of egg development (Supplemental Movie S1), indicating that the abnormal phenotypes did not merely result from osmotic abnormality.
FIGURE 5:
Abnormal egg morphology and eggshell permeability of the piga-1(tm2939) embryos. (A) Wild-type embryo. (B–F) piga-1(tm2939) embryos. (B) Apparently normal embryo. (C) Spherical embryo. (D) Embryo with protruding eggshell (arrow). (E) Embryo with constriction in the middle. (F) Embryo bent in the middle. (G) Eggshell permeability of the wild-type and the piga-1(tm2939) embryos. The lipophilic dye FM4-64 stains cell membranes. In wild-type N2 and rescued piga-1(tm2939) embryos, only eggshells and polar bodies were stained. In the piga-1(tm2939) embryos, their eggshells were permeable to FM4-64, and the cell membranes of the embryos were stained. Bars, 20 μm.
TABLE 3:
Eggshell permeability to the lipophilic dye FM4-64.
| Strain | Permeable eggshell (%) | n |
|---|---|---|
| N2 | 0 | 70 |
| piga-1(tm2939) | 25.4 | 67 |
| piga-1(tm2939) Ex[Plag-2::piga-1::egfp] | 0 | 74 |
| piga-1(tm2939) Ex[Ppie-1::piga-1::egfp] | 0 | 70 |
In addition to the osmotic phenotypes, abnormalities of gonads and intestine were detected in the null worms. The length of the gonadal arms was decreased, and in some animals, the morphology of the gonad was distorted, with wrinkled gonadal arms (Supplemental Figure S3A). The width of the intestine and the number of intestinal granules also decreased (Supplemental Figure S3B). Thus the piga-1 deletion–mutant worm tm2939 is deficient in GPI-anchored proteins in the cell membrane and shows severe germline abnormalities, including embryonic lethality, as well as various somatic abnormalities.
Somatic or germline expression of piga-1 rescues abnormal phenotypes of the null worms
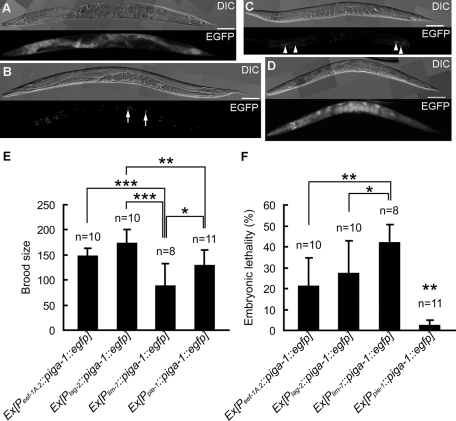
To confirm that these phenotypes were due to deletion of piga-1, we injected the wild-type piga-1 gene (D2085.6 genomic sequence) driven by the eef-1A.2 promoter (Peef-1A.2::piga-1::egfp) into tm2939 worms. The eef-1A.2 gene (eft-4) codes for translational elongation factor 1α, and its promoter drives its expression in almost all cell types. The wild-type piga-1 gene driven with this promoter rescued all of the germline and embryonic defects observed in the tm2939 homozygous animals (Table 2 and Figure 6A). The EGFP fluorescence of the transgene products indicated that the PIGA-1 protein was expressed in somatic cells, and the lack of EGFP fluorescence and EGFP immunoreactivity in the germline indicated that PIGA-1 expression was suppressed in the germline (Figure 6A). The germline expression of transgenes is silenced by a germline silencing mechanism (Green et al., 2008). Thus we were not sure whether PIGA-1 expression in the germline rescued the abnormal phenotypes. To circumvent the germline silencing of the piga-1 gene, we next injected the wild-type piga-1 gene driven by pie-1 promoter with cut DNA to rescue the piga-1–null allele (Kelly et al., 1997). The worms were cultured at 25°C to suppress germline silencing. As shown in Figures 5G and 6D and Table 2, all abnormalities of the null allele, including eggshell permeability defects, were rescued by expressing the wild-type piga-1 gene in the germline. The PIGA-1 fluorescence was solely detected in the germline, and no EGFP-tagged PIGA-1 was detected in somatic cells (Figure 6D). The embryonic lethality of the null allele was almost completely rescued (Figure 6F). From these two rescue experiments, we conclude that synthesis of GPI-anchored proteins either in the somatic cells or in the germline is sufficient for the development of germline cells and normal embryogenesis.
TABLE 2:
Transgenic rescue experiments of piga-1(tm2939) mutants with wild-type piga-1 genes driven by different promoters.
| Transgene | Expression tissue | Obtained transgenic lines | Rescue lines |
|---|---|---|---|
| Peef-1A.2::piga-1::egfp | Almost all cells | 2 | 2 |
| Plag-2::piga-1::egfp | Distal tip cells | 8 | 8 |
| Plim-7::piga-1::egfp | Gonadal sheath cells | 7 | 5 |
| Ppie-1::piga-1::egfp | Germline and embryos | 2 | 2 |
| Psth-1::piga-1::egfp | Spermatheca and uterus | 3 | 0 |
| Pmyo-3::piga-1::egfp | Muscles | 7 | 0 |
| Prgef-1::piga-1::egfp | Pan-neurons | 6 | 0 |
FIGURE 6:
Tissue-specific rescue of the piga-1(tm2939) mutant. (A–D) Differential interference contrast and fluorescence images of the piga-1(tm2939) rescue lines 24 h after the L4 stage. (A) piga-1(tm2939) Ex[Peef-1A.2::piga-1::egfp, pRF4]. EGFP expression was observed in all cells except for germline cells and early embryos. (B) piga-1(tm2939) Ex[Plag-2::piga-1::egfp, pRF4]. EGFP expression was detected in DTCs (arrows). Nonspecific gut granule fluorescence was also observed in the intestine. (C) piga-1(tm2939) Ex[Plim-7::piga-1::egfp, pRF4]. EGFP expression was observed in gonadal sheath cells (arrowheads). (D) piga-1(tm2939) Ex[Ppie-1::piga-1::egfp, pRF4]. EGFP expression was detected in embryos in utero. Autofluorescence was also observed in the pharynx and intestine. Bars, 100 μm. (E, F) Brood size and embryonic lethality in the piga-1(tm2939) rescue lines. Brood size was reduced (E) and embryonic lethality was increased (F) in the Plim-7::piga-1::egfp rescue line compared with the Peef-1A.2::piga-1::egfp, Plag-2::piga-1::egfp, and Ppie-1::piga-1::egfp rescue lines. The Ppie-1::piga-1::egfp transgene rescued the defects most effectively. Embryonic lethality is indicated by the percentage of unhatched eggs. The data for the most effectively rescued line are displayed. Bar graphs represent the mean values of brood size and embryonic lethality. Error bars indicate SD. *p < 0.05, **p < 0.01, ***p < 0.001 (Holm's multiple-comparison test).
Expression of piga-1 in somatic gonad is sufficient to rescue the null allele
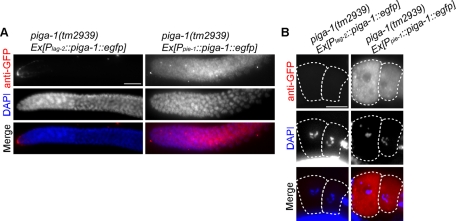
Although the wild-type piga-1 gene driven by the eef-1A.2 promoter rescued all of the abnormalities in germline formation and embryogenesis, germline expression of the gene seemed to be suppressed by germline silencing. Thus, in this rescue experiment, the piga-1 gene expressed in somatic tissues might have been responsible for the complete rescue of the GPI-anchor–deficient phenotypes in germline cells and embryos. To identify somatic tissue and cells responsible for this successful rescue of the null allele, we generated a wild-type piga-1::egfp gene linked to several different tissue-specific promoters and microinjected them to rescue the tm2939 strain (Table 2). To our surprise, piga-1 driven by the DTC-specific promoter Plag-2 rescued the germline phenotypes most effectively. The osmotic permeability phenotype was also rescued completely (Table 3 and Figure 5G). lag-2 expression from this promoter in adult gonad is limited to the DTCs and is excluded from the germline (Henderson et al., 1994), and the lag-2 promoter specifically drives the expression of the introduced transgenes in DTCs. We examined the presence of EGFP fluorescence, as well as immunoreactive EGFP protein, in the rescued animals by fluorescence microscopy and by immunostaining with anti-GFP antibody. Only DTCs were positive for PIGA-1 (Figure 6B and Figure 7A, left column). Neither the EGFP signal nor EGFP-tagged protein was observed in the germline of the rescued worms. Thus there was little possibility of the expression of the wild-type piga-1 transgene in the germline, and we concluded that the transgene Plag-2:::piga-1::egfp was expressed only in DTCs. In contrast, the piga-1 transgene expressed in the spermatheca and the uterus (Psth-1::piga-1::egfp) did not rescue the phenotypes. The muscle-specific promoter (Pmyo-3) and the pan-neuronal promoter (Prgef-1) were also unable to rescue the defects. Punc-119, another pan-neuronal promoter, successfully rescued the phenotype, but we detected weak GFP expression in DTCs as well as in neuronal cells (unpublished data), confirming the importance of piga-1 expression in DTCs. The expression of the transgene in gonadal sheath cells (Plim-7::piga-1::egfp) provided weak rescue activity (Figure 6C and Table 2). These rescued worms grew slowly (Supplemental Figure S4), and brood size recovery was poor (Figure 6E). Furthermore, the embryonic lethality of this rescue line was higher than that of other three rescue lines (Figure 6F).
FIGURE 7:
Immunofluorescence staining of the rescued piga-1(tm2939) worms using the anti-GFP antibody. (A) In the distal gonad, the fluorescence signal was detected only in DTCs in piga-1(tm2939) Ex[Plag-2::piga-1::egfp] (left column). The signal was observed all over the germline in piga-1(tm2939) Ex[Ppie-1::piga-1::egfp] (right column). (B) In oocytes, the fluorescence signal was detected in piga-1(tm2939) Ex[Ppie-1::piga-1::egfp] but not in piga-1(tm2939) Ex[Plag-2::piga-1::egfp]. Dotted lines indicate outlines of oocytes. Bars, 20 μm.
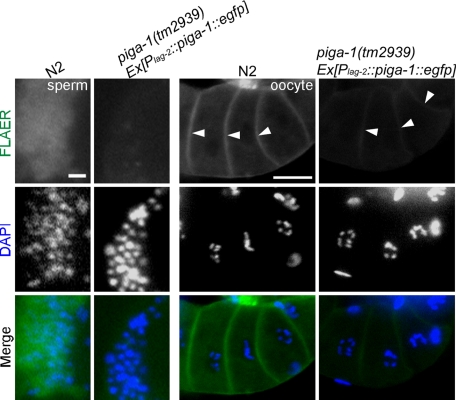
In mammalian cells, intercellular transfer of GPI-anchored proteins occurs (Dunn et al., 1996). Thus we examined whether GPI-anchored proteins synthesized in DTCs could be transferred to the germline of the transgenically rescued worms. As shown in Figure 8, we detected weak but positive FLAER staining on the oocyte cell surface of the Plag-2:::piga-1::egfp rescued worms. The FLAER signal in mitotic germline cells was too weak to detect, and only oocyte signals were detected in the experiment. The presence of GPI-anchored proteins was detected in oocytes but not in sperm (Figure 8). Because the piga-1 gene is active in DTCs and DTCs extend processes proximally, the GPI-anchored proteins synthesized in the DTCs could be transferred to the germline mitotic cells by direct contact or transferred by extensive secretion, which is characteristic of DTCs (Hall et al., 1999). The results show that the expression of the piga-1 gene in DTCs accompanied by the transfer of GPI-anchored proteins to the germline was sufficient for the complete rescue of the germline defects.
FIGURE 8:
Weak FLAER staining was observed in the oocyte membrane of the rescued piga-1(tm2939) mutant compared with that of the wild-type animal. No FLAER signal was detected in sperm of the rescued mutants. Arrowheads indicate oocyte membranes. Bars, 5 μm (left column) and 20 μm (right column).
Loss of GPI-anchor synthesis affects germline mitotic cell number
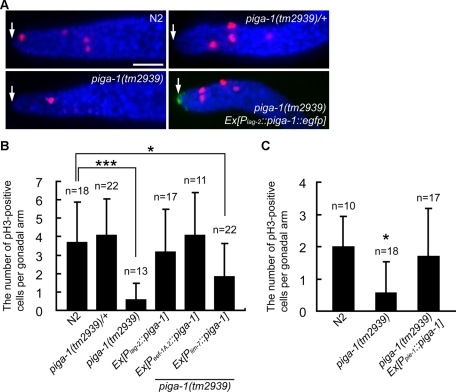
DTCs express the Notch ligand LAG-2, which maintains germline stem cells at the distal end of gonadal arms in the undifferentiated state. DTCs are linked to oocyte precursor mitotic cells (including germline stem cells) and provide the LAG-2 signal. DTCs therefore have been believed to form the germline stem cell niche (Byrd and Kimble, 2009; Cinquin et al., 2010). Decreasing the LAG-2 signal induces germline stem cells to switch from mitosis to meiosis, resulting in oocyte maturation. As shown in Figure 1B, GPI-anchored proteins are present on DTCs. In addition, our data demonstrate that germline cell formation was severely affected by the loss of genes involved in GPI-anchor synthesis. To examine the effects of piga-1 deficiency on mitotic germline cells, we labeled the mitotic cells in dissected gonads by immunostaining with anti–phosphohistone H3 (anti-pH3) antibody, which is a marker of mitosis (Hendzel et al., 1997). The number of mitotic prometaphase nuclei (pH3 positive) was significantly reduced in piga-1(tm2939) worms (Figure 9, A and B). The average number of pH3-positive cells of the piga-1(tm2939) worm was 0.6 ± 0.9 (n = 13), compared with 3.7 ± 2.2 (n = 18) in the wild-type worm. Induction of DTC-specific expression of the piga-1 gene led the prometaphase nucleus number to recover to normal levels. The average number of pH3-positive cells in the rescued mutant (Plag-2::piga-1 worm) was 3.2 ± 2.3 (n = 17). Expression of piga-1 from the eef-1A.2 promoter also restored the pH3-positive cell number to the normal level, whereas the lim-7 promoter was less effective. When piga-1 was expressed in the germline from the pie-1 promoter, the number of pH3-positive cells returned to the normal level (Figure 9C). These results indicate that PIGA-1 synthesis in somatic gonad and/or germline cells is sufficient to restore the number of prometaphase mitotic cells to the normal level, showing the importance of GPI-anchored proteins in germline mitotic cell number maintenance. From these results, we conclude that GPI-anchor synthesis in the germline and/or DTCs (somatic gonad) is sufficient for maintaining normal numbers of mitotic germline cells.
FIGURE 9:
The number of mitotic germ cells at the distal gonad was decreased in piga-1(tm2939) worms. Normal numbers of mitotic germ cells were detected in N2 and piga-1(tm2939)/+ worms and in worms rescued with the transgene. (A) Immunofluorescence staining using the anti-pH3 antibody. Red, cells stained with the anti-pH3 antibody; blue, DAPI-stained DNA; green, PIGA-1::EGFP. Arrows indicate DTCs. Bar, 20 μm. (B, C) Bar graphs represent the mean numbers of pH3-positive cells at 20°C (B) and 25°C (C). Error bars indicate SD. *p < 0.05, ***p < 0.001 (Holm's multiple-comparison test).
DISCUSSION
Here we demonstrated that GPI-anchor synthesis in the somatic gonad and/or in the germline is indispensable in the germline formation and embryonic development of the nematode C. elegans. Abnormal germline phenotypes of the GPI-anchor–deficient worm were successfully rescued by expressing wild-type piga-1 (orthologue of human PIGA) in the somatic gonad (DTCs or gonadal sheath cells) or in the germline. Total inhibition of GPI-anchor synthesis by piga-1 knockout resulted in severe germline malformation beginning at the distal end of the gonad, as well as eggshell malformation at the proximal end of the gonad.
GPI-anchored proteins are embedded in the outer leaflet of the plasma membrane, and they are generally more highly concentrated at the membrane rafts than transmembrane proteins. This high concentration seems to be essential for signal transduction (Yamashita and Fukushima, 2007; Lingwood and Simons, 2010). In the present experiments, the recruitment of proteins to the cell membrane by GPI-anchor modification was blocked, as shown in Figure 4D. Thus inhibition of GPI-anchor synthesis could affect various signal transduction pathways that are important in germline formation, and the total lack of GPI-anchored proteins on the cell surface seems to be the major cause of the germline and eggshell abnormalities described in this study. Synthesis of GPI-anchored proteins seems to be essential for reproduction of this species.
Our transgenic rescue experiment using wild-type piga-1::egfp driven by the lag-2 promoter demonstrates that expression of PIGA-1 in DTCs is sufficient for the total rescue of the germline phenotypes. Because no PIGA-1 was detected in the germline cells, the FLAER staining of proximal oocytes strongly indicated that the GPI-anchored proteins in the oocytes originated from DTCs. The transfer of GPI-anchored proteins from DTCs to the germline is highly probable, whereas the possibility that nondetectable amounts of PIGA-1 are present in the germline and in other somatic cells cannot be excluded altogether. It is possible that the GPI-anchor moiety from one protein is recycled and attached to another protein. Thus the GPI-anchored proteins found in oocytes are not necessarily identical to the GPI-anchored proteins in DTCs. As shown in Figure 8, oocytes were stained with FLAER, but sperm were not stained at all. The sperm are separated from other germline cells by the spermatheca, and they are situated at the most proximal position of the gonad. The physical separation of sperm from DTCs may be the reason for the loss of FLAER staining in these cells. We have no evidence that GPI-anchored proteins are normally transferred from the DTCs to the germline in wild-type worms, and we cannot exclude the possibility that this transfer is an emergency response of the somatic gonad to the lack of PIGA-1 synthesis in the germline. Further experiments are needed to resolve this question.
Recently GPI-anchor modification has been used to deliver GPI-linked therapeutic proteins to target cells with a transcellular transfer mechanism. GPI-anchored therapeutic proteins added from outside of the cells are incorporated into lipid rafts, after which they leave the rafts and move to cytoplasmic lipid droplets. The lipid droplets can be secreted as exosomes or microvesicles and incorporated into the membranes of nearby cells (Müller, 2010). In the present study, the transgenic expression of piga-1 in DTCs rescued all of the germline phenotypes of the piga-1–null worms. The results strongly indicate that intercellular transfer of GPI-anchored proteins from DTCs to germline cells rescued the germline phenotypes completely, and the transfer seems to be very efficient. Our results suggest the intriguing possibility that intercellular transport of GPI-anchored proteins could be used to deliver drugs to nearby tissues and cells. The transgenically rescued worm described in this article could be used as a model system to examine the utility of drug delivery systems based on GPI-anchor modification.
Proteome analyses of the transgenically rescued worms and wild-type N2 worms were carried out to determine what kinds of GPI-anchored proteins are synthesized in these worms and which proteins are important for germline and eggshell development (Supplemental Tables S2 and S3). GPI-anchored proteins were identified with Triton X-114 phase partitioning followed by PI-PLC digestion. The isolated proteins were separated by SDS–PAGE, silver stained, and analyzed by LC/MS/MS. Supplemental Tables S2 and S3 show all of the GPI-anchored proteins predicted by the PredGPI and FragAnchor programs that we identified in three different experiments. Most of the proteins listed in Supplemental Table S1 were also identified in these experiments, and many new candidate GPI-anchored proteins were identified.
In Drosophila melanogaster, male hub cells and female cap cells play similar roles to nematode DTCs in forming the Drosophila germline stem cell niche (Fuller and Spradling, 2007). Dally is a GPI-anchored heparan sulfate proteoglycan of the glypican family in Drosophila. Dally is expressed in the female cap cells, and another glypican, Dally-like, is expressed in the male hub cells. Dally and Dally-like are required for maintaining the germline stem cell niches in female and male Drosophila, respectively (Guo and Wang, 2009; Hayashi et al., 2009). C. elegans has two members of the glypican family, LON-2 and GPN-1. We did not detect either protein in our proteome analyses. The knockout/knockdown phenotypes and the expression patterns of these worm glypican genes are different from the germline phenotypes described in this article, indicating different roles of these genes in development (Foehr et al., 2006; Hudson et al., 2006; Gumienny et al., 2007). C. elegans glypicans may not be involved in the maintenance of the germline stem cell niche.
The Notch signaling pathway plays important roles in germline stem cell niche formation at the distal end of the gonad of the nematode. Because inhibition of GPI-anchor synthesis resulted in decreased mitotic cell number at the distal end of the gonad, we suspect the involvement of GPI-anchored proteins in Notch signaling. In the present proteome analyses, however, canonical Notch ligands such as LAG-2 and 10 other DSL ligands (apx-1, arg-2, lag-2, and dsl-1 to dsl-7) were not detected in the GPI-anchored protein fraction. Both PredGPI and FragAnchor predict that these ligands are not GPI anchored. An examination of the identified GPI-anchored protein candidates in Supplemental Table S3 shows that Jagged-like GPI-anchored protein (C02B10.3) was present in the GPI-anchored protein fraction from lag-2 promoter–rescued worms. This protein has sequence homology to human tanascins, Jagged-1, Notch2, and Notch3. Although the GPI-anchored protein RIG-6, which is a noncanonical Notch ligand and a homologue of contactin, was not detected in the present proteome analysis, RNAi against rig-6 affected germline development (our unpublished result). Therefore the involvement of the noncanonical Notch signaling pathway in germline development is an intriguing possibility for future studies.
The possible GPI-anchored proteins identified in this study include the neuronal IgCAM homologue RIG-3, an integrin β subunit, cathepsin A homologue Y40D12A.2, growth arrest–specific protein homologue PHG-1 (PHAS-1), ephrin-B2 homologue VAB-2, and WRK-1. WRK-1 is the prototypical GPI-anchored protein expressed in the DTCs. The deletion mutant alleles of wrk-1 (tm1099 and ok695) show neuronal but not germline abnormalities (Boulin et al., 2006). This implies that WRK-1 may not be the GPI-anchored protein required for normal germline development. WRK-1 interacts with an ephrin (VAB-2) and an ephrin receptor (VAB-1). VAB-2/EFN-1 is a predicted GPI-anchored ephrin ligand for VAB-1, and RNAi against vab-1 results in embryonic lethality (Partridge et al., 2008). VAB-2/EFN-1 is predominantly expressed in neurons, but serial analysis of gene expression analysis shows that the gene is also expressed in the germline (Wang et al., 2009). VAB-2 was detected in our proteome analysis of lag-2 promoter–rescued worms. These results suggest that the VAB-2 expressed in germline cells or DTCs is one of the GPI-anchored proteins responsible for the germline phenotypes described in this study. No single gene knockdown results in the aforementioned germline phenotypes (Wormbase 220; www.wormbase.org). One plausible explanation for this is that there is no single GPI-anchored protein species responsible for the phenotypes, but the synthesis of several different GPI-anchored proteins is necessary for the normal development of the germline and eggshells. On the basis of the results presented in this article, we are planning to carry out multiple knockouts/knockdowns of these GPI-anchored proteins and to examine the expression timing and localization of these GPI-anchored molecules. Genetic analysis and reverse-genetic analysis will also play important roles in resolving the molecular mechanisms of the functions of GPI-anchored proteins in germline and egg development.
MATERIALS AND METHODS
C. elegans strains
All strains were cultured at 20°C as described previously (Brenner, 1974). N2 Bristol was used as the wild-type strain. The following allele was used in this study: OD70 (unc-119(ed3) III;ltIs44 [Ppie-1::mCherry::PH + unc-119(+)] V; Kachur et al., 2008). Prior to our experiments, we crossed the OD70 strain with N2 twice. A piga-1(tm2939) allele was isolated from pools of worms mutagenized by the TMP/UV method described previously (Gengyo-Ando and Mitani, 2000) with appropriate primers. The piga-1 deletion mutant was backcrossed four times with N2. The piga-1 mutant was balanced with mIn1[mIs14 dpy-10(e128)] II and maintained as a heterozygote (piga-1(tm2939/+)) because the homozygous mutant (piga-1(tm2939)) was not viable. The piga-1(tm2939/+) worm was crossed with the OD70 strain to produce the piga-1(tm2939/+);ltIs44 worms.
RNAi methodology
All RNAi experiments in this study were performed using the feeding method essentially as described previously (Timmons et al., 2001). C. elegans RNAi library clones (Kamath and Ahringer, 2003) were used for RNAi of most of genes listed in Table 1, except for Y48E1B.2, T27F7.4, C27A12.9, T22C1.3, and dpm-1 (Y66H1A.2). The cDNA fragments corresponding to these genes, except dpm-1, were amplified from total cDNA of N2 worms by PCR and cloned into the L4440 (pPD129.36) vector. The dpm-1 genomic sequence was amplified from N2 genomic DNA by PCR and cloned into the L4440 vector. The plasmids were transformed into Escherichia coli HT115 (DE3). A single colony of HT115 carrying the plasmids was cultured in LB medium containing 100 μg/ml carbenicillin overnight. The cultured E. coli was seeded onto NGM plates and incubated at 37°C overnight. The plates were treated with 2 mM isopropyl-β-d-thiogalactoside and incubated at 37°C for 6 h to allow HT115 to express double-strand RNA. L4 hermaphrodites were transferred onto the feeding plates and maintained at 25°C. We observed the phenotypes of F1 generation after 72 and 96 h. HT115 carrying the L4440 vector without any insert was used as a control. The PCR primers used in this study are listed in Supplemental Table S4.
Phenotypic analysis
To observe phenotypes of piga-1 (tm2939) mutants, L4 larvae of piga-1(tm2939/+) and piga-1(tm2939) worms were transferred onto separate nematode growth medium (NGM) plates seeded with E. coli OP50 and maintained at 20°C. After 24 h, the animals were observed with a Leica DMRXA full automatic microscope (see Microscopy).
To count the brood size, L4 larvae were placed onto fresh NGM plates seeded with OP50 (one worm per plate) and maintained at 20°C. The P0 worms were transferred onto fresh NGM plates every 24 h for 3 d. We counted the hatched larvae and unhatched eggs on the plates.
Plasmid construction
All plasmids generated in this study were constructed in a pFX_EGFPT vector backbone (Gengyo-Ando et al., 2006). For the rescue experiments, we obtained the following promoter sequences for each gene by PCR using N2 genomic DNA as a template: a lag-2 promoter Plag-2 containing 5 kb of upstream sequence, with the first codon specifically expressed in DTCs; a lim-7 promoter Plim-7 containing 3 kb of upstream sequence, with the first exon and intron predominantly expressed in gonadal sheath cells (Voutev et al., 2009); and a sth-1 promoter Psth-1 containing 1.3 kb of upstream sequence, with the first codon expressed in the spermatheca and uterus (Bando et al., 2005). Promoter fragments were inserted into the pFX_EGFPT vector using the TA cloning method described by Gengyo-Ando et al. (2006) to generate Plag-2::egfp, Plim-7::egfp, and Psth-1::egfp plasmids. A Peef-1A.2::egfp plasmid was generated by replacing the venus sequence of the Peef-1A.2::venus vector (Kitagawa et al., 2007) with the egfp sequence. Pmyo-3::egfp, Prgef-1::egfp and Punc-119::egfp plasmids were generated by Dejima et al. (2010). To generate rescue plasmids expressing the wild-type piga-1 gene under the control of each promoter, the piga-1 genomic sequence was amplified from N2 genomic DNA by PCR using appropriate primers tagged with the NotI restriction site. The piga-1 fragment was digested with NotI and cloned into the NotI restriction site of each plasmid to generate Peef-1A.2::piga-1::egfp, Plag-2::piga-1::egfp, Plim-7::piga-1::egfp, Psth-1::piga-1::egfp, Pmyo-3::piga-1::egfp, Prgef-1::piga-1::egfp, and Punc-119::piga-1::egfp.
For the germline rescue experiment, we constructed the pFX vector containing the promoter, the third intron, and the 3′ sequence of the pie-1 gene. First, the 2.4-kb fragment of the pie-1 promoter was amplified by PCR and inserted into the pFX_EGFPT vector using the TA cloning method to generate the pFX_Ppie-1::egfp vector. Second, the third intron sequence (1 kb) of the pie-1 gene was amplified by PCR and inserted into the BamHI site located upstream the pie-1 promoter sequence of the pFX_Ppie-1::egfp vector. This addition of the third intron to the upstream of the pie-1 promoter enhances gene expression (Strome et al., 2001). Third, the pie-1 3′ sequence (1.4 kb) was amplified by PCR and inserted into the BglII/PciI sites located downstream the egfp coding sequence of the pFX_Ppie-1::egfp vector. Finally, the piga-1 genomic sequence was inserted into the NotI site located between the pie-1 promoter and the egfp coding sequence to generate the Ppie-1::piga-1::egfp plasmid.
For expression analysis of wrk-1, the coding sequence was amplified from N2 genomic DNA by PCR using appropriate primers tagged with the NotI and BglII restriction sites. The wrk-1 fragment was digested with NotI and BglII and cloned into the NotI and BglII double-digested pFX vector (pFX_wrk-1 vector). To insert the coding sequence of EGFP into the pFX_wrk-1 vector, the coding sequence of EGFP was amplified from the pFX_EGFPT vector by PCR using appropriate primers tagged with the BamHI restriction site. The egfp fragment was digested with BamHI and inserted into the BamHI restriction site of the wrk-1 sequence (pFX_egfp-tagged wrk-1 vector; Figure 1, D and E). Five kilobases of the wrk-1 promoter were amplified by PCR using primers tagged with the NotI restriction site. The wrk-1 promoter fragment was digested with NotI and inserted into the NotI restriction site of the pFX_egfp-tagged wrk-1 vector to generate the Pwrk-1::egfp-tagged wrk-1 plasmid. DNA sequence analysis was performed with a 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA). The PCR primers used in this study are listed in Supplemental Table S4.
Microinjection
Microinjections were performed as described by Mello and Fire (1995). For expression analysis of the egfp-tagged wrk-1, the Pwrk-1::egfp-tagged wrk-1 plasmid was injected at 15–20 ng/μl with a coinjection marker pRF4 (rol-6(su1006)) at 80 ng/μl into the gonads of adult N2 and the piga-1(tm2939/+) hermaphrodites.
For rescue experiments of the piga-1(tm2939), the rescue plasmids were injected into the gonads of the adult piga-1(tm2939/+) hermaphrodites. The Peef-1A.2::piga-1::egfp plasmid was injected at 5 ng/μl with pRF4 at 100 ng/μl. The Plag-2::piga-1::egfp, Pmyo-3::piga-1::egfp and Prgef-1::piga-1::egfp plasmids were injected at 10 ng/μl with pRF4 at 90 ng/μl. The Plim-7::piga-1::egfp, Psth-1::piga-1::egfp and Punc-119::piga-1::egfp plasmids were injected at 20 ng/μl with pRF4 at 80 ng/μl.
For the germline rescue experiment, microinjections were performed as previously described (Kelly et al., 1997). For the generation of complex extrachromosomal arrays, DNAs were digested and linearized with restriction enzymes, and then the digested DNAs were injected into the gonads of the adult piga-1(tm2939/+) hermaphrodites. The rescue plasmid (pFX_Ppie-1::piga-1::egfp), pRF4, and N2 genomic DNA were digested with PvuII, EcoRI, and XbaI, respectively. XbaI fragments the piga-1 gene in the N2 genomic DNA. The PvuII-digested pFX_Ppie-1::piga-1::egfp was injected at 0.5 ng/μl with EcoRI-digested pRF4 at 0.5 ng/μl and XbaI-digested N2 genomic DNA fragment at 31 ng/μl. To circumvent germline silencing of the transgenes, injected animals were maintained at 25°C. Transgenic worms were selected and maintained based on their EGFP expression and roller phenotype.
Selection of the piga-1(tm2939) rescue lines
We injected the rescue plasmid into the gonads of piga-1(tm2939/+) hermaphrodites to obtain piga-1(tm2939/+) transgenic lines (see Microinjection). To examine whether the introduced transgene could rescue the piga-1(tm2939) mutants, 10 piga-1(tm2939) mutants expressing the transgene were transferred onto a fresh NGM plate seeded with OP50 and cultured at 20°C, except for the germline rescue experiment at 25°C. After several days, we determined whether the mutants with the transgene could produce offspring on the plate. Homozygous mutant lines that carried the transgene and were capable of fertility were counted as rescue lines. Three independent experiments were performed for each line.
FLAER staining
Alexa 488 proaerolysin (FLAER) was obtained from Protox Biotech (Victoria, Canada). For FLAER staining, gonads were dissected and covered with an 18 × 18 mm coverslip. The gonads were then freeze-cracked and fixed in methanol at –20°C for 10 min. They were immediately transferred to phosphate-buffered saline (PBS) to prevent drying. The slides were washed twice in fresh PBS for 5 min and blocked with 1% bovine serum albumin (BSA) in PBS for 30 min at room temperature in a humid chamber. FLAER diluted in PBS (final concentration of 20–50 nM) was applied, and the specimens were incubated for 2 h at room temperature in a dark, humid chamber. After incubation, the slides were washed twice in PBS for 5 min and stained with 4,6-diamidino-2-phenylindole (DAPI) diluted in PBS (final concentration of 5 μg/ml) for 10 min. Slides were then washed twice in PBS for 10 min and mounted in VECTASHIELD Mounting Medium (Vector Laboratories, Burlingame, CA).
Immunofluorescence staining
Adult hermaphrodites 24 h after L4 stage were used for immunofluorescence staining. Gonads were dissected and covered with an 18 × 18 mm coverslip. The gonads were freeze-cracked and fixed in methanol at –20°C for 5 min and then immediately transferred to PBS. The slides were washed twice in fresh PBS for 5 min and once in PBS containing 0.05% Tween 20 (PBST) for 5 min. Slides were blocked with 1% BSA/PBST for 30 min at room temperature in a humid chamber. Specimens were incubated with primary antibody diluted 1:1000 in 1% BSA/PBST at 4°C overnight in a humid chamber. The following primary antibodies were used: rabbit polyclonal immunoglobulin G (IgG) anti–phosphohistone H3 (Ser10) antibody (Millipore, Billerica, MA) and mouse monoclonal IgG anti-GFP antibody mFX73 (Wako, Osaka, Japan). The slides were then washed twice in PBS and incubated with secondary antibody diluted 1:200 in PBS at room temperature for 2 h in a dark, humid chamber. The secondary antibodies were as follows: Alexa Fluor 594 goat anti–rabbit IgG and Alexa Fluor 594 goat anti–mouse IgG (Invitrogen Molecular Probes, Eugene, OR). After incubation, the slides were washed twice in PBS for 5 min and stained with DAPI (final concentration of 5 μg/ml in PBS) for 10 min. Finally, the slides were washed twice in PBS for 10 min and mounted in VECTASHIELD Mounting Medium.
Eggshell permeability assay
For eggshell permeability assay, embryos were dissected in egg buffer (118 mM NaCl, 48 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.3) containing 8 μM FM4-64 (Invitrogen Molecular Probes). The osmolality of egg buffer (340 mOsm) was measured with OSMOMAT 030-D Osmometer (Gonotec, Berlin, Germany).
Microscopy
Differential interference contrast and fluorescent images were obtained with a Leica DMRXA full automatic microscope (Leica, Wetzlar, Germany). The acquired images were processed using MetaMorph software (version 6.1r5; Universal Imaging, West Chester, PA). Confocal images were acquired with an LSM 510 META (Carl Zeiss, Jena, Germany). Worms were placed on an eight-well printed microscope slide glass (Matsunami Glass, Osaka, Japan) and were anesthetized with M9 buffer containing 16 mM sodium azide.
Precondensation of Triton X-114
Precondensation of Triton X-114 (Sigma-Aldrich, St. Louis, MO) was performed according to the previously described method (Hooper, 1992). In preparation, 50 ml of Tris-buffered saline (TBS; 10 mM Tris-HCl, 150 mM NaCl, pH 7.4) was mixed with 0.8 mg of butylated hydroxytoluene. This solution was gently mixed with 1 ml of Triton X-114 and placed on ice for 3 h. The solution was incubated at 30°C overnight to induce condensation of Triton X-114. After the incubation, the upper phase (aqueous phase) was removed. The remaining lower phase (detergent-rich phase) was mixed with an equal volume of fresh TBS and placed on ice for 3 h. The condensation procedure was then repeated twice. The resulting detergent-rich phase was used as a precondensed Triton X-114.
Fractionation of GPI-anchored proteins
Fractionation of GPI-anchored proteins was performed according to the previously described method (Hooper, 1992). The lyophilized fraction of total proteins was dissolved in TBS containing 2 mM EDTA and 1 mM phenylmethylsulfonyl fluoride. The solution was gently mixed with precondensed Triton X-114 on ice and incubated at 4°C overnight. The sample was centrifuged at 15,000 × g at 4°C for 20 min to remove detergent-insoluble materials. The supernatant was then incubated at 37°C for 10 min and centrifuged at 13,000 × g for 5 min at room temperature to induce Triton X-114 phase partition. The upper phase (aqueous phase) was removed, and the lower phase (detergent-rich phase) was gently mixed with fresh TBS on ice. The phase partition method was repeated twice more. The resulting detergent-rich phase was mixed with four volumes of ice-cold acetone and incubated at –20°C overnight to remove the detergent and concentrate proteins. The sample was centrifuged at 13,000 × g for 30 min at 4°C. The supernatant was removed, and the precipitate was dried to evaporate remaining acetone. The precipitate was then dissolved in 200 μl of Tris-HCl buffer (10 mM Tris-HCl, pH 7.4) and gently mixed with 1 μl of PI-PLC (Invitrogen Molecular Probes) on ice. After incubation at 37°C for 90 min, the sample was gently mixed with 200 μl of TBS and 60 μl of precondensed Triton X-114 on ice. After the phase partition procedure, the upper phase (aqueous phase) was recovered and mixed with fresh precondensed Triton X-114 on ice. This phase partition was then repeated. The upper phase underwent acetone precipitation at –20°C overnight as described. After centrifugation at 13,000 × g for 30 min at 4°C, the supernatant was removed. The resulting precipitate was dissolved in fresh TBS, and this solution was used as a GPI-anchored protein-enriched fraction.
Identification of proteins by LC/MS/MS
The extracted GPI-anchored proteins were separated by SDS–PAGE and stained with SYPRO Ruby (Invitrogen Molecular Probes) or with Silver Stain MS Kit (Wako) according to the manufacturer's protocols. Noticeable bands were cut into pieces and incubated in 30 mM dithiothreitol at 56°C for 1 h. The gel pieces were subsequently incubated in 69 mM sodium iodoacetate at 28°C for 45 min in the dark and then divided into two tubes. To one tube was added sequence grade–modified trypsin (wt:wt 1:50; Promega, Madison, WI) in 25 mM ammonium bicarbonate, and the mixture was incubated at 37°C for 16 h. After the reaction was quenched by acidification with 1% trifluoroacetic acid (Wako), the digest was subjected to SpeedVac concentration (Thermo Fisher Scientific, Waltham, MA) and reconstituted in 0.1% formic acid.
The peptides were separated in a C18 column (Magic C18, 0.2 × 150 mm, 3 μ; Bruker-Michrom, Auburn, CA) at a flow rate of 3 μl/min in a high-performance liquid chromatography system (Paradigm MS4; Bruker-Michrom). The eluents were 2% acetonitrile/0.1% formic acid (pump A) and 90% acetonitrile/0.1% formic acid (pump B). The peptides were eluted with a linear gradient of 5–65% in pump B for 20 min and analyzed using an online linear ion trap mass spectrometer (Finnigan LTQ; Thermo Fisher Scientific) with a single mass scan (m/z 450–2000) and a data-dependent MS/MS scan in the positive-ion mode. All product ions were submitted to the computer database search analysis with the Mascot search engine (Matrix Science, Boston, MA) and TurboSEQUEST search engine (Thermo Fisher Scientific) using the National Center for Biotechnology Information database (C. elegans).
Statistical analysis
Statistical analyses, including one-way analysis of variance, followed by Holm's multiple-comparison test, were performed using R statistical package (R, version 2.11.0; www.r-project.org).
Supplementary Material
Acknowledgments
We thank Katsuko Yamashita and Keiko Fukushima (Tokyo Institute of Technology, Yokohama, Japan) for critical comments on the manuscript. We also thank Masaki Matsumoto from the Division of Proteomics of the Medical Institute of Bioregulation (Kyushu University, Fukuoka, Japan) and Mizuho Oda and Emiko Koba from the Laboratory for Technical Support of the Medical Institute of Bioregulation (Kyushu University) for the liquid chromatography–tandem mass spectrometry analysis. We acknowledge the Caenorhabditis Genetics Center for the strains used in this study, which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by aid from the Core Research for Evolutional Science and Technology Program of the Japan Science and Technology Corp. to K.N. This work was also supported partially by a Grant-in-Aid from the Japan Society for the Promotion of Science Fellows (to K.D.), a Grant-in-Aid for Young Scientists (B) (to S.M.), and a Grant-in-Aid for Challenging Exploratory Research (to K.N.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- DIC
differential interference contrast
- DTC
distal tip cell
- EGFP
enhanced green fluorescent protein
- GPI
glycosylphosphatidylinositol
- LC
liquid chromatography
- MS
mass spectrometry
- NGM
nematode growth medium
- PH
pleckstrin homology
- pH3
phosphohistone H3
- PIGA
phosphatidylinositol glycan-class A
- PI-PLC
phosphatidylinositol-specific phospholipase C
- PNH
paroxysmal nocturnal hemoglobinuria
- RNAi
RNA interference
- TMP/UV
trimethylpsoralen/ultraviolet
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-10-0855) on February 1, 2012.
REFERENCES
- Agostoni E, Gobessi S, Petrini E, Monte M, Schneider C. Cloning and characterization of the C. elegans gas1 homolog: phas-1. Biochim Biophys Acta. 2002;1574:1–9. doi: 10.1016/s0167-4781(01)00321-9. [DOI] [PubMed] [Google Scholar]
- Almeida AM, et al. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat Med. 2006;12:846–851. doi: 10.1038/nm1410. [DOI] [PubMed] [Google Scholar]
- Almeida A, Layton M, Karadimitris A. Inherited glycosylphosphatidyl inositol deficiency: a treatable CDG. Biochim Biophys Acta. 2009;1792:874–880. doi: 10.1016/j.bbadis.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Bando T, Ikeda T, Kagawa H. The homeoproteins MAB-18 and CEH-14 insulate the dauer collagen gene col-43 from activation by the adjacent promoter of the spermatheca gene sth-1 in Caenorhabditis elegans. J Mol Biol. 2005;348:101–112. doi: 10.1016/j.jmb.2005.01.045. [DOI] [PubMed] [Google Scholar]
- Boulin T, Pocock R, Hobert O. A novel Eph receptor-interacting IgSF protein provides C. elegans motoneurons with midline guidepost function. Curr Biol. 2006;16:1871–1883. doi: 10.1016/j.cub.2006.08.056. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky RA, Mukhina GL, Li S, Nelson KL, Chiurazzi PL, Buckley JT, Borowitz MJ. Improved detection and characterization of paroxysmal nocturnal hemoglobinuria using fluorescent aerolysin. Am J Clin Pathol. 2000;114:459–466. doi: 10.1093/ajcp/114.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd DT, Kimble J. Scratching the niche that controls Caenorhabditis elegans germline stem cells. Semin Cell Dev Biol. 2009;20:1107–1113. doi: 10.1016/j.semcdb.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinquin O, Crittenden SL, Morgan DE, Kimble J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc Natl Acad Sci USA. 2010;107:2048–2053. doi: 10.1073/pnas.0912704107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejima K, et al. Two Golgi-resident 3′-phosphoadenosine 5′-phosphosulfate transporters play distinct roles in heparan sulfate modifications and embryonic and larval development in Caenorhabditis elegans. J Biol Chem. 2010;285:24717–24728. doi: 10.1074/jbc.M109.088229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn DE, Yu J, Nagarajan S, Devetten M, Weichold FF, Medof ME, Young NS, Liu JM. A knock-out model of paroxysmal nocturnal hemoglobinuria: Pig-a(-) hematopoiesis is reconstituted following intercellular transfer of GPI-anchored proteins. Proc Natl Acad Sci USA. 1996;93:7938–7943. doi: 10.1073/pnas.93.15.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhaber F, Eisenhaber B, Kubina W, Maurer-Stroh S, Neuberger G, Schneider G, Wildpaner M. Prediction of lipid posttranslational modifications and localization signals from protein sequences, big-Pi, NMT and PTS1. Nucleic Acids Res. 2003;31:3631–3634. doi: 10.1093/nar/gkg537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser N, Maser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics. 2005;21:1846–1852. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- Foehr ML, Lindy AS, Fairbank RC, Amin NM, Xu M, Yanowitz J, Fire AZ, Liu J. An antagonistic role for the C. elegans Schnurri homolog SMA-9 in modulating TGFβ signaling during mesodermal patterning. Development. 2006;133:2887–2896. doi: 10.1242/dev.02476. [DOI] [PubMed] [Google Scholar]
- Fuller MT, Spradling AC. Male and female Drosophila germline stem cells: Two versions of immortality. Science. 2007;316:402–404. doi: 10.1126/science.1140861. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K, Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;269:64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- Gengyo-Ando K, Yoshina S, Inoue H, Mitani S. An efficient transgenic system by TA cloning vectors and RNAi for C. elegans. Biochem Biophys Res Commun. 2006;349:1345–1350. doi: 10.1016/j.bbrc.2006.08.183. [DOI] [PubMed] [Google Scholar]
- Green RA, Audhya A, Pozniakovsky A, Dammermann A, Pemble H, Monen J, Portier N, Hyman A, Desai A, Oegema K. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 2008;85:179–218. doi: 10.1016/S0091-679X(08)85009-1. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, MacNeil LT, Wang H, de Bono M, Wrana JL, Padgett RW. Glypican LON-2 is a conserved negative regulator of BMP-like signaling in Caenorhabditis elegans. Curr Biol. 2007;17:159–164. doi: 10.1016/j.cub.2006.11.065. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang Z. The glypican Dally is required in the niche for the maintenance of germline stem cells and short-range BMP signaling in the Drosophila ovary. Development. 2009;136:3627–3635. doi: 10.1242/dev.036939. [DOI] [PubMed] [Google Scholar]
- Hall DH, Winfrey VP, Blaeuer G, Hoffman LH, Furuta T, Rose KL, Hobert O, Greenstein D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: relations between the germ line and soma. Dev Biol. 1999;212:101–123. doi: 10.1006/dbio.1999.9356. [DOI] [PubMed] [Google Scholar]
- Hayashi Y, Kobayashi S, Nakato H. Drosophila glypicans regulate the germline stem cell niche. J Cell Biol. 2009;187:473–480. doi: 10.1083/jcb.200904118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Gao D, Lambie EJ, Kimble J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Hong Y, Ohishi K, Inoue N, Kang JY, Shime H, Horiguchi Y, van der Goot FG, Sugimoto N, Kinoshita T. Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicumα-toxin. EMBO J. 2002;21:5047–5056. doi: 10.1093/emboj/cdf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper NM. Identification of a glycosylphosphatidylinositol anchor on membrane proteins. In: Hooper NM, Turner AJ, editors. In: Lipid Modification of Proteins: A Practical Approach. Oxford: IRL Press; 1992. pp. 89–115. [Google Scholar]
- Hudson ML, Kinnunen T, Cinar HN, Chisholm AD. C. elegans Kallmann syndrome protein KAL-1 interacts with syndecan and glypican to regulate neuronal cell migrations. Dev Biol. 2006;294:352–365. doi: 10.1016/j.ydbio.2006.02.036. [DOI] [PubMed] [Google Scholar]
- Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol Pharm Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- Johnston WL, Krizus A, Dennis JW. The eggshell is required for meiotic fidelity, polar-body extrusion and polarization of the C. elegans embryo. BMC Biol. 2006;4:35. doi: 10.1186/1741-7007-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachur TM, Audhya A, Pilgrim DB. UNC-45 is required for NMY-2 contractile function in early embryonic polarity establishment and germline cellularization in C. elegans. Dev Biol. 2008;314:287–299. doi: 10.1016/j.ydbio.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Kaitna S, Schnabel H, Schnabel R, Hyman AA, Glotzer M. A ubiquitin C-terminal hydrolase is required to maintain osmotic balance and execute actin dependent processes in the early C. elegans embryo. J Cell Sci. 2002;115:2293–2302. doi: 10.1242/jcs.115.11.2293. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kawagoe K, Kitamura D, Okabe M, Taniuchi I, Ikawa M, Watanabe T, Kinoshita T, Takeda J. Glycosylphosphatidylinositol-anchor-deficient mice: implications for clonal dominance of mutant cells in paroxysmal nocturnal hemoglobinuria. Blood. 1996;87:3600–3606. [PubMed] [Google Scholar]
- Keller P, Tremml G, Rosti V, Bessler M. X inactivation and somatic cell selection rescue female mice carrying a Piga-null mutation. Proc Natl Acad Sci USA. 1999;96:7479–7483. doi: 10.1073/pnas.96.13.7479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Murakami Y, Morita YS. Diseases associated with GPI anchors. In: Kamerling JP, editor. Comprehensive Glycoscience. Chapter 4.21. Vol. 4. Amsterdam: Elsevier; 2007. pp. 393–419. [Google Scholar]
- Kitagawa H, et al. Expression of rib-1, a Caenorhabditis elegans homolog of the human tumor suppressor EXT genes, is indispensable for heparan sulfate synthesis and embryonic morphogenesis. J Biol Chem. 2007;282:8533–8544. doi: 10.1074/jbc.M611107200. [DOI] [PubMed] [Google Scholar]
- Krawitz PM, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Gen. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Müller G. Oral delivery of protein drugs: driver for personalized medicine? Curr Issues Mol Biol. 2010;13:13–24. [PubMed] [Google Scholar]
- Nishimura J, Murakami Y, Kinoshita T. Paroxysmal nocturnal hemoglobinuria: an acquired genetic disease. Am J Hematol. 1999;62:175–182. doi: 10.1002/(sici)1096-8652(199911)62:3<175::aid-ajh7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Nomura KH, Kobayashi R, Hirabayashi Y, Fujisue-Sakai M, Mizuguchi S, Nomura K. Involvement of blood-group-B-active trisaccharides in Ca2+-dependent cell-cell adhesion in the Xenopus blastula. Dev Genes Evol. 1998;208:9–18. doi: 10.1007/s004270050148. [DOI] [PubMed] [Google Scholar]
- Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- Partridge FA, Tearle AW, Gravato-Nobre MJ, Schafer WR, Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- Pierleoni A, Martelli PL, Casadio R. PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 2008;9:392. doi: 10.1186/1471-2105-9-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisson G, Chauve C, Chen X, Bergeron A. FragAnchor: a large-scale predictor of glycosylphosphatidylinositol anchors in eukaryote protein sequences by qualitative scoring. Genomics Proteomics Bioinformatics. 2007;5:121–130. doi: 10.1016/S1672-0229(07)60022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao W, Isaac RE, Keen JN. An analysis of the Caenorhabditis elegans lipid raft proteome using geLC-MS/MS. J Proteomics. 2011;74:242–253. doi: 10.1016/j.jprot.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Rappleye CA, Paredez AR, Smith CW, McDonald KL, Aroian RV. The coronin-like protein POD-1 is required for anterior-posterior axis formation and cellular architecture in the nematode Caenorhabditis elegans. Genes Dev. 1999;13:2838–2851. doi: 10.1101/gad.13.21.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosti V, Tremml G, Soares V, Pandolfi PP, Luzzatto L, Bessler M. Murine embryonic stem cells without pig-a gene activity are competent for hematopoiesis with the PNH phenotype but not for clonal expansion. J Clin Invest. 1997;100:1028–1036. doi: 10.1172/JCI119613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Grant BD, Harada A, Sato K. Rab11 is required for synchronous secretion of chondroitin proteoglycans after fertilization in Caenorhabditis elegans. J Cell Sci. 2008;121:3177–3186. doi: 10.1242/jcs.034678. [DOI] [PubMed] [Google Scholar]
- Strange K, Christensen M, Morrison R. Primary culture of Caenorhabditis elegans developing embryo cells for electrophysiological, cell biological and molecular studies. Nat Protoc. 2007;2:1003–1012. doi: 10.1038/nprot.2007.143. [DOI] [PubMed] [Google Scholar]
- Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Spindle dynamics and the role ofγ-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tsukahara K, et al. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol Microbiol. 2003;48:1029–1042. doi: 10.1046/j.1365-2958.2003.03481.x. [DOI] [PubMed] [Google Scholar]
- Voutev R, Keating R, Hubbard EJA, Vallier LG. Characterization of the Caenorhabditis elegans Islet LIM-homeodomain ortholog, lim-7. FEBS Lett. 2009;583:456–464. doi: 10.1016/j.febslet.2008.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita K, Fukushima K. Carbohydrate recognition by cytokines and its relevance to their physiological activities. In: Kamerling JP, editor. Comprehensive Glycoscience. Chapter 3.25. Vol. 3. Amsterdam: Elsevier; 2007. pp. 539–562. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.