cAMP/PKA stimulation elicited posttranslational increases in CFTR expression and the interaction of specific 14-3-3 proteins with phosphorylated sites within the R region. This improved the efficiency of nascent CFTR biogenesis and reduced its interaction with the COPI retrograde retrieval mechanism, making more CFTR available for anion secretion.
Abstract
Cystic fibrosis transmembrane conductance regulator (CFTR) is a cAMP/protein kinase A (PKA)–regulated chloride channel whose phosphorylation controls anion secretion across epithelial cell apical membranes. We examined the hypothesis that cAMP/PKA stimulation regulates CFTR biogenesis posttranslationally, based on predicted 14-3-3 binding motifs within CFTR and forskolin-induced CFTR expression. The 14-3-3β, γ, and ε isoforms were expressed in airway cells and interacted with CFTR in coimmunoprecipitation assays. Forskolin stimulation (15 min) increased 14-3-3β and ε binding to immature and mature CFTR (bands B and C), and 14-3-3 overexpression increased CFTR bands B and C and cell surface band C. In pulse-chase experiments, 14-3-3β increased the synthesis of immature CFTR, reduced its degradation rate, and increased conversion of immature to mature CFTR. Conversely, 14-3-3β knockdown decreased CFTR B and C bands (70 and 55%) and elicited parallel reductions in cell surface CFTR and forskolin-stimulated anion efflux. In vitro, 14-3-3β interacted with the CFTR regulatory region, and by nuclear magnetic resonance analysis, this interaction occurred at known PKA phosphorylated sites. In coimmunoprecipitation assays, forskolin stimulated the CFTR/14-3-3β interaction while reducing CFTR's interaction with coat protein complex 1 (COP1). Thus 14-3-3 binding to phosphorylated CFTR augments its biogenesis by reducing retrograde retrieval of CFTR to the endoplasmic reticulum. This mechanism permits cAMP/PKA stimulation to make more CFTR available for anion secretion.
INTRODUCTION
Cystic fibrosis transmembrane conductance regulator (CFTR) is an agonist-regulated anion channel expressed at the apical membranes of epithelial cells. CFTR-dependent anion secretion establishes the driving forces for salt and water secretion to clear the apical surface of secreted macromolecules, for example, airway mucins and pancreatic enzymes. The enabling step in CFTR channel activation involves phosphorylation of the regulatory region (R region), an intrinsically disordered region mediating protein interactions that receives regulatory input from protein kinase A (PKA), protein kinase C (PKC), and AMP-activated protein kinase (AMPK). It contains nine PKA consensus phosphorylation motifs (Gadsby and Nairn, 1999). Phosphorylation at multiple sites in the R region is believed to evoke a change in CFTR conformation that permits the nucleotide-binding domains (NBD1 and 2) to associate, an interaction that forms sites for the binding and hydrolysis of ATP to drive channel gating (opening and closing) activity (Vergani et al., 2005).
A central issue in cystic fibrosis (CF) disease is the low efficiency with which the common CFTR variant ΔF508 achieves the native, folded state that permits its exit from the endoplasmic reticulum (ER) and its trafficking to, and stability at, the apical membrane of epithelial cells (Cheng et al., 1990; Okiyoneda et al., 2010). Many therapeutic approaches to the basic defect in CF are directed toward improving CFTR folding and/or interfering with its ER-associated degradation. Either of these strategies may benefit from conditions that increase the amount of CFTR in the biosynthetic pipeline (Hutt et al., 2010).
In addition to the control of channel gating, increases in cellular cAMP have been shown to augment CFTR biogenesis by interacting with a cAMP response element (CRE) identified 48 nucleotides upstream of the start site for gene transcription (McDonald et al., 1995). The interaction of this regulatory region with CRE-binding protein and transcription factors has been demonstrated, making CFTR transcription a cAMP- and stimulation-responsive process (Matthews and McKnight, 1996).
At the posttranscriptional level, a number of studies implicated protein phosphorylation and interactions with 14-3-3 proteins in the processing and stability of channels and receptors (Mrowiec and Schwappach, 2006). Although the 14-3-3 protein family was first identified in 1967 (Moore and Perez, 1967), we now appreciate that these regulatory proteins generally interact with phosphorylated motifs that surround phosphoserine or phosphothreonine residues to control a diverse array of cellular functions (Yuan et al., 2003). As such, they often stabilize a protein's phosphorylated state (Kjarland et al., 2006). 14-3-3 proteins promote the plasma membrane expression of integral membrane proteins such as the nicotinic acetylcholine and γ -aminobutyric acid receptors (Zerangue et al., 1999; Jeanclos et al., 2001), potassium channels (TASK-1, TASK-3, and KCNK3; Rajan et al., 2002), and the epithelial Na+ channel, ENaC (Liang et al., 2006, 2008).
In the present study, the levels of CFTR in cells expressing the gene from an exogenous promoter were found to be augmented by stimulation of the cAMP/PKA pathway. The mechanism of this posttranslational regulation of CFTR expression was linked to the selective interaction of the channel with specific 14-3-3 protein isoforms at known PKA phosphorylation sites within the R region. The effect of this pathway was demonstrated in 14-3-3 protein knockdown experiments in which steady-state expression of CFTR was markedly reduced. Thus, in addition to stimulation-dependent transcriptional regulation of CFTR expression, these posttranslational regulatory events also increase CFTR biogenesis during stimulation. Their manipulation is expected to augment the pool of CFTR available for therapeutic interventions.
RESULTS
PKA stimulation increases CFTR expression level
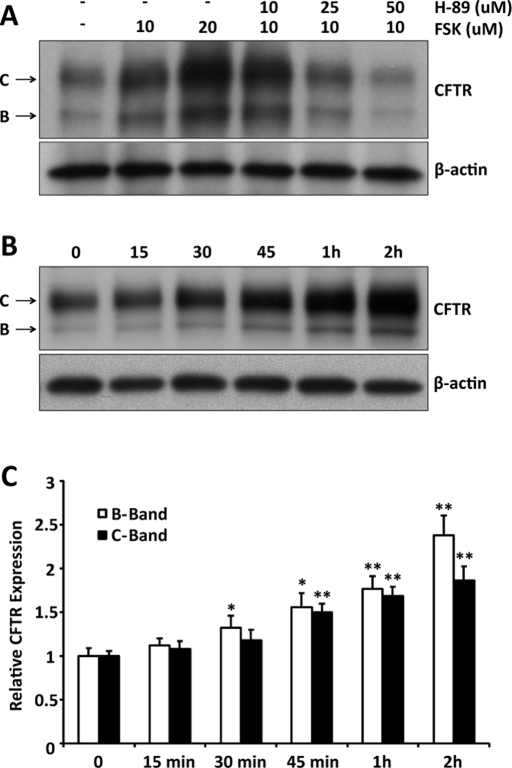
The effect of cAMP/PKA stimulation on steady-state CFTR expression was examined initially by immunoblot (IB) using human embryonic kidney (HEK) 293 cells transiently transfected with CFTR and treated overnight with forskolin (FSK). Forskolin stimulation increased CFTR expression in a dose-related manner, and addition of the PKA inhibitor H-89 decreased CFTR levels (Figure 1A). The forskolin result was not due to stimulation of CFTR's promoter, since expression from the plasmid is driven by the cytomegalovirus promoter. In addition, this effect on expression was not observed for other proteins expressed transiently from the same vector, including green fluorescent protein (GFP) and a truncated version of CD4 (Supplemental Figure S1). At the highest dose of H-89, CFTR expression fell below the control level (−/−), suggesting that endogenous kinase activity might play a role in CFTR expression under basal conditions.
FIGURE 1:
CFTR expression is increased by cAMP/PKA stimulation. (A) HEK293 cells transiently transfected with CFTR were treated for 16 h with forskolin, with or without the PKA inhibitor H-89, at the indicated concentrations and processed for immunoblot to assess the effect of stimulation on steady-state CFTR expression levels. (B) Time course of the increase in CFTR expression during forskolin (20 μM) stimulation. (C) Quantitation of immature (band B) and mature (band C) CFTR expression from all experiments like that shown in B (n = 3). *p < 0.05; **p < 0.01.
We also used immunoblot analyses to examine the expression of CFTR as a function of time during the stimulation of HEK293 cells by FSK (20 μM). As shown in Figure 1B, the adenylyl cyclase activator increased the expression of both mature and immature CFTR. The time course of CFTR levels from all experiments was quantified, normalized to β-actin expression, and is shown in Figure 1C. Increased CFTR, observed at early stimulation times, was also consistent with a nontranscriptional regulatory process, and similar experiments performed in the presence of actinomycin D supported this conclusion (unpublished data). Although CFTR expression is regulated in part by upstream cAMP response elements (McDonald et al., 1995), the present data are consistent with stimulation-related control of CFTR expression at the posttranslational level. Our prior studies indicated that phosphorylation-dependent 14-3-3 protein interactions increase the stability of the epithelial Na channel in response to Nedd4-2 and AS160 phosphorylation (Liang et al., 2008, 2010). This and work on K channel biogenesis (reviewed later) led us to the hypothesis that 14-3-3 proteins participate in the phosphorylation-dependent control of CFTR expression level.
Expression of 14-3-3 isoforms in HEK293 and airway cells
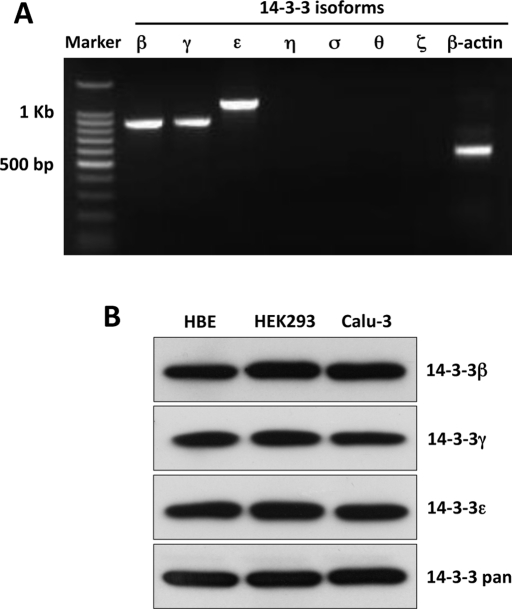
It is increasingly apparent that the seven identified isoforms of 14-3-3 proteins frequently show selectivity of function and cell type expression pattern (Fu et al., 2000). Therefore we used reverse transcriptase (RT)-PCR and IB to examine 14-3-3 isoform expression in HEK293 cells, in the Calu-3 airway cell line, and in differentiated primary cultures of human bronchial epithelia (HBE). cDNA from these cells was used with isoform-specific primers to perform semiquantitative PCR, as previously discussed (Liang et al., 2006). In HEK293 and Calu-3 cells, significant PCR products of appropriate size and sequence corresponded to the 14-3-3 isoforms β, γ, and ε, whereas other isoforms were weak or absent (Supplemental Figure S2). Of importance, primary HBE cultures produced similar findings (Figure 2A). The lack of signal or the weak signals observed for some isoforms was not due to issues of primer adequacy or PCR conditions, as adequate signals were obtained in positive control lines (unpublished data). For protein detection, cell lysates were obtained from HEK293, Calu-3 cells, and HBE primaries, resolved by SDS–PAGE, and probed for 14-3-3β, γ, and ε using isoform-specific antibodies (Figure 2B). Similar to the PCR data, the IB results indicate that 14-3-3β, γ, and ε were abundantly expressed in airway and HEK293 cells at the protein level.
FIGURE 2:
The 14-3-3 isoforms β, γ, and ε are expressed in HEK293 and airway cells. (A) The expression of 14-3-3 protein isoforms was determined by RT-PCR using cDNA from differentiated, polarized, primary cultures of HBE. Similar patterns of isoform expression were obtained from HEK293 and Calu-3 cells (Supplemental Figure S1, A and B). Products of the appropriate size and sequence for 14-3-3β, γ, and ε were obtained. (B) Immunoblots performed using isoform-specific antibodies and cell lysates from HEK293 or Calu-3 cell lines or HBE primary cultures showed a similar expression pattern at the protein level. The PCR and IB data are representative of the results from three experiments.
Phosphorylation augments the interaction of CFTR with 14-3-3β
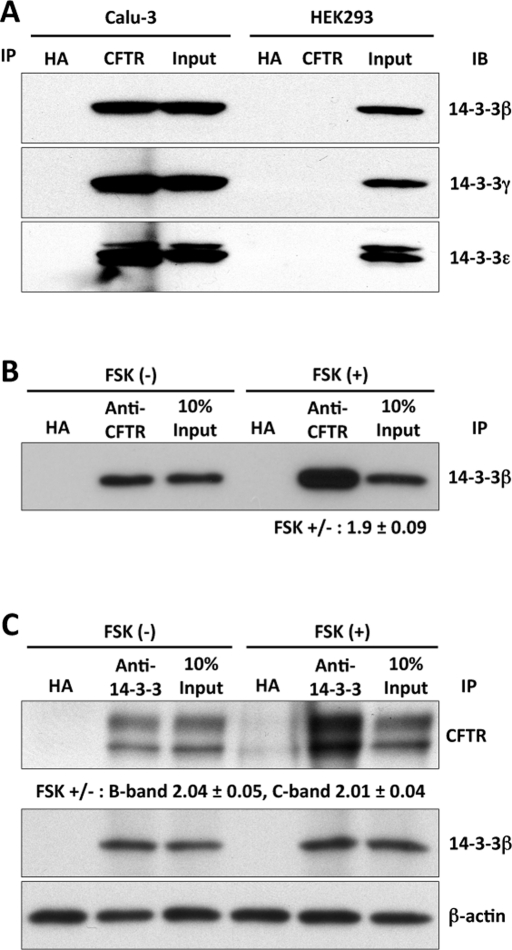
To determine whether interactions between 14-3-3 proteins and CFTR could be detected in vivo, we performed coimmunoprecipitation (coIP) experiments using lysates from the Calu-3 cell line. The initial precipitation was performed with antibodies against CFTR, and the immunoprecipitates were then blotted with antibodies specific for 14-3-3β, γ, or ε. Physical interactions of 14-3-3β, γ, and ε with CFTR were detected in Calu-3 cells as shown in Figure 3A. The absence of signal in HEK293 cells not expressing CFTR confirms the specificity of these antibodies.
FIGURE 3:
CFTR interacts with 14-3-3β, γ, and ε. (A) In vivo CFTR-14-3-3 isoform interactions were evaluated in coIP experiments using lysates from Calu-3 or HEK293 cells. IPs were performed using antibodies to CFTR (M3A7) or the HA epitope as control. HEK293 cells were not transfected with CFTR cDNA. (B) Forskolin stimulation (15 min) increased the interaction of 14-3-3β with CFTR. CFTR-transfected HEK293 cells; CFTR IP followed by 14-3-3β IB. Immunoglobulin G against the HA epitope served as IP control. (C) Reverse IP/IB protocol performed using the same cells and conditions; 14-3-3β was associated with both immature (band B) and mature (band C) CFTR, and their interaction was increased significantly by forskolin stimulation.
The dependence of 14-3-3 binding on CFTR phosphorylation was examined in similar coIP experiments performed with HEK293 cells transiently expressing CFTR and stimulated with 10 μM forskolin for 15 min. The results show that the interaction between 14-3-3β and CFTR increased approximately twofold in response to cAMP/PKA stimulation (Figure 3B). Using a reverse IP/IB protocol and CFTR-transfected HEK293 cells, we found that 14-3-3β brought down both the mature (band C) and immature (band B) forms of CFTR. The IP of 14-3-3β was similar in the presence and absence of forskolin, but its interaction with CFTR was increased significantly, approximately twofold, by forskolin treatment (Figure 3C). These augmented interactions were observed with endogenous 14-3-3β and only 15 min of forskolin treatment and are therefore not due to increased CFTR expression.
14-3-3β and ε selectively increase CFTR expression in response to forskolin
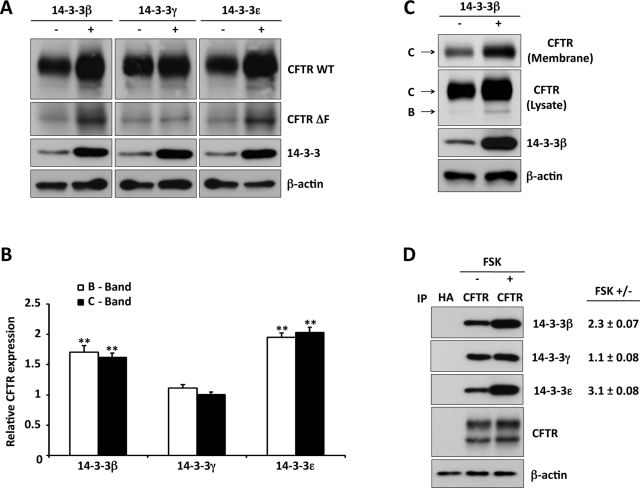
The significance of 14-3-3 proteins in the regulation of CFTR expression by cAMP/PKA was evaluated by overexpressing the airway-specific isoforms 14-3-3β, γ, and ε with CFTR in HEK293 cells. As shown in Figure 4A, coexpression of 14-3-3β and ε increased the steady-state levels of mature and immature wild-type (WT) CFTR (bands C and B), as well as the immature form of the common CFTR variant ΔF508 (Figure 4A). A significant effect of 14-3-3γ on CFTR expression level was not observed. Quantification of these data are presented in Figure 4B, which demonstrates specificity among the expressed isoforms in their ability to posttranslationally augment the expression of CFTR, which again was approximately twofold. Cell surface biotinylation was used to determine whether the augmented steady-state CFTR in response to 14-3-3 overexpression translated into increased plasma membrane CFTR. Biotinylated proteins were isolated by streptavidin pulldown, as described in Materials and Methods. As shown in Figure 4C, 14-3-3β expression increased cell surface CFTR, which averaged twofold and corresponded to a 1.6-fold increase in cell lysate CFTR expression (n = 3).
FIGURE 4:
CFTR interactions with 14-3-3 proteins are selective and increased by cAMP/PKA stimulation. (A) 14-3-3 isoforms (as indicated) were coexpressed with WT or ΔF508 CFTR in HEK293 cells, followed by CFTR IB. Expression of 14-3-3β or ε increased immature and mature WT CFTR and immature ΔF508 but expression of 14-3-3γ did not. (B) Quantitation of the effect of 14-3-3 isoform expression on WT CFTR B and C bands relative to that observed in the absence of 14-3-3 coexpression; data from three experiments as in A. **p < 0.01. (C) 14-3-3β coexpression increases cell surface CFTR, as determined by biotinylation and streptavidin pulldown. The data are typical of three experiments. (D) Stimulation-induced interactions of CFTR with 14-3-3 isoforms. HEK293 cells transiently expressing CFTR; CFTR IP followed by 14-3-3 isoform-specific IB under basal or stimulated (forskolin, 10 μM, 15 min) conditions. Values to the right provide the relative increase in 14-3-3 interaction with stimulation from three experiments. CFTR interactions with 14-3-3β and ε were selectively increased by stimulation.
Next we examined the interaction of the three predominant 14-3-3 isoforms with CFTR in IP/IB experiments performed by using lysates from HEK293 cells. Protein complexes were isolated from cells under basal conditions or after 15 min of forskolin stimulation. As illustrated in Figure 4D, IPs performed with anti-hemagglutinin (HA) immunoglobulin G (IgG) as control yielded no 14-3-3 signal. Under basal conditions, CFTR interacted with the 14-3-3β, γ, and ε isoforms, consistent with the data of Figure 3A. Relative to nonstimulated conditions, forskolin increased CFTR interaction with 14-3-3β and 14-3-3ε by 2.3- and 3.1-fold, respectively (Figure 4D), whereas its interaction with the 14-3-3γ isoform was not affected by stimulation. Together with the data of Figure 4A, these findings indicate that the increases in total and plasma membrane CFTR expression level evoked by cAMP/PKA stimulation or 14-3-3 overexpression involve selective interactions with 14-3-3β and ε.
The fate of newly synthesized CFTR is modulated by 14-3-3 protein interactions
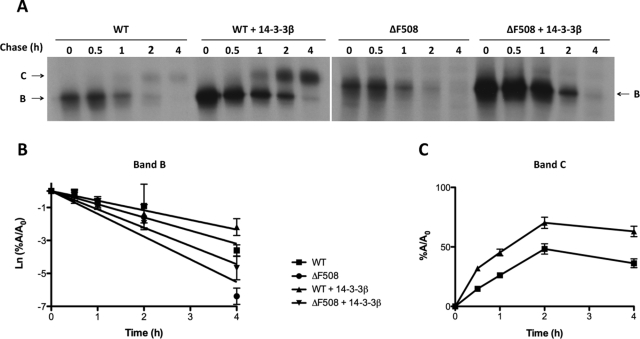
The effect of 14-3-3 proteins on the fate of nascent CFTR was examined using pulse-chase experiments performed in HEK293 cells coexpressing 14-3-3β and either WT or ΔF508 CFTR. We examined the synthesis of CFTR during the 30-min pulse interval, the turnover rate of the immature (band B) forms, and the efficiency of conversion of the immature to the mature form (band C). Representative autoradiographs are shown in Figure 5A; quantification of all experiments is provided in Figure 5, B and C, for WT and mutant CFTR, respectively. As illustrated in Figure 5A, 14-3-3β coexpression significantly increased the formation of the immature forms of both WT and mutant CFTR during the 30-min pulse. In addition, 14-3-3β slowed the rate at which immature CFTR disappeared during the chase. Both of these effects may be explained by reduced CFTR degradation, which is known to occur early, as cotranslational ubiquitylation has been detected (Sato et al., 1998). Nevertheless, an increase in CFTR folding efficiency may also contribute to the increase in CFTR biosynthetic rate, at least for WT CFTR, and this possibility is consistent with the improved efficiency of conversion of immature to mature protein observed at 2 h of chase (48% control vs. 70% 14-3-3β, p < 0.005). Despite these actions of expressed 14-3-3β on WT CFTR, significant maturation of ΔF508 CFTR was not apparent. Taken together, these data indicate that a substantial contribution to the effect of 14-3-3 proteins on steady-state CFTR levels results from their ability to reduce CFTR degradation and enhance the biogenesis of the nascent protein. Whether the 14-3-3s also affect the turnover of post-Golgi CFTR, as suggested by their interaction with the mature protein, requires additional study.
FIGURE 5:
14-3-3β expression enhances nascent CFTR biogenesis. (A) Pulse-chase experiments were performed using HEK293 cells transiently expressing WT or ΔF508 CFTR and 14-3-3β or empty vector (control) were radiolabeled with [35S]methionine–cysteine mix (PerkinElmer) for 30 min and then chased at the indicated times before lysis with RIPA buffer and IP with anti-CFTR antibodies 596 and 217. The positions of bands B and C are indicated by arrows. (B) Rate of turnover of the immature form of CFTR (band B) shown as the natural logarithm of the amount of band B at a given time of the chase (A) relative to the amount at the beginning of the chase (A0). (C) Efficiency of conversion of immature (band B) to mature (band C) CFTR, shown as the percentage of band C detected at a given time of the chase (A) relative to the amount at the beginning of the chase (A0). In B and C, data are mean ± SEM (n = 3), and lines are the fit of first-order regression to the data.
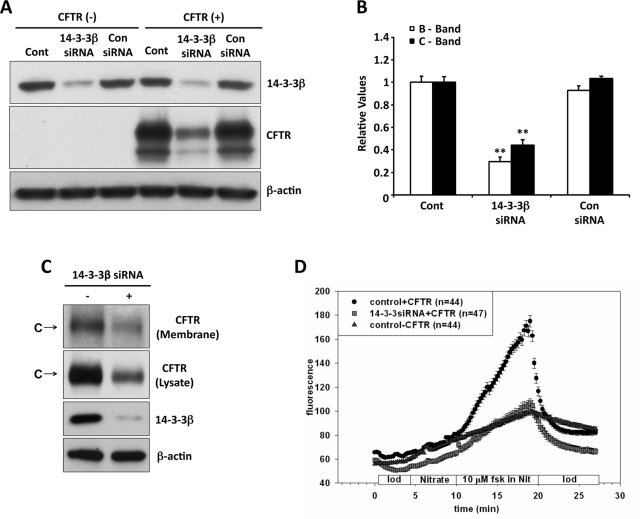
14-3-3β knockdown decreases CFTR expression and function
We used 14-3-3β–targeted small interfering RNAs (siRNAs) to knock down this 14-3-3 isoform in HEK293 cells transfected with or without CFTR and compared the results with those obtained with a control, scrambled siRNA. The RNA interference–induced reduction in 14-3-3β expression was ∼90%, whereas the control siRNA did not affect 14-3-3β expression (Figure 6A). The corresponding expression level of CFTR is illustrated by the blots of Figure 6A; the data from all experiments are quantified in Figure 6B. Decreasing 14-3-3β reduced the expression of both immature and mature CFTR bands (B and C) on average by ∼70 and ∼55%, respectively. The expression of β-actin was monitored as control. The large effect of 14-3-3β knockdown, despite endogenous expression of the γ and ε isoforms, is likely due to two factors: 1) 14-3-3γ did not participate in CFTR regulation—unlike β and ε, its expression did not increase CFTR level, and phosphorylation did not enhance its interaction with CFTR—and 2) 14-3-3β and ε form an obligatory heterodimer (Liang et al., 2008), a property that will be discussed in more detail later.
FIGURE 6:
14-3-3β knockdown decreases CFTR expression and stimulated anion efflux. (A) CFTR expression level. Control and 14-3-3β siRNAs were obtained as SMART-pool reagents (Dharmacon, Lafayette, CO), as described in Liang et al. (2006). (B) Quantitation of data from three experiments, normalized to the control CFTR levels. Reduced 14-3-3β expression (∼90%) decreased steady-state CFTR band B (∼70%) and band C (∼55%); the control siRNA had no effect. (C) 14-3-3β knockdown reduced CFTR detected by biotinylation of the surface membranes of Calu-3 cells, performed as described in Materials and Methods. The data are representative of two such experiments. (D) 14-3-3β knockdown reduced CFTR-mediated anion efflux. HEK293 cells seeded onto glass coverslips were transiently transfected with CFTR, and 14-3-3β siRNAs or empty vector (control). After 48 h, SPQ assays of forskolin plus IBMX-stimulated iodide efflux were performed as described in Material and Methods. The stimulation-induced increase in slope was reduced ∼70% by 14-3-3β knockdown, similar to the reduction in CFTR expression level. Data are mean ± SE from two independent experiments performed on 44–47 cells for each.
Next we evaluated the effect of 14-3-3β knockdown on total and cell surface CFTR in the airway cell line, Calu-3, shown earlier to express the β, γ, and ε isoforms (Supplemental Figure S2B). As shown in Figure 6C, reduced 14-3-3β decreased both total and plasma membrane CFTR, consistent with the findings from HEK293 cells.
To determine whether the reduction in cell surface CFTR affects channel function, we measured the effect of 14-3-3β knockdown on CFTR- and cAMP-dependent anion efflux across the plasma membranes of HEK293 cells, using the halide-sensitive fluorescence indicator 6-methoxy-N-(3-sulfopropyl)quinolinium (SPQ; Figure 6D). In cells expressing CFTR alone, we noted the anticipated upward deflection in the time course of fluorescence intensity upon addition of forskolin plus 3-isobutyl-1-methylxanthine (IBMX). This response reflects dequenching of SPQ, which is associated with cAMP/PKA-stimulated iodide efflux from iodide-preloaded cells. This response was reduced ∼70% in cells following 14-3-3β knockdown. These findings are consistent with the similar effect of reduced 14-3-3β on the expression of CFTR protein and indicate that the mature CFTR detected biochemically and at the plasma membrane (Figure 6D) is functional.
14-3-3β interacts with the R region and N-terminus of CFTR
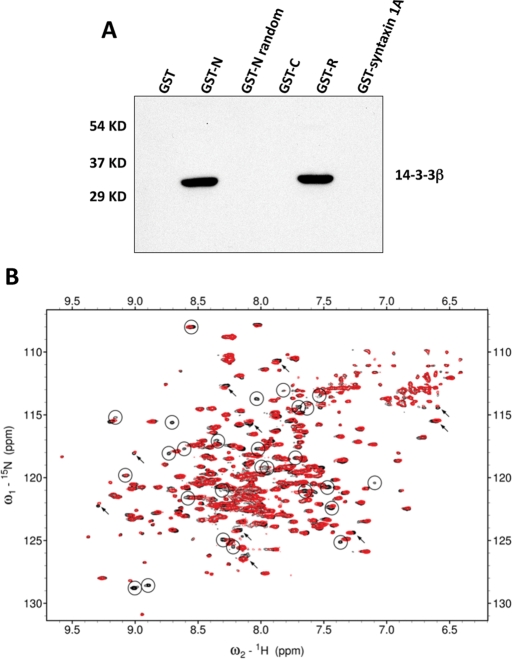
The results from coimmunoprecipitation experiments indicated that CFTR and 14-3-3 interact in vivo (Figure 3). To evaluate CFTR domain interactions with 14-3-3 proteins, we performed pulldown experiments using glutathione-S-transferase (GST alone or GST-CFTR domain fusion proteins incubated with Calu-3 cell extracts; Figure 7A). In this study, the GST fusion proteins included CFTR N-terminus, C-terminus, and R region; GST-syntaxin 1A served as a control. The results show that 14-3-3β interacted significantly with the CFTR N-terminus and the nonphosphorylated R region, whereas no significant binding was detected between the 14-3-3β isoform and GST, GST-CFTR C-terminus (GST-C), GST-CFTR N-terminus (GST-N) random, or GST-syntaxin 1A.
FIGURE 7:
14-3-3β interaction with the CFTR R region. (A) In vitro pairwise binding of 14-3-3β with CFTR domains. The indicated bead-immobilized GST-CFTR domains or GST-syntaxin 1A fusion proteins were prepared as described in Material and Methods and incubated with Calu-3 cell extracts. After washing, samples were resolved on 12% SDS–PAGE and detected using anti–14-3-3β, which interacted directly with the N-terminus and nonphosphorylated R region of CFTR. (B) 1H15N correlation spectra of 15N-labeled human 14-3-3β protein in the absence (black) and in the presence (red) of unlabeled PKA phosphorylated R region measured at 50°C. Many peaks with significant broadening are highlighted with circles (demonstrating reduced intensity, with fewer contours seen in the contour plot), and selected peaks with substantial peak shifts are marked with arrows. The differences between the two spectra provide evidence for interaction of the R region with 14-3-3β.
Interactions of 14-3-3 protein with multiple motifs within the R region
To further probe the interaction between the 14-3-3 and the R region, we monitored binding using nuclear magnetic resonance (NMR) experiments. Peaks in the 1H15N correlation spectrum for 14-3-3β are perturbed in the presence of phosphorylated R region, with some chemical shift changes and significant peak broadening observed, confirming the binding (Figure 7B). The sequences of the nine PKA phosphorylation sites in the R region have some similarity to the two broadly defined consensus 14-3-3 recognition motifs (Johnson et al., 2010; Table 1). To investigate the hypothesis that each of these PKA sites could interact with 14-3-3β, we compared the spectra of the R region in free and 14-3-3β bound states (Bozóky and Forman-Kay, unpublished data) and calculated the estimated relative occupancy of the bound state for each site based on the broadening of resonances for the central three residues of the binding segment (Table 1). Comparison of intensities in free and bound states is a useful approach to monitoring interactions of disordered proteins such as the R region (Baker et al., 2007). The results of this analysis suggest that most of the PKA sites in the phosphorylated R region have reasonable affinity for 14-3-3β, with the most broadened being the pS768, pS795, and pS813 PKA sites (Table 1A). The broadening observed for each site provides evidence for a dynamic complex involving an equilibrium exchange of interactions of the various sites with the 14-3-3β interface. The extent of PKA phosphorylation is nearly 100% at these sites, as determined by both NMR and mass spectrometry analysis (unpublished data). For the nonphosphorylated R region, we observed significant but reduced binding to 14-3-3β compared with the phosphorylated state, with the greatest broadening largely at residues not aligned with PKA sites or consensus 14-3-3 recognition motifs (Table 1B).
TABLE 1:
Analysis of R region 14-3-3β binding sites.
| Central residue of the binding motif | Sequence | Estimated relative occupancy of the bound state |
|---|---|---|
| A. Interacting segments of phosphorylated R region | ||
| Mode I | RXXpSXP | |
| Mode II | RX[SYFWTQAD]XpSX[PLM] | |
| pS660 | SAERRNpSILTETL | 0.85 |
| pS670 | ETLHRFpSLEGDAP | 0.39 |
| pS700 | GEKRKNpSILNPIN | 0.58 |
| pS712 | NSIRKFpSIVQKTP | 0.83 |
| pS737 | PLERRLpSLVPDSE | 0.72 |
| pS753 | AILPRIpSVISTGP | 0.83 |
| pS768 | QARRRQpSVLNLMT | 1 |
| pS795 | ASTRKVpSLAPQAN | 0.98 |
| pS813 | IYSRRLpSQETGLE | 0.93 |
| B. Interacting segments of nonphosphorylated R regiona | ||
| S670 | ETLHRFSLEGDAP | 0.54 |
| N706 | SILNPINSIRKFS | 0.36 |
| R764 | GPTLQARRRQSVL | 0.47 |
| L770 | RRRQSVLNLMTHS | 0.56 |
| S809 | TELDIYSRRLSQE | 0.40 |
Part A shows binding motifs surrounding the phosphorylation sites at which 14-3-3β interactions were observed for phosphorylated R region, compared with the consensus mode I and II 14-3-3 interaction motifs (Johnson et al., 2010). Part B shows binding motifs at which 14-3-3β interactions were observed for non-phosphorylated R region. The values in both parts of the table provide estimates of the relative population of the 14-3-3 bound state for each site based on broadening of R region resonances for the three central residues of the motif (pS − 1, pS, and pS + 1 in the case of the phosphorylated motifs) due to 14-3-3 protein binding (Bozóky and Forman-Kay, unpublished data), normalized to pS768, at which the greatest broadening is observed.
aNote that these nonphosphorylated segments likely also bind in the context of the PKA phosphorylated state of the R region, but they compete with the stronger-binding phosphorylated segments.
14-3-3 binding competes with CFTR–coat protein complex interactions
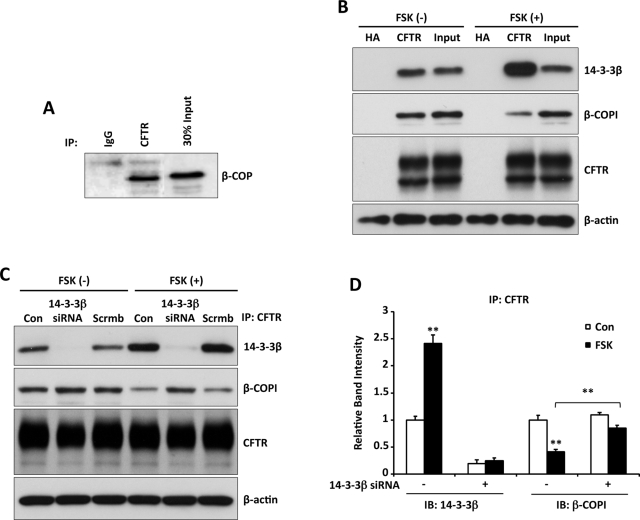
The coat protein complex (COPI) was implicated in protein transport between the ER and Golgi (Lee et al., 2004; Rennolds et al., 2008), and prior studies of CFTR suggested an interaction with the coat protein β-COP (Rennolds et al., 2008). Studies of the trafficking of the dual-pore potassium channel K(2P)3.1 demonstrated that its interaction with 14-3-3β suppressed its COPI-mediated ER retention, permitting forward trafficking to the plasma membrane (O'Kelly et al., 2002). To determine whether altered CFTR interactions with the COPI coat contribute to 14-3-3–mediated CFTR trafficking, we first asked whether CFTR and the coat complex component β-COP interact in Calu-3 cells. Coimmunoprecipitation experiments showed that these proteins were associated in vivo (Figure 8A).
FIGURE 8:
14-3-3β competes for β-COPI binding to CFTR in response to cAMP/PKA stimulation. (A) β-COP interacts with CFTR. IP from Calu-3 cell lysates using HA control or CFTR immunoglobulin G, followed by IB for β-COP, demonstrates an interaction of CFTR with this COPI coat protein. (B) CoIP experiments were performed using CFTR-transfected HEK293 cells with or without forskolin stimulation (10 μM, 15 min), followed by CFTR/14-3-3β or CFTR/β-COPI coIP. The interaction of CFTR with 14-3-3β increased with stimulation, whereas that with β-COPI decreased. (C) Recovery of the β-COP interaction with CFTR, induced by 14-3-3β knockdown. Lysates from HEK293 cells transiently expressing CFTR and either 14-3-3β or scrambled siRNAs (Figure 6) and treated with forskolin (10 μM, 15 min) as indicated were subjected to CFTR IP, followed by IB for 14-3-3β, β-COP, or CFTR. (D) Quantitation of the IB data from three experiments as in C. Stimulation reproduces a reciprocal relation between 14-3-3β and β-COP binding to CFTR, whereas β-COP binding returns toward control values when 14-3-3β expression is reduced. The data were normalized to 14-3-3β and β-COP intensities observed under basal conditions in the absence of knockdown. **p < 0.01.
Next we asked whether the interaction with β-COP was competitive with phosphorylation-dependent 14-3-3 protein binding (Figure 3, B and C). CFTR coIP experiments were performed using CFTR-transfected HEK293 cells maintained under basal conditions or treated with forskolin (10 μM) for 15 min, followed by 14-3-3β or β-COPI immunoblot. The results of Figure 8B show that the interaction between 14-3-3β and CFTR increased significantly with forskolin stimulation, whereas the association of CFTR with β-COPI was reduced.
Another test of this reciprocal interaction was performed using 14-3-3β knockdown experiments. As earlier, 14-3-3β expression was reduced in HEK293 cells transduced to express CFTR, and the cells were either treated with forskolin for 15 min or maintained in the basal state. CFTR was immunoprecipitated from cell lysates and the IPs blotted for 14-3-3β or β-COP. Under nonstimulated conditions, the interaction of CFTR with β-COP was not influenced by 14-3-3β knockdown. Stimulation by forskolin again reduced the interaction of CFTR with β-COPI, but this interaction returned to control levels when 14-3-3β expression was reduced. These findings support the idea that CFTR is retained in the ER via COPI-mediated retrograde trafficking and that phosphorylation-dependent binding of 14-3-3 proteins suppresses CFTR's interaction with COPI to facilitate its forward transport.
DISCUSSION
The results of this study indicate that the posttranscriptional biogenesis of CFTR is phosphorylation dependent. Thus they imply a role for the regulatory factors that control CFTR channel activity in determining also the amount of CFTR available for anion and fluid secretion. CFTR levels were increased by cAMP/PKA stimulation and blocked by the PKA inhibitor H-89. These effects were associated with PKA-mediated phosphorylation of sites within CFTR's R region and their interaction with 14-3-3 proteins. Because CFTR transcription is regulated by upstream cAMP response elements (McDonald et al., 1995; Pittman et al., 1995; Matthews and McKnight, 1996; Li et al., 1999), both transcriptional and posttranslational mechanisms provide for stimulation-dependent CFTR production.
14-3-3 protein specificity
Recent findings highlight the significance of isoform specificity in the actions of 14-3-3 proteins, which interact with their substrates as dimers with two potential binding sites for target protein(s) (Aitken et al., 2002; Wilker and Yaffe, 2004). The basis of 14-3-3 substrate selectivity may lie in the preference of the dimer binding cleft for specific phosphopeptide motifs (Wilker et al., 2005) or in structures peripheral to the binding core, the least conserved regions among the 14-3-3 isoforms (Cardasis et al., 2007). 14-3-3β, γ, and ε were the predominant isoforms expressed in HEK293 cells and in Calu-3 and primary HBE airway cells. A selective role of 14-3-3β and ε in CFTR biogenesis emerged from experiments in which isoform overexpression increased steady-state CFTR protein levels (Figure 4, A and B), and this specificity was preserved when their phosphorylation-dependent interactions with CFTR were examined (Figure 4D). Although 14-3-3γ was expressed at levels similar to those for β and ε, this isoform did not play a role in enhancing CFTR expression.
The physiological significance of these interactions was verified by 14-3-3β knockdown, which reduced total and cell surface CFTR and led to a corresponding reduction in cAMP/PKA-stimulated anion efflux. In view of the similar actions of 14-3-3β and ε, it may seem curious that the knockdown of 14-3-3β alone markedly reduced CFTR expression level and regulated anion efflux. However, this finding is consistent with the solved crystallographic structures of 14-3-3β and ε mixtures, which contained >95% β/ε heterodimers (Yang et al., 2006), and with our prior coIP studies, showing that the IP of either isoform quantitatively depleted the other isoform from lysates of cells endogenously expressing β and ε (Liang et al., 2008). Thus heterodimer formation accounts for the effect on CFTR levels of 14-3-3β knockdown alone, as we observed previously for the regulation of apical membrane ENaC density (Liang et al., 2008).
Phosphorylation-dependent 14-3-3 interactions in CFTR biogenesis
Both phosphorylation-dependent and -independent modes of 14-3-3 binding have been implicated in protein progression along the early secretory pathway (Smith et al., 2011). CFTR was associated with 14-3-3 proteins under both basal and stimulated conditions, and the NMR data shows binding of 14-3-3β to both phospho and nonphospho states of the R region, implying that both types of interactions may be involved in determining the fate of CFTR. The present study focuses primarily on the regulation of CFTR expression by its PKA-mediated phosphorylation. Prior evidence of phosphorylation-regulated forward trafficking of cargo proteins includes the presence of PKA in the ER and its stimulation of hERG K channel progression to the cell surface (Sroubek and McDonald, 2011). PKA also plays a role in the ER exit of other channels and receptors (Kuwana et al., 1998; Jeanclos et al., 2001; Mrowiec and Schwappach, 2006), including the KCNK and ROMK1 potassium channels (O'Kelly et al., 2002; O'Connell et al., 2005).
Physical interactions with 14-3-3 proteins formed the basis of the stimulation-induced increase in CFTR levels (Figure 1), since 1) cAMP/PKA stimulation increased the ability of 14-3-3β to coIP CFTR (Figure 3C), 2) stimulation enhanced the ability of CFTR to associate with 14-3-3β and ε (Figure 4D), and 3) NMR studies showed that PKA-induced phosphorylation of known phospho sites within the R region increased the interaction of these sites with 14-3-3β (Table 1). Although this analysis implicated most of the recognized R region phospho sites in phosphorylation-dependent 14-3-3 binding, the relative population of the 14-3-3 bound state was particularly high for binding sequences in its C-terminus (Table 1). Mutation analysis of CFTR's phospho sites demonstrated their redundant contributions to the channel activation process (Chang et al., 1993; Wilkinson et al., 1997; Gadsby and Nairn, 1999), and a similar phenomenon is likely to apply in attempts to define the sites responsible for the phosphorylation-dependent 14-3-3 interactions in CFTR biogenesis. Therefore a mutation analysis of these sites was not undertaken at this time.
The exchanging engagement of multiple CFTR R region PKA sites on the two binding surfaces of the 14-3-3 dimer is an example of a dynamic complex involving intrinsically disordered proteins (Tompa and Fuxreiter, 2008; Mittag et al., 2010a). These flexible proteins, which lack stable folded structure, including the CFTR R region, are highly charged, are depleted in hydrophobic residues, and contain numerous sites of posttranslational modification, particularly phosphorylation. Intrinsically disordered proteins are increasingly recognized to play critical roles in mediating regulatory protein interactions, in which they can undergo disorder-to-order transition upon binding in some cases but in others can remain highly mobile within the context of dynamic complexes such as proposed for the R region: 14-3-3 interaction. The interaction of the R region with NBD1 was also suggested to share this dynamic character, with multiple segments of the R region binding to NBD1 in a manner believed to transiently stabilize helical structure (Baker et al., 2007). The interaction of 14-3-3 with the R region, in contrast, is likely to involve extended structure for the R region PKA sites bound to each of the 14-3-3 interfaces, as observed for numerous crystal structures of peptide complexes of 14-3-3 proteins (Yang et al., 2006; Molzan et al., 2010; Schumacher et al., 2010). Such plasticity in binding is typical of disordered proteins, as observed for a disordered segment of p53 (Oldfield et al., 2008).
The interacting PKA sites within the R region have a phosphoserine that matches the 14-3-3 consensus binding motifs (Table 1); however, they do not otherwise agree well with the consensus sequences, suggesting that each site would interact weakly. The significant binding observed, with a Kd value of ∼5 μM (Bozóky and Forman-Kay, unpublished data), is thus likely due to synergistic interactions of the nine sites with two sites interacting at any given time, reminiscent of the dynamic interaction of multiple phosphorylated Sic1 motifs (Mittag et al., 2008, 2010b). The dynamic character of R region binding to 14-3-3 likely facilitates its rapid interactions with other partners or domains. Of importance, the binding of R region to 14-3-3 shifts the equilibrium away from its intramolecular partners, including NBD1, an interaction believed to be inhibitory to NBD heterodimer formation. Although critical for channel function, NBD heterodimer formation may also enhance folding efficiency, synergizing with other roles of 14-3-3 in processing.
The R region is believed to play a pivotal role in the stability of CFTR during its biogenesis. On the basis of the use of C-terminal CFTR truncations to mimic translation intermediates, N-terminal CFTR fragments lacking the R region were unstable, and they were associated with chaperones, reducing their aggregation (Strickland et al., 1997; Meacham et al., 1999). Once the R region was added, however, chaperone interactions were reduced, and the nascent protein became more stable.
The present findings suggest that 14-3-3 protein interactions contribute to this stability. Indeed, newly synthesized WT and ΔF508 CFTR disappeared more slowly in cells expressing 14-3-3β, consistent with protection from degradation and an increased efficiency of immature-to-mature CFTR conversion (Figure 5). The significant increases in WT and ΔF508 CFTR synthesis observed during pulse labeling suggest that 14-3-3 proteins affect CFTR biogenesis at an early stage, likely during translation and/or domain folding and assembly.
Non–phosphorylation-dependent interactions with 14-3-3 proteins
Protein interactions and increased CFTR levels with 14-3-3 coexpression were observed also in the absence of cAMP/PKA stimulation, as evidenced by 1) increases in steady-state levels of CFTR (Figure 4A), 2) pulse-chase studies, demonstrating enhanced CFTR biogenesis (at steps discussed above), 3) 14-3-3 interactions with full-length CFTR in vivo and with its R region and N-terminus in vitro, and 4) NMR-based identification of 14-3-3β association with sites in the nonphosphorylated R region. In addition, 14-3-3 knockdown reduced steady-state CFTR expression and regulated anion efflux, implying an interaction between these proteins in the basal state.
14-3-3β and the CFTR N-terminus interacted in vitro. Previous studies implicate the N-terminus as a region important for normal, ER-based CFTR processing, and they suggest that its interactions with the R region facilitate CFTR biogenesis and function (Skach, 2000; Fu et al., 2001). The N-terminal truncations of CFTR were poorly processed, and several N-terminus disease mutations are processing mutants. This region may contribute to non–phosphorylation-dependent 14-3-3 protein interactions that affect CFTR expression and stability.
Riordan, Amaral, and coworkers (Hegedus et al., 2006; Roxo-Rosa et al., 2006) described four arginine-framed tripeptide (AFT) motifs (RXR) that contributed to the ER retention of CFTR. Two of these lie in NBD1, one in the R region, and one at the N-terminus. Concerted mutations of all four AFT motifs in ΔF508 CFTR led to its export from the ER, suggesting that these sites play a role in CFTR retention and degradation. These retention motifs were discovered originally in another ABC protein-related ion channel, the Kir6.2/SUR (sulfonylurea receptor) complex, which comprises the ATP-sensitive K channel of pancreatic beta cells. In this system, 14-3-3 proteins interact with RXR motifs without a requirement for cargo protein phosphorylation (Zerangue et al., 1999; Heusser et al., 2006).
In addition to the RXR site at the N-terminus, the AFT motif in the R region overlaps with the phosphorylation site at S768, which showed 14-3-3β–dependent broadening of the NMR spectrum in both the presence and absence of cAMP/PKA-induced phosphorylation. S768 has been characterized as a phosphorylation site that is inhibitory to CFTR activity (Wilkinson et al., 1997; Vais et al., 2004; Csanady et al., 2005), and it has been linked to inhibition of CFTR by AMP-activated protein kinase (King et al., 2009). The role of this site in CFTR progression and whether 14-3-3 antagonizes the inhibitory action of AMPK require additional study.
Mechanisms for 14-3-3–dependent regulation of CFTR biogenesis
Many misfolded or incompletely assembled multidomain proteins are retained in the ER via interactions with COPI components (Smith et al., 2011). The K channel literature indicates that strategies to avoid retrograde transport and degradation elicited by interactions with COPI include 1) the burying of arginine-framed ER retention motifs (RXR) by intramolecular shielding, due to protein folding or subunit assembly, and 2) the shielding of retention motifs due to 14-3-3 protein binding, with or without cargo protein phosphorylation (Bonifacino et al., 1990; Zerangue et al., 1999).
In addition to AFT motifs, retrograde trafficking to the ER may involve recognition of cytoplasmic dibasic or tribasic “retrieval” motifs (i.e., KKXX, K(X)KXX, or KRK) on cargo proteins by the COPI coat (Teasdale and Jackson, 1996; Andersson et al., 1999), and they also involve mutually exclusive COPI or 14-3-3 protein binding (Kuwana et al., 1998; Vivithanaporn et al., 2006). O'Kelly et al. (2002) identified 14-3-3 binding motifs in a number of proteins that are subject to ER retention via dibasic signals, suggesting that this is a general method for regulating protein exit from the ER. CFTR contains sites that could function as dibasic retrieval signals.
Our findings suggest that CFTR forward transport is regulated, at least in part, by competitive 14-3-3 protein and COPI subunit interactions. The mechanism involves CFTR phosphorylation, which leads to 14-3-3 protein binding at sites within the R region and competition with COPI coat protein binding to reduce CFTR retrieval to the ER. This presumably accounts for cAMP/PKA-mediated stimulation of CFTR biogenesis. An alternative mechanism by which 14-3-3 proteins may regulate CFTR expression involves phosphorylation-independent 14-3-3 binding, perhaps to CFTR's AFT motifs, and this would resemble the process of Kir6.2 forward transport.
These processes also influence the production of ΔF508 CFTR, and yet modulation of this pathway was not sufficient to produce mutant CFTR maturation. Despite the increase in throughput, downstream quality control elements ultimately prevented maturation of the mutant protein. Nevertheless, manipulation of these processes, perhaps via activation of the cAMP/PKA pathway, might increase the efficacy of small-molecule correctors designed to improve the transit of ΔF508 CFTR to the cell surface.
MATERIALS AND METHODS
Antibodies
Polyclonal antibodies specific for 14-3-3 isoforms were purchased from Santa Cruz Biotechnology (Santa Cruz, CA), as follows: β (A-15), γ (C-16), and ε (T-16); their isoform specificity was previously validated (Liang et al., 2006). The polyclonal pan–14-3-3 (K-19) and anti-CD4 (sc-7219) antibodies were also from Santa Cruz Biotechnology. Mouse monoclonal antibodies to CFTR were obtained from Millipore (M3A7; Billerica, MA) or (596 and 217 from Cystic Fibrosis Foundation Therapeutics, Bethesda, MD). Antibodies to β-COPI, β-actin, the HA epitope, and GFP were obtained from Sigma-Aldrich (St. Louis, MO). Secondary antibodies against mouse or rabbit were obtained from GE Healthcare (Piscataway, NJ).
Cell culture
Calu-3 cells were cultured as described (Sun et al., 2000). HEK293 cells were cultured in DMEM (Sigma-Aldrich) with 10% fetal bovine serum, 4 mm l-glutamine, and penicillin–streptomycin and passaged every 3–4 d. Human bronchial epithelia (HBE) cells were obtained as excess pathological tissue following transplantation and organ donation under a protocol approved by the University of Pittsburgh Institutional Review Board. Polarized epithelia were cultured on human placental collagen-coated Costar Transwell filters (3470; 0.33 cm2, 0.4-μm pore; Lowell, MA) in 2% Ultroser G medium as previously described (Myerburg et al., 2010) and harvested after culture at an air–liquid interface for 4–6 wk.
RNA expression
Total RNA was extracted from Calu-3 and HEK293 cells and reverse transcribed to single-stranded cDNA as described previously (Liang et al., 2006). Semiquantitative RT-PCR was performed to analyze gene expression using 20 pmol of specific primers for 14-3-3 isoforms or β-actin. Primer sequences and PCR conditions are provided in our previous work (Liang et al., 2006).
Transient transfections
HEK293 cells grown in 60-mm dishes were transiently transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) with the indicated pcDNA3 or pcDNA3.1 expression plasmids, 4 μg of cDNA/dish. For the transfections with 14-3-3 or control siRNAs, 100 pmol of siRNA was used for 5 × 105 cells in 2 ml of culture medium. After 24 h, the cells were rinsed with phosphate-buffered saline and either prepared for pulse-chase assays or lysed in RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, and 0.1% SDS) or Nonidet P-40 lysis buffer (0.09% NP-40, 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 10 mM NaMoO4). Samples were incubated for 2 h in the appropriate lysis buffer and centrifuged at 16,000 × g for 30 min at 4°C. Cell extracts were used for immunoblot analysis.
Plasmid constructs and protein expression
cDNAs encoding GST-CFTR R-domain, GST-N, and GST-C were described previously (Zhang et al., 2002). GST-CFTR-N random was constructed as GST-N, except for a one-nucleotide shift in the reading frame, which yields a different amino acid composition; its net charge and size are comparable to those of CFTR-N. Purification and dialysis of GST fusion proteins were performed as described (Sun et al., 2000).
Proteins for NMR were expressed in BL21(DE3) CodonPlus (RIL) cells (Stratagene, Santa Clara, CA). The human CFTR R region (654–838; F833L) was expressed from a pPROEX HTb vector (Invitrogen) with an N-terminal hexahistidine tag and purified as described previously (Baker et al., 2007). (Note that the F833L polymorphism yields an R region that is significantly more soluble.) Human 14-3-3β protein encoded on a pET–small ubiquitin-related modifier (SUMO) plasmid (Invitrogen) was purified from the soluble fraction using a buffer of 50 mM Tris-HCl, pH 7.5, 200 mM NaCl, 2% (wt/vol) arginine, and 10 mM mercapthoethan-2-ol. The purification involves Ni2+ affinity chromatography, followed by SUMO protease cleavage and a second Ni2+ affinity chromatography, with a final size exclusion chromatography on a Superdex 200 column (Pharmacia, GE Healthcare Bio-Sciences, Piscataway, NJ). R region phosphorylation was performed using 40 μM R region with 100 U/ml cAMP-dependent protein kinase catalytic subunit (New England BioLabs, Ipswich, MA) in a buffer of 50 mM Tris-HCl, pH 7.5, 20 mM MgCl2, 10 mM ATP, and 2 mM dithiothreitol for 2 h at 37°C, followed by HPLC purification on a C4 prep column (Phenomenex, Torrance, CA) to separate fully phosphorylated R region from partially phosphorylated protein. The phosphorylation state of the R region was verified by mass spectrometry.
Immunoblot analyses
Equal amounts of protein from either FSK-treated or nontreated HEK293 cells, or the immunoprecipitates described previously, were resolved by 10% SDS–PAGE and transferred to polyvinylidene fluoride membranes. Unbound sites were blocked for 1 h at room temperature with 5% (wt/vol) skim milk powder in TBST (10 mM Tris, pH 8.0, 150 mM NaCl, 0.05% Tween 20). The blots were incubated with primary antibodies (anti–14-3-3 isoform, each 1:2000; anti-CFTR 217, 596, each 1:5000; anti-GFP, 1:4000; anti-CD4, 1:2000; anti–β-COPI, 1:1000; or anti–β-actin, 1:3000) at room temperature for 2 h. The blots were then washed three times for 10 min each with TBST and incubated for 1 h with 2 μg/ml horseradish peroxidase–conjugated secondary antibodies (1:1000; Amersham-Pharmacia Biotech, GE Healthcare Bio-Sciences, Piscataway, NJ) in TBST with 5% milk, followed by three TBST washes. The reactive bands were visualized with enhanced chemiluminescence (PerkinElmer Life Sciences, Wellesley, MA) and exposed to x-ray film (Eastman Kodak, Rochester, NY). β-Actin expression provided an internal control.
Cell surface biotinylation
Plasma membrane protein biotinylation was performed on HEK293 cells transiently cotransfected with CFTR and 14-3-3β or 14-3-3β siRNA. Prior to Calu-3 cell biotinylation by the same procedure, the siRNA was introduced by electroporation (BTX ECM 830; Harvard Apparatus, Holliston, MA). All assay procedures were performed on ice using ice-cold solutions. Cells were washed with phosphate-buffered saline with agitation, to remove growth media. The membranes were biotinylated using 1.5 mg/ml S-S-biotin (Pierce Chemical Co., Rockford, IL) in borate buffer (85 mM NaCl, 4 mM KCl, 15 mM Na2B4O7, pH 9) for 30 min. Labeling was quenched by adding a double volume of fetal bovine serum–containing medium. The cells were then washed three times with PBS with agitation and harvested. Cells were lysed in 0.4% deoxycholic acid, 1% NP-40, 50 mM ethylene glycol tetraacetic acid, and 10 mM Tris-Cl, pH 7.4, for 15 min. The protein concentration of the postnuclear supernatant was determined, and 300 μg of protein was combined with a streptavidin bead slurry (Pierce Chemical Co.) and incubated overnight at 4°C. Samples from the streptavidin beads were carefully washed three times in lysis buffer, solubilized with Laemmli sample buffer, separated on 5% SDS–PAGE, and blotted for CFTR.
Pulldown assays and coimmunoprecipitation
A 10-μg amount of GST fusion protein was incubated with 20 μl of preequilibrated glutathione–Sepharose 4B beads in 200 μl of DIGNAM-D buffer containing 0.1% bovine serum albumin at 4°C for 1 h (Zhang et al., 2002). Calu-3 membrane fractions were added, and the incubation was continued for an additional 2 h at 4°C. After five washes with DIGNAM-D buffer, samples were resuspended in 30 μl of 2× SDS sample buffer, boiled for 2 min, resolved on 12% SDS–PAGE, and probed with anti–14-3-3β antibody (1:2000). Coimmunoprecipitation was performed as described (Sun et al., 2000). In brief, protein assays (bicinchoninic acid; Pierce Biotechnology, Rockford, IL) ensured that equivalent amounts of protein were used for Western blot analysis and immunoprecipitation. Precleared cell lysates (∼1 mg of protein) were mixed with the appropriate primary antibodies for 1.5 h at 4°C in lysis buffer (0.4% deoxycholate acid, 1% NP-40, 50 mM EDTA, 10 mM Tris–HCl at pH 7.4). Twenty-five microliters of washed protein A– or G–Sepharose beads was added to each sample and incubated 1 h at 4°C with gentle rotation. Immunocomplexes were washed with lysis buffer four times and precipitated by centrifugation at 12,000 × g for 10 s. The immunocomplexes were resuspended in SDS sample buffer and subjected to immunoblotting (see later discussion). Controls for the immunoprecipitations were performed using a concentration of HA antibody equal to that of the primary precipitating antibody. Ten percent of the protein extract used in the immunoprecipitation was loaded for subsequent immunoblot.
Pulse-chase and immunoprecipitation
Cells were starved for 30 min in methionine-free, α-modified Eagle's medium (α-MEM; Invitrogen) before being radiolabeled for 30 min in the same medium supplemented with 150 μCi/ml EasyTag Express 35S protein labeling mix (PerkinElmer). During the chase periods (0, 0.5, 1, 2, and 4 h), the labeling medium was replaced by α-MEM supplemented with nonradioactive methionine (1 mM; Sigma-Aldrich). Cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer (1 ml) containing deoxycholic acid (1% wt/vol; Sigma-Aldrich), Triton X-100 (1% vol/vol; GE Healthcare Bio-Sciences), SDS (0.1% wt/vol; Invitrogen), Tris (50 mM, pH 7.4; Sigma-Aldrich), and NaCl (150 mM). CFTR protein was immunoprecipitated as described (Farinha et al., 2004) after centrifugation of samples at 14,000 × g for 30 min at 4°C. To detect CFTR, the supernatant was incubated overnight with 1 μg of each of the anti-CFTR monoclonal antibodies 596 and 217 at 4°C with protein G–agarose beads (25 μg, Roche, Indianapolis, IN). Beads were washed four times using RIPA buffer (1 ml) and protein eluted for 1 h at room temperature with cracking buffer (40 μl) containing dithiothreitol (0.5 mM, Sigma-Aldrich), bromophenol blue (0.001% wt/vol), glycerol (5% vol/vol), SDS (1.5% wt/vol), and Tris (31.25 mM), pH 6.8. Samples were separated electrophoretically on 7% vol/vol polyacrylamide gels. Then, gels were prefixed (methanol [30% vol/vol] and acetic acid [10% vol/vol]) for 30 min, washed thoroughly in water, and soaked in salicylic acid (1 M) for 1 h. After drying at 65°C under vacuum for 1 h, gels were exposed to x-ray film (Kodak BioMax MR film). Fluorograms of gels were digitized, and integrated peak areas were determined using ImageQuant 7.0 software (GE Healthcare Bio-Sciences).
SPQ fluorescence assays
SPQ halide efflux assays were performed on HEK293 cells as described previously (Chao et al., 1989; Silvis et al., 2009). In brief, the iodide-sensitive fluorescent indicator SPQ (Molecular Probes, Eugene, OR) was introduced into HEK293 cells in a hypotonic solution of iodide buffer (in mM: 130 NaI, 4 KNO3, 1 Ca(NO3)2, 1 Mg(NO3)2, 10 glucose, and 20 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, diluted 1:1 with water) and containing a final concentration of 10 mM SPQ. Cells were loaded for 20 min at 37°C in a humidified chamber with 5% CO2. The SPQ-loaded cells were then mounted on a Diaphot 300 inverted microscope (Nikon, Melville, NY) with a 37°C heated stage and perfused with iodide buffer while regions of interest were for chosen for GFP and non-GFP–expressing cells using Simple PCI, version 5.1, software (Compix Imaging Systems, Cranberry, PA). The GFP signal intensity varied from cell to cell; therefore, for SPQ fluorescence data collection, three cell populations were chosen, based on GFP expression levels as estimated by visual inspection of the cells. Changes in CFTR-mediated SPQ fluorescence were monitored at 445 nm in response to excitation at 340 nm during perfusion at 37°C in nitrate buffer (NaI replaced with 130 mM NaNO3) for 3 min without forskolin and then for 8 min with 10 μM forskolin added. The slopes or single-exponential rate constants were calculated using SigmaPlot, version 7.1 (Systat Software, Chicago, IL), for each mean fluorescence trace generated from the >50 cells examined per population per coverslip.
Statistical analysis
Data were obtained from experiments performed two to four times, and values are presented as mean ± SD or SEM as indicated. The p values were calculated by analysis of variance, followed by unpaired t test, as appropriate. Pulse-chase data were analyzed by comparing degradation rates (slopes of regression lines) by Student's t test. On a regression modeling procedure, the slope follows a t distribution (Kleinbaum et al., 1988; Helliwell and Jackson, 1994). Therefore the slopes of two straight lines can be compared using a t distribution with n1 + n2 − 4 degrees of freedom, where n1 and n2 are the numbers of points used on the regression procedure in groups 1 and 2, respectively. Differences between groups of data were considered statistically significant when p < 0.05.
NMR measurements
All NMR data (heteronuclear single-quantum correlation and HNCO triple resonance experiments) were collected on a Varian Inova 800-MHz spectrometer at 10 or 50°C with a triple-resonance probe. Recorded data were processed using NMRPipe (Delaglio et al., 1995) and analyzed using Sparky (www.cgl.ucsf.edu/home/sparky). Experiments were carried out at 300 μM concentration in the absence and presence of the partner, from the R region perspective, in a buffer of 50 mM Tris-HCl, pH 7.50, 150 mM NaCl, 2 mM dithiothreitol, and 10% (vol/vol) D2O and, from the 14-3-3β perspective, in a buffer of 125 mM KH2PO4, pH 6.80, 125 mM KCl, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride, and 10% (vol/vol) D2O.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health Grants DK68196 and DK72506, Cystic Fibrosis Foundation Therapeutics Grant FRIZZE05XX0, and fellowship support from the Cystic Fibrosis Foundation (LIANG09F0). Additional support from the Cystic Fibrosis Foundation Therapeutics (FORMAN05XX0) and Cystic Fibrosis Canada (J.D.F.-K.) and a fellowship from the Canadian Institutes of Health Research Strategic Training Program in Protein Folding and Interaction Dynamics (Z.B.) are acknowledged. We thank Fei Sun (Wayne State University, Detroit, MI) for providing the GST fusion proteins used in this work.
Abbreviations used:
- CFTR
cystic fibrosis transmembrane conductance regulator
- ER
endoplasmic reticulum
- GST
glutathione-S-transferase
- GST-C
GST-CFTR C-terminus
- GST-N
GST-CFTR N-terminus
- HEK
human embryonic kidney
- NBD
nucleotide-binding domain
- R region
regulatory region
- RT-PCR
reverse transcriptase-PCR
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-08-0662) on January 25, 2012.
REFERENCES
- Aitken A, Baxter H, Dubois T, Clokie S, Mackie S, Mitchell K, Peden A, Zemlickova E. Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem Soc Trans. 2002;30:351–360. doi: 10.1042/bst0300351. [DOI] [PubMed] [Google Scholar]
- Andersson H, Kappeler F, Hauri HP. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J Biol Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- Baker JM, Hudson RP, Kanelis V, Choy WY, Thibodeau PH, Thomas PJ, Forman-Kay JD. CFTR regulatory region interacts with NBD1 predominantly via multiple transient helices. Nat Struct Mol Biol. 2007;14:738–745. doi: 10.1038/nsmb1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Cardasis HL, Sehnke PC, Laughner B, Eyler JR, Powell DH, Ferl RJ. FTICR-MS analysis of 14-3-3 isoform substrate selection. Biochim Biophys Acta. 2007;1774:866–873. doi: 10.1016/j.bbapap.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Chang XB, Tabcharani JA, Hou YX, Jensen TJ, Kartner N, Alon N, Hanrahan JW, Riordan JR. Protein kinase A (PKA) still activates CFTR chloride channel after mutagenesis of all 10 PKA consensus phosphorylation sites. J Biol Chem. 1993;268:11304–11311. [PubMed] [Google Scholar]
- Chao AC, Dix JA, Sellers MC, Verkman AS. Fluorescence measurement of chloride transport in monolayer cultured cells. Mechanisms of chloride transport in fibroblasts. Biophys J. 1989;56:1071–1081. doi: 10.1016/S0006-3495(89)82755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Csanady L, et al. Preferential phosphorylation of R-domain serine 768 dampens activation of CFTR channels by PKA. J Gen Physiol. 2005;125:171–186. doi: 10.1085/jgp.200409076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Farinha CM, et al. Biochemical methods to assess CFTR expression and membrane localization. J Cyst Fibros. 2004;3(Suppl 2):73–77. doi: 10.1016/j.jcf.2004.05.017. [DOI] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Fu J, Ji HL, Naren AP, Kirk KL. A cluster of negative charges at the amino terminal tail of CFTR regulates ATP-dependent channel gating. J Physiol. 2001;536:459–470. doi: 10.1111/j.1469-7793.2001.0459c.xd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadsby DC, Nairn AC. Control of CFTR channel gating by phosphorylation and nucleotide hydrolysis. Physiol Rev. 1999;79:S77–S107. doi: 10.1152/physrev.1999.79.1.S77. [DOI] [PubMed] [Google Scholar]
- Hegedus T, Aleksandrov A, Cui L, Gentzsch M, Chang XB, Riordan JR. F508del CFTR with two altered RXR motifs escapes from ER quality control but its channel activity is thermally sensitive. Biochim Biophys Acta 1758. 2006:565–572. doi: 10.1016/j.bbamem.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Helliwell PS, Jackson S. Relationship between weakness and muscle wasting in rheumatoid arthritis. Ann Rheum Dis. 1994;53:726–728. doi: 10.1136/ard.53.11.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heusser K, Yuan H, Neagoe I, Tarasov AI, Ashcroft FM, Schwappach B. Scavenging of 14-3-3 proteins reveals their involvement in the cell-surface transport of ATP-sensitive K+ channels. J Cell Sci. 2006;119:4353–4363. doi: 10.1242/jcs.03196. [DOI] [PubMed] [Google Scholar]
- Hutt DM, et al. Reduced histone deacetylase 7 activity restores function to misfolded CFTR in cystic fibrosis. Nat Chem Biol. 2010;6:25–33. doi: 10.1038/nchembio.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanclos EM, Lin L, Treuil MW, Rao J, DeCoster MA, Anand R. The chaperone protein 14-3-3eta interacts with the nicotinic acetylcholine receptor alpha 4 subunit. Evidence for a dynamic role in subunit stabilization. J Biol Chem. 2001;276:28281–28290. doi: 10.1074/jbc.M011549200. [DOI] [PubMed] [Google Scholar]
- Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427:69–78. doi: 10.1042/BJ20091834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King JD, Jr, Fitch AC, Lee JK, McCane JE, Mak DO, Foskett JK, Hallows KR. AMP-activated protein kinase phosphorylation of the R domain inhibits PKA stimulation of CFTR. Am J Physiol Cell Physiol. 2009;297:C94–C101. doi: 10.1152/ajpcell.00677.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjarland E, Keen TJ, Kleppe R. Does isoform diversity explain functional differences in the 14-3-3 protein family? Curr Pharm Biotechnol. 2006;7:217–223. doi: 10.2174/138920106777549777. [DOI] [PubMed] [Google Scholar]
- Kleinbaum DG, Kupper LL, Muller KE. Applied Regression Analysis and Other Multivariated Methods. Pacific Grove, CA: Duxbury Press; 1988. [Google Scholar]
- Kuwana T, Peterson PA, Karlsson L. Exit of major histocompatibility complex class II-invariant chain p35 complexes from the endoplasmic reticulum is modulated by phosphorylation. Proc Natl Acad Sci USA. 1998;95:1056–1061. doi: 10.1073/pnas.95.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, LeLeiko NS, Walsh MJ. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- Liang X, Butterworth MB, Peters KW, Frizzell RA. AS160 modulates aldosterone-stimulated epithelial sodium channel forward trafficking. Mol Biol Cell. 2010;21:2024–2033. doi: 10.1091/mbc.E10-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Butterworth MB, Peters KW, Walker WH, Frizzell RA. An obligatory heterodimer of 14-3-3beta and 14-3-3epsilon is required for aldosterone regulation of the epithelial sodium channel. J Biol Chem. 2008;283:27418–27425. doi: 10.1074/jbc.M803687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Peters KW, Butterworth MB, Frizzell RA. 14-3-3 isoforms are induced by aldosterone and participate in its regulation of epithelial sodium channels. J Biol Chem. 2006;281:16323–16332. doi: 10.1074/jbc.M601360200. [DOI] [PubMed] [Google Scholar]
- Matthews RP, McKnight GS. Characterization of the cAMP response element of the cystic fibrosis transmembrane conductance regulator gene promoter. J Biol Chem. 1996;271:31869–31877. doi: 10.1074/jbc.271.50.31869. [DOI] [PubMed] [Google Scholar]
- McDonald RA, Matthews RP, Idzerda RL, McKnight GS. Basal expression of the cystic fibrosis transmembrane conductance regulator gene is dependent on protein kinase A activity. Proc Natl Acad Sci USA. 1995;92:7560–7564. doi: 10.1073/pnas.92.16.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Kay LE, Forman-Kay JD. Protein dynamics and conformational disorder in molecular recognition. J Mol Recognit. 2010a;23:105–116. doi: 10.1002/jmr.961. [DOI] [PubMed] [Google Scholar]
- Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure. 2010b;18:494–506. doi: 10.1016/j.str.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci USA. 2008;105:17772–17777. doi: 10.1073/pnas.0809222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molzan M, et al. Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol Cell Biol. 2010;30:4698–4711. doi: 10.1128/MCB.01636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore BW, Perez VJ. Specific acidic proteins of the nervous system. In: Carlson FD, editor. In: Physiological and Biochemical Aspects of Nervous Integration. Englewood Cliffs, NJ: Prentice-Hall; 1967. pp. 343–359. [Google Scholar]
- Mrowiec T, Schwappach B. 14-3-3 proteins in membrane protein transport. Biol Chem. 2006;387:1227–1236. doi: 10.1515/BC.2006.152. [DOI] [PubMed] [Google Scholar]
- Myerburg MM, King JD, Jr, Oyster NM, Fitch AC, Magill A, Baty CJ, Watkins SC, Kolls JK, Pilewski JM, Hallows KR. AMPK agonists ameliorate sodium and fluid transport and inflammation in cystic fibrosis airway epithelial cells. Am J Respir Cell Mol Biol. 2010;42:676–684. doi: 10.1165/rcmb.2009-0147OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell AD, Leng Q, Dong K, MacGregor GG, Giebisch G, Hebert SC. Phosphorylation-regulated endoplasmic reticulum retention signal in the renal outer-medullary K+ channel (ROMK) Proc Natl Acad Sci USA. 2005;102:9954–9959. doi: 10.1073/pnas.0504332102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kelly I, Butler MH, Zilberberg N, Goldstein SA. Forward transport. 14-3-3 binding overcomes retention in endoplasmic reticulum by dibasic signals. Cell. 2002;111:577–588. doi: 10.1016/s0092-8674(02)01040-1. [DOI] [PubMed] [Google Scholar]
- Okiyoneda T, Barriere H, Bagdany M, Rabeh WM, Du K, Hohfeld J, Young JC, Lukacs GL. Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science. 2010;329:805–810. doi: 10.1126/science.1191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman N, Shue G, LeLeiko NS, Walsh MJ. Transcription of cystic fibrosis transmembrane conductance regulator requires a CCAAT-like element for both basal and cAMP-mediated regulation. J Biol Chem. 1995;270:28848–28857. doi: 10.1074/jbc.270.48.28848. [DOI] [PubMed] [Google Scholar]
- Rajan S, et al. Interaction with 14-3-3 proteins promotes functional expression of the potassium channels TASK-1 and TASK-3. J Physiol. 2002;545:13–26. doi: 10.1113/jphysiol.2002.027052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennolds J, et al. Cystic fibrosis transmembrane conductance regulator trafficking is mediated by the COPI coat in epithelial cells. J Biol Chem. 2008;283:833–839. doi: 10.1074/jbc.M706504200. [DOI] [PubMed] [Google Scholar]
- Roxo-Rosa M, Xu Z, Schmidt A, Neto M, Cai Z, Soares CM, Sheppard DN, Amaral MD. Revertant mutants G550E and 4RK rescue cystic fibrosis mutants in the first nucleotide-binding domain of CFTR by different mechanisms. Proc Natl Acad Sci USA. 2006;103:17891–17896. doi: 10.1073/pnas.0608312103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J Biol Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- Schumacher B, Mondry J, Thiel P, Weyand M, Ottmann C. Structure of the p53 C-terminus bound to 14-3-3: implications for stabilization of the p53 tetramer. FEBS Lett. 2010;584:1443–1448. doi: 10.1016/j.febslet.2010.02.065. [DOI] [PubMed] [Google Scholar]
- Silvis MR, Bertrand CA, Ameen N, Golin-Bisello F, Butterworth MB, Frizzell RA, Bradbury NA. Rab11b regulates the apical recycling of the cystic fibrosis transmembrane conductance regulator in polarized intestinal epithelial cells. Mol Biol Cell. 2009;20:2337–2350. doi: 10.1091/mbc.E08-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach WR. Defects in processing and trafficking of the cystic fibrosis transmembrane conductance regulator. Kidney Int. 2000;57:825–831. doi: 10.1046/j.1523-1755.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- Smith AJ, Daut J, Schwappach B. Membrane proteins as 14-3-3 clients in functional regulation and intracellular transport. Physiology (Bethesda) 2011;26:181–191. doi: 10.1152/physiol.00042.2010. [DOI] [PubMed] [Google Scholar]
- Sroubek J, McDonald TV. Protein kinase A activity at the endoplasmic reticulum surface is responsible for augmentation of human ether-a-go-go-related gene product (HERG) J Biol Chem. 2011;286:21927–21936. doi: 10.1074/jbc.M110.201699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland E, Qu BH, Millen L, Thomas PJ. The molecular chaperone Hsc70 assists the in vitro folding of the N-terminal nucleotide-binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1997;272:25421–25424. doi: 10.1074/jbc.272.41.25421. [DOI] [PubMed] [Google Scholar]
- Sun F, Hug MJ, Lewarchik CM, Yun CH, Bradbury NA, Frizzell RA. E3KARP mediates the association of ezrin and protein kinase A with the cystic fibrosis transmembrane conductance regulator in airway cells. J Biol Chem. 2000;275:29539–29546. doi: 10.1074/jbc.M004961200. [DOI] [PubMed] [Google Scholar]
- Teasdale RD, Jackson MR. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu Rev Cell Dev Biol. 1996;12:27–54. doi: 10.1146/annurev.cellbio.12.1.27. [DOI] [PubMed] [Google Scholar]
- Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Vais H, Zhang R, Reenstra WW. Dibasic phosphorylation sites in the R domain of CFTR have stimulatory and inhibitory effects on channel activation. Am J Physiol Cell Physiol. 2004;287:C737–C745. doi: 10.1152/ajpcell.00504.2003. [DOI] [PubMed] [Google Scholar]
- Vergani P, Lockless SW, Nairn AC, Gadsby DC. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 2005;433:876–880. doi: 10.1038/nature03313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivithanaporn P, Yan S, Swanson GT. Intracellular trafficking of KA2 kainate receptors mediated by interactions with coatomer protein complex I (COPI) and 14-3-3 chaperone systems. J Biol Chem. 2006;281:15475–15484. doi: 10.1074/jbc.M512098200. [DOI] [PubMed] [Google Scholar]
- Wilker E, Yaffe MB. 14-3-3 proteins—a focus on cancer and human disease. J Mol Cell Cardiol. 2004;37:633–642. doi: 10.1016/j.yjmcc.2004.04.015. [DOI] [PubMed] [Google Scholar]
- Wilker EW, Grant RA, Artim SC, Yaffe MB. A structural basis for 14-3-3sigma functional specificity. J Biol Chem. 2005;280:18891–18898. doi: 10.1074/jbc.M500982200. [DOI] [PubMed] [Google Scholar]
- Wilkinson DJ, Strong TV, Mansoura MK, Wood DL, Smith SS, Collins FS, Dawson DC. CFTR activation: additive effects of stimulatory and inhibitory phosphorylation sites in the R domain. Am J Physiol. 1997;273:L127–L133. doi: 10.1152/ajplung.1997.273.1.L127. [DOI] [PubMed] [Google Scholar]
- Yang X, Lee WH, Sobott F, Papagrigoriou E, Robinson CV, Grossmann JG, Sundstrom M, Doyle DA, Elkins JM. Structural basis for protein-protein interactions in the 14-3-3 protein family. Proc Natl Acad Sci USA. 2006;103:17237–17242. doi: 10.1073/pnas.0605779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Michelsen K, Schwappach B. 14-3-3 dimers probe the assembly status of multimeric membrane proteins. Curr Biol. 2003;13:638–646. doi: 10.1016/s0960-9822(03)00208-2. [DOI] [PubMed] [Google Scholar]
- Zerangue N, Schwappach B, Jan YN, Jan LY. A new ER trafficking signal regulates the subunit stoichiometry of plasma membrane K(ATP) channels. Neuron. 1999;22:537–548. doi: 10.1016/s0896-6273(00)80708-4. [DOI] [PubMed] [Google Scholar]
- Zhang H, Peters KW, Sun F, Marino CR, Lang J, Burgoyne RD, Frizzell RA. Cysteine string protein interacts with and modulates the maturation of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2002;277:28948–28958. doi: 10.1074/jbc.M111706200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.