Abstract
Background
Cryptococcus neoformans is a pathogenic yeast that causes cryptococcosis, a life threatening disease. The prevalence of cryptococcosis in Asia has been rising after the onset of the AIDS epidemic and estimates indicate more than 120 cases per 1,000 HIV-infected individuals per year. Almost all cryptococcal disease cases in both immunocompromised and immunocompetent patients in Asia are caused by C. neoformans var. grubii. Epidemiological studies on C. neoformans in pan-Asia have not been reported. The present work studies the genetic diversity of the fungus by microsatellite typing and susceptibility analysis of approximately 500 isolates from seven Asian countries.
Methodology/Principal Findings
Genetic diversity of Asian isolates of C. neoformans was determined using microsatellite analysis with nine microsatellite markers. The analysis revealed eight microsatellite complexes (MCs) which showed different distributions among geographically defined populations. A correlation between MCs and HIV-status was observed. Microsatellite complex 2 was mainly associated with isolates from HIV-negative patients, whereas MC8 was associated with those from HIV-positive patients. Most isolates were susceptible to amphotericin B, itraconazole, voriconazole, posaconazole, and isavuconazole, but 17 (3.4%) and 10 (2%) were found to be resistant to 5-flucytosine and fluconazole, respectively. Importantly, five Indonesian isolates (approximately 12.5% from all Indonesian isolates investigated and 1% from the total studied isolates) were resistant to both antifungals. The majority of 5-flucytosine resistant isolates belonged to MC17.
Conclusions
The findings showed a different distribution of genotypes of C. neoformans var. grubii isolates from various countries in Asia, as well as a correlation of the microsatellite genotypes with the original source of the strains and resistance to 5-flucytosine.
Introduction
Cryptococcus neoformans and C. gattii are encapsulated basidiomycetous yeasts that can cause life-threatening infections in humans. According to the current classification, C. neoformans consists of two varieties, namely variety grubii (serotype A) and variety neoformans (serotype D). C. neoformans and C. gattii differ in ecology, biochemistry, molecular characteristics, and their ability to cause disease [1], [2]. C. neoformans is known as an opportunistic pathogen because it mainly infects immunocompromised patients, whereas C. gattii is considered a primary pathogen that infects otherwise healthy individuals [3]. Clinical characteristics of C. neoformans and C. gattii differ as well. The latter species causes more frequently cryptococcomas and has a lower susceptibility to antifungal agents, resulting in prolonged treatment and a higher mortality rate when compared to C. neoformans [4].
Cryptococcus neoformans continues to be the most important cause of fungal meningitis in immunocompromised patients. The global burden of cryptococcal infections among HIV-infected patients is estimated at nearly one million new cases per year [5]. In South Asia and Southeast Asia, the incidence of HIV infection is the second-highest with over 4.5 million HIV-infected patients. The prevalence of cryptococcal meningitis was estimated to be 13.6 and 120 per thousand HIV-infected individuals per year among HIV-infected patients in these two regions, respectively [5]. In contrast, most cases of cryptococcosis in East Asia, especially in China and Japan, where HIV prevalence is low, have been reported from apparently immuncompetent patients and were mostly caused by C. neoformans var. grubii [6]–[9]. Similarly, Vietnamese patients with cryptococcal meningitis were usually infected by this variety, but here it manifests in both immunocompromised and immunocompetent patients [10].
Most cryptococcal meningitis cases in Asia are caused by C. neoformans [7]. Notwithstanding the clinical importance of the fungus in this part of the world, a systematic survey on the genetic diversity of the pathogen has not been performed. Genotyping of isolates may reveal differences in host range and clinical symptoms. Recently, a genotyping study of Vietnamese clinical isolates using amplified fragment length polymorphism (AFLP) revealed two genotypes, VNIγ and VNIδ with the former as the major genotype for isolates originating from non-HIV-infected patients [10]. For epidemiological studies of the C. neoformans/C. gattii complex, several molecular methods have been used, such as AFLP, M13-based PCR fingerprinting, Multi-locus Sequence Typing (MLST) and analysis of the intergenic spacer (IGS) ribosomal DNA sequences [11]–[15].
Microsatellite analysis is a genotyping technique that is becoming increasingly popular for molecular typing of medically important fungi [16]–[18]. Microsatellites, also referred to as short tandem repeats (STRs), are genomic sequences that consist of tandem repeated short motifs [19]. Mutations in microsatellites lead to a change in the number of repeats creating genotypic diversity. In some fungi, e.g. Aspergillus fumigatus and A. flavus, molecular typing using microsatellites proved to be more discriminatory than MLST [18], [19]. In our study, microsatellite typing was applied to estimate the extent of genetic diversity and the epidemiological relationships of a collection of clinical cryptococcal isolates that originated from East, South and Southeast Asia, and the Middle East. Moreover, a set of environmental isolates from Japan and Thailand was included. In vitro antifungal susceptibility was determined for amphotericin B, flucytosine, fluconazole, itraconazole, voriconazole, posaconazole and isavuconazole. The aims of this study were: (i) to analyze the genotypic diversity as well as the distribution of C. neoformans var. grubii from different geographical regions in Asia, (ii) to relate the genetic background of the cryptococcal isolates to disease status and origin from the human body, (iii) to test the in vitro antifungal susceptiblity of the isolates against seven antifungal drugs, and (iv) to determine if differences in susceptibility correlate with the observed genotypic diversity.
Materials and Methods
Isolates and media
A total of 426 clinical isolates of Cryptococcus neoformans var. grubii isolates were obtained from the collections of the Chinese Cryptococcus Reference Centre at the Second Military Medical University, Shanghai, China (n = 115); Department of Microbiology, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand (n = 79); Department of Medical Microbiology, Postgraduate Institute of Medical Education and Research, Chandigarh, India (n = 61); Sappasitthiprasong Hospital, Ubon Ratchathani, Thailand (n = 59); Department of Parasitology, Faculty of Medicine, University of Indonesia, Jakarta, Indonesia (n = 40); Department of Microbiology, Meiji Pharmaceutical University, Tokyo, Japan (n = 28); Department of Microbiology, Faculty of Medicine, Khon Kaen University, Khon Kaen, Thailand (n = 20); Department of Microbiology, Faculty of Medicine, Health Sciences Centre, Kuwait University, Jabriya, Kuwait (n = 10); Mycology Unit, Microbiology Division, Department of Laboratory Medicine and Pathology, Hamad Medical Corporation, Doha, Qatar (n = 5) and Department of Pathology, Faculty of Medicine, Prince of Songkla University, Hat Yai , Thailand (n = 9) (Table S1). Furthermore, 67 environmental isolates were obtained from the Department of Microbiology, Faculty of Medicine, Chiang Mai University, Thailand (n = 58) and the Department of Microbiology, Meiji Pharmaceutical University, Tokyo, Japan (n = 9) (Table S2). Two-hundred and thirty-six isolates originated from HIV-infected patients and 156 isolates were obtained from HIV-negative patients. Thirty-four out of 426 clinical isolates were from patients with unknown HIV status.
Species identification was initially performed by standard mycological methods [20]. l-Canavanine-glycine-bromothymol blue (CGB) medium was used to distinguish between C. neoformans and C. gattii isolates [21]. Cryptococcus isolates were stored in sterile 2 ml screw-capped tubes containing porous beads (Microbank, ProLab Diagnostics, Richmond Hill, ON, Canada) at −80°C until further use.
Mating- and serotype analysis by PCR, and microsatellite typing
Genomic DNA extraction was performed as previously described [22]. All PCR amplifications for mating- and serotyping were carried out in a total volume of 20 µl containing 0.1 mM dNTPs, 0.5 U of Taq DNA polymerase (Gentaur, Brussels, Belgium), 1 µl of template genomic DNA (100 ng/µl) and 0.5 µM of the forward and reverse primers, as described by Bovers et al. [23] in 1× PCR reaction buffer containing 50 mM MgCl2 [24].
Microsatellite analysis was performed using nine microsatellite markers [17]. However, instead of three multiplex PCRs, each locus was amplified in a separate PCR reaction and reaction products containing different fluorescent labels were pooled prior to analysis. One microliter of combined reaction product was mixed with 8.75 µl water and 0.25 µl ET-550R ROX Size Standard (GE Healthcare, Diegem, Belgium). Samples were analyzed on a MegaBACE 500 automated DNA analysis platform (GE Healthcare) equipped with a 48 capillary array according to the instructions of the manufacturer. Repeat numbers were assigned using Fragment Profiler v1.2 (GE Healthcare), imported into BioNumerics v6.0 software (Applied Maths, Sint Martens-Latem, Belgium) and analyzed using the multistate categorical similarity coefficient. According to previously described criteria, microsatellite complexes (MCs) were defined as groups of two or more microsatellite genotypes that differ by a maximum of two loci [17].
Antifungal susceptibility testing
In vitro antifungal susceptibility testing of amphotericin B (AMB; Bristol Myers Squibb, Woerden, The Netherlands), flucytosine (5FC; Valeant Pharmaceuticals, Zoetermeer, The Netherlands), fluconazole (FLU; Pfizer Central Research, Sandwich, Kent, United Kingdom), itraconazole (ITR; Janssen Cilag, Tilburg, The Netherlands), posaconazole (POS; Schering-Plough Corp., Kenilworth, NJ, USA), voriconazole (VOR; Pfizer Central Research) and isavuconazole (ISA; Basilea Pharmaceutica, Basel, Switzerland) was performed using the standard broth microdilution method as described in CLSI document M27-A3 [25]. The minimal inhibitory concentrations (MIC) were read optically after 72 h of incubation at 35°C. For AMB, the MIC was defined as the lowest concentration of drug showing no yeast growth. For the other antifungal compounds, the MIC was defined as the lowest concentration that caused a prominent reduction of yeast growth (≥50%). Candida krusei ATCC6258 and Candida parapsilosis ATCC22019 were used as quality controls.
The resistance breakpoints of FLU and 5FC were taken from CLSI document M27-A3 [25] and Perfect et al. [26] as follows: ≥16 µg/ml for FLU; ≥32 µg/ml for 5FC. According to Nguyen and Yu [27] the resistance breakpoint of AMB is ≥2 µg/ml. Interpretive criteria of ITR and the new azoles have not been proposed yet for C. neoformans. However, a MIC≤1 µg/ml was suggested as the susceptibility breakpoint for ITR, VOR, POS and ISA [28]–[31].
Data Analysis
The Simpson's index of diversity was calculated to assess the genotypic diversity of microsatellite genotypes among the different populations [32]. The value obtained is scaled from zero to one, where a value of one indicates that all isolates are different, and a value of zero means that all belong to the same genotype.
Chi-square and Fisher exact tests were applied to examine the correlation between MCs and isolate categories, including HIV status, sample sources (environmental or clinical), and geographical origin. In vitro antifungal susceptibility testing results were statistically analyzed using the t-test method. P values less than 0.05 were considered significant for all statistical methods. The statistics were analyzed using StatsDirect v2.7.8 (StatsDirect, Cheshire, United Kingdom).
Results
Mating-type and serotype analysis
Of the 426 clinical C. neoformans var. grubii isolates obtained from HIV-positive patients (n = 236), HIV-negative patients (n = 156), and those with an unknown HIV status (n = 34), 425 isolates (99.8%) belonged to mating-type α and serotype A (αA), and one isolate (0.2%) from a HIV-negative patient in China belonged to mating-type α and serotype AD (αAαD). All 67 environmental isolates of C. neoformans var. grubii obtained from avian droppings from Chiang Mai, Thailand (n = 58) and Tokyo, Japan (n = 9) were mating type α and serotype A (Table S1 and Table S2).
Microsatellite genotyping
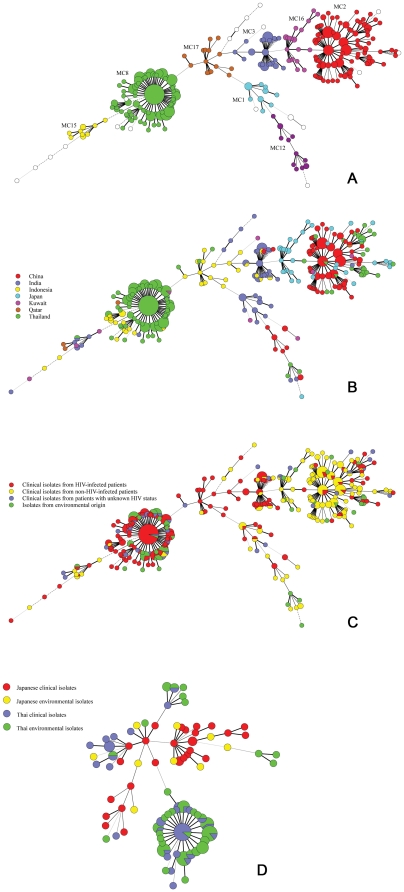
The genetic diversity of 492 isolates of C. neoformans var. grubii (αA) and one αAαD hybrid isolate was analyzed by microsatellite typing. Within this collection of 493 isolates, 265 different genotypes were found. These 265 genotypes were distributed over eight microsatellite complexes (MCs), which were defined as described previously [17]. Of the eight observed MCs, five were previously found in a collection of cryptococcal isolates from Cuba (i.e., MC1, MC2, MC3, MC8 and MC12), while three novel MCs were observed in the Asian collection presented here. These were assigned MC15, MC16, and MC17 (Figure 1A, Table S1 and Table S2). Twenty-four of the 493 isolates, which belonged to 19 microsatellite genotypes, could not be assigned to a MC. MC8 was the predominant MC among Asian isolates (n = 206, 41.8%), followed by MC2 (n = 151, 30.6%), MC3 (n = 42, 8.5%), MC16 (n = 21, 4.3%), MC17 (n = 15, 3.1%), MC1 (n = 14, 2.8%), MC12 (n = 13, 2.6%) and MC15 (n = 11, 2.2%). The distribution of MCs was found to differ in each country (Figures 1A and B, and Table 1). MC2 was predominant among the Chinese, Japanese, and Thai cryptococcal populations, whereas MC8 was the major type among isolates of patients from Southeast Asian countries (i.e., Indonesia and Thailand). In India, the majority of isolates belonged to MC1 and MC3. In addition, MC16 and MC17 were the predominant MCs containing Japanese and Indonesian isolates, respectively. The 17 isolates studied from the Middle East (Kuwait and Qatar) occur more scattered and belonged to MC2, MC3, MC8, and MC15 (Figures 1A and B, and Table 1). The relationship between most MCs is not well resolved. However, MC2 and MC16 that contained isolates from the Chinese, Thai, and Japanese populations are well interconnected.
Figure 1. Genotypic variation of C. neoformans isolates from different Asian countries by microsatellite typing.
(A) Minimum spanning tree based on a multistate categorical analysis representing 429 C. neoformans var. grubii isolates from different countries. Each circle represents a unique genotype. The size of the circle corresponds to the number of isolates within that genotype. Numbers and connecting lines correspond to the number of different markers between genotypes. Genotypes with identical colors are part of a microsatellite complex (MC). Circles without color are unique genotypes that are not part of a MC.; (B) Same as A, but now showing the genotypes from different geographic locations. Different colors correspond to different countries.; (C) Same as A and B, but now showing the genotypes from clinical and environmental sources.; (D) Same as A, B and C, but now showing the genotypes of Thai and Japanese population from clinical and environmental sources.
Table 1. Distribution of microsatellite complexes (MCs) between different countries.
| MCs | Chinan (%) | Japann (%) | Indian (%) | Indonesian (%) | Thailandn (%) | Kuwaitn (%) | Qatarn (%) | Total number of isolatesn (%) |
| MC1 | 0 | 0 | 14 (23) | 0 | 0 | 0 | 0 | 14 (2.8) |
| MC2 | 102 (88.7) | 14 (37.8) | 1 (1.6) | 0 | 30 (13.3) | 2 (20) | 2 (40) | 151 (30.6) |
| MC3 | 2 (1.7) | 0 | 30 (49.2) | 9 (22.5) | 0 | 1 (10) | 0 | 42 (8.5) |
| MC8 | 0 | 0 | 3 (4.9) | 17 (42.5) | 183 (81.3) | 2 (20) | 1 (20) | 206 (41.8) |
| MC12 | 8 (7) | 0 | 2 (3.3) | 0 | 3 (1.3) | 0 | 0 | 13 (2.6) |
| MC15 | 0 | 0 | 6 (9.8) | 0 | 1 (0.5) | 2 (20) | 2 (40) | 11 (2.2) |
| MC16 | 0 | 20 (54.1) | 0 | 0 | 1 (0.5) | 0 | 0 | 21 (4.3) |
| MC17 | 0 | 0 | 0 | 12 (30) | 3 (1.3) | 0 | 0 | 15 (3.1) |
| None MCs | 3 (2.6) | 3 (8.1) | 5 (8.2) | 2 (5) | 4 (1.8) | 3 (30) | 0 | 20 (4.1) |
| Total | 115 | 37 | 61 | 40 | 225 | 10 | 5 | 493 |
The predominant MCs in each country are indicated in bold.
The genotypic diversity of C. neoformans var. grubii was found to be most diverse (D = 1.000) in the Kuwait and Qatar populations, followed by the Japanese (D = 0.998), Indonesian (D = 0.994), Indian (D = 0.983), Chinese (D = 0.975), and Thai populations (D = 0.968) (Table 2). The genetic diversity within MCs showed values of Simpson's index of diversity close to 1, thus indicating that each MC contained a high level of genetic diversity. A significant correlation between MCs and HIV-status was observed by the Chi-square (p<0.0001) and Fisher exact tests (p<0.0001) (Figures 1A and C, and Table 3). The majority of isolates from HIV-negative patients belonged to MC2 that accounted for 104 out of 156 isolates from HIV-negative patients (66.6%), followed by MC16 (n = 13, 8.3%). In contrast, MC8 was the predominant MC containing isolates from HIV-positive patients (138 of 236, 58.5%), followed by MC2 (n = 30, 12.7%), MC3 (n = 33, 14%) and MC17 (n = 13, 5.5%).
Table 2. Simpson's index of diversity (D) in geographically different populations and for different microsatellite complexes (MCs).
| Country | DCountry | MCs | DMCs |
| China | 0.975 | MC1 | 0.912 |
| Japan | 0.998 | MC2 | 0.983 |
| India | 0.983 | MC3 | 0.945 |
| Indonesia | 0.994 | MC8 | 0.960 |
| Thailand | 0.968 | MC12 | 0.974 |
| Kuwait | 1.000 | MC15 | 0.964 |
| Qatar | 1.000 | MC16 | 0.990 |
| MC17 | 1.000 |
Table 3. Distribution of microsatellite complexes (MCs) from patients according to HIV status.
| MCs | HIV positiven (%) | HIV negativen (%) | Unknown statusn (%) | Total number of isolatesn (%) |
| MC1 | 6 (2.5) | 6 (3.9) | 2 (5.9) | 14 (3.3) |
| MC2 | 30 (12.7) | 104 (66.6) | 6 (17.7) | 140 (32.9) |
| MC3 | 33 (14) | 8 (5.1) | 1 (2.9) | 42 (9.9) |
| MC8 | 138 (58.5) | 4 (2.6) | 19 (55.9) | 161 (37.8) |
| MC12 | 2 (0.9) | 8 (5.1) | 0 | 10 (2.3) |
| MC15 | 5 (2.1) | 6 (3.9) | 0 | 11 (2.6) |
| MC16 | 1 (0.4) | 13 (8.3) | 3 (8.8) | 17 (4) |
| MC17 | 12 (5.1) | 1 (0.6) | 1 (2.9) | 14 (3.3) |
| None MCs | 9 (3.8) | 6 (3.9) | 2 (5.9) | 17 (4) |
| Total | 236 | 156 | 34 | 426 |
The predominant MC in each HIV status category is indicated in bold.
The distribution of environmental and clinical isolates among MCs was studied using two populations from Tokyo, Japan and Chiang Mai, Thailand. In both cases, the clinical and environmental isolates belonged to the same MCs (Figure 1D and Table 4). MC2 and MC8 were the common MCs in the Thai population. MC8 contained 26 out of 40 clinical isolates and 45 out of 58 environmental isolates, whereas MC2 contained 12 out of 40 clinical isolates and 8 out of 58 environmental isolates. Among the Japanese isolates, MC16 contained 16 out of 28 clinical isolates and 4 out of 9 environmental isolates, and MC2 contained 11 out of 28 clinical isolates and 3 out of 9 environmental isolates. The combined results of the Thai and Japanese populations showed a correlation between MCs and the clinical or environmental origin (p = 0.0004, Chi-square; p = 0.0001, Fisher exact test). However, no such correlation was observed within the Thai population from Chiang Mai (p = 0.0963, Chi-square; p = 0.0729, Fisher exact test) nor within the Japanese population from Tokyo (p = 0.9192, Chi-square; p>0.9999, Fisher exact test).
Table 4. Distribution of C. neoformans isolates from clinical and environmental samples from Thailand and Japan in microsatellite complexes (MCs).
| MCs | Chiang Mai, Thailand | Tokyo, Japan | |||
| Clin. Isolatesn (%) | Env. Isolatesn (%) | Clin. Isolatesn (%) | Env. Isolatesn (%) | Total number of isolatesn (%) | |
| MC1 | 0 | 0 | 0 | 0 | 0 |
| MC2 | 12 (30) | 8 (13.8) | 11 (39.3) | 3 (33.3) | 34 (25.2) |
| MC3 | 0 | 0 | 0 | 0 | 0 |
| MC8 | 26 (65) | 45 (77.6) | 0 | 0 | 71 (52.6) |
| MC12 | 0 | 3 (5.2) | 0 | 0 | 3 (2.2) |
| MC15 | 0 | 1 (1.7) | 0 | 0 | 1 (0.7) |
| MC16 | 0 | 0 | 16 (57.1) | 4 (44.4) | 20 (14.8) |
| MC17 | 0 | 0 | 0 | 0 | 0 |
| None MCs | 2 (5) | 1 (1.7) | 1 (3.6) | 2 (22.2) | 6 (4.5) |
| Total | 40 | 58 | 28 | 9 | 135 |
The predominant MCs in each sample type in these countries are indicated in bold.
In vitro antifungal susceptibility testing
The MIC values of all C. neoformans var. grubii isolates for the seven antifungal compounds tested are listed in Table 5. Almost all cryptococcal isolates were susceptible to AMB, ITR, FLU, VOR, POS and ISA. Notably, 18 clinical isolates (3.7%) from Indonesia (n = 13), Thailand (n = 4) and China (n = 1) were resistant to 5FC with MIC ≥32 µg/ml (Table S3). Most of 5FC resistant isolates occurred in MC3 (n = 4), MC8 (n = 8) and MC17 (n = 5), and one belonged to MC2 (Table S4). Approximately 33.3% of all MC17 isolates were resistant to 5FC and this was found to be highly significant (p<0.0001, Chi-square; p = 0.0005, Fisher exact test) when compared to the presence of 3.9 and 9.5% of resistant isolates in MC8 and MC3, respectively. Ten FLU-resistant isolates (2%) occurred in different countries, including China (n = 2), India (n = 1), Indonesia (n = 5), and Thailand (n = 2) and belonged to MC2 (n = 3), MC3 (n = 2), MC8 (n = 2), MC17 (n = 2) and one isolate from India (number 25_17) that could not be assigned to any MC (n = 1) (Table S4). Five isolates from Indonesia (strain numbers 676, 1206, 1571, 3400, and 1048) that belonged to MC3 (n = 2), MC8 (n = 1) and MC17 (n = 2) were resistant to both 5FC and FLU. Isolates obtained from HIV-negative patients and HIV-positive patients did not differ in the susceptibility to three antifungal compounds, including 5FC (p = 0.0298, t-test), FLU (p = 0.0108, t-test) and VOR (p<0.0001, t-test). No significant differences were observed for ITR (p = 0.0002, t-test), POS (p<0.0001, t-test), and ISA (p<0.0001, t-test) when clinical and environmental isolates were compared. When all isolates were considered, the broadest ranges and highest MIC values were those of 5FC (<0.063 to >64 µg/ml), followed by FLU (0.125 to 32 µg/ml). The lowest MIC range was observed for ISA (<0.016 to 0.125 µg/ml), followed by POS, VOR and ITR (<0.016 to 0.5 µg/ml) (Table 5). Among the azoles, FLU showed the lowest activity (MIC90 = 4 µg/ml) and ISA the highest (MIC90 = 0.063 µg/ml) (Table 5).
Table 5. The MIC range, MIC50, MIC90, and geometric mean for all 493 C. neoformans isolates for seven antifungals according to clinical and environmental origin of isolates.
| Isolates | Antifungal agent | MIC | |||
| Range | MIC50 | Geometric Mean | MIC90 | ||
| All C. neoformans isolates (n = 493) | Amphotericine B | 0.063–1 | 0.25 | 0.251 | 0.5 |
| 5-Flucytosine | <0.063–>64 | 4 | 3.483 | 8 | |
| Fluconazole | 0.125–32 | 2 | 2.294 | 4 | |
| Itraconazole | <0.016–0.5 | 0.063 | 0.063 | 0.25 | |
| Voriconazole | <0.016–0.5 | 0.063 | 0.049 | 0.125 | |
| Posaconazole | <0.016–0.5 | 0.063 | 0.061 | 0.125 | |
| Isavuconazole | <0.016–0.125 | 0.031 | 0.027 | 0.063 | |
| Isolates from HIV-positive patients (n = 236) | Amphotericine B | 0.063–1 | 0.25 | 0.236 | 0.5 |
| 5-Flucytosine | <0.063–>64 | 4 | 3.816 | 8 | |
| Fluconazole | 0.125–32 | 2 | 2.532 | 4 | |
| Itraconazole | <0.016–0.5 | 0.063 | 0.062 | 0.25 | |
| Voriconazole | <0.016–0.5 | 0.063 | 0.055 | 0.125 | |
| Posaconazole | <0.016–0.5 | 0.063 | 0.062 | 0.125 | |
| Isavuconazole | <0.016–0.125 | 0.031 | 0.025 | 0.063 | |
| Isolates from HIV-negative patients (n = 156) | Amphotericine B | 0.063–1 | 0.25 | 0.253 | 0.5 |
| 5-Flucytosine | 0.25–32 | 4 | 3.091 | 8 | |
| Fluconazole | 0.125–16 | 2 | 2.072 | 4 | |
| Itraconazole | <0.016–0.25 | 0.063 | 0.057 | 0.125 | |
| Voriconazole | <0.016–0.25 | 0.063 | 0.036 | 0.125 | |
| Posaconazole | <0.016–0.25 | 0.063 | 0.054 | 0.125 | |
| Isavuconazole | <0.016–0.125 | 0.031 | 0.024 | 0.063 | |
| Clinical isolates from Thailand and Japan (n = 68) | Amphotericine B | 0.063–0.5 | 0.25 | 0.291 | 0.5 |
| 5-Flucytosine | 0.5–8 | 4 | 3.433 | 8 | |
| Fluconazole | 0.25–32 | 2 | 2.452 | 4 | |
| Itraconazole | <0.016–0.25 | 0.063 | 0.058 | 0.125 | |
| Voriconazole | <0.016–0.5 | 0.063 | 0.070 | 0.125 | |
| Posaconazole | <0.016–0.125 | 0.063 | 0.057 | 0.125 | |
| Isavuconazole | <0.016–0.125 | 0.031 | 0.026 | 0.063 | |
| Environmental isolates from Thailand and Japan | Amphotericine B | 0.125–0.5 | 0.25 | 0.286 | 0.5 |
| (n = 67) | 5-Flucytosine | 0.063–8 | 4 | 2.933 | 4 |
| Fluconazole | 0.125–8 | 2 | 2.106 | 4 | |
| Itraconazole | <0.016–0.5 | 0.125 | 0.095 | 0.25 | |
| Voriconazole | <0.016–0.125 | 0.063 | 0.055 | 0.125 | |
| Posaconazole | <0.016–0.25 | 0.125 | 0.090 | 0.125 | |
| Isavuconazole | <0.016–0.063 | 0.063 | 0.039 | 0.063 | |
Discussion
Microsatellites are an excellent tool to discriminate among C. neoformans var. grubii isolates [17], [33]. Indeed, 265 different genotypes were observed in this Asian collection of 493 isolates. Most isolates belonged to only eight microsatellite complexes (MCs), including four new ones. Simpson's index of diversity, however, showed that genetic diversity within each individual population and each MCs was very high.
The genotypic structure of the C. neoformans var. grubii isolates differed widely among the populations from the different countries. This finding supports the hypothesis that local geographic differences occur in Asia between C. neoformans var. grubii populations that could be the result from different founder effects and/or regional factors such as differences in the local environment and climate [34]–[36]. Recently, a population biology study using MLST analysis of C. neoformans var. grubii compared cryptococcal isolates from Thailand with a globally collected set of isolates. This study showed that the Asian population was genetically less diverse than those occurring in Africa and North America [37]. Furthermore, 15 of the 115 Chinese isolates (Table S1) were investigated previously by M13 PCR fingerprinting and identified as genotype VNIc [6]. All these isolates clustered in MC2, thus supporting the relatively limited genetic diversity among Chinese cryptococcal strains as seen by AFLP. Thus, it seems that the genetic diversity of cryptococcal isolates from Asian countries is lower than that of populations occurring in other parts of the world. The minimum spanning tree based on microsatellite analysis showed that MC8 contained mostly Thai isolates (Figure 1B), thus supporting the presence of limited genetic diversity among this population as has been described before [36]–[39]. However, other Thai isolates from HIV-infected patients and bird excreta from the North and Northeast occurred in MCs that comprised also isolates from China and Japan (i.e., MC2 and MC16). Can these findings be explained by assuming a relation with bird migration, especially the East Asia-Australian flyway, through which pathogenic microorganisms such as C. neoformans var. grubii may disperse between China and Thailand [40]? The scattered distribution of isolates from Kuwait and Qatar may be due to migration of foreign workers from Southeast Asia who may have carried isolates that were obtained from their country of origin, similar as has been demonstrated for African immigrants in France [41]. Further support for this hypothesis is that young children in USA acquired the organism from their surrounding environments [42] and we think that this also happens elsewhere.
Isolates belonging to MC8 contained mainly cryptococcal isolates from HIV-infected patients, whereas MC2 and MC16 comprised mostly isolates from non-HIV-infected patients (Figure 1C). A recent report from Vietnam revealed two genotypic clusters based on AFLP analysis, viz., VNIγ and VNIδ, that contained isolates from HIV-negative and positive patients, respectively [10]. Thus genotypic differences were seen between both patient categories based on AFLP analysis and this observation is reminiscent to the differences that we observed in the current microsatellite data. However, no comparison could be made between the AFLP- and microsatellite typing because, unfortunately, none of the Vietnamese isolates was available for our studies.
In Japan and Thailand, environmental isolates co-occurred with clinical isolates in MC2, MC8, and MC16 (Figure 1A, C and D, and Table 3), and these results suggest a genetic relatedness between environmental and clinical cryptococcal isolates in these countries. Such a relationship has been suggested before [34], [43]–[46], but is at stake with a recent analysis from Cuba where the clinical isolates largely belonged to a different MC than those obtained from the environment [17]. At present, the observed differences between the genetic relationship of environmental and clinical isolates in different parts of the globe are not easily explained, but it may be that different environmental niches are occupied in various locales.
Susceptibility analysis of 493 C. neoformans isolates to seven antifungals yielded no change in MIC ranges and MIC50 and MIC90 values of AMB and FLU when compared to previous studies [47]–[49]. The MIC ranges and values for ITR and the three novel antifungal agents POS, ISA, and VOR were slightly changed to those reported previously [22], [48]–[49]. The MIC50 and MIC90 values for ISA were lower than those observed for the other antifungal agents, thus corroborating previous observations [48], [49]
Importantly, resistance in approximately 4% of the isolates to 5FC was observed. Most 5FC-resistant isolates came from Indonesia and Thailand where this drug is not in use [50]–[51]. In our study, we found that approximately 35% of all MC17 isolates were resistant to 5FC. These findings suggest that intrinsic resistance to 5FC occurs among Southeast Asian isolates of C. neoformans, especially in MC17 and in Indonesia. Intrinsic resistance to 5FC in C. neoformans is uncommon and occurs with a low incidence of approximately 1–2% as reported from the USA in the 1990s [52]–[53]. The reason for this relative high number of 5FC resistant isolates in the Southeast Asian region is not clear, and needs further study. Furthermore, resistance to FLU was observed. The FLU-resistant isolates occurred in different countries, including China, India, Indonesia, and Thailand. C. neoformans may develop resistance to FLU after treatment [52], [54], but the molecular origin of the resistance mechanism among the Asian strains remains unknown.
Five out of 10 FLU resistant isolates (2 from MC3, 1 from MC8, and 2 from MC17) from Indonesia were also not susceptible to 5FC. Our findings suggested that isolates in certain MCs, especially MC17, may be prone to a decreased susceptibility to antifungals, especially FLU and 5FC. The observed double resistance to 5FC and FLU has not been reported before, and may pose a risk for the patients infected with such isolates. Isolates from HIV-infected and non-HIV-infected patients did not differ in MIC values of 5FC, FLU, and VOR, but the MIC ranges of isolates from HIV-infected patients were broader than those of isolates from non-HIV-infected patients. The highest MIC values of isolates from HIV-infected patients were 16 times higher for 5FC and FLU and two to four times as high as those from non-HIV-infected patients, respectively.
This is the first extensive report on in vitro antifungal susceptibility and genotyping of clinical and environmental isolates of C. neoformans from several Asian countries. Based on the in vitro susceptibility test results, the patients in the region seem to receive appropriate antifungal therapy with AMB plus 5FC for induction therapy followed by consolidation/maintenance therapy of FLU. AMB is still the most effective agent to treat an infection with C. neoformans [4]. A Chinese study showed that the risk of death in patients with cryptococcosis who did not receive AMB-based initial therapy was about 7–9 times higher than those given AMB [54]. Another study from Thailand used different combinations of antifungal therapies, i.e., AMB alone, AMB plus 5FC, AMB plus FLU, or triple antifungals (AMB plus 5FC and FLU) to treat cryptococcal meningitis showed that treatment of AMB combined with 5FC remains a powerful treatment strategy that results in rapid clearance of the pathogen [55]. Our study did not reveal a significant change in the MICs of AMB for C. neoformans, and therefore, initial treatment with AMB remains the recommended choice. However, given the well-tolerated nature and a excellent activity against Cryptococcus strains, the new generation of triazoles may become an important addition to the currently used antifungals.
In summary, genotypic differences in microsatellite patterns occur between C. neoformans populations from the Asian countries studied. Most of the countries had a unique distribution of MCs but the overall genetic diversity estimated by Simpson's index of diversity was high. Good in vitro antifungal activity was observed, but 5FC and FLU resistant, as well as 5FC/FLU double resistant isolates occurred.
Supporting Information
Origin of Cryptococcus neoformans var. grubii isolates and clinical background information of the patients.
(DOC)
Environmental Cryptococcus neoformans var. grubii isolates from Thailand and Japan.
(DOC)
The MIC range, MIC50, MIC90, and geometric mean for clinical isolates of C. neoformans in each country.
(DOC)
The MIC range, MIC50, MIC90, and geometric mean for the different microsatellite complexes (MCs) of C. neoformans.
(DOC)
Acknowledgments
The opinions expressed by authors contributing to this journal do not necessarily reflect the opinions of the Chinese Centers for Disease Control and Prevention or the institutions with which the authors are affiliated. We thank S.P. Simwami and M.C. Fisher for kindly donating cryptococcal isolates and W.H.M. Chew for excellent technical assistance.
Footnotes
Competing Interests: JFM has been a consultant to Astellas, Basilea, Merck and Schering-Plough and received speaker's fees from Gilead, Janssen Pharmaceutica, Merck, Pfizer, and Schering-Plough. CHK received a grant from Pfizer. RW is currently receiving a grant from IIR-Pfizer for doing research on Cryptococcus. RW is a speaker for Pfizer and Astellas Pharma. All other authors: no potential conflicts of interest relating to employment, consultancy, patents, products in development or marketed products. The sponsors of the research played no decision-making role in the design, execution, analysis and reporting of the research. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported by grants (no. 30970130 and no. 80171335) from the National Natural Science Foundation of China. FH was funded by the Odo van Vloten Foundation, The Netherlands. KK was granted by University of Phayao, Thailand. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bovers M, Hagen F, Boekhout T. Diversity of the Cryptococcus neoformans-Cryptococcus gattii species complex. Revista Iberoamericana de Micologia. 2008;25:S4–12. doi: 10.1016/s1130-1406(08)70019-6. [DOI] [PubMed] [Google Scholar]
- 2.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annual Review of Microbiology. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 3.Springer DJ, Chaturvedi V. Projecting global occurrence of Cryptococcus gattii. Emerging Infectious Diseases. 2010;16:14–20. doi: 10.3201/eid1601.090369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clinical Infectious Diseases. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Varma A, Diaz MR, Litvintseva AP, Wollenberg KK, et al. Cryptococcus neoformans strains and infection in apparently immunocompetent patients, China. Emerging Infectious Diseases. 2008;14:755–762. doi: 10.3201/eid1405.071312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Manosuthi W, Banerjee U, Zhu LP, Chen J, et al. Cryptococcosis in Asia. In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Cryptococcus from human pathogen to model yeast. Washington DC: ASM press; 2011. pp. 287–297. [Google Scholar]
- 8.Shen YZ, Qi TK, Ma JX, Jiang XY, Wang JR, et al. Invasive fungal infections among inpatients with acquired immune deficiency syndrome at a Chinese university hospital. Mycoses. 2007;50:475–480. doi: 10.1111/j.1439-0507.2007.01421.x. [DOI] [PubMed] [Google Scholar]
- 9.Choi YH, Ngamskulrungroj P, Varma A, Sionov E, Hwang SM, et al. Prevalence of the VNIc genotype of Cryptococcus neoformans in non-HIV-associated cryptococcosis in the Republic of Korea. FEMS Yeast Research. 2010;10:769–778. doi: 10.1111/j.1567-1364.2010.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day JN, Hoang TN, Duong AV, Hong CTT, Diep PT, et al. Most cases of cryptococcal meningitis in HIV-uninfected patients in Vietnam are due to a distinct amplified fragment length polymorphism-defined cluster of Cryptococcus neoformans var. grubii VNI. Journal of Clinical Microbiology. 2011;49:658–664. doi: 10.1128/JCM.01985-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz MR, Boekhout T, Kiesling T, Fell JW. Comparative analysis of the intergenic spacer regions and population structure of the species complex of the pathogenic yeast Cryptococcus neoformans. FEMS Yeast Research. 2005;5:1129–1140. doi: 10.1016/j.femsyr.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 12.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Medical Mycology. 2009;47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boekhout T, Theelen B, Diaz M, Fell JW, Hop WC, et al. Hybrid genotypes in the pathogenic yeast Cryptococcus neoformans. Microbiology. 2001;147:891–907. doi: 10.1099/00221287-147-4-891. [DOI] [PubMed] [Google Scholar]
- 15.Meyer W, Aanensen DM, Boekhout T, Cogliati M, Diaz MR, et al. Consensus multi-locus sequence typing scheme for Cryptococcus neoformans and Cryptococcus gattii. Medical Mycology. 2009;47:561–570. doi: 10.1080/13693780902953886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Valk HA, Meis JFGM, Klaassen CHW. Microsatellite based typing of Aspergillus fumigatus: strengths, pitfalls and solutions. Journal of Microbiological Methods. 2007;69:268–272. doi: 10.1016/j.mimet.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Illnait-Zaragozi M-T, Martinez-Machin GF, Fernandez-Andreu CM, Boekhout T, Meis JF, et al. Microsatellite typing of clinical and environmental Cryptococcus neoformans var. grubii isolates from Cuba shows multiple genetic lineages. PloS One. 2010;5:e9124–e9124. doi: 10.1371/journal.pone.0009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rudramurthy SM, de Valk HA, Chakrabarti A, Meis JFGM, Klaassen CHW. High resolution genotyping of clinical Aspergillus flavus isolates from India using microsatellites. PloS One. 2011;6:e16086–e16086. doi: 10.1371/journal.pone.0016086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaassen CHW. MLST versus microsatellites for typing Aspergillus fumigatus isolates. Medical Mycology. 2009;47(Suppl 1):S27-33-S27-33. doi: 10.1080/13693780802382244. [DOI] [PubMed] [Google Scholar]
- 20.Hazen KC, Baron EJ, Colombo AL, Girmenia C, Sanchez-Sousa A, et al. Comparison of the susceptibilities of Candida spp. to fluconazole and voriconazole in a 4-year global evaluation using disk diffusion. Journal of Clinical Microbiology. 2003;41:5623–5632. doi: 10.1128/JCM.41.12.5623-5632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon-Chung KJ, Polacheck I, Bennett JE. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). Journal of Clinical Microbiology. 1982;15:535–537. doi: 10.1128/jcm.15.3.535-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagen F, Illnait-Zaragozi M-T, Bartlett KH, Swinne Dl, Geertsen E, et al. In vitro antifungal susceptibilities and amplified fragment length polymorphism genotyping of a worldwide collection of 350 clinical, veterinary, and environmental Cryptococcus gattii isolates. Antimicrobial Agents and Chemotherapy. 2010;54:5139–5145. doi: 10.1128/AAC.00746-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bovers M, Hagen F, Kuramae EE, Diaz MR, Spanjaard L, et al. Unique hybrids between the fungal pathogens Cryptococcus neoformans and Cryptococcus gattii. FEMS Yeast Res. 2006;6:599–607. doi: 10.1111/j.1567-1364.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- 24.Barreto de Oliveira MT, Boekhout T, Theelen B, Hagen F, Baroni FA, et al. Cryptococcus neoformans shows a remarkable genotypic diversity in Brazil. Journal of Clinical Microbiology. 2004;42:1356–1359. doi: 10.1128/JCM.42.3.1356-1359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical Laboratory Standards Institute. M27-A3. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard. 2008. Wayne, PA.
- 26.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clinical Infectious Diseases. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen MH, Yu CY. In vitro comparative efficacy of voriconazole and itraconazole against fluconazole-susceptible and -resistant Cryptococcus neoformans isolates. Antimicrobial Agents and Chemotherapy. 1998;42:471–472. doi: 10.1128/aac.42.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaller MA, Boyken L, Hollis RJ, Messer SA, Tendolkar S, et al. In vitro susceptibilities of clinical isolates of Candida species, Cryptococcus neoformans, and Aspergillus species to itraconazole: global survey of 9,359 isolates tested by clinical and laboratory standards institute broth microdilution methods. Journal of Clinical Microbiology. 2005;43:3807–3810. doi: 10.1128/JCM.43.8.3807-3810.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfaller MA, Messer SA, Boyken L, Hollis RJ, Rice C, et al. In vitro activities of voriconazole, posaconazole, and fluconazole against 4,169 clinical isolates of Candida spp. and Cryptococcus neoformans collected during 2001 and 2002 in the ARTEMIS global antifungal surveillance program. Diagnostic Microbiology and Infectious Disease. 2004;48:201–205. doi: 10.1016/j.diagmicrobio.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Pfaller MA, Messer SA, Boyken L, Rice C, Tendolkar S, et al. Global trends in the antifungal susceptibility of Cryptococcus neoformans (1990 to 2004). Journal of Clinical Microbiology. 2005;43:2163–2167. doi: 10.1128/JCM.43.5.2163-2167.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souza LKH, Souza Junior AH, Costa CR, Faganello J, Vainstein MH, et al. Molecular typing and antifungal susceptibility of clinical and environmental Cryptococcus neoformans species complex isolates in Goiania, Brazil. Mycoses. 2010;53:62–67. doi: 10.1111/j.1439-0507.2008.01662.x. [DOI] [PubMed] [Google Scholar]
- 32.Simpson EH. Measurement of Diversity. Nature. 1949;163:688–688. [Google Scholar]
- 33.Illnait-Zaragozi MT, Martinez-Machin GF, Fernandez-Andreu CM, Hagen F, Boekhout T, et al. Microsatellite typing and susceptibilities of serial Cryptococcus neoformans isolates from Cuban patients with recurrent cryptococcal meningitis. BMC Infectious Diseases. 2010;10:289–289. doi: 10.1186/1471-2334-10-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Franzot SP, Hamdan JS, Currie BP, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for both local genetic differences and a global clonal population structure. Journal of Clinical Microbiology. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain N, Wickes BL, Keller SM, Fu J, Casadevall A, et al. Molecular epidemiology of clinical Cryptococcus neoformans strains from India. Journal of Clinical Microbiology. 2005;43:5733–5742. doi: 10.1128/JCM.43.11.5733-5742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meyer W, Marszewska K, Amirmostofian M, Igreja RP, Hardtke C, et al. Molecular typing of global isolates of Cryptococcus neoformans var. neoformans by polymerase chain reaction fingerprinting and randomly amplified polymorphic DNA-a pilot study to standardize techniques on which to base a detailed epidemiological survey. Electrophoresis. 1999;20:1790–1799. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1790::AID-ELPS1790>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Simwami SP, Khayhan K, Henk DA, Aanensen DM, Boekhout T, et al. Low diversity Cryptococcus neoformans variety grubii multilocus sequence types from Thailand are consistent with an ancestral African origin. PLoS Pathogens. 2011;7:e1001343–e1001343. doi: 10.1371/journal.ppat.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sriburee P, Khayhan S, Khamwan C, Panjaisee S, Tharavichitkul P. Serotype and PCR-fingerprints of clinical and environmental isolates of Cryptococcus neoformans in Chiang Mai, Thailand. Mycopathologia. 2004;158:25–31. doi: 10.1023/b:myco.0000038435.14281.f4. [DOI] [PubMed] [Google Scholar]
- 39.Sukroongreung S, Lim S, Tantimavanich S, Eampokalap B, Carter D, et al. Phenotypic switching and genetic diversity of Cryptococcus neoformans. J Clin Microbiol. 2001;39:2060–2064. doi: 10.1128/JCM.39.6.2060-2064.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, et al. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 41.Garcia-Hermoso D, Janbon G, Dromer F. Epidemiological evidence for dormant Cryptococcus neoformans infection. Journal of Clinical Microbiology. 1999;37:3204–3209. doi: 10.1128/jcm.37.10.3204-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski L, et al. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics. 2001;107:E66–E66. doi: 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- 43.Casali AK, Goulart Lc, Rosa e Silva LvK, Ribeiro AM, Amaral AA, et al. Molecular typing of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. FEMS Yeast Research. 2003;3:405–415. doi: 10.1016/S1567-1356(03)00038-2. [DOI] [PubMed] [Google Scholar]
- 44.Currie BP, Freundlich LF, Casadevall A. Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. Journal of Clinical Microbiology. 1994;32:1188–1192. doi: 10.1128/jcm.32.5.1188-1192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delgado ACN, Taguchi H, Mikami Y, Myiajy M, Villares MCB, et al. Human cryptococcosis: relationship of environmental and clinical strains of Cryptococcus neoformans var. neoformans from urban and rural areas. Mycopathologia. 2005;159:7–11. doi: 10.1007/s11046-004-9618-4. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto Y, Kohno S, Koga H, Kakeya H, Tomono K, et al. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. Journal of Clinical Microbiology. 1995;33:3328–3332. doi: 10.1128/jcm.33.12.3328-3332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandt ME, Pfaller MA, Hajjeh RA, Hamill RJ, Pappas PG, et al. Trends in antifungal drug susceptibility of Cryptococcus neoformans isolates in the United States: 1992 to 1994 and 1996 to 1998. Antimicrobial Agents and Chemotherapy. 2001;45:3065–3069. doi: 10.1128/AAC.45.11.3065-3069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Illnait-Zaragozi M-T, Martinez GF, Curfs-Breuker I, Fernandez CM, Boekhout T, et al. In vitro activity of the new azole isavuconazole (BAL4815) compared with six other antifungal agents against 162 Cryptococcus neoformans isolates from Cuba. Antimicrobial Agents and Chemotherapy. 2008;52:1580–1582. doi: 10.1128/AAC.01384-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson GR, 3rd, Wiederhold NP, Fothergill AW, Vallor AC, Wickes BL, et al. Antifungal susceptibilities among different serotypes of Cryptococcus gattii and Cryptococcus neoformans. Antimicrobial Agents and Chemotherapy. 2009;53:309–311. doi: 10.1128/AAC.01216-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chottanapund S, Singhasivanon P, Kaewkungwal J, Chamroonswasdi K, Manosuthi W. Survival time of HIV-infected patients with cryptococcal meningitis. Journal of the Medical Association of Thailand. 2007;90:2104–2111. [PubMed] [Google Scholar]
- 51.Ganiem AR, Parwati I, Wisaksana R, van der Zanden A, van de Beek D, et al. The effect of HIV infection on adult meningitis in Indonesia: a prospective cohort study. AIDS. 2009;23:2309–2316. doi: 10.1097/QAD.0b013e3283320de8. [DOI] [PubMed] [Google Scholar]
- 52.Perea S, Patterson TF. Antifungal resistance in pathogenic fungi. Clinical Infectious Diseases. 2002;35:1073–1080. doi: 10.1086/344058. [DOI] [PubMed] [Google Scholar]
- 53.Vermes A, Guchelaar HJ, Dankert J. Flucytosine: a review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. Journal of Antimicrobial Chemotherapy. 2000;46:171–179. doi: 10.1093/jac/46.2.171. [DOI] [PubMed] [Google Scholar]
- 54.Perfect JR, Cox GM. Drug resistance in Cryptococcus neoformans. Drug Resistance Updates. 1999;2:259–269. doi: 10.1054/drup.1999.0090. [DOI] [PubMed] [Google Scholar]
- 55.Brouwer AE, Rajanuwong A, Chierakul W, Griffin GE, Larsen RA, et al. Combination antifungal therapies for HIV-associated cryptococcal meningitis: a randomised trial. Lancet. 2004;363:1764–1767. doi: 10.1016/S0140-6736(04)16301-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Origin of Cryptococcus neoformans var. grubii isolates and clinical background information of the patients.
(DOC)
Environmental Cryptococcus neoformans var. grubii isolates from Thailand and Japan.
(DOC)
The MIC range, MIC50, MIC90, and geometric mean for clinical isolates of C. neoformans in each country.
(DOC)
The MIC range, MIC50, MIC90, and geometric mean for the different microsatellite complexes (MCs) of C. neoformans.
(DOC)