Abstract
The recent crystal structure of two monoferric human serum transferrin (FeNhTF) molecules bound to the soluble portion of the homodimeric transferrin receptor (sTFR) has provided new details of this binding interaction which dictates iron delivery to cells. Specifically, substantial rearrangements in the homodimer interface of the sTFR occur as a result of the binding of the two FeNhTF molecules. Mutagenesis of selected residues in the sTFR highlighted in the structure was undertaken to evaluate the effect on function. Elimination of Ca2+ binding in the sTFR by mutating two of four coordinating residues ([E465A,E468A]) results in low production of an unstable and aggregated sTFR. Mutagenesis of two histidines ([H475A,H684A]) at the dimer interface had little effect on the kinetics of iron release at pH 5.6 from either lobe, reflecting the inaccessibility of this cluster to solvent. Creation of a H318A sTFR mutant allows assignment of a small pH dependent initial decrease in the fluorescent signal to His318. Removal of the four C-terminal residues of the sTFR, Asp757-Asn758-Glu759-Phe760, eliminates pH-stimulated iron release from the C-lobe of the Fe2hTF/sTFR Δ757–760 complex. The loss is accounted for by the inability of this sTFR mutant to bind and stabilize protonated hTF His349 (a pH-inducible switch) in the C-lobe of hTF. Collectively, these studies support a model in which a series of pH-induced events involving both TFR residue His318 and hTF residue His349 occurs in order to promote receptor-stimulated iron release from the C-lobe of hTF.
The efficient delivery of iron to mammalian cells relies on the transferrin/transferrin receptor system. Iron (in the form of Fe3+) circulates in the blood bound to the ~80 kDa bilobal glycoprotein human serum transferrin (hTF). The two lobes of hTF (termed N- and C-lobes) are further divided into two subdomains (N1 and N2, C1 and C2). As with most other members of the transferrin superfamily, hTF is capable of binding two Fe3+ atoms deep within a cleft formed between the two subdomains of each lobe.1, 2 The sequestration of Fe3+ within the cleft of hTF is critical to preventing its hydrolysis or reduction to Fe2+ which promotes the production of harmful radicals via the Fenton series of reactions.3, 4 Iron-containing hTFs (diferric and the two monoferric hTF species) bind with nM affinity to the homodimeric transferrin receptor (TFR) located on the cell surface of all cells.5 Following clathrin-dependent endocytosis, a decrease in pH within the early endosome (to ~5.5–6.0) aids in the release of Fe3+ (to an as of yet unidentified biological chelator) from the hTF/TFR complex in a receptor-mediated manner.6, 7 Still within the endosome, the Fe3+ must be reduced to Fe2+ by a ferrireductase, such as Steap38, before being transported out of the endosome by the divalent metal transporter, DMT1.9 Iron-free hTF, referred to as apohTF, remains tightly bound to the TFR at endosomal pH, facilitating its proper sorting and recycling back to the cell surface.10 Upon exposure to the more alkaline pH of the blood (~7.4), apohTF is released or displaced by Fe2hTF from the TFR11 and becomes available to sequester more Fe3+.
The TFR plays a critical role throughout the entire process of cellular iron delivery. Iron release from hTF requires opening of the cleft and is accompanied by large conformational changes within each lobe (opening ~59° and 50° for the N- and C-lobes, respectively).1, 2 These significant conformational changes in hTF must be accommodated and compensated for by the TFR within the hTF/TFR complex. As well as being critical to the discrimination between iron-containing and apohTF at both neutral and endosomal pH, the TFR also significantly affects iron release from hTF at endosomal pH.12 Clearly, the interactions controlling this finely tuned system of cellular iron delivery must be elucidated to understand this process completely.
The crystal structure of the TFR ectodomain (PDB ID: 1CX8) 13 revealed that the homodimeric receptor is comprised of three distinct domains: a protease-like domain, an apical domain and a helical domain. Importantly, the TFR crystal structure was used to create a cryo-EM model of the hTF/TFR complex (PDB ID: 1SUV).14 The 7.5 Å resolution cryo-EM model provided the first structural insight into the hTF-TFR interaction. In the model, the N-lobe of hTF is situated between the protease-like domain of the TFR and the cell membrane, while a large portion of the C1 subdomain interacts with the helical domain of the TFR. However, given the relatively low resolution, the model lacked the precision needed to identify specific molecular interactions between hTF and the TFR.
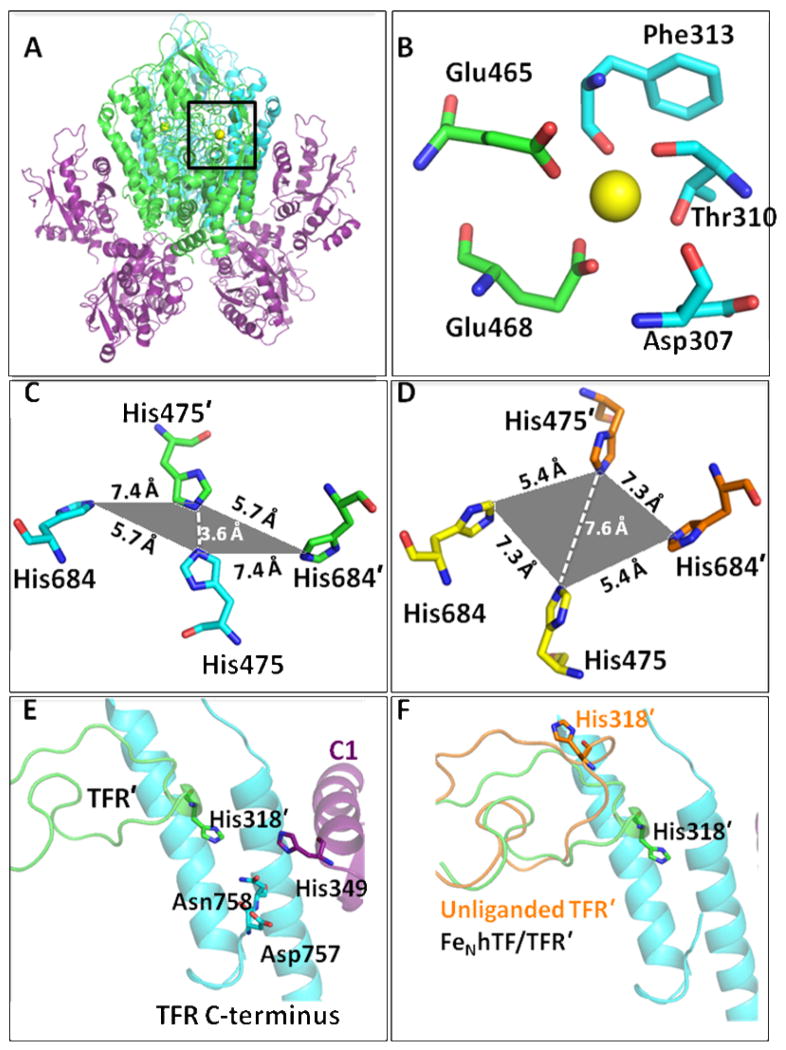
The recently solved 3.22 Å crystal structure of recombinant monoferric N-lobe hTF (FeNhTF) bound to the soluble portion of the TFR (sTFR, residues 121–760, PDB ID: 3S9L)15 has provided more detailed information with regard to the binding interactions between hTF and the TFR (Figure 1A). As previously predicted from the crystal structure of the TFR bound to another ligand (the HFE protein),16 the FeNhTF/sTFR structure shows that the structure of the TFR changes significantly as a result of hTF binding. Since Cheng et al. 14 utilized the crystal structure of the unliganded TFR, these structural changes in the TFR as a result of hTF binding were not accounted for in the cryo-EM model. As observed in the HFE/TFR crystal structure,16 the geometry of a set of four histidine residues (His475 and His684 from each TFR monomer) is altered in the FeNhTF/sTFR structure (Figures 1C and D).15 Specifically, His475 and His475′ (which are 7.6 Å apart in the unliganded sTFR structure)13 are brought to within 3.6 Å of each other when hTF binds to the TFR.15 Given the physiologically relevant pKa of His residues (~6.0), it was suggested that repulsion of this histidine cluster at low pH would promote receptor-mediated iron release and/or release of the HFE protein at endosomal pH.15, 16 Additionally, an intersection formed between the apical and protease-like domains of one TFR monomer (TFR), the C-terminus (helical domain) of the other TFR monomer (TFR′) and the C1 subdomain of hTF was identified in the FeNhTF/sTFR crystal structure (Figure 1E).15 A number of interesting structural elements are located within this intersection including a metal binding site, previously identified in other TFR structures. 13, 16 In the FeNhTF/sTFR structure, a Ca2+ atom in this metal binding site is coordinated by two residues from the protease-like domain (Glu465 and Glu468) and three residues from the apical domain of the TFR (Asp307, Thr310 and Phe313, Figure 1B). Another significant feature of this TFR-TFR′-C1 intersection is the large movement of a long loop (residues 275–338) in the apical domain. Specifically, the movement of this loop causes residue His318 to move nearly ~18 Å in the FeNhTF/sTFR structure in comparison to the unliganded TFR structure (Figure 1F). Additionally, the location of α-helix 1 in the C-lobe of hTF, on which a number of residues involved in both the binding of hTF to the TFR (Asp356) 17 and pH-dependent receptor-mediated iron release from the C-lobe (His349)15, 18 are located, is shifted ~5 Å (nearly one full helical turn) in the FeNhTF/sTFR structure compared to the cryo-EM model.15 Although the last two amino acids (Glu759 and Phe760) of the TFR could not be placed in the FeNhTF/sTFR structure due to a lack of sufficient density at pH 7.5, it is plausible that α-helix 1 in the C-lobe of hTF could interact with the C-terminus of the TFR monomer at endosomal pH (Figure 1E).
Figure 1.
(A) The FeNhTF/sTFR crystal structure (PDB ID: 3S9L).15 One TFR monomer (TFR) is shown in cyan, the other TFR monomer (TFR′) is shown in green and the two molecules of FeNhTF are shown in purple. (B) Ca2+ binding site located between the protease-like (green) and apical domains (blue) of the TFR (A, black box). (C) The histidine cluster formed at the dimer interface. Two His residues, His475 and His684, from one TFR monomer (cyan) converge with the same two His residues, His475′ and His684′, from the other TFR monomer (green) as a result of hTF binding (in comparison to their position in the unliganded TFR structure, PDB ID: 1CX8 13 as shown in yellow and orange in (D)). (E) The TFR-TFR′-C1 intersection formed when hTF binds to the sTFR. One TFR monomer is shown in cyan, the other TFR monomer (TFR′) is shown in green and the FeNhTF is shown in purple. (F) The long loop containing His318 moves ~18 Å upon hTF binding, as shown by overlaying the structure of the unliganded TFR (orange) with the sTFR in the FeNhTF/sTFR structure (green).
Iron release from Fe2hTF can proceed via two pathways: N-lobe first followed by C-lobe (k1N→ k2C, where k1N is the rate constant for release of the first iron from Fe2hTF coming from its N-lobe and k2C the rate constant for release of second iron coming from its C-lobe) or alternatively iron release from the C-lobe first followed by the N-lobe (k1C →k2N). A comprehensive kinetic scheme of iron removal from diferric hTF (Fe2hTF) to apohTF was determined by monitoring the increase in intrinsic tryptophan fluorescence as iron is released from recombinant hTF constructs at pH 5.6.12, 19 In the absence of the sTFR, essentially all iron is released first from the N-lobe followed by the C-lobe.12 As first suggested by Aisen et al.,20 a switch in the order of iron release is observed in the presence of the TFR, such that iron is preferentially removed from the C-lobe first followed by the N-lobe (k1C → k2N).12 However, since this is only the case ~66% of the time,12 both pathways must be taken into account when fitting kinetic data from the Fe2hTF/sTFR complex. Under our defined conditions, the sTFR enhances iron release from the C-lobe ~7–11 fold and retards iron release from the N-lobe ~6–15 fold.12 Hence, binding to the TFR not only switches the order of iron release from hTF, but also balances the rates of iron release from the two lobes to maximize efficient delivery of iron to cells during the endocytic cycle.
Intrinsic tryptophan fluorescence allows iron release rates from each lobe of the hTF/sTFR complexes to be monitored, as well as protein conformational changes. A rapid initial drop in the fluorescence has been a puzzling feature of the kinetic profile of all hTF/sTFR complexes.21, 22 Since this initial decrease in fluorescence is observed even when the sTFR homodimer alone (without hTF) is exposed to putative endosomal conditions (pH 5.6) and is not observed in any hTF (without the sTFR), it was previously attributed to a pH-sensitive change in the sTFR.21 The time points for this initial quench in the tryptophan fluorescence are routinely removed prior to fitting kinetic data for iron release from the hTF/sTFR complexes.12
Based on the rearrangements within the sTFR observed in the structure of the FeNhTF/sTFR complex as discussed above, a number of sTFR mutants have been produced: H318A sTFR, [H475A,H684A] sTFR, [E465A,E468A] sTFR and a truncated form of the sTFR in which the last four amino acids, Asp757-Asn758-Glu759-Phe760, have been removed, namely sTFR Δ757–760. Using our established kinetic scheme of iron removal from hTF/sTFR complexes, 12 we have measured the iron release kinetics from various recombinant hTF constructs (non-glycosylated Fe2hTF, FeNhTF, and monoferric C-lobe or FeChTF) bound to the sTFR mutants to assess the effect of these mutations on function.
MATERIALS AND METHODS
Materials
Dulbecco’s modified Eagle’s medium-Ham F-12 nutrient mixture (DMEM-F12) and fetal bovine serum (FBS) were obtained from the GIBCO-BRL Life Technologies Division of Invitrogen. Novex 6% TBE urea mini-gels, TBE running buffer (5X) and TBE-urea sample buffer (2X), and the iBlot dry blot transfer system were also from Invitrogen. Antibiotic-antimycotic solution (100X) solution and trypsin were from Mediatech, Inc. The QuikChange site-directed mutagenesis kit was from Stratagene. Pro293A-CDM serum-free medium, L-glutamine, and 4–20% acrylamide gels were from Lonza. Methotrexate from Bedford Laboratories was obtained at a hospital pharmacy. All tissue culture dishes, Corning flasks, and expanded surface roller bottles were from local distributors as were Amicon Ultracel 30 kDa molecular weight cutoff membrane microconcentrator devices. Ni-NTA Superflow resin came from Qiagen. Hi-prep 26/60 Sephacryl S-200HR and S-300HR columns were from GE Healthcare. Ethylenediaminetetraacetic acid (EDTA), Ponceau stain and Orange G were from Fisher. Nitrilotriacetic acid (NTA) and ferrous ammonium sulfate were from Sigma. The chemiluminescence detection kit was from Thermo Scientific.
Expression and Purification of sTFR Mutants
Recombinant N-terminally His-tagged glycosylated sTFR and various mutants were produced as previously described.23 Briefly, mutations were introduced into the pNUT vector using the QuikChange site-directed mutagenesis kit. The mutagenic primers are shown in Table S2. Following transfection and selection with methotrexate, adherent baby hamster kidney (BHK) cells containing the mutant N-His tagged sTFR pNUT vector were transferred into expanded surface roller bottles.23 The culture medium (~200 mL/roller bottle) was collected every 2–4 days. The first two or three batches contain Dulbecco’s modified Eagle medium/F12 with antibiotic-antimycotic solution and 10% fetal bovine serum, were collected and discarded. Subsequent batches (generally 4–6) containing Pro293A-CDM serum-free medium with L-glutamine and 1 mM butyric acid, were collected, pooled and saved at 4°C until purification. The amount of each sTFR mutant produced was determined by a solid-phase competitive immunoassay as previously described.23
Purification of the sTFR mutants followed the same protocol developed for the sTFR.23 Briefly, purification entailed concentration followed by the addition of 5X buffer to yield a final concentration of 1X Qiagen start buffer (50 mM Tris, pH 7.5, containing 300 mM NaCl, 20 mM imidazole, 10% glycerol, and 0.05% NaN3) before passage over a Ni-NTA column (1 × 10 cm) at a flow rate of 2 mL/min. Each sTFR sample was displaced from the column by the addition of 250 mM imidazole to the start buffer. Peak fractions were pooled, reduced using microconcentrators to less than 2 mL, filtered and loaded onto a Sephacryl S300HR 26/60 column equilibrated and run in 100 mM NH4HCO3 at a flow rate of 1.5 mL/min (V0 = ~93 mL). Fractions containing the sTFR (or mutant) were pooled and stored at 4°C in 100 mM NH4HCO3.
All recombinant non-glycosylated hTFs (diferric hTF, Fe2hTF; FeNhTF, monoferric N-lobe hTF in which mutation of iron binding ligands, Y426F/Y517F, prevents iron binding in the C-lobe); FeChTF monoferric C-lobe hTF in which mutation of iron binding ligands, Y95F/Y188F, prevents iron binding in the N-lobe) were produced as previously described.24
Immunoblotting
Non-reduced sTFR samples were separated by SDS-PAGE on 4–20% acrylamide gels pre-electrophoresed (20 min at 100 V) with Orange G buffer (0.25% Orange G, 30% glycerol). Samples were then loaded and electrophoresed on the gel (1.75 h at 120V). Proteins were transferred to nitrocellulose using the iBlot dry blot transfer system. Transfer of proteins to the membrane was confirmed by staining with Ponceau stain. The immunoblot was analyzed using the mouse IgG1 monoclonal antibody to the TFR, A4A6 (1 μg/mL, a generous gift from the laboratory of Dr. James Cook at the University of Kansas Medical Center). Bound antibody was detected using horseradish peroxidase conjugated goat anti-mouse IgG and a chemiluminescence detection kit.
hTF/sTFR Complex Formation and Purification
The hTF/sTFR complexes were prepared by adding a small molar excess (~20%) of hTF (Fe2hTF or FeChTF) to 1.5 mg of each mutant sTFR. Following equilibration at room temperature for ~5 min, hTF/mutant sTFR complexes were purified by passage over a Sephacryl S300HR gel filtration column in 100 mM NH4HCO3 to remove excess hTF. Fractions containing the complex were concentrated to 15 mg/mL with respect to hTF.
Kinetics of Iron Release from hTF/sTFR Complexes at pH 5.6
Iron release from the hTF/mutant sTFR complexes was monitored at 25° C using an Applied Photophysics SX.20MV stopped-flow spectrofluorimeter as previously described.12, 18 One syringe contained the hTF/sTFR complex (375 nM) in 300 mM KCl and the other syringe contained MES buffer (200 mM, pH 5.6), KCl (300 mM) and EDTA (8 mM). Rate constants were determined by fitting the increase in fluorescence intensity versus time using Origin software (version 7.5) to standard models as described in detail previously.12, 18 When determining rate constants for iron release, the initial quench in the tryptophan fluorescence, attributed to a pH-inducible conformational change in the sTFR, was removed prior to fitting. All data were corrected to zero fluorescence intensity before fitting.
Analysis of the initial quench in tryptophan fluorescence required the derivation of a new model that is similar to the previously described A → B model, but also includes an initial decay term (Figure 2 of the Supporting Information). The equation used to fit the initial decay data, as well as the complete derivation and program code for Origin, are provided in the Supplemental Data. Again, all data were corrected to zero at the fluorescent minimum before fitting. Because the fluorescent increase following the minimum affects the fit, an equal number of data points on each side of the fluorescent minimum were included in the fitting process. The half-life (t1/2) was calculated by the following equation: t1/2 = ln(2)/k2.
Urea gel analysis of hTF/mutant sTFR Complexes
The iron status of hTF bound to the sTFR mutants was examined by urea gel electrophoresis using Novex 6% TBE-urea mini-gels in 90 mM Tris–borate, pH 8.4, containing 16 mM EDTA as previously described.12, 18 Iron-containing complexes were mixed 1:1 with 2X TBE-urea gel sample buffer (final concentration 0.5 μg/μL). To determine the extent of iron removal from the various hTF/mutant sTFR complexes, an aliquot of each was added to iron removal buffer (100 mM MES buffer, pH 5.6, containing 300 mM KCl and 4 mM EDTA) and incubated at room temperature for 5 min. The iron removal process was halted by addition of 2X TBE-urea gel sample buffer. Samples (3.0 μg) were loaded and the gel was electrophoresed for 2.25 h at 125 V. Protein bands were visualized by staining with Coomassie blue.
RESULTS
Recombinant sTFR production
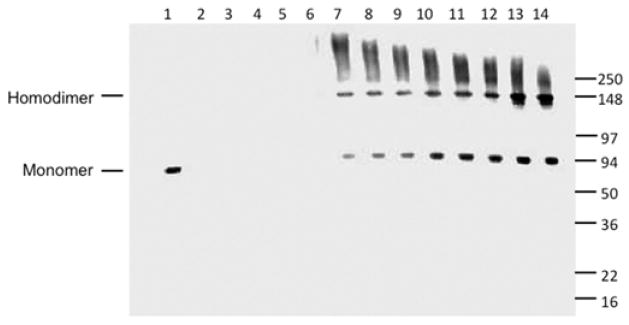
As reported previously, the expression of the glycosylated sTFR in our BHK system generally produces between 30–40 mg of protein per liter of tissue culture medium (Table S1).23 Production of the H318A sTFR and sTFR Δ757–760 mutants was comparable to production of the wild type sTFR, while an ~50% decrease was observed in the production of the [H475A,H684A] sTFR mutant. However, mutation of the two glutamate residues in the protease-like domain of the sTFR ([E465A,E468A] sTFR mutant) significantly decreased the yield of receptor to <1.0 mg/L.
In the final step of purification, homodimeric wild type sTFR typically elutes from an S300HR size exclusion column as a broad peak centered at 165 mL.23 However, the [E465A,E468A] sTFR mutant (as indicated by monitoring the A280) eluted from the column immediately following the void volume (V0) of 93 mL (data not shown), indicating that it exists largely in an oligomeric state. While the wild type sTFR migrates as a monomer (Figure 2, lane 1) even under non-reducing SDS-PAGE conditions, immunoblot analysis confirmed the presence of homodimers as well as a range of higher order oligomers in the S300HR fractions of the [E465A,E468A] sTFR mutant (Figure 2, lanes 7–14). Unfortunately, the poor yield and oligomeric state of the [E465A,E468A] sTFR mutant precluded further experiments with this mutant.
Figure 2.
Ca2+ coordination by Glu465 and Glu468 is critical to structural integrity of the sTFR. Purified sTFR (100 ng, lane 1) served as a standard. Following purification, aliquots from tissue culture batches 4, 5 and 6 (lanes 2–4) as well as the pool of batches 4–6 (lane 5). Fractions 3–11 (corresponding to elution volumes between 102 and 126 mL) from the S300HR column of the [E465A,E468A] sTFR mutant were analyzed SDS-PAGE followed by immunoblotting (lanes 6–14, respectively). Note that homodimeric wild type sTFR typically elutes as a broad peak centered at 165 mL.
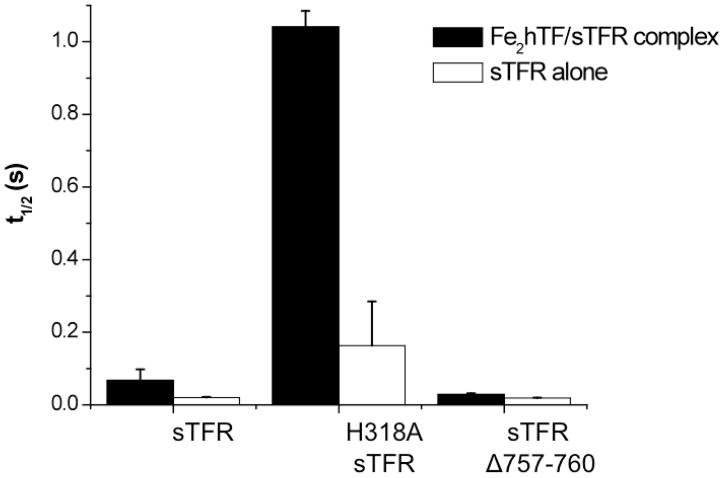
pH-inducible changes in the sTFR
As described in the Introduction, for all sTFR containing samples, an initial rapid drop in tryptophan fluorescence precedes the increase associated with iron release from Fe2hTF. Interestingly, the half-life of this pH-mediated rapid decrease in fluorescence of sTFR depends on whether Fe2hTF is bound (Fe2hTF/sTFR complex versus sTFR alone, Figure 3). The H318A sTFR and sTFR Δ757–760 mutations significantly affect the duration of this pH-sensitive change in the sTFR. Specifically, this pH-sensitive decrease in fluorescence is considerably lengthened in the presence of the H318A sTFR mutation, the half-life being ~8–15 fold longer, depending on whether Fe2hTF is bound (Figure 3). Conversely, the half-life of the pH-sensitive decrease in the fluorescence is slightly shorter in the presence of the sTFR Δ757–760 truncation and is unaffected by the presence of Fe2hTF (Figure 3). No effect on the pH-sensitive fluorescent decrease was observed in the [H475A,H684A] sTFR mutant, with or without Fe2hTF, in comparison to the wild type sTFR (data not shown).
Figure 3.
The half life (t1/2) of a pH-inducible conformational change in the sTFR is altered by the H318A sTFR mutation and the sTFR Δ757–760 truncation. Average half lives ± 95% confidence intervals are shown for the sTFR, H318A sTFR and sTFR Δ757–760 alone (white) and in complex with Fe2hTF (black).
Iron release kinetics from hTF/H318A sTFR complexes
Rate constants were determined by fitting the increase in fluorescence intensity versus time. This increase in intrinsic tryptophan fluorescence has been previously attributed to Trp residues in hTF and not the sTFR.21 Since no Trp or Tyr residues in hTF were mutated, no differences in the amplitude of the fluorescent signal in any of the hTF/mutant sTFR complexes in comparison to the hTF/wt sTFR complex were observed. TFR residue His318, part of a long loop in the apical domain, flips into the TFR-TFR′-C1 intersection upon hTF binding, moving nearly 18 Å relative to its position in the unliganded TFR structure (Figure 1F). Kinetic rate constants for conformational changes and iron release from various hTF/H318A sTFR complexes are presented in Table 1. In fitting the Fe2hTF/H318A sTFR kinetic data, rate constants for both pathways, k1N→ k2C and k1C→ k2N, were allowed to vary (Table 1). The rate constants, k1C, k1N and k2C are smaller in the Fe2hTF/H318A sTFR complex by 67%, 39% and 18%, respectively, relative to the control while the rate constant k2N is unaffected. The ratio, k1C/(k1C+ k1N,) corresponds to the fraction following the k1C → k2N pathway. For the wild-type Fe2hTF/sTFR complex, 66% of the iron is removed by the k1C → k2N pathway and 34% by the k1N → k2C pathway. In contrast, the rate constants for the Fe2hTF/H318A sTFR complex indicate that neither pathway is favored over the other, i.e., 51% of the iron is released via k1C → k2N pathway and 49% via the k1N → k2C pathway.
Table 1.
Iron release kinetics from H318A sTFR complexes.a
| Complex | k1C (min−1) | k2N (min−1) | k1N (min−1) | k2C (min−1) |
|---|---|---|---|---|
| Fe2hTF/sTFRb | 5.5 ± 0.9 | 1.4 ± 0.2 | 2.8 | 7.2 |
| Fe2hTF/H318A sTFR | 1.8 ± 0.7 | 1.5 ± 0.2 | 1.7 ± 0.1 | 5.9 ± 0.9 |
|
| ||||
| Complex |
k1 (min−1) Conformational Change |
k2C (min−1) Fe3+ Release |
||
|
| ||||
| FeChTF/sTFRb | 20.6 ± 1.2 | 7.2 ± 0.4 | ||
| FeChTF/H318A sTFR | - | 5.5 ± 0.6 | ||
|
| ||||
| Complex |
k1 (min−1) Conformational Change |
k2N (min−1) Fe3+ Release |
||
|
| ||||
| FeNhTF/sTFRb | 22.0 ± 0.7 | 1.7 ± 0.6 | ||
| FeNhTF/H318A sTFR | 17.4 ± 2.4 | 1.1 ± 0.1 | ||
Averages and 95% confidence intervals for kinetic runs performed on N=3–5 different days. Each day 3 kinetic traces were averaged before fitting.
From reference 12.
Normally, a rapid conformational change precedes iron release from the two monoferric complexes FeNhTF/sTFR and FeChTF/sTFR, exhibiting conformational rate constants k = 22.0 min−1 and 20.6 min−1, respectively. In the FeNhTF/H318A sTFR complex, a small decrease in the rate of the conformational change (k = 17.4 vs. 22.0 min−1) is observed, whereas the conformational change is completely absent in the FeChTF/H318A sTFR complex. The rate constant for iron release from the FeNhTF/H318A sTFR complex is decreased somewhat (k2N = 1.1 vs. 1.7 min−1, Table 1). Although the preceding conformational change is absent in the FeChTF/H318A sTFR complex, iron release from this mutant is relatively unaffected, i.e., k2C is similar to the k2C value obtained from fitting the Fe2hTF/H318A sTFR complex (5.5 ± 0.6 vs. 5.9 ± 0.9 min−1) and only 23% lower than the value of k2C = 7.2 min−1 for FeChTF/sTFR (Table 1).
Iron release kinetics from hTF/[H475A,H684A] sTFR complexes
Rate constants for iron release and conformational changes from the various hTF/[H475A,H684A] sTFR complexes are reported in Table 2. Relatively small differences are observed between the hTF/[H475A,H684A] mutant sTFR complexes and the respective control complexes except for k1C for the Fe2hTF complex which is reduced by ~ 45%.
Table 2.
Iron release kinetics from [H475A,H684A] sTFR complexes.a
| Complex | k1C (min−1) | k2N (min−1) |
|---|---|---|
| Fe2hTF/sTFRb | 5.5 ± 0.9 | 1.4 ± 0.2 |
| Fe2hTF/[H475A,H684A] sTFR | 3.1 ± 0.1 | 1.2 ± 0.1 |
|
| ||
| Complex |
k1 (min−1) Conformational Change |
k2C (min−1) Fe3+ Release |
|
| ||
| FeChTF/sTFRb | 20.6 ± 1.2 | 7.2 ± 0.4 |
| FeChTF/[H475A,H684A] sTFR | 16.7 ± 2.2 | 7.5 ± 0.9 |
|
| ||
| Complex |
k1 (min−1) Conformational Change |
k2N (min−1) Fe3+ Release |
|
| ||
| FeNhTF/sTFRb | 22.0 ± 0.7 | 1.7 ± 0.6 |
| FeNhTF/[H475A,H684A] sTFR | 15.3 ± 0.3 | 1.7 ± 0.1 |
Averages and 95% confidence intervals for kinetic runs performed on N=3–5 different days. Each day 3 kinetic traces were averaged before fitting.
From reference 12.
Iron release kinetics from hTF/sTFR Δ757–760 complexes
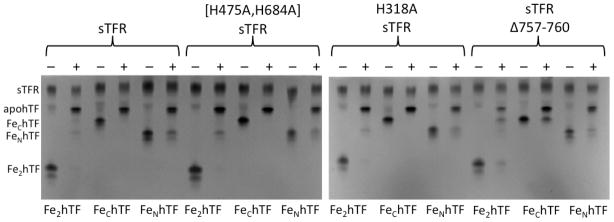
The C-terminal residues of one TFR monomer are positioned between and make contact with both the C1 subdomain of hTF and the other TFR monomer (designated TFR′, Figure 1E) in the TFR-TFR′-C1 intersection. Kinetic rate constants for conformational changes and iron release from various hTF/sTFR Δ757–760 complexes are shown in Table 3. We were unable to fit the kinetic data for the Fe2hTF/sTFR Δ757–760 complex using the rate constants and two pathway model used for the Fe2hTF/sTFR control complex. Instead, iron release from the Fe2hTF/sTFR Δ757–760 complex was preceded by a very rapid conformational change (k1), followed by iron release from the N-lobe (k2=k1N) and very slow iron release from the C-lobe of hTF (k3=k2C). The specific assignment of these rate constants is established by urea gel analysis of the Fe2hTF/sTFR Δ757–760 complex (Figure 4), which indicates that, because of the small rate constant, a k2C=1.0 min−1, a population of FeChTF remains after subjecting the Fe2hTF/sTFR Δ757–760 complex to iron removal buffer. Thus, iron release from this construct proceeds by way of a single pathway (k1N→k2C), eliminating the need to include the other pathway (k1C→k2N) in fitting the kinetic data for the Fe2hTF/sTFR Δ757–760 complex.
Table 3.
Iron release kinetics from sTFR Δ757–760 complexes.a
| Complex | k1C (min−1) | k2N (min−1) | |
|---|---|---|---|
| Fe2hTF/sTFRb | 5.5 ± 0.9 | 1.4 ± 0.2 | |
|
| |||
|
k1 (min−1) Conformational Change |
k2=k1N (min−1) | k3=k2C (min−1) | |
|
| |||
| H349A Fe2hTF/sTFRc | 23.7 ± 4.6 | 6.7 ± 0.3 | 0.61 ± 0.02 |
| Fe2hTF/sTFR Δ757–760 | 89.4 ± 2.0 | 5.2 ± 0.2 | 1.0 ± 0.1 |
|
| |||
| Complex |
k1 (min−1) Conformational Change |
k2C (min−1) Fe3+ Release |
|
|
| |||
| FeChTF/sTFRa | 20.6 ± 1.2 | 7.2 ± 0.4 | |
| H349A FeChTF/sTFRd | 9.2 ± 0.8 | 1.4 ± 0.5 | |
| FeChTF/sTFR Δ757–760 | 48.9 ± 3.7 | 3.2 ± 0.1 | |
|
| |||
| Complex |
k1 (min−1) Conformational Change |
k2N (min−1) Fe3+ Release |
|
|
| |||
| FeNhTF/sTFRa | 22.0 ± 0.7 | 1.7 ± 0.6 | |
| FeNhTF/sTFR Δ757–760 | 31.9 ± 0.5 | 3.4 ± 0.1 | |
Figure 4.
Urea gel analysis of hTFs in the presence of the sTFR and sTFR mutants. Samples were electrophoresed before (−) and after (+) incubation with iron removal buffer (100 mM MES, pH 5.6, containing 300 mM KCl and 4 mM EDTA) for 5 min.
As mentioned, iron release from the FeChTF/sTFR complex (k2C) is preceded by a conformational change (k1). While the rate constant for the conformational change is markedly increased (48.9 vs. 20.6 min−1) in the FeChTF/sTFR Δ757–760 complex relative to the control FeChTF/sTFR, the rate constant k2C for iron release is significantly decreased (3.2 vs. 7.2 min−1, Table 3). Additionally, in the FeNhTF/sTFR Δ757–760 complex, the rate constant for the initial conformational change (k1) is also increased (31.9 vs. 22.0 min−1), but the rate constant for iron release (k2N) is doubled in comparison to the FeNhTF/sTFR control (3.4 vs. 1.7 min−1) (Table 3). Thus, opposite effects of the Δ757–760 mutation on iron release are seen for the two monoferric hTFs.
Kinetic analysis of the FeChTF/sTFR Δ757–760 complex as a function of pH
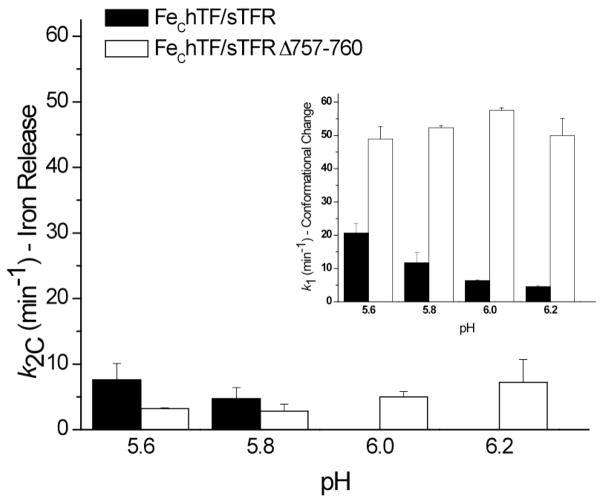
The pH sensitivity of the FeChTF/sTFR Δ757–760 complex was examined by monitoring the iron release kinetics of the complex between pH 5.6 to 6.2. Both rate constants, k1 and k2C, of the FeChTF/sTFR complex are sensitive to pH: the rate constant reporting the conformational change (k1) is decreased as the pH increases from 5.6–6.2 (Figure 5, inset), while iron release does not occur at pH 6.0 and above (Figure 5). The pH profile of the FeChTF/sTFR Δ757–760 complex differs drastically in comparison to the control: the already rapid rate constant for conformational change increases slightly from pH 5.6 to 6.0, as does the rate constant for iron release (Figure 5).
Figure 5.
Effect of pH on rate constants from FeChTF/sTFR and FeChTF/sTFR Δ757–760 complexes. Rate constants (k1 = Conformational Change (inset) and k2C = Iron Release) ± 95% confidence intervals as a function of pH are shown for the FeChTF/sTFR control (black) and the FeChTF/sTFR Δ757–760 mutant (white).
DISCUSSION
The homodimeric TFR ectodomain is comprised of three distinct domains in each monomer. The protease-like domain (domain I, 121–188 and 384–606) contains two of the four Ca2+ binding residues (Glu465 and Glu468, Figure 1B) mutated in the present work, as well as, one of the histidines (His475, Figure 1C) comprising half of the histidine cluster that forms as a result of hTF binding. The apical domain (domain II, 189–383), in which a long loop (residues 275 to 338) containing His318 resides (Figure 1E), is not directly involved in binding hTF. When hTF binds to the ectodomain of the TFR, part of this loop markedly changes its position moving into proximity of the C-terminal residues of the other TFR monomer (Figure 1F).15 Specifically, Phe316′ moves 8 Å, while His318′ flips directly into the TFR-TFR′-C1 intersection (a movement of nearly 18 Å) bringing it to within 5 Å of the TFR C-terminus.15 Lastly, the helical domain (domain III, 607–760) responsible for dimerization, contains the other histidine (His684, Figure 1C) in the interface histidine cluster and the four C-terminal amino acids of the TFR (Asp757, Asn758, Glu759 and Phe760, Figure 1E).
Glu465 and Glu468 in the protease-like domain of the sTFR, and Asp307, Thr310 and Phe313 in the apical domain, participate in the octahedral coordination of a metal ion (identified as Ca2+ in BHK derived sTFR)(Figure 1B). The essential role of the bound metal in stabilizing the structure of the TFR is demonstrated by the poor production of the [E465A,E468A] sTFR mutant and the inability to isolate any non-aggregated sTFR, preventing further experimentation with this mutant. Although the exact role of the metal in vivo remains unclear, our results suggest that the coordination of this Ca2+ is important to the structural integrity of the sTFR.
The histidine cluster (His475 and His684 from each TFR monomer) formed when either HFE or hTF binds to the TFR (Figure 1C) has been suggested to be a pH-inducible motif which may trigger a conformational event in the TFR involved in the release of HFE or of iron from hTF within the endosome.15, 16 However, as previously noted,15 this histidine cluster, buried deep within the interface between the two TFR monomers, may be inaccessible to changes in pH. The relatively small differences in most of the kinetic parameters in the [H475A,H684A] sTFR complexes (Table 2) are inconsistent with the suggestion that this cluster is involved in the mechanism of pH-induced iron release, at least under the tested conditions (pH 5.6 with 300 mM KCl and 4 mM EDTA).
As mentioned, due to the inherent flexibility of the C-terminus, at neutral pH the final two amino acids of the TFR (Glu759 and Phe760) are not observed in the FeNhTF/sTFR crystal structure.15 Significantly, the C-terminus of one TFR monomer is located in the center of the TFR-TFR′-C1 intersection, directly between TFR′ residue His318 and hTF residue His349 shown to be a pH-inducible switch required for TFR stimulated iron release from the C-lobe (Figure 1E).15, 18 We propose that the proximity of these two pH-sensitive histidine residues to the C-terminus of the other TFR monomer provides an explanation for the many rather subtle kinetic effects discussed below.
A curious feature of the kinetic profiles of all hTF/sTFR complexes is the initial drop in the fluorescent signal as a result of the decrease in pH to 5.6. We have previously reported this phenomenon 21 and because it is only observed in sTFR containing samples and in sTFR alone, assigned it to a pH-induced conformational change in the sTFR. The significantly longer duration of this feature in the H318A sTFR mutant alone or complexed with Fe2hTF is clearly shown in Figure 3 and is consistent with protonated His318 promoting and reporting this change. When hTF binds to the ectodomain of the TFR, His318 moves into the interface, close to TFR residues Trp641 and Trp740 of the TFR′ monomer (~3.6 and 4.1 Å, respectively, Figure 1 of the Supporting Information). Because protonated histidine residues quench tryptophan fluorescence by electron transfer,25 it is reasonable to assign the pH-induced decrease in fluorescence of sTFR largely to His318 quenching of nearby Trp641 and/or Trp740 at low pH.
A new and interesting finding is that the binding of Fe2hTF significantly prolongs the duration of the fluorescent quenching event in the sTFR (~3.4 fold), a phenomenon that is even more pronounced in the presence of the H318A sTFR mutation (~6.4 fold, Figure 3). In contrast, binding of Fe2hTF does not increase the duration of the pH-inducible change in the truncated sTFR (Δ757–760) (Figure 3). Based on these results, we suggest that the C-terminus of the TFR interacts with His318′ of the TFR′ at neutral pH either through a hydrogen bond with Asn758 or a weak hydrophobic interaction with Phe760, but only when iron containing hTF is bound. This interaction between the C-terminus and His318 is likely destabilized at low pH (5.6), thereby freeing the C-terminus to interact with hTF via residue His349. Further support for this suggestion is provided by the kinetics of iron release from the H318A sTFR containing complexes (Table 1). Specifically, the conformational change that is normally found prior to iron release from the C-lobe is not observed in the FeChTF/H318A sTFR complex. In a manner reminiscent of the H349W or H349Y hTF FeChTF mutant/sTFR complexes, which also lack the conformational change,18 iron release from the FeChTF/H318A sTFR complex can occur without the pH-induced conformational change. Thus, elimination of the His318:C-terminus interaction as in the H318A sTFR mutant enables the C-terminus to interact with His349 hTF even at neutral pH.
Whereas the conformational change preceding iron release from the FeChTF/sTFR complex is not observed in the FeChTF/H318A sTFR complex, it is accelerated more than 2-fold in the FeChTF/sTFR Δ757–760 complex, k1 = 20.6 vs. 48.9 min−1 (Table 3). This finding supports the link between His318 of TFR′ and the C-terminus of the other TFR monomer and provides further evidence that the two interact and have an effect on the conformational change.
In contrast to the FeChTF/H318A sTFR complex, the conformational change is observed and little affected in the other monoferric complex, FeNhTF/H318A sTFR complex (Table 1). These results imply that the conformational change preceding iron release in the two monoferric complexes has a different physical basis despite having nearly identical rate constants (20.6 min−1 for the FeChTF/sTFR complex versus 22.0 min−1 for the FeNhTF/sTFR complex, Table 1).
Interestingly, since the C-terminus of the sTFR (residues 757–760) is only proposed to interact with the C1 subdomain of the C-lobe of hTF and residues of the protease-like domain of the other TFR monomer, it was expected that truncation of the sTFR C-terminus would not affect iron release from the N-lobe of FeNhTF. However, the rate constant for the initial conformational change (k1) is increased by 45% and the rate constant for iron release (k2N) is doubled in the FeNhTF/sTFR Δ757–760 complex in comparison to the FeNhTF/sTFR control (Table 3). These results indicate communication between the two lobes of hTF within the hTF/TFR complex. Given that His318 interacts with the C-terminus of the other TFR monomer at pH 7.5, removal of the C-terminus eliminates this interaction, allowing His318 to interact with Trp641 and/or Trp740 even at neutral pH (Figure 1 of the Supporting Information). Of possible significance, Trp641 is located on αIII-3 in the helical domain of the TFR which interacts with both the N1 and C1 subdomains in the FeNhTF/sTFR crystal structure.14 Thus, we postulate that the absence of the interaction between His318 and the C-terminus in the FeNhTF/sTFR Δ757–760 complex accelerates both the rate of conformational change and iron release from the N-lobe in comparison to the FeNhTF/sTFR control via the interaction between His318 and Trp641 within αIII-3.
It is well established that a pH-induced conformational change involving hTF residue His349 drives iron release from the C-lobe.14; 17 A previous in silico model26 of the hTF/TFR complex suggested that hTF C1-subdomain residue His349 interacts with hTF C2-subdomain residue Lys511 through a weak electrostatic interaction between the lone pair of electrons on the ND1 of His349 and the cation of Lys511. However, due to the ~4.0–5.0 Å distance between His349 and Lys511 in the recent crystal structure of diferric hTF (PDB ID: 3V83), this interaction is unlikely. Interestingly, mutation of Lys511 to alanine increases the rate of iron release from the C-lobe, by an unknown mechanism.16 Closer examination of the crystal structure of diferric hTF reveals that Lys511 lies within 3.2 Å of and likely forms a salt bridge with hTF C1-subdomain residue Glu372, possibly explaining the observed effects of the K511A mutation.
Notably, the rate constant for iron release from the control FeChTF/sTFR complex titrates with an apparent pKa of 5.8–5.9, consistent with titration of one or more histidine residues (specifically hTF residue His349).18 Intriguingly, the pH profile of the rate constants for the FeChTF/sTFR Δ757–760 complex (Figure 5) follows the same trend as the pH profile for the H349A FeChTF/sTFR complex.18 Moreover, the kinetics of iron release from the Fe2hTF/sTFR Δ757–760 complex (Table 3) are very similar to the published kinetic data for the H349A Fe2hTF/sTFR complex15: both exclusively follow the k1N→ k2C pathway preceded by a rapid conformational change (Table 3). Additionally, the rate constant for iron release from the C-lobe, k2C, is markedly reduced in the Fe2hTF/sTFR Δ757–760 complex. Hence, the inability to anchor the protonated His349 (as in the H349A hTF or the truncated sTFR) has a pronounced effect on iron release. These findings strongly support the suggestion that the TFR C-terminus stabilizes protonated His349 and promotes iron release from the C-lobe of hTF at pH 5.6 either through a cation-π interaction with the C-terminal Phe760 of the sTFR, or through formation of a salt bridge with Glu759 or Asp757. Individual mutation of the four C-terminal residues is required to establish the contribution of each to the TFR facilitated mechanism of iron release from the C-lobe of hTF.
In summary, the present data suggest that iron release from the hTF/TFR complex is controlled by a relay within the TFR-TFR′-C1 intersection (Figure 1E). The absence of the coordinated Ca2+ ion (Figures 1B and 2) or a glycosylation site in this region (Asn317)23 both have a negative effect on the structural stability of this region. At pH 7.5, His318′ (TFR′) interacts with the C-terminus of the other TFR monomer (Figure 1E). Upon exposure to the acidic environment within the endosome, the interaction between His318′ and the C-terminus is severed by the protonation of His318′, allowing His318′ to interact with and quench nearby tryptophan residues 641 and/or 740 (Figure 3 and Figure 1 of the Supporting Information). A conformational change which promotes receptor-stimulated iron release from the C-lobe of hTF (Figures 4 and 5) is then triggered by the interaction formed between protonated hTF residue His349 and the TFR C-terminus. Communication of these kinetic events to the N-lobe of hTF, possibly through helix αIII-3 of the TFR to which both the N1 and C1 subdomains are bound, prompts iron release from the N-lobe. Collectively, these results help to establish a molecular basis for the pH-induced events that dictate efficient release of iron from each lobe within the endosome in a physiologically relevant timeframe.
Supplementary Material
Acknowledgments
This work was supported by U. S. Public Service Grant R01 DK 21739 to A.B.M. and the National Institute of General Medical Sciences Grant R37-GM-20194 to N.D.C. A.N.S. is funded by an AHA Predoctoral Fellowship (10PRE4200010).
We would like to thank Joe Klaus for technical assistance with the immunoblot and Dr. Brian E. Eckenroth for helpful discussions.
ABBREVIATIONS
- hTF
human serum transferrin
- TFR
transferrin receptor
- apohTF
iron-free hTF
- Fe2hTF
recombinant N-terminal hexa-His tagged non-glycosylated diferric hTF
- sTFR
glycosylated N-terminal hexa-His tagged soluble recombinant transferrin receptor (residues 121–760)
- FeNhTF
recombinant N-terminal hexa-His tagged non-glycosylated monoferric hTF that binds iron only in the N-lobe (Y426F/Y517F mutations prevent iron binding in C-lobe)
- FeChTF
recombinant N-terminal hexa-His tagged non-glycosylated monoferric hTF that binds iron only in the C-lobe (Y95F/Y188F mutations prevent iron binding in N-lobe)
- BHK cells
baby hamster kidney cells
- t1/2
half-life
- UG
Ultroser G
- V0
void volume
Footnotes
SUPPORTING INFORMATION AVAILABLE
Derivation of an equation for an A→B model including initial decay, the recombinant production levels of all sTFR mutants, oligonucleotide primers used to prepare the sTFR mutants, a representation of the FeNhTF/sTFR crystal structure showing the location of the TFR-TFR′-C1 intersection including residues Trp641 and Trp740, as well as an example of the analysis of the initial quench in tryptophan fluorescence (fit to the A → B model which also includes the initial decay term). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hall DR, Hadden JM, Leonard GA, Bailey S, Neu M, Winn M, Lindley PF. The crystal and molecular structures of diferric porcine and rabbit serum transferrins at resolutions of 2.15 and 2.60 Å, respectively. Acta Crystallogr D Biol Crystallogr. 2002;58:70–80. doi: 10.1107/s0907444901017309. [DOI] [PubMed] [Google Scholar]
- 2.Wally J, Halbrooks PJ, Vonrhein C, Rould MA, Everse SJ, Mason AB, Buchanan SK. The crystal structure of iron-free human serum transferrin provides insight into inter-lobe communication and receptor binding. J Biol Chem. 2006;281:24934–24944. doi: 10.1074/jbc.M604592200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fenton HJH. On a new reaction of tartaric acid. Chem News. 1876;33:190. [Google Scholar]
- 4.Fenton HJH. The oxidation of tartaric acid in presence of iron. Chem Soc Proc. 1893;9:113. [Google Scholar]
- 5.Aisen P, Enns C, Wessling-Resnick M. Chemistry and biology of eukaryotic iron metabolism. Int J Biochem Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- 6.Klausner RD, Van Renswoude J, Ashwell G, Kempf C, Schechter AN, Dean A, Bridges KR. Receptor-mediated endocytosis of transferrin in K562 cells. J Biol Chem. 1983;258:4715–4724. [PubMed] [Google Scholar]
- 7.Bali PK, Zak O, Aisen P. A new role for the transferrin receptor in the release of iron from transferrin. Biochemistry. 1991;30:324–328. doi: 10.1021/bi00216a003. [DOI] [PubMed] [Google Scholar]
- 8.Ohgami RS, Campagna DR, Greer EL, Antiochos B, McDonald A, Chen J, Sharp JJ, Fujiwara Y, Barker JE, Fleming MD. Identification of a ferrireductase required for efficient transferrin-dependent iron uptake in erythroid cells. Nat Genet. 2005;37:1264–1269. doi: 10.1038/ng1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA. Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature. 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 10.Dautry-Varsat A, Ciechanover A, Lodish HF. pH and the recycling of transferrin during receptor-mediated endocytosis. Proc Natl Acad Sci U S A. 1983;80:2258–2262. doi: 10.1073/pnas.80.8.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leverence R, Mason AB, Kaltashov IA. Noncanonical interactions between serum transferrin and transferrin receptor evaluated with electrospray ionization mass spectrometry. Proc Natl Acad Sci U S A. 2010;107:8123–8128. doi: 10.1073/pnas.0914898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrne SL, Chasteen ND, Steere AN, Mason AB. The unique kinetics of iron-release from transferrin: The role of receptor, lobe-lobe interactions and salt at endosomal pH. J Mol Biol. 2010;396:130–140. doi: 10.1016/j.jmb.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence CM, Ray S, Babyonyshev M, Galluser R, Borhani DW, Harrison SC. Crystal structure of the ectodomain of human transferrin receptor. Science. 1999;286:779–782. doi: 10.1126/science.286.5440.779. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Y, Zak O, Aisen P, Harrison SC, Walz T. Structure of the human transferrin receptor-transferrin complex. Cell. 2004;116:565–576. doi: 10.1016/s0092-8674(04)00130-8. [DOI] [PubMed] [Google Scholar]
- 15.Eckenroth BE, Steere AN, Chasteen ND, Everse SJ, Mason AB. How the binding of human transferrin primes the transferrin receptor potentiating iron release at endosomal pH. Proc Natl Acad Sci U S A. 2011;108:13089–13094. doi: 10.1073/pnas.1105786108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett MJ, Lebron JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46–53. doi: 10.1038/47417. [DOI] [PubMed] [Google Scholar]
- 17.Steere AN, Miller BF, Roberts SE, Byrne SL, Chasteen ND, Smith VC, MacGillivray RTA, Mason AB. Ionic residues of human serum transferrin affect binding to the transferrin receptor and iron release. Biochemistry. 2012;51:686–694. doi: 10.1021/bi201661g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steere AN, Byrne SL, Chasteen ND, Smith VC, MacGillivray RT, Mason AB. Evidence that His349 acts as a pH-inducible switch to accelerate receptor-mediated iron release from the C-lobe of human transferrin. J Biol Inorg Chem. 2010;15:1341–1352. doi: 10.1007/s00775-010-0694-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steere AN, Byrne SL, Chasteen ND, Mason AB. Kinetics of iron release from transferrin bound to the transferrin receptor at endosomal pH. Biochim Biophys Acta. 2012;1820:326–333. doi: 10.1016/j.bbagen.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bali PK, Aisen P. Receptor-induced switch in site-site cooperativity during iron release by transferrin. Biochemistry. 1992;31:3963–3967. doi: 10.1021/bi00131a011. [DOI] [PubMed] [Google Scholar]
- 21.James NG, Byrne SL, Mason AB. Incorporation of 5-hydroxytryptophan into transferrin and its receptor allows assignment of the pH induced changes in intrinsic fluorescence when iron is released. Biochim Biophys Acta. 2008;1794:532–540. doi: 10.1016/j.bbapap.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.James NG, Byrne SL, Steere AN, Smith VC, MacGillivray RT, Mason AB. Inequivalent contribution of the five tryptophan residues in the C-lobe of human serum transferrin to the fluorescence increase when iron is released. Biochemistry. 2009;48:2858–2867. doi: 10.1021/bi8022834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrne SL, Leverence R, Klein JS, Giannetti AM, Smith VC, MacGillivray RT, Kaltashov IA, Mason AB. Effect of glycosylation on the function of a soluble, recombinant form of the transferrin receptor. Biochemistry. 2006;45:6663–6673. doi: 10.1021/bi0600695. [DOI] [PubMed] [Google Scholar]
- 24.Mason AB, Halbrooks PJ, Larouche JR, Briggs SK, Moffett ML, Ramsey JE, Connolly SA, Smith VC, MacGillivray RT. Expression, purification, and characterization of authentic monoferric and apo-human serum transferrins. Protein Expr Purif. 2004;36:318–326. doi: 10.1016/j.pep.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 25.Callis PR, Liu T. Quantitative prediction of fluorescence in proteins. J Phys Chem B. 2004;108:4248–4259. [Google Scholar]
- 26.Sakajiri T, Yamamura T, Kikuchi T, Yajima H. Computational structure models of apo and diferric transferrin-transferrin receptor complexes. Protein J. 2009;28:407–414. doi: 10.1007/s10930-009-9208-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.