Summary
Nutrient sensing and the capacity to respond to starvation is tightly regulated as a means of cell survival. Among the features of the starvation response are induction of both translational repression and autophagy. Despite the fact that intracellular parasite like Toxoplasma gondii within a host cell predicted to be nutrient rich, they encode genes involved both in translational repression and autophagy. We therefore examined the consequence of starvation, a classic trigger of autophagy, on intracellular parasites. As expected, starvation results in the activation of the translational repression system as evidenced by elevation of phosphorylated TgIF2α (TgIF2α-P). Surprisingly, we also observe a rapid and selective fragmentation of the single parasite mitochondrion that leads irreversibly to parasite death. This profound effect was dependent primarily on the limitation of amino acids and involved signaling by the parasite TOR homolog. Notably, the effective blockade of mitochondrial fragmentation by the autophagy inhibitor 3-methyl adenine (3-MA) suggests an autophagic mechanism. In the absence of a documented apoptotic cascade in T. gondii, the data suggest that autophagy is the primary mechanism of programmed cell death in T. gondii and potentially other related parasites.
Keywords: Toxoplasma, starvation, autophagy, mitophagy, eIF2α, TOR
Introduction
Autophagy is a catabolic process in eukaryotic cells involved in the targeted degradation of cellular organelles and the cytoplasm (Inoue et al., 2010, Mizushima et al., 2010). Autophagy is widely regarded to be critical for cell survival under starvation conditions and for the turnover of defunct organelles (Kroemer et al., 2010, Kristensen et al., 2008). The molecular machinery driving this process exhibits a remarkable level of conservation from yeast to mammals (Yorimitsu et al., 2005). The process of autophagy is triggered by the formation of unique double membranes that mature into autophagosomes and eventually fuse with lysosomes, resulting in the degradation of captured components (Weidberg et al., 2011). The execution of autophagy is dependent on 31 ATG proteins described in yeast that are largely conserved through mammals (Klionsky et al., 2010). Initial bioinformatic studies in several parasitic protozoa suggest that consistent with their evolutionary antiquity, homologs of many but not all the yeast ATG genes are evident (Duszenko et al., 2011, Herman et al., 2006). This suggests that parasitic protozoa may possess a progenitor of autophagy as defined in yeast, or, alternatively, use the machinery for parasite specific functions (Duszenko et al., 2011, Denninger et al., 2008, Luder et al., 2010). With this study we sought to establish the response of Toxoplasma gondii to starvation stress in order to characterize the response of the parasite to what is considered a classic trigger of autophagy.
The protozoan parasite Toxoplasma gondii is an important opportunistic pathogen in immune compromised individuals as well as a significant cause of morbidity and mortality when vertically transmitted during pregnancy (Tenter et al., 2000, Montoya et al., 2008). Within the mammalian host the parasite exhibits rapid growth as a tachyzoite defining the acute phase of the infection before transforming into the slow growing tissue cyst form (Tenter et al., 2000). The specific triggers for differentiation appear to be linked to the establishment of immunity although recent evidence points also to physiological and metabolic triggers (Weiss et al., 2000, Radke et al., 2006). Thus while the intracellular niche that T. gondii establishes is not typically considered to be nutrient limiting, specialized cases such as tryptophan limitation in activated macrophages or interferon stimulated fibroblasts are known to limit parasite growth (Pfefferkorn, 1984, Pfefferkorn et al., 1986, Silva et al., 2002). Under such conditions, induction of autophagy within the parasite could serve a protective role as seen in other systems (Kroemer et al., 2010). Alternatively, activation of autophagy may promote death by triggering the targeted degradation of critical cellular components (Kourtis et al., 2009, Levine et al., 2009). The notion of autophagy as a mechanism of both cell survival and death is emerging in studies from yeast to mammals (Kourtis et al., 2009, Levine et al., 2009, Gozuacik et al., 2007).
In this study we directly examine the impact of nutrient limitation on the induction of autophagy in T. gondii. We were specifically interested in determining whether potential autophagy-related processes contribute to parasite survival or death. The absence of a bona fide apoptotic death pathway in protozoa and yeasts suggests autophagy may play a role in programmed cell death (Luder et al., 2010). We also probe how nutrient limitation and consequent translational repression induce the cell biological consequences of starvation. We find that starvation induces the rapid and selective degradation of the single parasite mitochondrion culminating in parasite death. Our findings represent the first functional description of autophagy in an Apicomplexan parasite and integrate key elements of the parasite response to nutrient limitation. These findings effectively establish an autophagy mediated programmed cell death pathway in T. gondii, and potentially other protozoa. The study of autophagy in parasites as a potential mechanism of death addresses a vital question in the development of new drugs (Brennand et al., 2011, Paguio et al., 2011).
Results
Intracellular Starvation activates stress-induced translational arrest in T. gondii
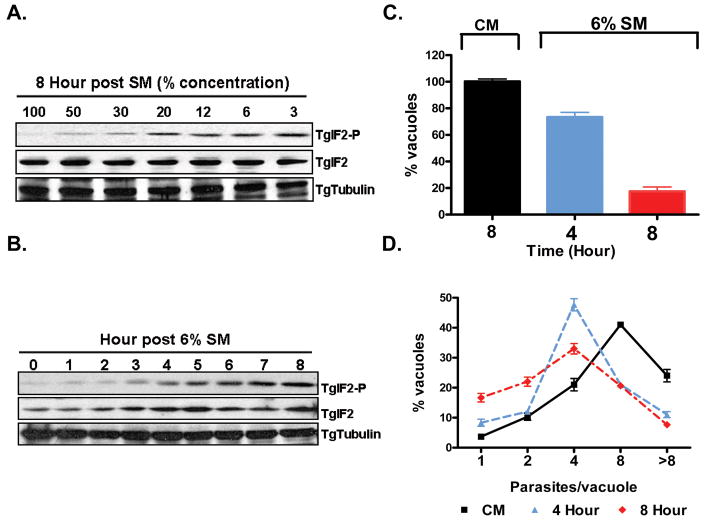
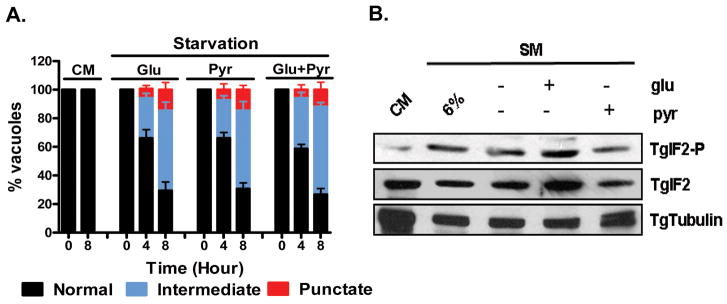
Extracellular T. gondii are reportedly able to activate the translational arrest response (Wek et al., 2006) defined by an increase in TgIF2α phosphorylation (Joyce et al., 2010, Sullivan et al., 2004). As this pathway is activated in diverse systems in response to nutrient limitation (Wek et al., 2006), we examined the effect of starvation on intracellular T. gondii by monitoring TgIF2α phosphorylation (TgIF2α-P) levels as complete media (CM, see experimental procedures) was progressively diluted with glucose free HBSS (see legend, Figure1). HFF cells infected for 24 hours in complete media were exposed to diluted media for 8 hours. Consistent with other cell types, levels of TgIF2α-P for parasite infected HFF cells increased in response to nutrient dilution (Figure 1A). While maximal TgIF2α-P levels are evident in media diluted to 2% (not shown), the loss of the cell monolayer at this dilution indicated toxicity to the host cell (Supplemental Figure 1). We therefore selected media diluted to 6% (6% Starvation Medium, 6%-SM) as our standard treatment (Supplemental Fig 1A). Furthermore, with 6 % SM, we do not observe activation of autophagy in host cells as defined by appearance of LCIII (lipid modified ATG8) (Mizushima et al., 2010) (data not shown). Finally, the level of host cell death, measured by the release of lactate dehydogenase (LDH) was identical for both complete and 6%-SM media in uninfected and infected cells respectively (Supplemental Figure1B).
Figure 1.
Nutrient limitation induces translational repression impacting both invasion and growth. (A) Dilution of complete media (CM) results in starvation-induced stress detected by the induction of increasing levels phosphorylated TglF2α (TgIF2-P), an indicator of translational repression. Cells were exposed for 8 hours to 100% CM (left), 50 % CM / 50 % glucose free HBSS (2nd lane) or 30 % / 70 %, 20% / 80 %, 12 % / 88 %, 6 % / 94 %, and 3 % / 97 %, respectively, western blots were run as described [methods], and samples probed with antibody specific to TgIF2α-P (top) TgIF2α (middle) or TgTubulin (bottom) (B) Kinetics of TgIF2α phosphorylation in 6% starvation media (6%-SM) reveal the onset of translational repression between 3 and 8 hours after transfer to nutrient deficient conditions. (C) Control parasites and parasites subjected to starvation for 4 or 8 hours were liberated from the host cells by syringe passage and used to infect new cell monolayers (2×105/coverslip). The infected monolayers were fixed and stained with SAG1 and GRA3 (see Figure2 following for representative images). The mean number of vacuoles (based on GRA3 staining of the PVM) per field was used as a measure of invasion potential (n=>20/sample). Starvation for as little as 4 hours was found to reduce the invasion capacity by 33% while a reduction of invasion efficiency of 85% was observed following 8 hours under nutrient limiting conditions. Mean number of vacuoles per field established by control parasites was 7.22 +/-0.23 (D) Previously starved parasites that productively infected a fresh monolayer exhibit growth inhibition proportional to the duration of starvation. The distribution of parasite number per vacuole, a reliable measure of intracellular growth, was determined in untreated parasites (CM, solid squares) and parasites previously exposed to 6%-SM for 4 (blue triangles) and 8 (red diamonds) hours. A minimum of 100 vacuoles per experiment were counted for each sample. The data in C and D are presented as the mean ± S.D. from three independent experiments.
The kinetics of phospho-TgIF2α generation were established following exposure of parasite infected cells as above in 6%-SM. TgIF2α-P levels are apparent as early as 3-hour post starvation induction and continued to increase in a time-dependent manner (Figure 1B).
Starved parasites exhibit defective invasion and growth retardation
To assess the consequence of starvation in terms of parasite infection and growth ability, we examined the competence of starved parasites to infect new host cells and their growth thereafter. HFF cells infected for 24 hours in complete media were incubated in 6%-SM for either 4 or 8 hours. The intracellular parasites were liberated by syringe passage and enumerated. Equal numbers of parasites were added to fresh monolayers grown on cover slips and allowed to infect for 24 hours in CM. The number of vacuoles per field was used as a measure of the invasive capacity. In addition, the distribution of parasite number per vacuole served as a measure of parasite growth.
Infection of fresh HFF monolayers with parasites maintained throughout in CM resulted in the establishment of on average 7.2 vacuoles per field (defined as 100%; Figure 1C). The effect of 6%-SM treatment for 4 and 8 hours results in 25% and 80% reduced vacuole counts, respectively (Figure 1C). In addition, vacuoles established by starved parasites exhibited growth retardation relative to those grown in CM throughout (Figure 1D). Both defects are likely due at least in part to translational arrest indicated by elevated TgIF2α-P (Figure 1B) (Joyce et al., 2010). Additionally, starvation may induce cellular and physiological changes outside of translational arrest that also promote these defects.
Starvation results in selective mitochondrial fragmentation in T. gondii
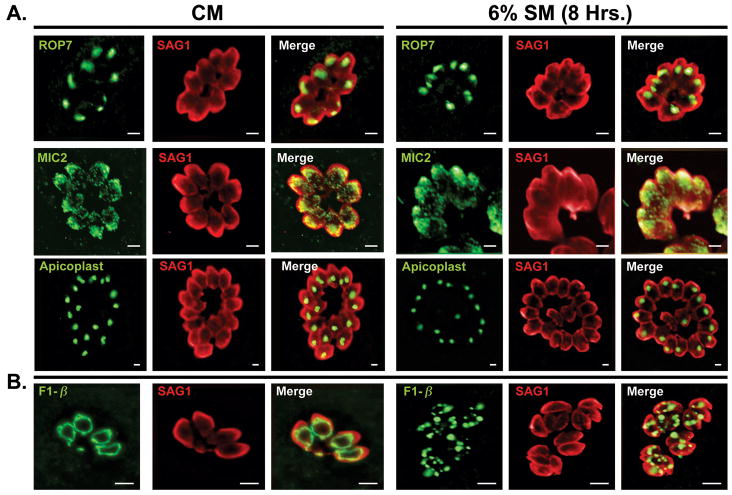
In addition to translational arrest, the selective degradation of expendable organelles provides a means of coping with starvation (Kanki et al., 2008, Kristensen et al., 2008). To address whether starvation triggered organelle degradation in T. gondii, we examined the morphology of key parasite organelles following 8 hours in 6%-SM. Since significant inhibition of parasite invasion was a consequence of intracellular starvation (Figure 1C), we examined the morphology of rhoptries and micronemes (Carruthers et al., 2007). Interestingly, the organization of these organelles were unaffected by exposure to 6%-SM for 8 hours (Figure 2A), even though they play a critical role in parasite attachment and invasion (Carruthers et al., 2007). Similarly the apicoplast, a relict plastid-derived organelle (Lim et al., 2010, Waller et al., 2005) appeared unaffected in starved parasites (Figure 2A).
Figure 2.
Impact of starvation on parasite organelle integrity. A) Incubation of established vacuoles in CM (left) or 6% SM (right) for 8 hours does not have any effect on the integrity or organization of the rhoptries (ROP7), micronemes (MIC2) or the apicoplast (anti-Atrx1) (B) Under normal growth conditions (CM) the tachyzoite mitochondrion possess a characteristic ring shape visualized using anti-F1β (far left). Treatment with 6% SM for 8 hours results in mitochondrial fragmentation (right hand side). The surface marker SAG1 is used as a counter-stain. Scale bars:(A-2 μm, B-5 μm).
The single parasite mitochondrion (Melo et al., 2000, Seeber et al., 1998, Vivier et al., 1972), detected with an antibody against the F1β–subunit of the mitochondrial H+ ATPase, normally assumes a ring-shaped appearance when T. gondii are grown in CM (Figure 2B, F1β). In contrast, a substantial number of vacuoles in 6%-SM displayed punctate or otherwise abnormal mitochondrial morphology (Figure 2B and see below). Mitochondrial fragmentation was observed to increase dramatically as a function of dilution of the culture media (Supplemental Figure1A). Increased fragmentation correlated with increased TgIF2α-P levels (Figure 1A) suggesting the effect was also a consequence of reduced nutrient availability. In addition, starvation induced mitochondrial starvation was seen not only in the Type I RHΔHX strain used here but also in the avirulent cyst forming Type II ME49 (data not shown). Finally, RH parasites expressing HXGPRT responded identically as does the deletion strain (data not shown).
Starvation induced mitochondrial fragmentation occurs sequentially
We next examined the kinetics of mitochondrial fragmentation (Figure. 3). Infected cell monolayers were fixed at hourly intervals and the morphology of the mitochondrion quantified (see legend, Figure 3). Progression from normal, ring-shaped morphology to puncta occurred via intermediate forms. These forms appeared, in turn, as linear mitochondria, patterns reminiscent of beads on a string, and then as dumbbell shapes with bulbous ends connected by thin segments (Figure 3A, F1β top to bottom).
Figure 3.
Sequential and progressive starvation induced mitochondrial fragmentation. (A) Mitochondrial fragmentation from normal ring morphology to punctate occurs via intermediate forms. Intermediate forms include linear mitochondria (2nd from top), a beads on a string appearance (3rd from top), and linear forms with bulbous ends (4th from top). Upon continued starvation parasites showed punctate mitochondria (5th from top) and then often lost their mitochondrion entirely (bottom). Scale bar each panel = 2 μm. (B) Quantification and kinetics of starvation induced mitochondrial fragmentation indicate that the intermediate forms precede the detection of punctate mitochondria which in turn precede the apparent loss of the organelle. Notably the mitochondrial morphology within individual parasites in a given vacuole all share the same morphology. (C) Parasites with abnormal mitochondrial are unable to invade a fresh monolayer. Parasites harvested from infected cells incubated in either complete medium (CM) or 6%-starvation medium (6% SM) were syringe passaged and served as the inoculum for a fresh monolayer. Roughly 75% of the 6% SM – incubated parasites possessed disrupted mitochondria. Notably, only parasites containing intact mitochondria were able to invade a fresh monolayer 2 hours following addition (see text). Establishment of the PV was confirmed using the PVM marker GRA3 a minimum of 50 vacuoles were scored for each of 3 replicates.
Normal (ring shaped), punctate, and intermediate morphologies were quantified. As shown in Figure 3B, the progression from normal morphology to puncta occurs via the intermediate forms. The changes in mitochondrial morphology are evident as early as 3 hours following transfer to 6%-SM, with the first appearance of punctate mitochondrial fragments at 4 hours post starvation. By 6 - 7 hrs even punctate forms disappear altogether (Figure3A bottom F1β, Figure3B diamond symbols),
We next tested whether parasites with morphologically abnormal mitochondria were invasion competent. Parasites exposed to 6%-SM for 8 hours harvested from the first monolayer, comprised the inoculum for a second fresh monolayer (Figure 3C). As expected, this inoculum exhibited a distribution of normal and altered mitochondrial morphologies essentially identical to those in the original monolayer (Figure 3C, cytospin). Notably, while 70% of the inoculum had abnormal mitochondria, all of the invasion competent parasites establishing vacuoles with GRA3 positive PVM’s had mitochondria with normal morphology following a 2 hour invasion assay (Figure 3C). Restoration of normal morphology for those observed to be fragmented prior to harvesting, was not observed upon the addition of CM, even after a 20 hour incubation (data not shown). Taken together, these data indicate that fragmentation is irreversible and that the integrity of the mitochondrion is vital for invasion. This is intriguing since localized glycolysis at the site of the motor complex is believed to power the invasion machinery (Pomel et al., 2008), although the ability of T. gondii to use glutamine to power the tricarboxilic cycle provides a bypass pathway in the absence of glucose (Blume et al., 2009).
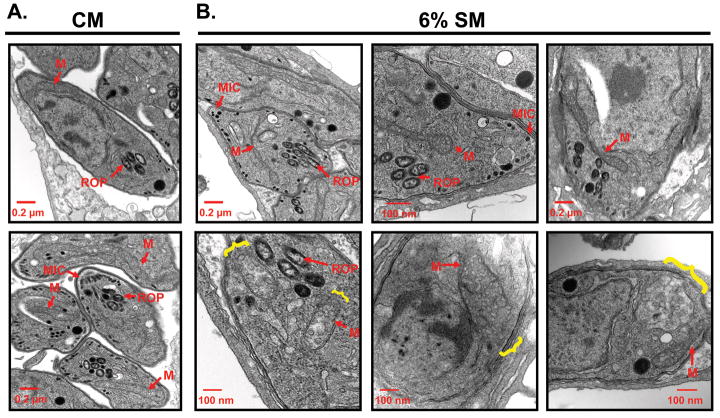
A limitation of immunofluorescence analysis to monitor the integrity of the mitochondrion is the possibility that starvation results in the reorganization and loss of the marker F1β but not the organelle itself. We therefore examined the organization of mitochondria by conventional transmission electron microscopy. Cross sections of the parasite mitochondrion are easily recognized based on their possession of the annular cristae (Melo et al., 2000) (Figure 4A). Also evident are rhoptries and micronemes (Dubey et al., 1998) (Figure 4A). Consistent with the IFA data, no significant changes were observed in the organization of the rhoptries or micronemes in 6%-SM treated cells (Figure 4B).
Figure 4.
Ultrastructural evidence for starvation induced selective mitochondrial fragmentation (A) Representative transmission electron microscopy (TEM) images of T. gondii maintained in CM exhibit normal morphology for all organelles including the mitochondrion (M), rhoptries (ROP), micronemes (MIC) (B) Representative TEM images of T. gondii starved for 8 exhibit an unusual “dumbbell” shaped mitochondrial morphology with club shaped ends and constricted linkers as well (B, top 3 panels). In addition, more profound morphological changes defined by the apparent disruption of the mitochondrial outer membrane accompanied by the “discharge” of cristae are consistent with the loss of mitochondrial integrity (B, bottom 3 panels). TEM analysis also demonstrated no change in the structural integrity of other parasite cellular organelles upon starvation as the rhoptries and micronemes are indistinguishable from control parasites. The yellow brackets outline the loss of mitochondrial integrity.
In contrast to the secretory organelles, dramatic changes consistent with the IFA results (Figure 3A) were noted for the mitochondrion by TEM (Figure 4B). Among these changes were mitochondrial profiles with marked constrictions and bulbous ends (Figure 4B, top 3 panels) as well as mitochondria that appear to have lost their integrity evident from the discontinuity in their limiting membranes as well as an apparent “discharge” of cristae into the cytoplasm (Figure 4B, brackets, lower panels). The ultrastructural data provide compelling evidence that starvation results in a selective fragmentation of the T. gondii mitochondrion but does not directly address the underlying mechanism for the phenotype.
Starvation-induced mitochondrial fragmentation occurs by an autophagic mechanism
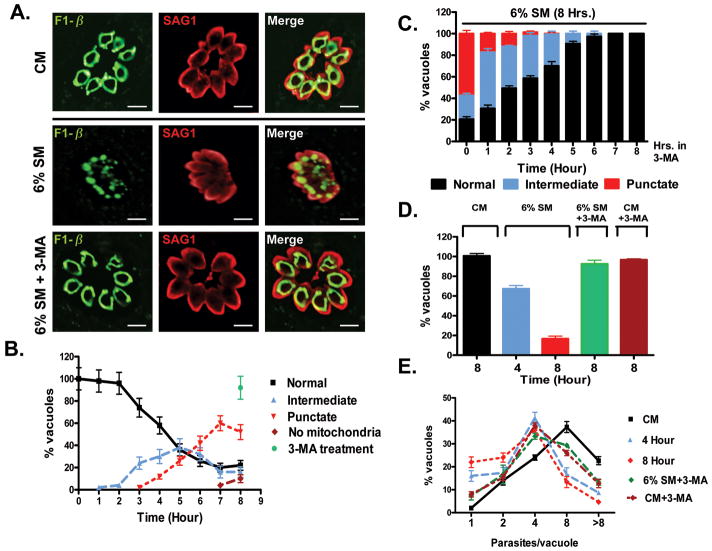
To test whether starvation induced mitochondrial fragmentation in T. gondii involves an autophagic mechanism we investigated the effect of 3-methyl adenine (3-MA). 3-MA is an established selective inhibitor of the Class III PI3 kinase Vps34 and is a reversible inhibitor of autophagy that blocks the recruitment of lipid to the autophagosome (Abeliovich et al., 2001, Seglen et al., 1982, Yue et al., 2010). The T. gondii genome encodes Vps34 homolog (TGGT1_126620) (Supplemental Table 1). 3-MA treatment of infected cells in CM had no effect on the integrity of mitochondria (Supplemental Figure 2A). In contrast, while 6%-SM resulted in mitochondrial fragmentation, the addition to 3-MA to 6%-SM effectively blocked fragmentation (Figure 5A,B). This blockade of starvation induced mitochondrial fragmentation by a selective inhibitor of autophagy indicates that mitochondrial fragmentation occurs by an autophagic mechanism. Notably, the presence of 3-MA had no impact on the morphology of the rhoptries, micronemes or apicoplast in either complete (data not shown) or starvation media (Supplemental Figure 3).
Figure 5.
Starvation-induced mitochondrial fragmentation occurs by an autophagic mechanism. (A) Addition of the autophagy inhibitor 3-methyl adenine (3-MA) to starvation media effectively blocks the starvation-induced fragmentation of the parasite mitochondrion. Scale bar = 5 μm. (B) Quantification of the kinetics of mitochondrial fragmentation in 6%-SM is largely blocked by 3-MA as virtually all vacuoles contain intact mitochondria even following 8 hours in 6%-SM supplemented with 3-MA (filled green circle). (C). Quantification and kinetics of 3-MA reversal experiment. RH infected HFF cells were starved for 8 hours with an addition of 3-MA at hourly intervals. Addition of 3-MA upto 3 hours post starvation completely blocked mitochondrial fragmentation. Addition of 3-MA 5 or 5 hours following the induction of starvation arrested mitochondrial fragmentation at the intermediate stage. Like wise addition of 3-MA at later time points blocked further fragmentation effectively freezing the extent of fragmentation to the levels at the time of the addition of the drug. Notably, addition of 3-MA at any time point did not trigger the reassembly of the mitochondrion indicating fragmentation is irreversible. (D) 3-MA mediated blockade of mitochondrial fragmentation is sufficient to rescue the starvation-induced defect in parasite invasion. Invasion of fresh monolayers by parasites released from infected cells incubated for 8 hours in CM, 6%-SM for 4 or 8 hours, or 6%-SM + 3-MA for 8 hours were scored for the number of vacuoles per field following infection with 2×105 parasites. Incubation of infected cells in 6%-SM supplemented with 3-MA (green bar) completely overcomes the invasion defect (red bar). (E) Partial rescue of the parasite growth (distribution of parasite load per vacuole) 24 hours following infection in parasites pre-treated with 6%-SM supplemented with 3-MA relative to those in starvation medium alone. Failure to completely reverse the growth defect may be associated with the documented block by 3-MA of the parasite cell cycle (Wang et al., 2010). All graphs were derived from at least 3 independent experiments with mean ± S.D. depicted. In D. the number of vacuoles per field for the CM control was 6.90+/- 0.28.
In order to establish the kinetics of the 3-MA mediated blockade of starvation induced mitophagy we staggered the addition of the drug to parasites in 6%SM. As expected, incubation of infected cells in 6%SM + 3-MA blocked for the 8 hour duration of the experiment completely blocked mitophagy. This blockade was effectively instituted even if 3-MA was added 3 hours into the 8-hour incubation. Notably, a 3-hour incubation with 6%SM does not significantly trigger the starvation-induced phosphorylation of TgIF2α (Figure 1B). In contrast, the addition of 3-MA following 4 or 5 hours of starvation results in the accumulation of the intermediate morphology without significant progression to the punctate form. Effectively, the addition of 3-MA at these time points freezes the progression of mitochondrial fragmentation at levels similar to what is seen at these time points (Figure 5B). This pattern is observed when 3-MA is added 6 and 7 hours post starvation as well (Figure 5C). Together these data indicate 2 key points: 1. 3-MA effectively blocks the progression of starvation induced mitophagy arresting further fragmentation at the time of its addition. 2. Both the initial formation of the “intermediate” forms and their progression to the punctate forms are sensitive to 3-MA suggesting an autophagic basis for both events.
While blocking mitophagy, 3-MA had no significant effect on the phosphorylation of TgIF2α in 6%-SM media indicating that it does not interfere with translational repression (Supplementary Figure 2B). Finally, the addition of 3-MA to 6%SM completely rescued the invasion defect observed in 6%SM starved parasites (Figure 5D). Taken together, these data indicate that loss of mitochondrial integrity, but not starvation itself or starvation induced translational repression are the key determinants responsible for the invasion defect which for T. gondii is a lethal event.
The effect of 3-MA on parasite growth also includes partial rescue of growth retardation following infection of a fresh monolayer in complete media relative to starvation media alone (Figure 5E). This is not entirely surprising as 3-MA has recently been shown to cause a reversible cell cycle arrest in T. gondii (Wang et al., 2010). In addition the potential for 3-MA mediated inhibition of host autophagy could further limit amino acid availability for the parasite further exacerbating the starvation condition (Wang et al., 2009).
Morphological evidence of autophagosomes in T. gondii
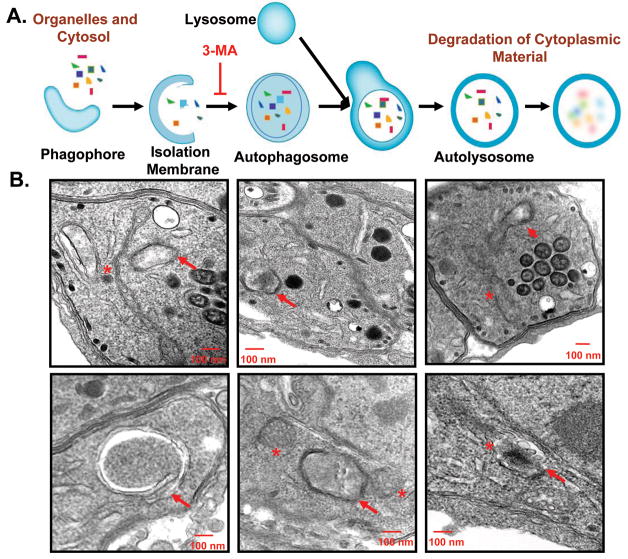
Autophagy progresses in a highly regulated manner that is initiated by the de novo formation of the phagophore and isolation membrane in the cytoplasm (Figure 6A) (Weidberg et al., 2011). The isolation membrane is comprised of 2 bilayers which form the autophagic cup that corrals both cytoplasm and organellar components into an enclosed autophagsome (Weidberg et al., 2011). These fuse with hydrolytic compartments resulting in the degradation of the contents and the generation of the building blocks to be recycled during nutrient limitation (Weidberg et al., 2011).
Figure 6.
Detection of putative starvation induced “autophagosome”-like structures in T. gondii. (A) Cartoon depicting the basic steps of autophagy in higher eukaryotes including the formation of the open isolation membrane and autophagosome formation. The specific step inhibited by 3-MA is indicated for a point of reference. (B) Double and multiple membrane containing structures consistent with the isolation membrane and autophagosomes (solid arrows) are observed in the parasites subjected to 6%-SM for 8 hours. These structures appear to engulf the cytoplasm and are often in the vicinity of altered and fragmented mitochondria (asterisk). The vacuolar compartment in the bottom right corner may contain a fragment of the mitochondrion as the internal tubules could be cristae. These structures represent putative autophagosomes since T. gondii possess a 4 membrane relict plastid, the apicoplast, which complicates definitive confirmation (see text). However, such structures were not typically observed in control parasites (data not shown).
We examined TEM sections of parasites incubated in 6%-SM to establish whether the characteristic double membrane autophagosomal intermediates could be detected. One confounding issue in identifying such structures is the presence in T. gondii of the 4-membrane containing apicoplast (Kohler, 2005) and the absence of specific autophagic markers for immune-electron microscopy. Notwithstanding these caveats, our ability to detect what appear to be double and/or multiple membrane containing structures, often in the vicinity of aberrant and fragmented mitochondria that appear to be engulfing the cytoplasm are highly suggestive of autophagosomes (Figure 6). While several of these structures appear to possess 3-4 membranes, a feature of the apicoplast, they could also represent a putative double membrane autophagosome that has engulfed a mitochondrial fragment, itself possessing 2 membranes. Confirmation of these structures as autophagosomes will require the generation of specific markers, as the molecular machinery is unraveled.
Loss of energy metabolites are not the primary triggers for mitophagy
Mitophagy in yeast occurs through a selective mechanism and is triggered by the limitation of both energy metabolites and amino acids (Kanki et al., 2008, Youle et al., 2011, Singh et al., 2011). Given the selective targeting of mitochondria by nutrient limitation we examined whether starvation for glucose and pyruvate were sufficient to trigger mitophagy in T. gondii. Cells infected for 24 hours in complete CM were shifted to glucose-free, pyruvate-free or glucose and pyruvate free media for 4 or 8 hours. We monitored the effect on the integrity of mitochondria using immunofluorescence. Starvation for energy metabolites did result in changes in the organization of the mitochondrion, which were considerably less extreme that treatment with 6%-SM (Figure 7A). Specifically, mitochondrial fragmentation was largely arrested at the intermediate stages and few vacuoles displayed punctate forms (Figure 7A).
Figure 7.
Energy metabolism is only partially responsible for starvation-induced mitophagy in T. gondii. (A) Quantification of the mitochondrial morphology phenotype following exposure to CM or media lacking glucose (glu), pyruvate (pyr) or both (glu+pyr) indicates induction of mitochondrial fragmentation with very few vacuoles possessing punctate mitochondria, thus, starvation for energy metabolites is not the primary trigger for mitochondrial fragmentation (see text). (B) Starvation of parasite infected cells for glu or pyr is sufficient to trigger the phosphorylation of TgIF2α (TgIF2-P) indicating the activation of the translational repression pathway. Panel A shows the mean ± S.D. from 3 independent experiments. At least 100 vacuoles were counted for each treatment in each experiment.
While the impact on mitochondrial integrity was modest, limitation of energy metabolites was a stress as TgIF2α-P levels were elevated suggesting translational inhibition (Figure 7B). The slower progression of mitophagy following starvation for energy metabolites suggests that the effect may be secondary due to an impact on ATP-dependent processes such as amino acid transport.
The primary mechanism involved in sensing energy flux involves the cyclic AMP-activated protein kinase (AMPK) an activity that has been extensively studied in higher eukaryotes (Hardie, 2008, Rutter et al., 2003) but not in parasitic protozoa. The Toxoplasma genome encodes a potential AMPK homolog (TGGT1_113440, a 46 KD putative serine/ threonine protein kinase with 58% identity to Human AMPK alpha). This led us to examine the effect of 2 AMPK agonists - metformin and AICAR (Hardie, 2008, Lee et al., 2011). Treatment with these drugs at high concentrations for up to 24 hours had a limited if any effect on mitochondrial integrity (Supplemental Figure 4), reinforcing the interpretation that the effect of glucose/pyruvate starvation on mitochondrial integrity is secondary as is likely energy metabolism in general.
Similar starvation experiment where we removed micronutrients (vitamins and lipoic acid) failed to trigger any significant changes in the morphology of the parasite mitochondrion (Supplementary Figure 5). The absence of a requirement for micronutrients (Supplementary Figure 5) and the modest impact of energy metabolites and drugs affecting energy metabolism (Figure 7, Supplemental Figure 4) suggest that starvation-induced mitophagy is linked to amino acid levels.
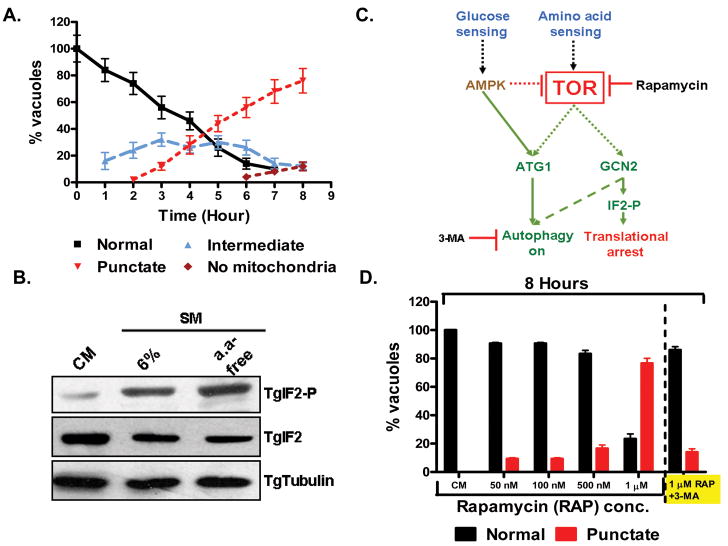
Mitophagy is triggered by amino acid starvation and is TgTOR dependent
Amino acid deprivation has been established as a primary trigger of autophagy, including mitophagy (Wang et al., 2011), in both yeast and mammalian cells (Mortimore et al., 1977, Meijer, 2008, Onodera et al., 2005). Addition of amino acid free media to HFF cells infected with T. gondii for 24 hours resulted in a rapid induction of mitophagy with changes in the mitochondrial architecture being evident as early as 1 hour following amino acid deprivation (Figure 8A). The progression to punctuate mitochondria was similarly rapid and accelerated (Figure 8A) relative to incubation in 6%-SM (Figure 3B). This rapid mitochondrial fragmentation occurred in all parasites within 8 hours following transfer to amino acid deprived media, with approximately 10% lacking a detectable mitochondrion (Figure 8A). Consistent with other systems (Wek et al., 2006, Kroemer et al., 2010), amino acid deprivation resulted in a robust induction of TgIF2α-P levels indicating the activation of the translational repression system (Figure 8B).
Figure 8.
Rapid mitophagy accompanies amino acid starvation in a TOR-dependent manner. (A) A time course depicting the distribution of mitochondrial morphologies following the addition of amino acid free media to an established T. gondii infection reveals rapid fragmentation of mitochondria with changes in morphology evident as early as 1 hour following exposure to amino acid free media. (B). Amino acid starvation potently activates the translation repression pathway based on the phosphorylation of TgIF2α (TgIF2-P). (C). Schematic describing the integration of the autophagic pathway with the mechanism for translational repression in higher eukaryotes (including S. cerevisiae) reveals a crucial role for the TOR complex. Rapamycin is a selective inhibitor of TOR. Repressing interactions are depicted with dashed arrows while activating reactions are depicted with solid arrows. Notably, amino acid starvation results in the inactivation of TOR a phenotype that is mimicked in complete media by rapamycin. (D). Rapamycin treatment of infected cells in complete media results in a dose dependent fragmentation of the parasite mitochondrion following an 8 hour treatment. Notably, treatment with rapamycin in the presence of 10 mM 3-MA (right) completely blocks the fragmentation of the mitochondrion indicating an autophagy-related process. Each experiment was performed at least 3 times with a minimum of a 100 vacuoles scored for each replicate in A and D.
Phosphorylation of TgIF2α is mediated by one or both of the two homologs of eIF2α-kinase, which are homologs of the yeast kinase GCN2 (Sullivan et al., 2004, Narasimhan et al., 2008). In yeast GCN2 itself is repressed by TOR mediated phosphorylation (Figure 8C) (Wek et al., 2006). The target of rapamycin (TOR) kinase is a vital component of the amino acid sensing mechanism in eukaryotic cells (Proud, 2007, Dann et al., 2006). The Toxoplasma genome (www.toxodb.org) is reported to encode a single TOR homolog (TgTOR, TGGT1_094250, a 263 KD putative fkbp-rapamycin associated protein with 44% identity with Human mTOR kinase (Brown et al., 1994)). Recent studies in mammalian cells and yeast establish TOR-kinase serves as a signaling hub that integrates both the autophagic response and translational repression in response to amino acid starvation (Figure 8C) (Kim et al., 2011b, Meijer, 2008, Egan et al., 2011, Kroemer et al., 2010, Kundu, 2011).
Rapamycin is a well established inhibitor of the TOR-complex both in yeast (Chang et al., 2009, Kunz et al., 1993) and mammalian cells (Huo et al., 2011). It has been shown to be growth inhibitory in the extracellular Trypanosoma brucei (Barquilla et al., 2008). We reasoned that if TgTOR was in fact an integral component of the amino acid sensing machinery in T. gondii, then rapamycin treatment should mimic amino acid starvation. We therefore incubated infected cells in CM with increasing levels of rapamycin for 8 hours. Treatment at < 500nM had a limited effect on mitochondrial integrity (Figure 8D), however, exposure to ≥ 1μm resulted in the detection of punctuate mitochondria in approximately 80% of the vacuoles (Figure 8D). Typically, 50 – 500nM rapamycin induces autophagy in mammalian cells, but this requires incubation times of 24-72 hours as opposed to the 8 hours used here (Huo et al., 2011). In neurons an absence of rapamycin toxicity at doses as high as 3μM for 72 hours has been reported (Pan et al., 2009). We were nonetheless concerned that the effects seen on the T. gondii mitochondrion 1μM may reflect non-specific toxicity and not autophagy. To address this point we tested whether 3-MA could block the rapamycin induced mitochondrial fragmentation (Figure 8D, right hand side). The observation that inhibition of autophagy with 3-MA effectively blocks rapamycin induced mitophagy confirms that the T. gondii TOR pathway directly integrates amino acid sensing and the parasite’s autophagic response.
Starvation induced mitochondrial fragmentation correlates with the loss of membrane potential
Involvement of autophagy in mitochondrial depolarization has been evident in both yeast and mammalian systems upon nutrient starvation (Rodriguez-Enriquez et al., 2009, Rodriguez-Enriquez et al., 2006, Okamoto et al., 2011, Zhang et al., 2007). To establish whether starvation-induced changes in mitochondrial mitochondrial morphology are associated with the loss of the membrane potential (ΔΨm) we used the membrane potential sensitive dye MitoTracker. Consistent with growth in complete media, accumulation of MitoTracker is evident in both the parasite mitochondrion and host mitochondria (Figure 9A,B, CM). The replacement of CM with 6%SM results in changes in the morphology of the parasite mitochondrion. While the ‘intermediate ”phenotype mitochondria continue to retain the ΔΨm noted by the accumulation of Mitotracker (Figure 9A second row), the punctate mitochondria exhibit only diffuse staining with the dye indicative of a loss of the membrane potential. This is not surprising as the TEM evidence indicates that while the intermediate-stage mitochondrion is morphologically altered, it still retains the integrity of its membranes (Figure 4). In contrast, the apparently punctate mitochondria exhibit a breakdown of membrane integrity precluding the establishment of the requisite electrochemical gradient. This is also evident in row 4 of Figure 9A where a vacuole with a normal mitochondrial morphology is adjacent to one with punctate mitochondria. Finally, the loss of host mitochondrial ΔΨm was observed in monolayers treated with 6%SM for 8 hours (Figure 9A, row 3,4). The loss of the host mitochondrial potential could be to the depletion of energy metabolites (glucose/pyruvate) or amino acids.
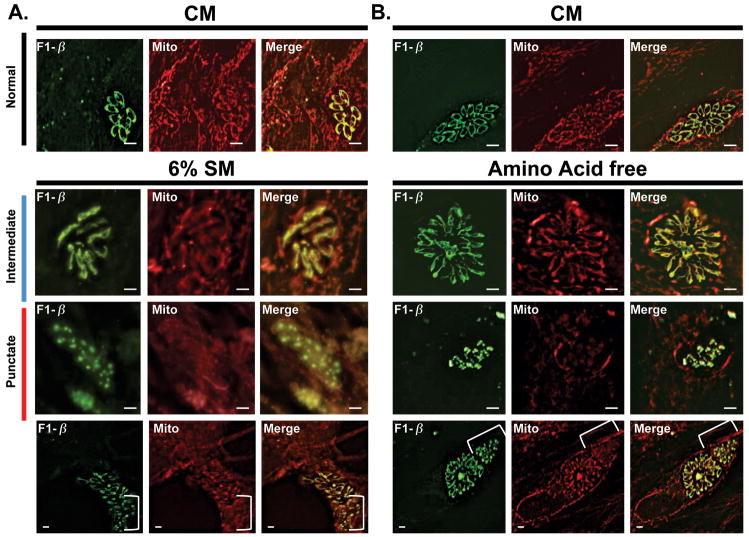
Figure 9.
Effect of starvation induced mitophagy on the mitochondrial membrane potential. (A) Accumulation of the membrane potential sensitive dye MitoTracker in both the parasite mitochondrion and host mitochondria is observed in CM (top panel). The 6%SM starvation mediated intermediate mitochondrial morphology does not result in the loss of the mitochondrial membrane potential (second panel). Notably, at 4-hour time point presented, host mitochondria retain their membrane potential (second panel). In contrast, mitochondrial fragmentation, seen here in 6%SM for 8 hours, results in a failure to accumulate MitoTracker which now appears as diffuse staining (third panel). Loss of the membrane potential is not a function of length of incubation in 6%SM but rather mitochondrial fragmentation as indicated by adjacent vacuoles at the 8 hour time point (fourth panel, punctate mitochondria bracketed). A loss of discrete MitoTracker labeling in host mitochondria following exposure to 6%SM for 8 hours is observed (third and fourth panel). (B) The effect of amino acid starvation on the membrane potential of parasite mitochondrion is similar to that seen with 6%SM whereby normal (top panel) and intermediate form (second panel) mitochondria retain the membrane potential while punctate mitochondria (third and fourth panel- bracketed) do not. In contrast to what was observed with regard to host mitochondria incubated for 8 hours in 6%SM (A), incubation in amino acid free medium does not result in the loss of the membrane potential of host mitochondria (third and fourth panels). (Scale bars in A and B: Top panel-5 μm, other panels-2μm).
In light of the mitophagy phenotype being linked to amino acid limitation we investigated the effect of amino acid starvation on the mitochondrial membrane potential. Consistent with the observations with 6%SM (Figure 9A), the membrane potential was retained in intermediate stage mitochondria (Figure 9B row 2) but depleted in parasites containing punctate mitochondria. In these cases, mitotracker staining was weak and diffuse within the parasites (Figure 9B rows 3,4). In contrast to what was observed in 6%SM however, amino acid deprivation had no impact on the ability of host mitochondria to accumulate MitoTracker even following an 8-hour incubation (Figure 9B row 3,4). These data suggest that the loss of host ΔΨm in 6%SM is a consequence of the depletion of energy metabolites while the loss of parasite ΔΨm is associated with the loss of mitochondrial integrity, a phenotype triggered by amino acid starvation.
Bioinformatic evidence reveals the presence and absence of ATG genes
The genes associated with autophagy have been studied in considerable detail in the yeast Saccharomyces cerevisiae (Klionsky et al., 2010, Mizushima, 2007). We interrogated the T. gondii genome (www.toxodb.org) to identify potential homologs of key autophagy related (Atg) genes. What is immediately evident is that while several homologs can be readily identified, others appear to be missing (Supplemental Table 1).
Several of the putative homologs, including TgATG1, TgATG3, TgATG7, TgATG8, TgATG18, TgATG20 and TgVPS34 have respectable identity (similarity) scores and may be considered to be true homologs (Supplemental Table 1). In contrast the assignment of several other genes with identity scores < 30% must be considered provisional assignments subject to experimental confirmation (Supplemental Table 1). Notably, a block of apparently missing activities include key members of the cytoplasm to vacuole (Cut) pathway (Lynch-Day et al., 2010, Teter et al., 2000) and components of the ATG12-ATG5 conjugation machinery (Inoue et al., 2010). While a potential homolog of TgATG5 was identified, the poor identity score complicates a definitive assignment (Supplemental Table 1).
The bioinformatic analysis does however indicate that genes for conventional autophagy are evident. Furthermore the finding that several of the putative TgATG genes that should possess key enzymatic activities (e.g. protein kinase –TgATG1, lipid kinase -TgVPS34, lipid conjugation systems -TgATG7, -TgATG5) are predicted to be substantially larger than their yeast counterparts (Supplemental Table 1) suggests additional parasite specific functions that could be independent of “classical” starvation induced autophagy. Future study of these important genes may therefore uncover novel aspects of parasite biology.
Discussion
Our understanding of autophagy and its role in cellular homeostasis is evolving (Mizushima, 2007, Kroemer et al., 2010). What was once seen primarily as a cellular catabolic mechanism to deal with nutrient limitation is now seen to be integrated into the central metabolic regulation of the cell (Kroemer et al., 2010). The role of autophagy in normal cellular homeostasis centers on the elimination of defective organelles (most notably mitochondria) and excess cytoplasm (Sakai et al., 2006, Wang et al., 2011, Weidberg et al., 2011, Kristensen et al., 2008). Under conditions of nutrient limitation, the autophagic cascade assumes a cell survival function. This is achieved by selectively targeting excess capacity in the cell for degradation thereby increasing pools of nutrients to sustain cellular functions (Weidberg et al., 2011, Kristensen et al., 2008). Cellular degradation pathways are under considerable control as degradation taken to an extreme results in the breakdown of critical components leading to cell death (Assuncao Guimaraes et al., 2004, Blommaart et al., 1997, Gozuacik et al., 2007). Thus autophagy, while a cell survival mechanism in certain contexts, is a mediator of cell death in others. An autophagy mediated cell death is classified as a Type II death pathway that has a distinct progression from that of the Type I apoptotic death (Gozuacik et al., 2007, Kourtis et al., 2009). In the absence of clear evidence for apoptotic pathways in early branching eukaryotes, autophagy presents a potential mechanism for programmed cell death (Luder et al., 2010, Duszenko et al., 2011). Such an assignment must however be made with caution as a death pathway caused by autophagy is often difficult to distinguish from a death mechanism that involves autophagy as a component of its execution (Galluzzi et al., 2009, Galluzzi et al., 2011).
The extent of mechanistic similarity of autophagy, between yeast and mammalian systems points to a high degree of functional conservation (Inoue et al., 2010, Mizushima et al., 2010). Much less is known about the autophagic pathways in truly early branching eukaryotes (Duszenko et al., 2011, Odronitz et al., 2007, Herman et al., 2006). We chose to functionally investigate whether or not the protozoan parasite Toxoplasma gondii possesses an autophagy related pathway by investigating the response of the parasite to nutrient limitation, a classical autophagic trigger.
Nutritional stress triggered by incubation of infected cells in diluted complete growth media (CM) results in the activation of the translational repression pathway defined by the increases in levels of TgIF2α-P (Figure 1A,B) (Joyce et al., 2010, Sullivan et al., 2004). Interestingly, starvation for as few as 4 hours, rather than establishing a state of stasis, results in parasite death. This is defined by a marked defect in their ability to invade a fresh monolayer (Figure 1C). Despite exhibiting an invasion defect, the integrity of the invasion related organelles (micronemes and rhoptries) (Carruthers et al., 2007) was not affected. Rather we observed a rapid and sequential fragmentation of the single parasite mitochondrion (Figure 3A). The kinetics of fragmentation proceeds from the normal ring shape (Melo et al., 2000), via linear intermediates, to puncta all the way to the loss of the organelle (Figure 3A,B). Evidence based on the incorporation of Mitotracker, a mitochondrial membrane potential (ΔΨm) sensitive dye, suggests that mitochondria retain their ΔΨm despite morphological alteration, losing it only when the mitochondrion is truly fragmented (Figure 9).
It is easy to envision the loss of viability in a parasite that has lost its mitochondrion, our inability to observe invasion by parasites exhibiting any mitochondrial defects was surprising (Figure 1C). This is remarkable given the fact that invasion is believed to be powered by glycolysis (Pomel et al., 2008, Polonais et al., 2010). Furthermore, our finding that once fragmentation is initiated, it is irreversible (Figure 5C) suggests loss of mitochondrial integrity is a marker of parasite death.
The systematic fragmentation and elimination of the mitochondrion is reminiscent of starvation-induced mitophagy observed in yeast and mammalian cells (Bhatia-Kissova et al., 2010, Kim et al., 2007, Youle et al., 2011). A critical difference lies in the fact that yeast and mammalian cells possess multiple mitochondria representing an excess capacity that can be regenerated upon the return of nutrient replete conditions (Bhatia-Kissova et al., 2010, Youle et al., 2011). With its single mitochondrion (Melo et al., 2000), T. gondii does not have the luxury of dispensing with this critical organelle.
Autophagy is a sequential process that is initiated by a cascade of events initiated by the de novo formation of a unique double membrane entity termed the phagophore which surrounds cytoplasm and organelles destined for clearance into a autophagosome that fuses with a lysosomal compartment resulting in the breakdown of the contents (Figure 6A) (Weidberg et al., 2011, Kristensen et al., 2008). A key activity in yeast (and other eukaryotes) is Vps34, a class III PI3 kinase that is involved in membrane recruitment to the autophagosome (Yue et al., 2010). This enzyme is selectively and competitively inhibited by 3-methyl adenine (3-MA), which is viewed as a selective inhibitor of autophagy (Yue et al., 2010, Abeliovich et al., 2001, Seglen et al., 1982). 3-MA is well tolerated by T. gondii as it is reversible and can be used to synchronize the parasite cell cycle (Wang et al., 2010). Our finding that the addition of 3-MA effectively blocked the starvation-induced fragmentation of the mitochondrion (Figure 5A,B) strongly implicates an autophagic mechanism in the selective mitophagic response. More importantly, while the presence of 3-MA did not impact the starvation-induced induction of translational repression (Supplemental Figure 2), it did rescue the starvation-induced inhibition of parasite invasion (Figure 5D).
A morphological signature of autophagy is a double (or multiple) membrane structure engulfing either organelles or the cytoplasm (Abeliovich et al., 2000, Hamasaki et al., 2010, Weidberg et al., 2011). A confounding issue in T. gondii and other Apicomplexa is the existence of the 4-membraned apicoplast, a relict non-photosynthetic plastid derived endosymbiont (Kohler, 2005). It should be noted, that the frequency of double/multi-membrane structures in starved cells was qualitatively higher by TEM (Figure 6, data not shown). However, the organization and number of apicoplasts in starved parasites was indistinguishable from those grown in complete media by IFA analysis (Figure 2A). While not confirmatory, these observations are suggestive of the double/multi-membrane structures seen by electron microcopy representing distinct stages of autophagosome formation. The fact that several of these entities are not entirely membrane enclosed further suggest they are not apicoplasts (Figure 6B).
The sheer size of the single mitochondrion evident both by IFA and TEM (Melo et al., 2000, Seeber et al., 1998) suggests that the intact organelle is an unlikely target for the autophagy-mediated breakdown. Consistent with this view, we observe a sequential breakdown in the course of starvation-induced mitochondrial fragmentation (Figure 3AB), which itself is blocked by 3-MA (Figure 5A). This suggests that activities involving PI3K/Vps34 play a role in mitochondrial fission consistent with what has been recently reported in mammalian cells (Kim et al., 2011a). In cells undergoing mitophagy, mitochondria that elongate evade autophagic destruction assuring a pool of protected organelles (Gomes et al., 2011). Given the fragmentation of the single parasite mitochondrion, a protective role during starvation does not appear to be likely.
The mitophagy response to starvation appeared to be directly linked to amino acid sensing (Figure 8) and not a response to energy metabolites (Figure 7) or micronutrients in the media (Supplemental Figure 5). Amino acid sensing in higher eukaryotes, including S. cerevisiae, is regulated by the TOR-complex (target of rapamycin) (Dann et al., 2006, Goberdhan et al., 2009, Noda et al., 1998) which recently has been shown to integrate glucose sensing (via AMPK) and amino acid sensing to autophagy and translational repression (Figure 8C) (Hardie, 2008, Kim et al., 2011b). A homolog of TOR has been identified in the Toxoplasma genome (TGGT1_094250) suggesting susceptibility to rapamycin. Indeed at a high concentration of rapamycin fragmentation of the parasite mitochondrion is evident, a phenotype that is largely blocked in the presence of 3-MA (Figure 8D), thereby linking the predicted TOR cascade to the autophagic response (Kim et al., 2011b).
The presence of a sensitive switch responsive to amino acid starvation that leads to a rapid death presents an enigma. The intracellular pools of amino acids derived from synthesis and turnover, coupled with transport from the extracellular environment suggest amino acids are not likely to be limiting under normal conditions (Sinai, 2008, Laliberte et al., 2008, Sinai et al., 1997a). Quite to the contrary, an increasing literature indicates that T. gondii and other intracellular pathogens actively induce autophagic pathways in the infected host cell, potentially as a means of increasing free amino acid pools (Wang et al., 2009, Subauste, 2009, Orlofsky, 2009, Halonen, 2009).
One amino acid, for which the parasite is auxotrophic is tryptophan (Pfefferkorn, 1984, Pfefferkorn et al., 1986). Tryptophan is known to become limiting in an activated macrophage where it contributes to parasite killing (Pfefferkorn, 1984, Pfefferkorn et al., 1986). This may provide a clue to why such a sensitive programmed cell death pathway, triggered by amino acid limitation may be retained within the parasite. Recent work from several laboratories has demonstrated that the capacity to present both endogenous as well as model antigens is dependent on the context in which the antigen is delivered (Jordan et al., 2010, Blanchard et al., 2010, Dzierszinski et al., 2007, Dzierszinski et al., 2008). The presentation of antigens detected only in the context of dead parasites (with epitopes in intracellular proteins) could present a selective advantage in the course of infection by distracting the immune response from away from viable organisms. Accelerating death by the activation of an autophagy based parasite death pathway may increase the pool of non-protective decoy epitopes.
Nutrient-limitation induced autophagy may also play a role in the transmission stages of the parasites, the tissue cyst and oocysts (Dubey, 1998, Dubey et al., 1998). One may postulate that nutrient limitation within a cyst may be alleviated by the activation of a bulk protein turnover mechanism like autophagy-although no direct evidence for this has been reported. Finally, programmed death mechanisms in protozoa have been suggested to be triggered as a means to controlling cell density in the host (Luder et al., 2010). Whether such a strategy applies to an intravacuolar pathogen is not known.
The pharmacological evidence for the presence of an autophagic pathway (Figure 5,8) is supported for by the bioinformatic analysis that reveals homologs of several key autophagy genes are encoded in the parasite genome, while others are not evident (Supplemental Table 1).
In summation, we find that autophagy related processes in T. gondii are activated in response to amino acid limitation with rapid kinetics leading to selective mitophagy and death. As an activatable and irreversible cell death pathway, the autophagic cascade presents a very attractive pathway for the development of drugs (Brennand et al., 2011, Paguio et al., 2011). Investigations into this pathway in an earlier branching eukaryote has the potential to reveal novel aspects in parasite biology while enriching our understanding of autophagy and its evolution into a highly specialized pathway.
Experimental Procedures
Reagents
Base cell culture media include α-MEM and Glucose and Pyruvate-free MEM (Invitrogen, Carlsbad, CA). MEM Vitamin solution was purchased from GIBCO (Life Technologies/ Invitrogen, Carlsbad, CA). 3-methyladenine (3-MA), Metformin (1, 1 – Dimethylbiguanide hydrochloride), Lipoic acid and AICAR (5-Aminoimidazole-4-carboxamide riboside) were purchased from Sigma (St. Louis, MO). Rapamycin was purchased from EMD Biosciences (Calbiochem, Gibbstown, NJ).
Cell Lines, Parasite and starvation medium
Low passage Human foreskin fibroblast (HFF) cells (ATCC) and Vero cells (ATCC CCL-81) were maintained in α-MEM supplemented with 7% heat inactivated FBS (Gemini Bio-Products, West Sacramento), 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM L-Glutamine (complete medium) (CM) (Gibco BRL, Rockville, MD). All cell lines were grown as monolayers at 37°C in an atmosphere of 5% CO2. The type I RHΔHX strain of Toxoplasma gondii (NIH-AIDS Resource Center) were used throughout the study and maintained by serial passage in Vero cells as previously described (Carmen et al., 2006). All experiments to investigate the role of starvation were conducted in infected HFF cells (passage <8) allowed to be confluent for at least 1 week. Starvation media (SM) comprised complete αMEM (CM) diluted in 1X Hanks Balanced Salts Solution (HBSS) to a final concentration from 2% - 50%. Glucose and pyruvate free (Gibco) was supplemented as above for complete αMEM with the exception of the use of dialyzed FBS. Reconstitution of glucose, pyruvate or both compounds was achieved by the addition of cell culture grade glucose (5.56mM) and/or pyruvate (1mM) respectively. Amino acid starvation medium was made by adding 1 mM sodium pyruvate, 5.56 mM cell culture grade glucose, vitamin solution (1X), and lipoic acid (0.2 mg/ L) to 1X HBSS solution, without supplemental L-Glutamine. Dialyzed serum was added as above.
Induction of starvation
A confluent HFF cell monolayer in either a 6-well plate (for immunoblot) or on a cover slip in a 24-well tissue culture dish (for IFA) (Falcon) was infected with 5×105 or 2×105 freshly passaged parasites for 24 hours. The infected cells were washed with 1X PBS, and either fresh complete media or SM added for up to 8 hours. For experiments involving pharmacological treatments (3-MA, Metformin, AICAR or rapamycin) the drug treatments were added in either CM (3-MA, Metformin, AICAR, rapamycin) or SM (3-MA) media for the time points and at the concentrations indicated in the figure legends. 3-MA was used at a concentration of 10mM.
Impact of starvation on invasion and growth
HFF cell monolayer in a 10 cm dish was infected with freshly passaged 1×107 parasites for 24 hours. Infected monolayers were washed with 1X PBS, and transferred to the 6%-SM for 4 and 8 hours with or without 3-MA. Control plates were returned to complete media. Intracellular parasites were harvested by syringe passage and enumerated. 2×105 parasites were allowed to infect fresh HFF monolayers grown on the cover slips for 24 hours in complete media. Infected HFF monolayers were fixed and the number of vacuoles per field (defined staining with the PVM marker GRA3) as well as the distribution of parasites per vacuole (defined using the surface marker SAG1) determined. These serve as measures of parasite invasive ability and replication respectively. In addition, a portion of the inoculum was pelleted onto a glass slide using a cytospin centrifuge, fixed and examined for the integrity of the mitochondrion as described below. In addition the integrity of the mitochondrion in the newly infected cell was established using anti-F1β. Specific conditions for immunofluorescence analyses are described below. At least 50 independent fields from the control, 4 and 8-hour starvation treatments were randomly chosen for the quantification of invasion efficiency while at least 100 PV’s were counted for their parasite burden- a measure of parasite growth.
Host viability assay
Necrosis was examined by measuring the LDH activity as a marker for cellular necrosis using LDH cytotoxicity assay kit (Cayman Chemical, Ann Arbor MI), according to manufacturer’s instructions. Briefly, HFF monolayers seeded in a 96-well plate were infected with 2 × 104 no. of RH parasites for 24 hours. Following infection, the complete media were switched to 6% starvation media for 8 hours, and subjected to the measurement of LDH activity in the supernatant. Non-infected HFF cells starved for 8 hours and cells treated with 0.1% Triton X-100 for 10 minutes were used as control.
Immunoblot analysis
Following induction of starvation and/ or drug treatment in a 6-well plate, both adherent and detached cells from the normal and starvation-induced population were collected by scraping and pelleted by centrifugation using a standard tabletop centrifuge at 1000×g for 5 minutes. The cell pellets were lysed directly in 4x SDS-PAGE sample buffer and subjected to SDS-PAGE followed by immunoblot analysis as previously described. immunoblots were probed overnight at 4°C using anti-TgIF2α-P (rabbit polyclonal, 1:500, Dr. W. Sullivan), anti-TgIF2α (rabbit polyclonal, 1:7000, Dr. W. Sullivan) (Joyce et al., 2010) and anti-Tg Tubulin (rabbit polyclonal 1: 2000, Dr. N. Morrissette) (Ma et al., 2010). Following washes, the blots were incubated with goat anti-rabbit-HRP (1:2000, JAX) and detected using enhanced chemiluminescence (Pierce) using X-ray film (Kodak). Immunoblot analyses experiments were performed at least 3 times with a representative experiment shown.
Immunofluorescence analysis and image processing
Confluent HFF cells growing on sterile 12 mm glass cover slips in 24-well plates were infected with freshly harvested parasites for 24 hours in complete medium (CM). The medium was replaced with either CM, or 6%-SM or media with the pharmacological treatments for the time points as noted in the figure legends. The infected cell monolayers were washed with PBS and fixed in methanol at -20°C. Primary antibodies used were mouse anti-TgROP7 (1:2000, rhoptry marker) (Bradley et al., 2005), mouse anti-TgF1β (1:2000, mitochondrial marker)(Bradley and Morrissette, submitted), mouse anti-TgAtrx1 (1:2000, apicoplast marker) (DeRocher et al., 2008), mouse anti-TgMIC2 (1:1000, microneme marker) (Huynh et al., 2006), rabbit anti-TgSAG1 (1:5000, parasite surface) (Lekutis et al., 2001) and mouse anti-GRA3 (1:3000, PVM) (Bermudes et al., 1994) as appropriate. Proteins were localized using species-specific Oregon Green or Texas Red conjugated secondary antibodies diluted at 1:2000 (Molecular Probes) and visualized using a Zeiss Axiovision Fluorescence Microscope with 100X oil immersion objective. Images were acquired as multiple Z-stacks and deconvoluted using the iterative algorithm using a high-resolution grayscale camera (Zeiss). The merged images were generated by pseudo-coloring the constituent grayscale images and merging them using Adobe Photoshop software. In all cases, adjustments for brightness and contrast, and for color balance in the merged images, were applied uniformly on the entire image. For quantification purposes at least three independent replicates were used. Data were plotted and subjected to statistical analyses Graph-pad Prism (version 5.0).
Mitochondrial membrane potential assay
Mitotracker ® Red CM – H2X Ros (Molecular Probes; Invitrogen detection technologies, Carlsbad CA) dye was used to detect the membrane potential of mitochondria as previously described (Sinai et al., 1997b). Infected cells were incubated in media as indicated in the figure legend. MitoTracker labeled cells, were formalin fixed, and subjected to immunofluorescence analysis. Mouse TgF1ß antibody was used as a marker for parasite mitochondrion.
Electron Microscopy
HFF monolayers growing on a 10cm dish were infected with freshly passaged RH parasites for 24 hours, and switched to CM, 6%SM or 3-MA containing 6%SM supplemented with 3-MA (data not shown). Infected monolayers were processed essentially as previously described (Sinai et al., 1997b). The EM-grids were visualized using a Phillips Tecnai 12 transmission electron microscope and images were digitally acquired.
Supplementary Material
(A) Effect of starvation medium on the integrity of the host cell monolayer (blue) and the appearance of altered T. gondii mitochondria (red). The mean number of host nuclei per field following incubation for 8 hours in 6%-SM. In addition the changes in starvation induced mitochondrial morphology were determined. Incubation of infected cells in 6%-SM resulted in an optimal balance of the 2 phenotypes. Nuclei were labeled with Hoechst stain while anti-F1β antibody was use to stain the parasite mitochondrion. A minimum of 50 fields was counted in each of 3 independent experiments. (B) Measurement of lactate dehydrogenase LDH activity in the culture supernatant in uninfected and infected cells incubated for 8 hours in CM or 6%SM. While LDH release was higher in infected cells, there was no effect of 6%SM on host cell viability relative to the appropriate CM condition. LDH release following incubation with 0.1% Triton X-100 for 10 minutes was used as positive control. The activity data are presented as ratio of the measured optical density relative to the 0.1% TX100 control.
(A) Treatment of infected cells with complete media (CM) with 10 mM 3-MA for 8 hours does not have any effect on the morphology of the parasite mitochondrion. Scale bar = 2μm. (B) The 3-MA mediated block of mitochondrial fragmentation is independent of translational repression as comparable to induction of TgIF2α-P levels are evident in the presence or absence of the compound. Notably 3-MA alone serves as a stress causing a small increase in TgIF2α-P above the untreated control.
Treatment of infected cells in 6%-SM supplemented with 10 mM 3-MA has no effect on the morphology of the rhoptries (ROP7), micronemes (MIC2) or the apicoplast (anti-ATrx1). Scale bar = 2μm.
The AMPK agonists Metformin and AICAR do not trigger significant levels of mitochondrial fragmentation suggesting that the manipulation of glucose sensing plays a limited role in the triggering of starvation induced mitophagy in T. gondii. Results shown are the mean ± S.D. from 3 replicates each.
Effect of starvation for micronutrients (vitamins and lipoic acid) on the mitophagy phenotype. HFF monolayers were infected for 24 hours prior to being switched to vitamin (A) or lipoic acid (B) free media for 8 hours. The impact of treatment on mitochondrial morphology was determined by immunofluorescence microscopy. Results shown are the mean ± S.D. from 3 replicates each. A minimum of 100 vacuoles were counted at random for each sample for each replicate.
Bioinformatic identification of potential T. gondii homologs of yeast Atg proteins with their basic characteristics and generalized biological function. Targets hits of >30% identity (50% similarity) are considered bonafide hits. Those with <30% identify (50% similarity) are considered provisional hits. References regarding the specific biological functions for the yeast genes are reviewed in (Klionsky et al., 2010). Genome Sources Saccharomyces: www.yeastgenome.org, Toxoplasma: www.toxodb.org.
Acknowledgments
The authors would like to thank Drs. John Boothroyd, Peter Bradley, Vern Carruthers Jean-Francois Dubremetz, Francesc Marti, Naomi Morrissette and William Sullivan for their gifts of antibodies and reagents. The assistance of the University of Kentucky College of Medicine Electron Microscopy and Imaging facility with the preparation of the EM samples is gratefully acknowledged. We would like to thank Dr. William Sullivan for critically reading the manuscript. Finally, we acknowledge members of the Sinai laboratory for their insights and Ms Sourita Das for assistance in putting together the figures. This work was supported by NIH RO1AI049367 to APS. Work in the Roepe laboratory is supported by NIH RO1 AI045957 awarded to PDR.
Note: While our work was in its final revision a manuscript making a link between the autophagy protein Atg3 and the maintenance of mitochondrial integrity was published in PLoS Pathogens. This work which is supportive of our study can be found at: Besteiro et. al. (2011), PLoS Pathog. 7(12): e1002416.
Literature Cited
- Abeliovich H, Dunn WA, Jr, Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol. 2000;151:1025–1034. doi: 10.1083/jcb.151.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assuncao Guimaraes C, Linden R. Programmed cell deaths. Apoptosis and alternative deathstyles. Eur J Biochem. 2004;271:1638–1650. doi: 10.1111/j.1432-1033.2004.04084.x. [DOI] [PubMed] [Google Scholar]
- Barquilla A, Crespo JL, Navarro M. Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci U S A. 2008;105:14579–14584. doi: 10.1073/pnas.0802668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermudes D, Dubremetz JF, Achbarou A, Joiner KA. Cloning of a cDNA encoding the dense granule protein GRA3 from Toxoplasma gondii. Mol Biochem Parasitol. 1994;68:247–257. doi: 10.1016/0166-6851(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Bhatia-Kissova I, Camougrand N. Mitophagy in yeast: actors and physiological roles. FEMS Yeast Res. 2010;10:1023–1034. doi: 10.1111/j.1567-1364.2010.00659.x. [DOI] [PubMed] [Google Scholar]
- Blanchard N, Shastri N. Topological journey of parasite-derived antigens for presentation by MHC class I molecules. Trends Immunol. 2010;31:414–421. doi: 10.1016/j.it.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Blommaart EF, Luiken JJ, Meijer AJ. Autophagic proteolysis: control and specificity. Histochem J. 1997;29:365–385. doi: 10.1023/a:1026486801018. [DOI] [PubMed] [Google Scholar]
- Blume M, Rodriguez-Contreras D, Landfear S, Fleige T, Soldati-Favre D, Lucius R, Gupta N. Host-derived glucose and its transporter in the obligate intracellular pathogen Toxoplasma gondii are dispensable by glutaminolysis. Proc Natl Acad Sci U S A. 2009;106:12998–13003. doi: 10.1073/pnas.0903831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PJ, Ward C, Cheng SJ, Alexander DL, Coller S, Coombs GH, et al. Proteomic analysis of rhoptry organelles reveals many novel constituents for host-parasite interactions in Toxoplasma gondii. J Biol Chem. 2005;280:34245–34258. doi: 10.1074/jbc.M504158200. [DOI] [PubMed] [Google Scholar]
- Brennand A, Gualdron-Lopez M, Coppens I, Rigden DJ, Ginger ML, Michels PA. Autophagy in parasitic protists: unique features and drug targets. Mol Biochem Parasitol. 2011;177:83–99. doi: 10.1016/j.molbiopara.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, Schreiber SL. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature. 1994;369:756–758. doi: 10.1038/369756a0. [DOI] [PubMed] [Google Scholar]
- Carmen JC, Hardi L, Sinai AP. Toxoplasma gondii inhibits ultraviolet light-induced apoptosis through multiple interactions with the mitochondrion-dependent programmed cell death pathway. Cell Microbiol. 2006;8:301–315. doi: 10.1111/j.1462-5822.2005.00622.x. [DOI] [PubMed] [Google Scholar]
- Carruthers V, Boothroyd JC. Pulling together: an integrated model of Toxoplasma cell invasion. Curr Opin Microbiol. 2007;10:83–89. doi: 10.1016/j.mib.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Chang YY, Juhasz G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK, Neufeld TP. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans. 2009;37:232–236. doi: 10.1042/BST0370232. [DOI] [PubMed] [Google Scholar]
- Dann SG, Thomas G. The amino acid sensitive TOR pathway from yeast to mammals. FEBS Lett. 2006;580:2821–2829. doi: 10.1016/j.febslet.2006.04.068. [DOI] [PubMed] [Google Scholar]
- Denninger V, Koopmann R, Muhammad K, Barth T, Bassarak B, Schonfeld C, et al. Kinetoplastida: model organisms for simple autophagic pathways? Methods Enzymol. 2008;451:373–408. doi: 10.1016/S0076-6879(08)03225-4. [DOI] [PubMed] [Google Scholar]
- DeRocher AE, Coppens I, Karnataki A, Gilbert LA, Rome ME, Feagin JE, et al. A thioredoxin family protein of the apicoplast periphery identifies abundant candidate transport vesicles in Toxoplasma gondii. Eukaryot Cell. 2008;7:1518–1529. doi: 10.1128/EC.00081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey JP. Advances in the life cycle of Toxoplasma gondii. Int J Parasitol. 1998;28:1019–1024. doi: 10.1016/s0020-7519(98)00023-x. [DOI] [PubMed] [Google Scholar]
- Dubey JP, Lindsay DS, Speer CA. Structures of Toxoplasma gondii tachyzoites, bradyzoites, and sporozoites and biology and development of tissue cysts. Clin Microbiol Rev. 1998;11:267–299. doi: 10.1128/cmr.11.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko M, Ginger ML, Brennand A, Gualdron-Lopez M, Colombo MI, Coombs GH, et al. Autophagy in protists. Autophagy. 2011;7:127–158. doi: 10.4161/auto.7.2.13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierszinski F, Pepper M, Stumhofer JS, LaRosa DF, Wilson EH, Turka LA, et al. Presentation of Toxoplasma gondii antigens via the endogenous major histocompatibility complex class I pathway in nonprofessional and professional antigen-presenting cells. Infect Immun. 2007;75:5200–5209. doi: 10.1128/IAI.00954-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierszinski FS, Hunter CA. Advances in the use of genetically engineered parasites to study immunity to Toxoplasma gondii. Parasite Immunol. 2008;30:235–244. doi: 10.1111/j.1365-3024.2007.01016.x. [DOI] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Aaronson SA, Abrams J, Alnemri ES, Andrews DW, Baehrecke EH, et al. Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death Differ. 2009;16:1093–1107. doi: 10.1038/cdd.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, et al. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2011 doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan DC, Ogmundsdottir MH, Kazi S, Reynolds B, Visvalingam SM, Wilson C, Boyd CA. Amino acid sensing and mTOR regulation: inside or out? Biochem Soc Trans. 2009;37:248–252. doi: 10.1042/BST0370248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gozuacik D, Kimchi A. Autophagy and cell death. Curr Top Dev Biol. 2007;78:217–245. doi: 10.1016/S0070-2153(06)78006-1. [DOI] [PubMed] [Google Scholar]
- Halonen SK. Role of autophagy in the host defense against Toxoplasma gondii in astrocytes. Autophagy. 2009;5:268–269. doi: 10.4161/auto.5.2.7637. [DOI] [PubMed] [Google Scholar]
- Hamasaki M, Yoshimori T. Where do they come from? Insights into autophagosome formation. FEBS Lett. 2010;584:1296–1301. doi: 10.1016/j.febslet.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMPK: a key regulator of energy balance in the single cell and the whole organism. Int J Obes (Lond) 2008;32(Suppl 4):S7–12. doi: 10.1038/ijo.2008.116. [DOI] [PubMed] [Google Scholar]
- Herman M, Gillies S, Michels PA, Rigden DJ. Autophagy and related processes in trypanosomatids: insights from genomic and bioinformatic analyses. Autophagy. 2006;2:107–118. doi: 10.4161/auto.2.2.2369. [DOI] [PubMed] [Google Scholar]
- Huo Y, Iadevaia V, Proud CG. Differing effects of rapamycin and mTOR kinase inhibitors on protein synthesis. Biochem Soc Trans. 2011;39:446–450. doi: 10.1042/BST0390446. [DOI] [PubMed] [Google Scholar]
- Huynh MH, Carruthers VB. Toxoplasma MIC2 is a major determinant of invasion and virulence. PLoS Pathog. 2006;2:e84. doi: 10.1371/journal.ppat.0020084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue Y, Klionsky DJ. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin Cell Dev Biol. 2010;21:664–670. doi: 10.1016/j.semcdb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan KA, Hunter CA. Regulation of CD8+ T cell responses to infection with parasitic protozoa. Exp Parasitol. 2010;126:318–325. doi: 10.1016/j.exppara.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce BR, Queener SF, Wek RC, Sullivan WJ., Jr Phosphorylation of eukaryotic initiation factor-2{alpha} promotes the extracellular survival of obligate intracellular parasite Toxoplasma gondii. Proc Natl Acad Sci U S A. 2010;107:17200–17205. doi: 10.1073/pnas.1007610107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J Biol Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Lemasters JJ. Mitochondrial degradation by autophagy (mitophagy) in GFP-LC3 transgenic hepatocytes during nutrient deprivation. Am J Physiol Cell Physiol. 2011a;300:C308–317. doi: 10.1152/ajpcell.00056.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011b;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Codogno P, Cuervo AM, Deretic V, Elazar Z, Fueyo-Margareto J, et al. A comprehensive glossary of autophagy-related molecules and processes. Autophagy. 2010;6 doi: 10.4161/auto.6.4.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S. Multi-membrane-bound structures of Apicomplexa: I. the architecture of the Toxoplasma gondii apicoplast. Parasitol Res. 2005;96:258–272. doi: 10.1007/s00436-005-1338-2. [DOI] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Autophagy and cell death in model organisms. Cell Death Differ. 2009;16:21–30. doi: 10.1038/cdd.2008.120. [DOI] [PubMed] [Google Scholar]
- Kristensen AR, Schandorff S, Hoyer-Hansen M, Nielsen MO, Jaattela M, Dengjel J, Andersen JS. Ordered organelle degradation during starvation-induced autophagy. Mol Cell Proteomics. 2008;7:2419–2428. doi: 10.1074/mcp.M800184-MCP200. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M. ULK1, mammalian target of rapamycin, and mitochondria: linking nutrient availability and autophagy. Antioxid Redox Signal. 2011;14:1953–1958. doi: 10.1089/ars.2010.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz J, Hall MN. Cyclosporin A, FK506 and rapamycin: more than just immunosuppression. Trends Biochem Sci. 1993;18:334–338. doi: 10.1016/0968-0004(93)90069-y. [DOI] [PubMed] [Google Scholar]
- Laliberte J, Carruthers VB. Host cell manipulation by the human pathogen Toxoplasma gondii. Cell Mol Life Sci. 2008;65:1900–1915. doi: 10.1007/s00018-008-7556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Kang R, Bae S, Yoon Y. AICAR, an activator of AMPK, inhibits adipogenesis via the WNT/beta-catenin pathway in 3T3-L1 adipocytes. Int J Mol Med. 2011;28:65–71. doi: 10.3892/ijmm.2011.674. [DOI] [PubMed] [Google Scholar]
- Lekutis C, Ferguson DJ, Grigg ME, Camps M, Boothroyd JC. Surface antigens of Toxoplasma gondii: variations on a theme. Int J Parasitol. 2001;31:1285–1292. doi: 10.1016/s0020-7519(01)00261-2. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in aging, disease and death: the true identity of a cell death impostor. Cell Death Differ. 2009;16:1–2. doi: 10.1038/cdd.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, McFadden GI. The evolution, metabolism and functions of the apicoplast. Philos Trans R Soc Lond B Biol Sci. 2010;365:749–763. doi: 10.1098/rstb.2009.0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luder CG, Campos-Salinas J, Gonzalez-Rey E, van Zandbergen G. Impact of protozoan cell death on parasite-host interactions and pathogenesis. Parasit Vectors. 2010;3:116. doi: 10.1186/1756-3305-3-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C, Tran J, Gu F, Ochoa R, Li C, Sept D, et al. Dinitroaniline activity in Toxoplasma gondii expressing wild-type or mutant alpha-tubulin. Antimicrob Agents Chemother. 2010;54:1453–1460. doi: 10.1128/AAC.01150-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer AJ. Amino acid regulation of autophagosome formation. Methods Mol Biol. 2008;445:89–109. doi: 10.1007/978-1-59745-157-4_5. [DOI] [PubMed] [Google Scholar]
- Melo EJ, Attias M, De Souza W. The single mitochondrion of tachyzoites of Toxoplasma gondii. J Struct Biol. 2000;130:27–33. doi: 10.1006/jsbi.2000.4228. [DOI] [PubMed] [Google Scholar]
- Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya JG, Remington JS. Management of Toxoplasma gondii infection during pregnancy. Clin Infect Dis. 2008;47:554–566. doi: 10.1086/590149. [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Schworer CM. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature. 1977;270:174–176. doi: 10.1038/270174a0. [DOI] [PubMed] [Google Scholar]
- Narasimhan J, Joyce BR, Naguleswaran A, Smith AT, Livingston MR, Dixon SE, et al. Translation regulation by eukaryotic initiation factor-2 kinases in the development of latent cysts in Toxoplasma gondii. J Biol Chem. 2008;283:16591–16601. doi: 10.1074/jbc.M800681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8:R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: Critical interplay between the two homeostats. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.08.001. in press. [DOI] [PubMed] [Google Scholar]
- Onodera J, Ohsumi Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem. 2005;280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- Orlofsky A. Toxoplasma-induced autophagy: a window into nutritional futile cycles in mammalian cells? Autophagy. 2009;5:404–406. doi: 10.4161/auto.5.3.7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paguio MF, Bogle KL, Roepe PD. Plasmodium falciparum resistance to cytocidal versus cytostatic effects of chloroquine. Mol Biochem Parasitol. 2011;178:1–6. doi: 10.1016/j.molbiopara.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T, Rawal P, Wu Y, Xie W, Jankovic J, Le W. Rapamycin protects against rotenone-induced apoptosis through autophagy induction. Neuroscience. 2009;164:541–551. doi: 10.1016/j.neuroscience.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Pfefferkorn ER. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc Natl Acad Sci U S A. 1984;81:908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn ER, Eckel M, Rebhun S. Interferon-gamma suppresses the growth of Toxoplasma gondii in human fibroblasts through starvation for tryptophan. Mol Biochem Parasitol. 1986;20:215–224. doi: 10.1016/0166-6851(86)90101-5. [DOI] [PubMed] [Google Scholar]
- Polonais V, Soldati-Favre D. Versatility in the acquisition of energy and carbon sources by the Apicomplexa. Biol Cell. 2010;102:435–445. doi: 10.1042/BC20100005. [DOI] [PubMed] [Google Scholar]
- Pomel S, Luk FC, Beckers CJ. Host cell egress and invasion induce marked relocations of glycolytic enzymes in Toxoplasma gondii tachyzoites. PLoS Pathog. 2008;4:e1000188. doi: 10.1371/journal.ppat.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Amino acids and mTOR signalling in anabolic function. Biochem Soc Trans. 2007;35:1187–1190. doi: 10.1042/BST0351187. [DOI] [PubMed] [Google Scholar]
- Radke JR, Donald RG, Eibs A, Jerome ME, Behnke MS, Liberator P, White MW. Changes in the expression of human cell division autoantigen-1 influence Toxoplasma gondii growth and development. PLoS Pathog. 2006;2:e105. doi: 10.1371/journal.ppat.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kai Y, Maldonado E, Currin RT, Lemasters JJ. Roles of mitophagy and the mitochondrial permeability transition in remodeling of cultured rat hepatocytes. Autophagy. 2009;5:1099–1106. doi: 10.4161/auto.5.8.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutter GA, Da Silva Xavier G, Leclerc I. Roles of 5’-AMP-activated protein kinase (AMPK) in mammalian glucose homoeostasis. Biochem J. 2003;375:1–16. doi: 10.1042/BJ20030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Oku M, van der Klei IJ, Kiel JA. Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Seeber F, Ferguson DJ, Gross U. Toxoplasma gondii: a paraformaldehyde-insensitive diaphorase activity acts as a specific histochemical marker for the single mitochondrion. Exp Parasitol. 1998;89:137–139. doi: 10.1006/expr.1998.4266. [DOI] [PubMed] [Google Scholar]
- Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva NM, Rodrigues CV, Santoro MM, Reis LF, Alvarez-Leite JI, Gazzinelli RT. Expression of indoleamine 2,3-dioxygenase, tryptophan degradation, and kynurenine formation during in vivo infection with Toxoplasma gondii: induction by endogenous gamma interferon and requirement of interferon regulatory factor 1. Infect Immun. 2002;70:859–868. doi: 10.1128/iai.70.2.859-868.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai AP. Biogenesis of and activities at the Toxoplasma gondii parasitophorous vacuole membrane. Subcell Biochem. 2008;47:155–164. doi: 10.1007/978-0-387-78267-6_12. [DOI] [PubMed] [Google Scholar]
- Sinai AP, Joiner KA. Safe haven: the cell biology of nonfusogenic pathogen vacuoles. Annu Rev Microbiol. 1997a;51:415–462. doi: 10.1146/annurev.micro.51.1.415. [DOI] [PubMed] [Google Scholar]
- Sinai AP, Webster P, Joiner KA. Association of host cell endoplasmic reticulum and mitochondria with the Toxoplasma gondii parasitophorous vacuole membrane: a high affinity interaction. J Cell Sci. 1997b;110(Pt 17):2117–2128. doi: 10.1242/jcs.110.17.2117. [DOI] [PubMed] [Google Scholar]
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subauste CS. Autophagy in immunity against Toxoplasma gondii. Curr Top Microbiol Immunol. 2009;335:251–265. doi: 10.1007/978-3-642-00302-8_12. [DOI] [PubMed] [Google Scholar]
- Sullivan WJ, Jr, Narasimhan J, Bhatti MM, Wek RC. Parasite-specific eIF2 (eukaryotic initiation factor-2) kinase required for stress-induced translation control. Biochem J. 2004;380:523–531. doi: 10.1042/BJ20040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenter AM, Heckeroth AR, Weiss LM. Toxoplasma gondii: from animals to humans. Int J Parasitol. 2000;30:1217–1258. doi: 10.1016/s0020-7519(00)00124-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter SA, Klionsky DJ. Transport of proteins to the yeast vacuole: autophagy, cytoplasm-to-vacuole targeting, and role of the vacuole in degradation. Semin Cell Dev Biol. 2000;11:173–179. doi: 10.1006/scdb.2000.0163. [DOI] [PubMed] [Google Scholar]
- Vivier E, Petitprez A. Donnes ultrastructurales complementaires, morphologiques et cytochimiques, sur Toxoplasma gondii. Protistologica. 1972;8:199–221. [Google Scholar]
- Waller RF, McFadden GI. The apicoplast: a review of the derived plastid of apicomplexan parasites. Curr Issues Mol Biol. 2005;7:57–79. [PubMed] [Google Scholar]
- Wang K, Klionsky DJ. Mitochondria removal by autophagy. Autophagy. 2011;7:297–300. doi: 10.4161/auto.7.3.14502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Karnataki A, Parsons M, Weiss LM, Orlofsky A. 3-Methyladenine blocks Toxoplasma gondii division prior to centrosome replication. Mol Biochem Parasitol. 2010;173:142–153. doi: 10.1016/j.molbiopara.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Weiss LM, Orlofsky A. Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J Biol Chem. 2009;284:1694–1701. doi: 10.1074/jbc.M807890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem. 2011;80:125–156. doi: 10.1146/annurev-biochem-052709-094552. [DOI] [PubMed] [Google Scholar]
- Weiss LM, Kim K. The development and biology of bradyzoites of Toxoplasma gondii. Front Biosci. 2000;5:D391–405. doi: 10.2741/weiss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Zhong Y. From a global view to focused examination: understanding cellular function of lipid kinase VPS34-Beclin 1 complex in autophagy. J Mol Cell Biol. 2010;2:305–307. doi: 10.1093/jmcb/mjq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Effect of starvation medium on the integrity of the host cell monolayer (blue) and the appearance of altered T. gondii mitochondria (red). The mean number of host nuclei per field following incubation for 8 hours in 6%-SM. In addition the changes in starvation induced mitochondrial morphology were determined. Incubation of infected cells in 6%-SM resulted in an optimal balance of the 2 phenotypes. Nuclei were labeled with Hoechst stain while anti-F1β antibody was use to stain the parasite mitochondrion. A minimum of 50 fields was counted in each of 3 independent experiments. (B) Measurement of lactate dehydrogenase LDH activity in the culture supernatant in uninfected and infected cells incubated for 8 hours in CM or 6%SM. While LDH release was higher in infected cells, there was no effect of 6%SM on host cell viability relative to the appropriate CM condition. LDH release following incubation with 0.1% Triton X-100 for 10 minutes was used as positive control. The activity data are presented as ratio of the measured optical density relative to the 0.1% TX100 control.
(A) Treatment of infected cells with complete media (CM) with 10 mM 3-MA for 8 hours does not have any effect on the morphology of the parasite mitochondrion. Scale bar = 2μm. (B) The 3-MA mediated block of mitochondrial fragmentation is independent of translational repression as comparable to induction of TgIF2α-P levels are evident in the presence or absence of the compound. Notably 3-MA alone serves as a stress causing a small increase in TgIF2α-P above the untreated control.
Treatment of infected cells in 6%-SM supplemented with 10 mM 3-MA has no effect on the morphology of the rhoptries (ROP7), micronemes (MIC2) or the apicoplast (anti-ATrx1). Scale bar = 2μm.
The AMPK agonists Metformin and AICAR do not trigger significant levels of mitochondrial fragmentation suggesting that the manipulation of glucose sensing plays a limited role in the triggering of starvation induced mitophagy in T. gondii. Results shown are the mean ± S.D. from 3 replicates each.
Effect of starvation for micronutrients (vitamins and lipoic acid) on the mitophagy phenotype. HFF monolayers were infected for 24 hours prior to being switched to vitamin (A) or lipoic acid (B) free media for 8 hours. The impact of treatment on mitochondrial morphology was determined by immunofluorescence microscopy. Results shown are the mean ± S.D. from 3 replicates each. A minimum of 100 vacuoles were counted at random for each sample for each replicate.
Bioinformatic identification of potential T. gondii homologs of yeast Atg proteins with their basic characteristics and generalized biological function. Targets hits of >30% identity (50% similarity) are considered bonafide hits. Those with <30% identify (50% similarity) are considered provisional hits. References regarding the specific biological functions for the yeast genes are reviewed in (Klionsky et al., 2010). Genome Sources Saccharomyces: www.yeastgenome.org, Toxoplasma: www.toxodb.org.