Abstract
In previous studies we showed that the replication of Cryptococcus neoformans in the lung environment is controlled by the glucosylceramide (GlcCer) synthase gene (GCS1), which synthesizes the membrane sphingolipid GlcCer from the C9-methyl ceramide. Here, we studied the effect of the mutation of the sphingolipid C9 methyltransferase gene (SMT1), which adds a methyl group to position 9 of the sphingosine backbone of ceramide. The C. neoformans Δsmt1 mutant does not make C9-methyl ceramide and, thus, any methylated GlcCer. However, it accumulates de-methylated ceramide and de-methylated GlcCer. The Δsmt1 mutant loses more than 80% of its virulence compared to the wild-type and the reconstituted strain. Interestingly, growth of C. neoformans Δsmt1 in the lung was decreased and C. neoformans cells were contained in lung granulomas, which significantly reduced the rate of their dissemination to the brain reducing the onset of meningoencephalitis. Thus, using fluorescent spectroscopy and atomic force microscopy we compared the wild-type and Δsmt1 mutant and found that the altered membrane composition and GlcCer structure affects fungal membrane rigidity, suggesting that specific sphingolipid structures are required for proper fungal membrane organization and integrity. Therefore, we propose that the physical structure of the plasma membrane imparted by specific classes of sphingolipids represents a critical factor for the ability of the fungus to establish virulence.
Introduction
Cryptococcosis is a chronic human disease caused by the fungus Cryptococcus neoformans that originates from the inhalation of either the encapsulated fungi or fungal spores from environmental sources. Once inhaled into the lung, C. neoformans can disseminate through the bloodstream to the central nervous system causing fungal meningo-encephalitis, which is a particularly life-threatening disease to immunocompromised patients (Casadevall et al., 1998, Datta et al., 2009, Litvintseva et al., 2005, Park et al., 2009, Mukherjee et al., 1993, Lin et al., 2006, Park et al., 2011). An important characteristic of this fungus that enables it to cause disease is its ability to grow in both neutral/alkaline and acidic environments inside the human body. The pathogen encounters neutral to alkaline environments in the extracellular space of the alveoli and in the bloodstream. Once the pathogen is engulfed by lung macrophages via the formation of the phagolysosome, C. neoformans finds itself in an acidic microenvironment.
Recently, studies have highlighted the importance of sphingolipids in the development of pathogenicity of this fungus by regulating growth of C. neoformans in acidic and neutral/alkaline environments. Certain sphingolipid metabolizing enzymes are required for intracellular growth (Garcia et al., 2008, Shea et al., 2006), whereas others are important for extracellular growth (Rittershaus et al., 2006). Intracellular growth appears to be controlled by inositol sphingophospholipid phospholipase C 1 (Isc1) through the production of very long chain phytoceramide species, which promotes C. neoformans growth at low pH via the regulation of Pma1 (Garcia et al., 2008, Shea et al., 2006). Interestingly, glucosylceramide (GlcCer) has also been previously implicated as a component that enables yeast to be alkali tolerant (Saito et al., 2006). Extracellular growth of C. neoformans is controlled by glucosylceramide synthase 1 (Gcs1) and thereby the production of glucosylceramide; the absence of glucosylceramide in fact causes a growth defect specifically when the fungus is in a neutral/alkaline environment similar to that found in the alveolar spaces (Kechichian et al., 2007, Rittershaus et al., 2006). How GlcCer regulates this extracellular growth is, at present, unclear.
Several studies over the last few years have established that sphingolipids are not only an integral part of the eukaryotic membranes (Lingwood et al., 2010, Sandhoff, 2010, Sonnino et al., 2010, van Meer et al., 2010, Giocondi et al., 2010, van der Meer-Janssen et al., 2010), but also they play a significant role in maintaining the physical and physiological functions of those membranes. In mammalian cells, sphingolipids serve as precursors for more than 300 species of glycosphingolipids (Pruett et al., 2008, Fahy et al., 2005, Merrill et al., 2001, Hakomori, 1981) and their synthesis and degradation contributes to control the levels of ceramides, which are bioactive lipids involved in the regulation of many biological processes (Leipelt et al., 2001). Cell membranes have a dynamic architecture that is the result of a delicate balance between the major lipid classes, namely sphingolipids, glycerophospholipids and sterols. In response to cellular needs and environmental signals, molecular clusters formed by specific lipids and proteins in the plasma membrane are continuously restructured in order to regulate essential cellular processes. In mammalian cells, these clusters are called lipid-microdomains or lipid rafts and consist of sphingolipids and cholesterol.
Fungal membranes also contain lipid microdomains and they too regulate many cellular functions (Brown et al., 1998). On the other hand, fungal lipid microdomains differ from the mammalian counterpart because they are composed of ergosterol instead of cholesterol as the main sterol and different sphingolipid structures (Hechtberger et al., 1994). Importantly, lipid raft formation is affected by ratios of sphingolipids/phospholipids/sterols, the degree of fatty acyl phospholipid chain unsaturation, and the sphingolipid structure of the head group or/and of the acyl chain (Xu et al., 2001). Model membranes are characterized by the presence of gel-phase domains, containing highly condensed and immobile lipids, co-existing with both liquid ordered (lo) and liquid disordered (ld)-phase domains (Brown, 2006). Under physiological conditions, the gel lipid phase is usually absent in living eukaryotic cells. However, a very recent study on yeast plasma membrane reported the presence of ‘gel-like’ domains highly ordered and enriched with sphingolipids (Aresta-Branco et al., 2011). In general, most biophysical evidence indicates that the rafts can be equated to a liquid ordered phase, which has a phase status in-between the very ordered or solid ordered gel phase and liquid disordered phase, with the rest of the membrane in a non-raft liquid disordered phase.
Rafts can be of different sizes and they can form clusters to function in cellular processes via oligomerization and scaffolding with other proteins (Brown et al., 1998). Raft clusters are dynamic, reversible, can be dissembled by removal of raft components (Harder et al., 1998, Pralle et al., 2000), and are especially important in host-pathogen interactions (Baorto et al., 1997, Tran et al., 1987). Siafakas et al. were the first to report that lipid rafts are important for cryptococcal pathogenicity (Siafakas et al., 2006) and that certain virulence determinants, such as the ceramide monohexoside that they isolated, are concentrated in lipid rafts.
GlcCer is an antigenic glycosphingolipid with a β-D-glucose moiety at the primary carbon and a 9-methyl-4,8-sphingadienine in an amide linkage to 2-hydroxyoctadecanoic acid (Rodrigues et al., 2000). Fungal GlcCer differs from plant and mammal GlcCer (Heinz, 1996, Lynch, 1993, Rodrigues et al., 2000) because of a methyl group present at the C9-position of the sphingoid base. In Pichia pastoris, sphingolipid C9 methylation of GlcCer occurs in the membrane, suggesting a role in membrane integrity (Ternes et al., 2006), whereas in Candida albicans (Noble et al., 2010) methylation of GlcCer is required for hyphal elongation (Oura et al., 2010), suggesting a role for virulence (Ramamoorthy et al., 2009). Importantly, fungal mutants lacking the sterol methyl transferase gene (erg6) have altered membrane structural features (Kleinhans et al., 1979, Lees et al., 1979). Together, these studies suggest that methylation of membrane lipids, and in particular sphingolipids, is important for fungal virulence and perhaps for membrane integrity.
Thus, we created a mutant of C. neoformans lacking the methyl group at the C9 position of GlcCer and performed pathogenesis, biochemical, and molecular studies that showed a clear correlation between the regulation of membrane conformational integrity regulated by GlcCer and C. neoformans pathogenicity.
Results
Disruption and reconstitution of C. neoformans SMT1 gene
To determine the role of the methyl group at the GlcCer C9 position in the pathophysiology of C. neoformans, we deleted the gene from C. neoformans var. grubii strain H99 wild-type (WT) and created the mutant C. neoformans Δsmt1 (Figure 1). The plasmid containing the deletion cassette had a NAT1 gene flanked upstream by 5′UTR and downstream by 3′UTR of the SMT1 gene. Details can be found in the supplementary material (Figure S1A). We re-introduced the SMT1 gene back into the Δsmt1 using the reconstitution cassette, which has the HYG allele as the selectable marker (Figure S1B). Homologous recombinants were screened by Southern hybridization using a fragment of SMT1 open reading frame and 5′UTR as probe (Figure 1). Recombinant # 2 was selected as the reconstituted strain and renamed as C. neoformans Δsmt1 + SMT1.
Figure 1. Deletion and reconstitution of SMT1 gene.
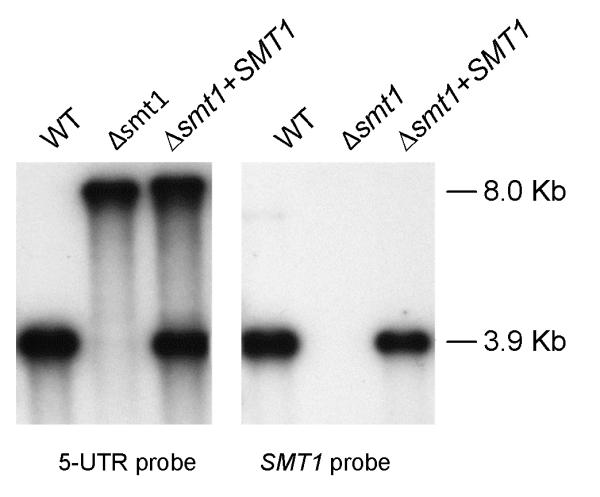
Southern hybridization analysis of genomic DNA of WT, Δsmt1, and Δsmt1+SMT1 digested with SalI and screened with the indicated probes. The Δsmt1 is negative when probed with SMT1 probe indicating the absence of the SMT1 gene. Whereas the Δsmt1+SMT1 shows the re-introduction of the SMT1 gene as illustrated.
Sphingolipid C9 methyltransferase is responsible for methylation at the glucosylceramide C9 position
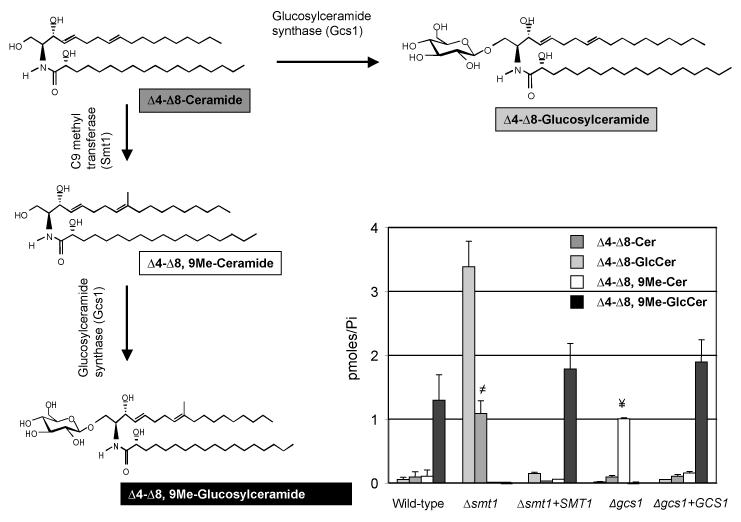
Lipid analysis of glycosylceramide and ceramide species from WT, Δsmt1, and Δsmt1 + SMT1 by mass spectrometry (Figure 2) showed that the C9 methyltransferase knockout mutant Δsmt1 produced only de-methylated GlcCer and it accumulated only de-methylated ceramide. The reconstituted strain Δsmt1 + SMT1 had lipid levels comparable to those observed in the WT strain, proving the successful re-introduction of the SMT1 gene. C. neoformans Δgcs1 and Δgcs1 + GCS1, previously described in (Rittershaus et al., 2006), were used as control strains (Figure 2). Therefore the changes in the lipid profile support the conclusion that the deleted SMT1 gene is indeed the sphingolipid C9 methyltransferase.
Figure 2. Deletion of Smt1 produces a strain that make only un-methylated GlcCer.
Analysis of specific lipids extracts of WT, Δsmt1, Δsmt1+SMT1, Δgcs1, and Δgcs1+GCS1 cells along with the structure illustration of α-OH-Δ4-Δ8-ceramide, α-OH-Δ4-Δ8,9-methyl-ceramide, α-OH-Δ4-Δ8,9-methyl-GlcCer and α-OH-Δ4-Δ8-GlcCer. The WT along with Δsmt1+SMT1 reveals almost identical amounts of α-OH-Δ4-Δ8,9-methy-GlcCer but negligible amounts of α-OH-Δ4-Δ8-ceramide and its methylated product α-OH-Δ4-Δ8,9-methyl-ceramide. However, the Δsmt1 does not produce methylated GlcCer and accumulates α-OH-Δ4-Δ8-ceramide. In contrast, Δgcs1 accumulated α-OH-Δ4-Δ8,9-methyl-ceramide and lacked methylated and unmethylated GlcCer. All results were means ± SD of three independent experiments. ≠P<0.05, Δsmt1 versus wild-type; ¥P<0.05, Δgcs1 versus WT.
To further prove the identity of the sphingolipid-specific enzyme C9 methyltransferase and its successful deletion, in vitro methyltransferase activity was performed using demethylated ceramide (α-OH-Δ4-Δ8-ceramide) as a substrate and adenosyl-L-methionine, S-[methyl-14C] (14C SAM), as the source of the radiolabelled methyl (CH3) group (Ternes et al., 2006). The methylated ceramide (α-OH-Δ4-Δ8,9-methylceramide) was used as a control substrate. These sphingolipids, which are not commercially available, were prepared in our laboratory (see supplementary materials for details). Briefly, WT type GlcCer [(E,E)-9-methylsphinga-4,8-dienine as sphingoid base in amide linkage to 2-hydroxyoctadecanoic acid] was isolated and purified from C. neoformans WT cells, whereas demethylated GlcCer [(E,E)-demethylated sphinga-4-8-dienine as a sphingoid base] was isolated and purified from Δsmt1. Corresponding ceramides were produced by treatment of the above GlcCer species with Cerezyme, which hydrolyses the glycosidic linkage of the β-D-glucose moiety with the 4-8-sphingadienine base (Rhome et al., 2011). These ceramide species were then purified using a thin layer chromatography (TLC) and used in the C9 methyltransferase assay.
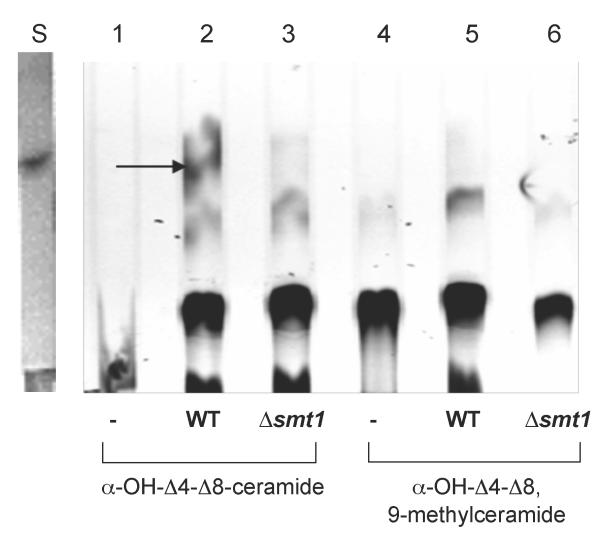
Sphingolipid C9 methyltransferase activity could only be detected using α-OH-Δ4-Δ8-ceramide incubated with cell homogenate from WT and not from Δsmt1 (Figure 3). As expected, C9 methyltransferase activity was not detected when using α-OH-Δ4-Δ8, 9-methylceramide as substrate (Figure 3). The corresponding quantification of the radiolabelled spot is shown in Figure S2.
Figure 3. In vitro enzyme assay of the sphingolipid C9 methyltransferase in C. neoformans.
Lipid extracts from the assay were separated by TLC and analyzed by radioscanning. The 14C-labelled methyl-ceramide can only be detected in WT lysates that have C9 methyltransferase, which acts upon the ceramide substrate-α-OH-Δ4-Δ8-ceramide and transfers a 14CH3 group from 14C SAM to form α-OH-Δ4-Δ8,9-methylated-ceramide represented by the radioactive spot in lane 2. In lanes 1 and 4, ‘-’ represent the respective substrate, ceramide or 9-methylceramide plus 14C SAM , but without any lysates. The ‘S’ lane represents 5 μg of purified and unlabeled 9-methylceramide as marker.
To determine whether other sphingolipids such as complex sphingolipids were altered in the mutant, we performed in vivo labeling studies with [3H]-palmitic acid and [3H]-myo-inositol (see supplementary materials and methods for details). Δsmt1 had no or negligible changes in complex sphingolipids compared to the WT (Figure S3).
Loss of the methyl group in position 9 of the ceramide backbone caused a growth arrest in an in vitro environment that mimics the lung extracellular alveolar space environment
Since the C. neoformans Δgcs1 mutant (which lacks GlcCer) undergoes growth arrest at neutral-alkaline pH, 37°C, 5% CO2, which mimics the alveolar micro-environment (Rittershaus et al., 2006), we studied the growth of the Δsmt1 mutant under these in vitro conditions in comparison to WT and Δgcs1 strains. The Δsmt1 mutant showed a similar phenotype to the Δgcs1 at pH 7.2 (Figure S4A) or pH 4.0 (Figure S4B). This growth defect is reversible because the growth was completely restored when the cells were washed and fresh acidic media were added (Figure S4C), suggesting that the deletion of the −CH3 group from the sphingadienine backbone of GlcCer itself can affect extracellular growth of this pathogenic fungus.
Deletion of the −CH3 group from GlcCer resulted in loss of virulence of C. neoformans
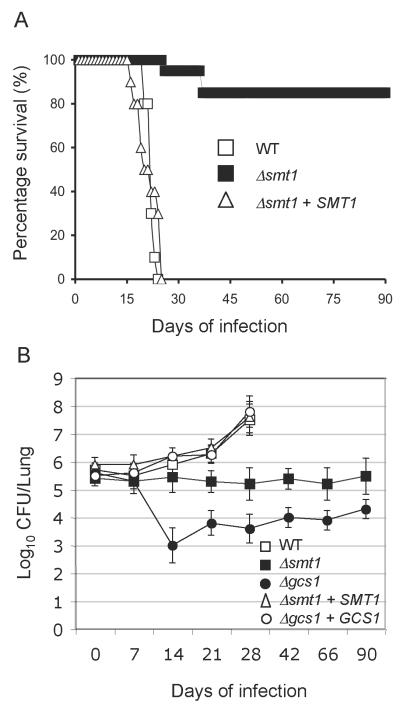
Virulence studies were performed in a murine model (CBA/J) of cryptococcosis. Mice were injected intranasally with either WT, mutant, or the reconstituted strain. Mice infected with WT and reconstituted strain survived an average of 22±1.0 and 21± 2.7 days, respectively. However, 85% of the mice infected with the Δsmt1 strain survived 90 days post infection (Figure 4A). Tissue burden culture of excised lungs were performed and illustrated in Figure 4B. They showed that the number of Δsmt1 cells recovered during the infection of 90 days is similar to the number of Δsmt1 cells (5×105 cells) injected at time 0. These results were similar of those obtained using the Δgcs1 mutant strain (Figure 4B). As expected, both Δsmt1+SMT1 and Δgcs1+GCS1 reconstituted strains showed a lung tissue burden similar to the WT strain (Figure 4B).
Figure 4. SMT1 is required for virulence of C. neoformans.
(A) ice infected intranasally with WT and the reconstituted strain Δsmt1+SMT1 have an average survival of 22.1±1.054 and 21.076± 2.77 days, respectively. Mice infected with Δsmt1 have ~ 85% survival at 90 days post infection (n=10 WT and Δsmt1+SMT1; n=15 for Δsmt1). (B) Tissue burden culture of mouse lungs infected with WT, Δsmt1, Δsmt1+SMT1, Δgcs1, and Δgcs1+GCS1 strains. Results show that a lower or similar CFU to the inoculum used to infect the mice, indicating that the mutants did not grow within the lung during observed period.
Of interest, at 90 days of infection, only one-third of the mice had Δsmt1 cells in the brain, with <3,000 fungal cells per organ (Figure S5), although all Δsmt1 infected mice did not show any sign of meningoencephalitis, such as tremor, irritability, increased respiration, or weight loss. The Δsmt1 strain slightly disseminated to other organs with an average tissue burden in the liver, kidney, and spleen of <1,000 fungal cells per organ (Figure S5), but this was only observed at 90 days after the injection (data not shown).
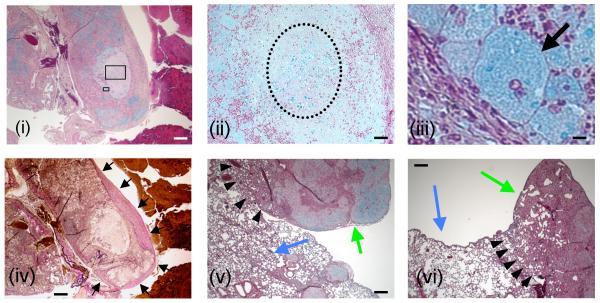
Lung histopathology performed at 90 days post-infection revealed an organized granulomatous response in all Δsmt1-infected mice, although the granuloma was not as well organized as in mice infected with the Δgcs1. A representative illustration of the granulomatous response to Δsmt1 is shown in Figure 5. In Figure 5i, a low magnification of the lung tissue stained with MOVAT shows a section of a lung lobe totally infiltrated with inflammatory cells (mostly neutrophils) surrounding a circular bright area corresponding to necrotic tissue. Many blue patches, corresponding to C. neoformans Δsmt1 cells, are present within the necrotic area as well as within the neutrophilic infiltrations. The necrotic area can be readily observed in Figure 5ii (dotted circle), higher magnification of Figure 5i. A ring of foamy and giant macrophages in which the cytoplasma is stained blue surrounds the necrotic area. The black arrow in Figure 5iii indicates one such macrophage in which the blue staining depicts fungal polysaccharides, suggesting that ingested C. neoformans cells that have been successfully killed. The ring of macrophages surrounding the necrotic tissue containing many C. neoformans cells is also illustrated in Figure S6, indicated by white arrows. This macrophage ring is similar to the one observed in lungs infected with the Δgcs1 mutant (white arrow in Figure S7).
Figure 5. Histopathological analysis of Δsmt1-infected lungs.
Lung sections of Δsmt1-infected mice were performed at 90 days of infection. They reveal distinct formation of granulomas within which Δsmt1 cells are contained. Panels i, ii, iii, v and vi are Movat stained sections, whereas panel iv is stained with VVG and depicts the same area illustrated in i. Boxed areas in 5i are magnified in 5ii and 5iii. C. neoformans Δsmt1 cells are stained blue in i, ii, iii, v and vi) are mostly contained within giant macrophage(s) (arrow in iii) or within a necrotic zone (dotted circle in ii). The granulomatous response is surrounded by collagen fibers stained pink-to-dark red (black arrows in iv). Normal lung tissues are indicated by blue arrows in v and vi, in close proximity of granuloma (green arrows in v and vi). Scale bars: 500 μm (i, iv, v and vi); 50 μm (ii); 10 μm (iii).
The area surrounding the granuloma is also infiltrated by fibroblasts and fibrotic tissue with significant amount of collagen deposition stained dark pink (Figure 5iv, black arrows). These granulomatous formations are in close proximity of normal lung tissue (compare green versus blue arrows in Figure 5v and 5vi). It should also be noted that the delimitation of the lung granuloma in Δsmt1-infected mice (arrowheads in Figure 5v and 5iv) was similar to the one found in the Δgcs1-infected mice (black arrowheads in Figure S7) but not identical as the host infiltration in the latter appears to be more pronounced.
The altered structure of GlcCer caused a defective cell membrane
Sodium dodecyl sulfate (an ionic detergent), Triton X-100 and Triton X-114 (non-ionic detergents) are reagents very commonly used to disrupt cell wall/membrane. We hypothesized that because fungal GlcCer is localized in the cell membrane, reduced or defective growth of the Δsmt1 mutant in presence of SDS in a spot-dilution assay would be preliminary evidence of a cell wall/cell membrane defect. The Δsmt1 strain was found to be susceptible to 0.05% SDS similarly to the Δgcs1 mutant (Figure S7). This susceptibility was more pronounced when cells were grown at 37°C, 5% CO2 compared to 30°C or in 0.04% CO2 (data not shown). Similar results were also obtained with non-ionic detergents Triton X-100 and Triton X-114 (data not shown). These results suggest that a structurally different GlcCer may have resulted in the formation of a defective cell membrane.
Enhanced membrane permeability of Δsmt1
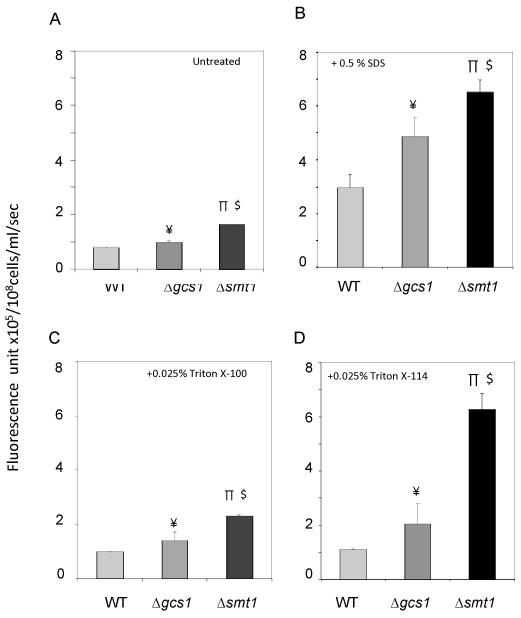
We next examined the membrane permeability of WT and Δsmt1 strains using Sytox green. A more permeable or defective membrane will allow the fluorophore to penetrate cells, bind to nucleic acids and intracellularly fluoresce (Thevissen et al., 1999). Δsmt1 cells grown in neutral/alkaline (pH 7.2) medium at 37°C, 5% CO2 showed increased uptake of Sytox green compared to WT (Figure 6A). Over time, Sytox green uptake increased, plateauing after 20 min (Figure S9). Cells treated with ionic (SDS) or non-ionic (Triton X-100, X-114) detergents showed even greater uptake of fluorescence at pH 7.2, 37°C, 5% CO2 compared to the WT strain (Figure 6B, 6C, and 6D respectively). Thus, a significantly greater permeabilization of the plasma membrane was observed in the Δsmt1 compared to the WT strain. This increased permeabilization was also conserved in the Δgcs1 strain (Figure 6). These results suggest that a relatively slight structural deformity of GlcCer due to the lack of the −CH3 moiety caused a leaky cell membrane lipid bed, which subsequently induced the increase in fluorescence uptake. Interestingly, this leaky phenotype is more pronounced in the Δsmt1 compared to the Δgcs1. To control for background fluorescence due to dead cells, fluorescent units were calculated taking into consideration cell viability, which was measured by serial dilutions, plating, and counting CFU/ml. Importantly, cell viability was identical among the different treatment groups (data not shown).
Figure 6. Sytox green (SG) uptake assays.
Untreated (A), 0.5% SDS-(B), 0.025% Triton X-100- (C) 0.025% Tritox X-114 treated (D) cells (density 108 cells/ml) were incubated in PBS and 2 μM SG in the dark for 10 min and then fluorescence (arbitrary unit) was quantified. Background corrections were made by subtracting the fluorescence in PBS (−/+ detergents) + SG for untreated and treated cells, respectively. Both Δsmt1 and Δgcs1 cells had greater Sytox green uptake than WT, whether or not cells were untreated or treated with detergents. The difference in fluorescence was most prominent at pH 7.2, 37 °C, 5% CO2, conditions (shown in the figure) which replicate the alveolar lung environment. All results were means ± SD of three independent experiments. ¥P<0.05, Δgcs1 versus WT; πP<0.05, Δsmt1 versus WT; $P<0.05, Δsmt1 versus Δgcs1;
Fluorescence spectroscopy confirmed altered membrane order in neutral to alkaline pH in Δsmt1
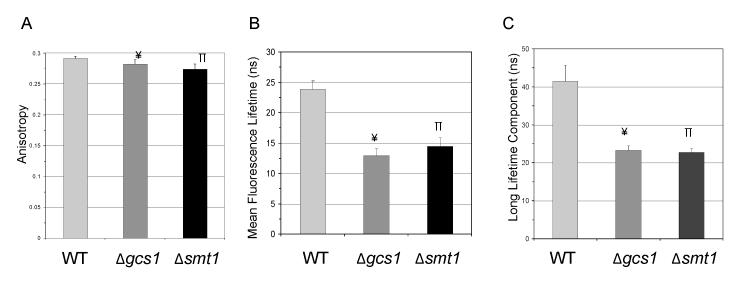
The biological membrane is a heterogeneous mixture in which the gel, liquid ordered (lo) and the liquid disordered (ld) phases co-exist and contribute to various cellular functions (Schuck et al., 2003). A common biophysical method to detect this heterogeneity or to probe alterations of the native membrane order is fluorescence spectroscopy because it is highly sensitive and multiple probes are available (Reyes Mateo et al., 1993). Here, we used 2 probes to study the fluidity of membrane microdomains. The probe TMA-DPH is expected to localize at the outer leaflet of the membrane and distribute equally among different membrane phases, presumably gel, l0, and ld (Barrow et al., 1985). Fluorescence anisotropy data for this probe suggested that at pH 7.2 there is a small but significant decrease in the global order of membranes in either Δsmt1 or Δgcs1 compared to WT (Figure 7A), due perhaps to a less ordered outer membranes. The t-PnA probe allows the measurement of the mean fluorescence lifetime (Figure 7B) and the long lifetime component (Figure 7C) of the fluorescence decay, which provides information about membrane order since these measurements are sensitive for the presence of different phases, particularly the gel phase (Sklar et al., 1977). Figure 7B shows that the global order is much higher in the WT. This higher global order in the WT is due to the presence of domains which have ‘gel-like’ phases, which is supported by the results obtained by the long lifetime component of the fluorescence decay of t-PnA illustrated in Figure 7C, showing a decay value of above 30 ns, typical of a gel phase (Castro et al., 2007). The long lifetime component of Δsmt1 and Δgcs1 is lower and typical of a lo phase (Figure 7C). Thus, these results suggest that the deletion of the CH3 group on GlcCer prevents the formation of domains that have gel phases and favors the formation of lo phases and by itself is sufficient to reproduce the membrane rearrangements caused by loss of GlcCer.
Figure 7. Analysis of membrane order by fluorescence spectroscopy study.
Fungal cells were grown at pH 7.2, 37 °C in the presence of 5% CO2. Two probes, TMA-DPH and t-PnA, were selected for fluorescence anisotropy studies. (A) Fluorescence anisotropy from probe TMA-DPH reveals a small but significant difference in anisotropy values of the three membranes. (B) Mean fluorescence lifetime studies with t-PnA indicate a less ordered membrane in mutants when compared to WT. (C) The long lifetime component of t-PnA fluorescence decay is the highest in WT and typical of a gel phase, thus demonstrating that WT membranes are more ordered than the two mutants. The long lifetime component values in Δsmt1 are typical of a lo phase. Results are means ± SD of three independent experiments. ¥P<0.05, Δgcs1 versus WT; πP<0.05, Δsmt1 versus WT.
Visualization of the membrane lipid topography by atomic force microscopy (AFM) illustrates that the −CH3 group in GlcCer helps in maintaining membrane lipid bilayer rigidity
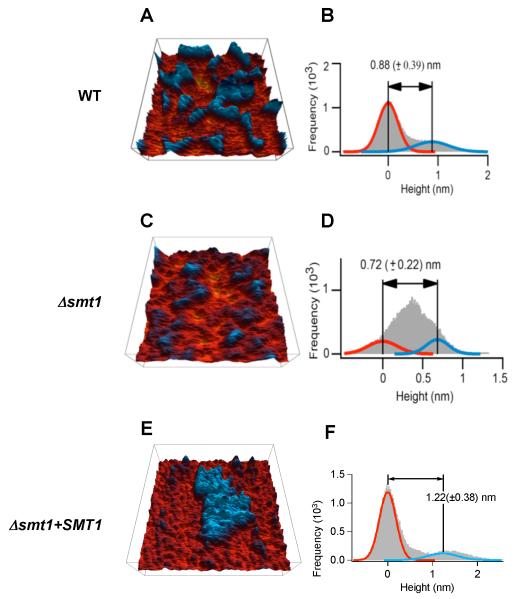
To confirm results obtained with fluorescent probes, we investigated the topography of the membrane lipids of WT, Δsmt1 and Δsmt1+SMT1 reconstituted strain using AFM. In these 3-D false-color images (Figure 8A, 8C and 8E), the taller domain is blue and the shorter one is red. Histogram analyses indicated that the two domains have a height difference of approximately 1 nm (Figure 8B, 8D and 8F). This height difference is explained by the relative abundance of total sphingolipids (including α-OH-Δ4-Δ8,9-methyl-GlcCer) and total phospholipids membranes. Indeed, the molecular size of complex sphingolipids (e.g. GlcCer or inositol containing sphingolipids) is approximately 0.5 nm longer than phospholipids whereas the molecular size of ceramide species (e.g. α-OH-Δ4-Δ8-ceramide which accumulates in the Δsmt1) is similar to those of phospholipids.
Figure 8. Atomic force microscopy of reconstituted membrane lipid bilayers.
A, C and E represent height images whereas B, D and E represent histograms of the height images of C. neoformans WT (A and B), Δsmt1 (C and D) and Δsmt1+SMT1 (E and F) strains. The sizes of the images are 750×750 nm2 for WT, and 500×500 nm2 for Δsmt1 and Δsmt1+SMT1 strains. In the 3-D height images (A, C and E), the sphingolipid domains are shown in blue in contrast to the phospholipid/ceramide in red. Using the histograms (B, D and F), the height differences between domains and backgrounds are computed by fitting two Gaussian curves to represent the two area types and calculating the differences of their means (a third Gaussian fit representing the transition of the two is not shown). All fungal cells used in these experiments were grown at pH 7.2, 37 °C, in the presence of 5% CO2. Data are representative of three separate experiments.
Analysis of the phase images of the topography results provide additional evidence to support the proposed domain assignments. In WT membrane the taller domains showed greater phase reduction and thus are more rigid than the shorter domains (data not shwon). This is consistent with sphingolipids being more rigid than phospholipids, since sphingolipid molecules have two straight acyl chains whereas phospholipid molecules have one straight and one tilted acyl chain. In the Δsmt1 membrane we found extra soft spots in the phase images (data not shown) perhaps due to the accumulation of α-OH-Δ4-Δ8-ceramide (shorter than GlcCer). The extra softness of ceramides may be due to the lack of hydrophilic head groups and the consequent lose packing of lipid bilayers. Although showing a slightly different pattern, Δsmt1+SMT1 reconstituted strain had an overall lipid topography similar to that of the WT strain (Figure 8E and 8F) further validating that the structural alteration in the GlcCer molecule is responsible for the changed membrane lipid topography in the mutant Δsmt1.
Discussion
Here we report that a structural variation in a membrane lipid-raft constituent glycosphingolipid molecule in the fungus C. neoformans changes the topography of the membrane lipid in which it is localized and is associated with a change in fungal patho-physiology. Specifically, the virulence of C. neoformans mutant Δsmt1 was significantly reduced and the mutant does not migrate from the lung environment to cause disease. Instead, the mutated fungus remains inside the lung trapped in a granuloma. This loss of virulence may be due to the less ordered global membrane-lipid environment of the mutant caused by the absence of domains with gel-like phases in Δsmt1 membranes, as suggested by permeability, fluorescence spectroscopy and AFM studies. The leaky mutant membrane is more pronounced in neutral-alkaline environments compared to acidic pH (at 37 °C, 5% CO2). Such a neutral-alkaline setting mimics physiological conditions encountered by the pathogen once it is lodged into the alveolar spaces, thus making the mutant highly susceptible to this environment.
It is well established that membrane order is a function of sphingolipid concentration and that cells regulate the synthesis of sphingolipids in a coordinated manner with the abundance and/or reduction in their levels depending on extracellular and cellular signals (Walther, 2010). At present, little is known about how the cell regulates sphingolipid levels and whether and how a specific structural sphingolipid feature can affect the molecular interactions among sphingolipids and also with other membrane components. However, each lipid, differing in molecular complexity, is expected to have a defined role in the membrane physiology of an organism, especially because lipids have highly selective molecular chaperonic abilities with respect to the chemical composition, physical properties, and the chirality of the molecule (Bogdanov et al., 1999). Thus, the structural preference and charge of the lipid can influence the folding and unfolding of protein along with the supramolecular organization of the lipo-protein complex, which makes up the functional membrane microdomains (Lang et al., 2001). Our observed alterations in specific sphingolipids (Figure 2) have obviously disrupted the delicate balance between sphingolipids and other membrane components, thereby leading to differential phase separation (Figure 8). In the Δsmt1 mutant, the fact that no significant increase in complex sphingolipids was observed from in vivo [3H]-palmitic acid labeling studies (Figure S3) or from in vivo labeling studies with [3H]-myo-inositol (data not shown) excludes a possible role of these lipids in regulating lipid phase topography in the Δsmt1. On the other hand, it is possible that different ceramide(s) species, intermediates in the GlcCer de novo biochemical pathway (Figure S10), might be involved in maintaining a balance which regulates lipid phase topography in this fungus. This hypothesis may be supported by the observation that the Δsmt1 (which lacks methylated ceramide and methylated GlcCer) has a more leaky and fluid membrane than the Δgcs1 mutant (which lacks only methylated GlcCer) (Figure 6). Thus, it is also possible that methylated ceramide, which accumulates in the Δgcs1 mutant, may compensate for the leaky membrane observed with Sytox green uptake assays. Irrespective of the accumulation of structurally different ceramide species in the mutants (de-methylated and methylated ceramide in Δsmt1 and Δgcs1, respectively) (Figure 2), both strains have attenuated virulence and a similar phenotype in a lung extracellular-like environment. Thus, it is possible that, in addition to the role of GlcCer per se, the methyl group on GlcCer in particular may be the key determinant of these phenotypes. Accumulation of ceramide, either methylated or de-methylated, may also be a critical player in the re-structuring of the membrane phase dynamics. In an attempt to investigate these possibilities, we performed experiments with the purified methylated or un-methylated ceramide species along with purified methylated or un-methylated GlcCer species, which were added back to fungal cultures to recapitulate WT phenotypes. Our attempts were not successful (data not shown), likely because these highly hydrophobic molecules cannot cross the capsule and the cell wall and be properly incorporated in the membranes to reconstitute the WT membrane topography. Another possibility may be that these sphingolipids can be properly assembled in the membranes only when synthesized endogenously.
Comparisons of the height images obtained from AFM between WT and the Δsmt1 mutant suggests that the sphingolipid microdomains in WT lipid bilayers are larger and are more tightly packed than their counterparts in Δsmt1. It is important to consider that the height differences among the strains may be due not only to the different lipid sizes but also to the phases formed by these lipids. In general, when the lipids are in the gel-like or lo phase, the membrane is thicker than when they are in the ld (fluid) phase. In the gel phase, lipid acyl chains are fully extended in an all-trans configuration, rendering a lo bilayer ~1 nm thicker than a ld bilayer.
In addition, the Δsmt1 mutant has additional presence of extra soft areas (data not shown) which are absent in the WT. Thus, this extra soft area may lead to the more permeable and more fluid, and therefore less rigid conformation of the membrane. The membrane topography obtained using the reconstituted strain is similar to the one obtained with the wild-type although not identical (Figure 8) and no soft areas were found in the reconstituted strain. It is important to consider that the reconstituted strain has more than one copy of SMT1 gene compared to WT (Figure 1) and produces slightly more GlcCer than the WT (Figure 2). Thus, the “extra” GlcCer may contribute to an increased the lipid packing and more rigid raft observed in the reconstituted strain.
Present studies are generally limited with regard to linking molecular interactions among sphingolipids (Gaber et al., 1989), our work is novel in that we observed that the molecular basis of the change in membrane-lipid topography could be directly linked to the methyl group loss from GlcCer. Perhaps this methyl group serves as “glue” in the membrane lipid network. Possibly, the GlcCer methyl group serves to anchor specific cryptococcal virulence factors. Notably, phospholipase B1 (Plb1) and superoxide dismutase 1 (Sod1) virulence factors were found to be associated with ceramide monohexosides in C. neoformans (Siafakas et al., 2006), or/and GlcCer may anchor key transmembrane transporter(s) at the plasma membrane which may get displaced when the sphingolipid loses its methyl group. Displacement of these virulence factors/trasmembrane transporters can contribute to the loss of virulence observed with the Δsmt1 strain.
The question remains as to how these sphingolipids promote growth at alkaline pH. In mammalian cells, it is well known that potassium (K+) channels are key regulators of transmembrane potential homeostasis and are regulated by extracellular pH (MacKinnon, 2003, Yellen, 2002). Some members of K+ channels, such as TASK-2, TALK-1 and TALK-2, are stimulated by extracellular alkalization. TASK-2 participates in ion fluxes necessary for cell volume regulation (Niemeyer et al., 2001), and its activity is coupled with HCO−3 transport in the kidney proximal tubule (Warth et al., 2004). Interestingly, the hydrophobic environment around transmembrane 4 (TM4) is essential for the alkaline-dependence of TASK-2 (Niemeyer et al., 2007), suggesting that this region acts as an extracellular pH sensor. This sensing mechanism is also common for the TALK subfamily channels (Niemeyer et al., 2007). Because TASK-2 and TALK-1 homologs are present in the C. neoformans genome database, it is worth speculating whether a proper ratio of methylated GlcCer and ceramide species is necessary to sustain the hydrophobicity of a transmembrane domain of these channels, thus regulating HCO−3 transport and the transmembrane potential. This hypothesis may be supported by our results showing that cell growth is arrested in the mutant, only in the presence of high CO2 concentrations. Thus, it is possible that the presence of methylated GlcCer is critical for the formation of the gel-like domains, which impart the required membrane rigidity for the proper localization or/and function of the K+ channels.
The growth defect in the mutant observed only at physiological concentrations of CO2 (5%), raises the question of whether GlcCer may regulate CO2 sensing mechanisms. The CO2 sensing system in C. neoformans is comprised of adenylyl cyclase (Cac1) and two carbonic anhydrases (Can1 and Can2) (Bahn et al., 2006, Idnurm et al., 2011). However, growth of C. neoformans Δcac1 or Δcan1 mutants is not inhibited by 5% CO2 as in the mutants, and the Δcan2 mutant has a growth defect at atmospheric (0.04%) CO2 but not at 5% CO2 (Alspaugh et al., 2002, Bahn et al., 2005, Mogensen et al., 2006, Schlicker et al., 2009). Thus, it is unlikely that GlcCer regulates growth of C. neoformans in the lung environment through the CO2 sensing system.
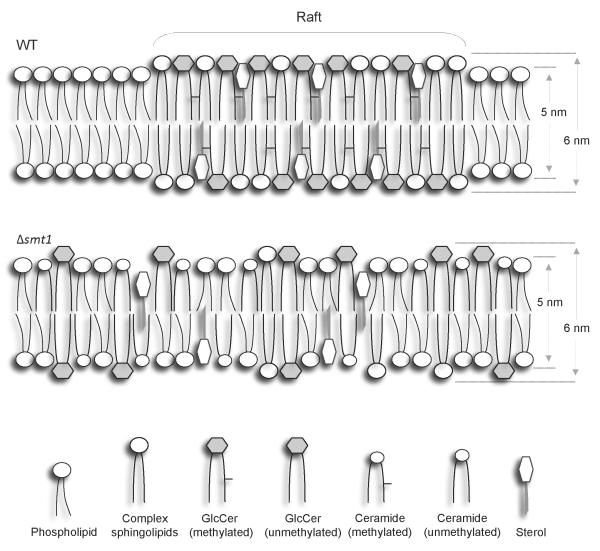
In Figure 9, we propose a schematic model of the fungal membrane lipid microenvironment. It is important to consider that in addition to the lack of methylated GlcCer, Δsmt1 strain accumulate different ceramide species, which, as discussed above, may contribute to regulate the phenotype(s) observed. The relative abundance of α-OH-Δ4-Δ8-ceramide in Δsmt1 may affect interaction between the raft lipids and different proteins, including K+ or/and additional channels. Clearly, the regulation of the membrane rigidity/structure may have important ties to virulence of C. neoformans by potentially regulating the pathogen survival in the host neutral/alkaline environment found in the lung alveoli. Future research on the cross talk between sphingolipids and K+ channels will provide new insights into the biophysical mechanism by which the GlcCer pathway regulates virulence of this important human pathogen.
Figure 9. Model for lipid micro-environments in WT and Δsmt1.
Δsmt1 cells have a less ordered lipid environment than WT cells and they are characterized by the presence of GlcCer and ceramide both unmethylated, suggesting that methylation of these sphingolipids might be critical for lipid raft formation.
Materials and Methods
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Medical University of South Carolina Institutional Animal Care and Use Committee (Permit Number: 2019). All animal procedures were performed according to the approved protocol, and all efforts were made to minimize suffering.
Strains, plasmids and culture conditions
Fungal strains used in this study were C. neoformans var. grubii serotype A strain H99 (WT), Δsmt1 mutant derived from H99, Δsmt1+SMT1 derived from Δsmt1, Δgcs1 also derived from H99 and the Δgcs1+GCS1 (Rittershaus et al., 2006). Escherichia coli strains DH-5α (Invitrogen and Stratagene) and TOP10 (Invitrogen) were used as competent cells. Plasmids pBluescript SK, pSC-A-amp/kan (Stratagene) and pCR II-TOPO 4.0 kb (Invitrogen) were used for cloning C. neoformans were routinely grown in yeast peptone dextrose (YPD) medium at 30 °C and 0.04% (atmospheric) CO2. Dulbecco’s modified eagle media (DMEM) (Gibco # 11995) buffered with 50 mM HEPES or Yeast Nitrogen Base (YNB) (Difco, Fisher Scientific # 239210) buffered with 25 mM HEPES and 2% glucose at pH 7.2 or pH 4.0 were used for growing C. neoformans strains at 37 °C and 5% CO2 as indicated. Bacterial strains for plasmid amplification were grown in Luria-Bertani (LB)) medium containing 50–75 mg/l ampicillin or kanamycin.
Stains, detergents, and reagents
Sytox green was from Molecular Probes, Invitrogen. Detergents SDS, Triton X-100 and all other reagents were from Sigma, Molecular Biology grade. Triton X-114 was from Thermo Scientific.
Fluorescence probes
t-PnA (trans-parinaric acid) and TMA-DPH (1-[4-trimethylammoniumphenyl]-6-phenyl-1,3,5-hexatriene p-toluenesulfonate]) were obtained from Molecular Probes (Leiden, The Netherlands).
Isolation and knockout of the sphingolipid C9 methyltransferase in C. neoformans
The putative homolog of the sphingolipid C9 methyltransferase (SMT1) gene was previously identified (Rhome et al., 2007) in the genome C. neoformans H99 at http://cneo.genetics.duke.edu/blast.html in chromosome 3, piece #23 using a blast search as containing the conserved domain sequence of the SMT1 gene (Ternes et al., 2006). Thus, a knockout cassette was generated as illustrated in the supplementary materials, containing a 1.294 kb of the 5′untranslated region (5′UTR) upstream the open reading frame (ORF) and a 1.385 kb of the 3′UTR. Primers and the restriction enzymes used are provided in supplementary materials. The 5′UTR and 3′UTR was PCR amplified using genomic DNA as a template. The amplified fragments were cloned in pCR TOPO vector (Invitrogen) and sequenced, resulting in plasmids pCR-5′UTR-TOPO and pCR-3′UTR-TOPO respectively. The nourseothricin acetyl transferase 1 (NAT1) gene, which confers resistance to the antibiotic nourseothricin (Werner BioAgents), under the control of C. neoformans actin promoter was restriction digested from the plasmid pCR-NAT1 (Heung et al., 2005, Heung et al., 2004) by HindIII and EcoRV, resulting in a 1.7 kb fragment. The 5′UTR and 3′UTR were spliced out with BamH1 and HindIII and EcoRV and Xba1, respectively. The TOPO vector was digested with BamH1 and Xba1. Then, the 4 fragments were simultaneously ligated in a single ligation reaction mix resulting in the pCR TOPO-5′UTR-NAT1-3′UTR and the resulting plasmid was named pΔsmt1 and used to delete the SMT1 gene. The deletion scheme is illustrated in Figure S1A. The WT was transformed with the plasmid pΔsmt1 biolistically, as described in (Toffaletti et al., 1993). Transformants were grown on YPD plates with 100 μg/ml of nourseothricin. Colonies were chosen randomly and genomic DNA was prepared for Southern blot analysis. Recombinants were screened by probing with a fragment of 5′UTR, NAT1 or SMT1 ORF fragment (Figure S1A). Transformant #1.2 showing deletion of the SMT1 gene by the insertion of the NAT1 fragment was chosen and designated Δsmt1 strain.
Reconstitution of the SMT1 gene
The SMT1 gene was reintroduced back into the Δsmt1 strain by generating a plasmid cassette pSK-SMT1-HYG containing the hygromycin B (HYG) gene under the control of the cryptococcal actin promoter. The reconstitution scheme is illustrated in Figure S1B. The resulting plasmid construct pSK-SMT1-HYG was biolistically delivered into the Δsmt1. Stable transformants growing on YPD plates containing 100 μg/ml Hygromycin B were selected. Genomic DNA extracted and Southern blot analysis of the transformants was performed. Out of a total of 11 transformants showing re-introduction of a single copy of the SMT1 gene by the insertion of the plasmid loop, transformants # 2, 4, 19, 25 and 36 were chosen for lipid analysis, and transformant #2 was designated Δsmt1+SMT1 reconstituted strain.
Mass spectrometry analysis
Total lipids were extracted as described in (Singh et al., 2011). In brief, single colony from WT, Δsmt1, Δsmt1+SMT1, Δgcs1 and Δgcs1+GCS1 were picked from freshly streaked YPD plates and grown in 15 ml YPD at 30°C for 20 hrs at 250 rpm. The cells were washed twice with sterile Phosphate Buffered Saline (PBS) and counted. Then, 5 × 108 cells were placed in a single glass tube, washed once with sterile DDW, and Mandala extraction was performed for extraction of inositol-containing phospholipids and phosphatidylcholine. Bligh and Dyer lipid extraction (Bligh et al., 1959) of neutral lipids was performed on the dried lipids obtained after Mandala lipid extraction (Mandala et al., 1995). A quarter of the samples were aliquoted for inorganic phosphate determination. The tubes were vacuum dried and used for mass spectrometry (MS) analysis. The MS and MS/MS scans were conducted using a Thermo Finnigan TSQ7000 triple quadruple mass spectrometer with electrospray ionization (Bielawski et al., 2009). The analyses included multiple reaction monitoring (MRM) for the characteristic product ions of m/z=276.2 (α-OH-Δ4-Δ8,9methyl-GlcCer at m/z 756.4), 262.0 (α-OH-Δ4-Δ8- GlcCer at m/z 742.4), 576.4 (α-OH-Δ4-Δ8,9methyl-ceramide at m/z 594.4), and 562.4 (α-OH-Δ4-Δ8-ceramide at m/z 580.4).
Purification of GlcCer
GlcCer was purified from WT and Δsmt1 strains as previously described (Rittershaus et al., 2006).
Preparation of ceramide substrate for in vitro C9 methyltransferase assay
Deglucosylation of purified α-OH-Δ4-Δ8-GlcCer (source Δsmt1) or α-OH-Δ4-Δ8,9methyl-GlcCer (source WT) was performed using purified sphingolipid (5 μg), incubated with Cerezyme (200 mU; Genzyme Corporation) in a total volume of 100 μl for 90 min at 37 °C, as described in (Rhome et al., 2011). The reaction was stopped by the addition of chloroform: methanol (1:2 by volume). Lipids were extracted by Bligh and Dyer and analyzed by Mass Spectrometry to assess the complete deglucosylation of the ceramide used and thereby formation of α-OH-Δ4-Δ8-ceramide and α-OH-Δ4-Δ8,9methyl-ceramide.
In vitro activity of C9 methyltransferase
For in vitro C9 methyltransferase activity in C. neoformans, fungal cells were grown in a 50 ml YPD broth in a shaker incubator for 20 h at 30 °C. Cells were harvested by centrifugation and then washed with sterile PBS and suspended in lysis buffer as described previously (Ternes et al., 2006) with some modifications. The cells were lysed by vortexing with 4 cups (1 cup holds 500 μl) (Singh et al., 2011) of glass beads at full speed for 5 times over a gap of 1 min on ice. This mixture was then ultrasonicated (5 pulses, each for 45 sec, with a gap of 45 sec on ice between each pulse). Cell-free homogenate was obtained by centrifuging at 1700 x g for 15 min at 4 °C. Proteins in the homogenate were quantified by Bradford assay (250 μg protein). Each assay was conducted in a 100 μl total volume containing 0.1% Triton X-100, 18 μM of Adenosyl-L-Methionine, S-[methyl-14C] solution in ethanol (Perkin-Elmer) (250,000 dpm at a specific activity of 2.2 GBq/mmol) and 5 μg of lipid (α-OH-Δ4-Δ8-ceramide and α-OH-Δ4-Δ8,9-methylceramide). The mixture was mixed gently by vortexing and sonicating for 5-10 sec twice and incubated for 1 h at 30 °C. The lipids were dried in a SPD 2010 Speedvac (Thermo Electron Corp) after the Bligh and Dyer lipid extraction and stored in −80 °C. When ready to spot, the lipids were dissolved in chloroform (40 μl): methanol (2:1 by volume) and analyzed by TLC (20 × 20 cm; 250 μM thickness from Whatman) in CHCl3/MeOH (80:20). Purified soy GlcCer was loaded on the TLC as a standard and the spot was marked with a pencil after the run was complete to identify corresponding GlcCer spots/bands in the appropriate lysate. Radioactivity was detected on X-ray film exposed for several days at −80 °C. The 14C-labeled spots were scraped off the TLC plates and analyzed for radioactivity in a liquid scintillation counter (Beckman).
Virulence studies and histology analysis in a murine mouse model of cryptococcosis
Four- to six-weeks-old CBA/J (Jackson laboratory) mice were used. Mice were anesthetized a xylazine/ketamine mixture (ip; 60 μl; 95 mg/kg ketamine, 5 mg/kg xylazine). WT, Δsmt1+SMT1 and Δsmt1 strains were grown in YPD for 20 h at 30 °C with constant shaking. The cells were harvested by washing twice in sterile PBS, counted and resuspended in sterile PBS at a concentration of 2.7 × 107 cells/ml. Ten mice each with WT and Δsmt1+SMT1 and 15 mice with Δsmt1 strains were infected with 5.4 × 105 cells (volume 20 μl) through nasal inhalation. The mice were fed ad libitum and inspected daily. Moribund mice or mice which appeared to be in pain were sacrificed with CO2 inhalation followed by cervical dislocation. All animal procedures were approved by the Medical University of South Carolina Institutional Animal Care and Use Committee and followed the guidelines of the American Veterinary Medical Association.
For tissue burden culture analysis, 12 mice were infected with C. neoformans wild type, Δsmt1+SMT1 or Δgcs1+GCS1 strains, whereas 21 mice were each infected with Δsmt1 or Δgcs1 mutants. At selected time points, 3 mice were sacrificed, lung, brain, liver, kidney, and spleen excised and homogenized in 10 ml PBS using Stomacher 80 (Lab System Fisher Scientific, Pittsburgh, Pennsylvania, USA) for 2 min at high speed. Serial dilutions were then plated on YPD plates and incubated for 48–72 h at 30 °C. CFU/organ were counted and compared.
For histopathology analysis, lung, brain, liver, kidney, and spleen of CBA/J mice infected with the Δsmt1 strain were collected after 90 days post infection of the survival studies. From a total of 15 (infected with Δsmt1), 13 mice survived 90 days post infection. Thus, 6 mice were used for additional tissue burden culture studies of lungs, brains, livers, kidneys and spleens whereas 7 mice were used for histopathological studies. For histopathology studies, organs were fixed in 3.7% formaldehyde in paraffin and stained as follows: hematoxylin and eosin was used to visualize the host inflammatory response (Lazcano et al., 1993); mucicarmine was used to staining the C. neoformans capsule; Russel’s modification of Movat’s pentachrome stain was used for staining mucin (alcian blue), fibrous tissue (intense red), and elastic tissue (black); and Verhoeff-van Gieson (VVG) stain was used to identify connective tissue such as elastic fibers (stains black) and collagen (stains red) (Lazcano et al., 1993, Luna, 1992, Sheehan, 1980).
Sytox uptake assay
Sytox green is a high affinity nucleic acid stain, which fluoresces upon nucleic acid binding (Roth et al., 1997, Thevissen et al., 1999), and we used this technique with some modifications. A starter culture of WT and Δsmt1 in YPD were grown at 30°C with constant shaking for 24 h. Cells were pelleted, washed twice with sterile PBS and inoculated into YNB supplemented with 25 mM HEPES, 2% glucose, pH 7.2, and were grown in a shaker incubator at 37°C in the presence of 5% CO2 for 20 h. Cells were harvested, washed, counted, and resuspended in the same media (pH 7.2) at a concentration of 108 cells/ml. From this suspension (100 μl) cells added to wells in a 96-well microplate containing YNB (100 μl) in the absence/presence of either 0.05% SDS or 0.025% Triton X-100 or 0.025% Triton X-114 and incubated in a shaker-incubator for 2 h at 37 °C and 5% CO2. For uptake assays for untreated cells, cells were suspended in PBS (pH 7.2) instead of YNB after harvesting, pelleting, and washing. After 2 h, cells were pelleted inside the microplate and medium/PBS was decanted. PBS (200 μl) with Sytox green (2 μM) were added to each well and incubated in the dark for 10 (for treated and untreated cells), 20 and 60 min for untreated cells. Emitted fluorescence was measured by a SpectroFluorimeter (Perkin Elmer) with Wallace 1420 multilabel counter, at an excitation wavelength 488 nm and emission wavelength of 535 nm (8-nm slit width). Fluorescence values of the sample were corrected by subtracting the fluorescence values of PBS + Sytox green (for untreated cells) and PBS + detergents + Sytox green for treated cells.
Fluorescence spectroscopy measurements
For fluorescence spectroscopy experiments, fungal cells (~2×106 cells/mL) grown in DMEM, pH 7.2 at 37 °C in the presence of 5% CO2 were harvested and incubated with 2 μM of either, TMA-DPH or t-PnA for 20 min. Fluorescence anisotropy was measured (at least 10 measurements for each analysis) in a SLM Aminco 8100 series 2 spectrofluorimeter. All measurements were performed in 1 cm × 0.4 cm quartz cuvettes under magnetic stirring. The excitation (λex)/emission (λem) wavelengths were 358/430 nm for TMA-DPH and 320/405 nm for t-PnA. The time-resolved fluorescence measurements with t-PnA were performed in a Fluorolog from Horiba Jobin Yvon (NJ, USA). The lifetime decays were obtained by the time Correlated Single Photon Counting (TCSPC) technique with fixed-wavelength “Plug and Play” interchangeable NanoLED pulsed laser-diodes of 315 nm. To obtain the fitting curves, the experimental decays were analyzed using the TRFA software (Scientific Software Technologies Center, Minsk, Belarus). For a decay described by a sum of exponentials where αi is the normalized pre-exponential and τi is the lifetime of the decay component i, the mean fluorescence lifetime is given by
Atomic force microscopic study of membranes
Differences in membrane microdomain formation between WT and the two mutants of C. neoformans were investigated by extracting their membrane lipids, reconstituting them on inert mica surfaces, and examining the reconstituted bilayers using atomic force microscopy (AFM) (Burns, 2007, Lin et al., 2007, Muller, 2008, Scheuring et al., 2007, Pelling et al., 2005). Total membranes were extracted from WT, Δsmt1, Δsmt1+SMT1 (Garcia et al., 2008) grown in DMEM supplemented with 50 mM HEPES, pH 7.2 at 37 °C in presence of 5% CO2. The cells were grown in 100 ml of the above mentioned media for 20 h, pelleted, washed, resuspended in homogenization buffer (100 mM Tris-HCl, pH 7.5, 5 mg/ml bovine serum albumin, 1X protease inhibitor cocktail) and lysed in a French Press (20,000 psi at 4 °C). The lysate was centrifuged at 10,000 x g at 4 °C for 10 min after adjusting the pH to 7.2. The supernatant was centrifuged at 1 × 105 x g for 1 h at 4 °C. The pellet was resuspended in 3 ml of extraction buffer (10 mM HEPES-KOH, pH 7.0, 100 mM KCl, 1 mM dithiothreitol, 0.45% (w/v) glycerol, and 0.5% w/v deoxycholate) by sonication (3 pulse at a gap of 45 sec over ice) and ultracentrifuged at 1.5 × 105 x g for 1 h. The previous step was repeated and the final pellet consisting of the total membranes was resuspended via sonication in 1 ml of the extraction buffer. Lipids were extracted from these membranes and reconstituted on mica (Structure Probe, Inc.) by dropping 0.2 μl of sample solution directly on the mineral surface covered with 2 μl DI water. The mica plate was incubated at 60 °C overnight in a closed chamber that kept the surface moisturized but without visible water droplet. The lipid bilayers were then examined by using AFM (Asylum Research MFP-3D). The microscope was operated in the tapping mode following standard procedures.
By rastering a probing tip (Olympus AC240TS) across an area, AFM provides a height image that records the topography on top of the lipid bilayers (i.e., variation in height). Because the fungal sphingolipids are approximately 0.5 nm longer than most of the fungal phospholipids, the microdomains formed by sphingolipids in reconstructed lipid bilayers are expected to be 1 nm taller than the phospholipid background. In addition to the height image, AFM also records a phase image that relates to the rigidity of the bilayers through the dissipation of the tapping energy (Cleveland et al., 1998, Connell et al., 2006, Vanegas et al., 2010). The sphingolipid bilayers are more rigid than their phospholipid counterparts and thus appear on the phase image as having more negative phase shifts.
For lipid reconstitution studies, methylated or demethylated GlcCer were purified from wild-type or Δsmt1 strain, respectively, as described above and, then, 5 μg of purified lipid was added to 95 μg of lipid membranes extracted from either Δgcs1 or Δsmt1 strain.
Statistical Analysis
Statistical analysis was performed by a 2 tailed Student’s T-test or ANOVA where applicable with MS Excel software. Mice survival was analyzed by Wilcoxon Signed-Rank test. A P<0.05 was considered to be statistically significant.
Supplementary Material
Author Summary.
In this paper, we describe the role of membrane lipids in the regulation of pathogenicity of the human fungal pathogen Cryptococcus neoformans. In particular, we show that the biochemical structure of certain membrane lipids, namely glucosylceramide, is important for maintaining the proper permeability and conformational rigidity of cryptococcal membranes. Alteration of the glucosylceramide biochemical structure using a genetic approach confers to the fungus certain characteristics similar to those observed when glucosylceramide is absent, suggesting that these fungal characteristics may be regulated by the specific structure of glucosylceramide. To our knowledge, this is the first study that investigates the structure-function relationship of glycosphingolipids to cryptococcal virulence.
Acknowledgments
The authors are grateful to all the members of the DNA Sequencing Facility and Lipidomics Core Facility at MUSC. A special thank to Margaret Romano (MUSC) for histological staining services. We also acknowledge Dr. Samarendra Kr. Singh for his invaluable help during cloning and Dr. Shriya Raj for her help in during the revision of the manuscript. We would also like to thank the members of Del Poeta’s and Luberto’s laboratory for guidance and helpful discussion. This work was performed in part by the National Institute of Health (NIH) awards AI056168, AI071142, AI078493, and AI087541 to M.D.P. and in part by by the University of Notre Dame Faculty Scholarship Award Program (FASP) to C.N. This work was conducted in a facility constructed with support from the National Institutes of Health, Grant Number C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. Dr. Maurizio Del Poeta is a Burroughs Welcome New Investigator in the Pathogenesis of Infectious Diseases.
References
- Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, et al. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aresta-Branco F, Cordeiro AM, Marinho HS, Cyrne L, Antunes F, de Almeida RF. Gel domains in the plasma membrane of Saccharomyces cerevisiae: highly ordered, ergosterol-free, and sphingolipid-enriched lipid rafts. The Journal of biological chemistry. 2011;286:5043–5054. doi: 10.1074/jbc.M110.154435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn YS, Cox GM, Perfect JR, Heitman J. Carbonic anhydrase and CO2 sensing during Cryptococcus neoformans growth, differentiation, and virulence. Curr. Biol. 2005;15:2013–2020. doi: 10.1016/j.cub.2005.09.047. [DOI] [PubMed] [Google Scholar]
- Bahn YS, Muhlschlegel FA. CO2 sensing in fungi and beyond. Curr Opin Microbiol. 2006;9:572–578. doi: 10.1016/j.mib.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Baorto DM, Gao Z, Malaviya R, Dustin ML, van der Merwe A, Lublin DM, Abraham SN. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature. 1997;389:636–639. doi: 10.1038/39376. [DOI] [PubMed] [Google Scholar]
- Barrow DA, Lentz BR. Membrane structural domains. Resolution limits using diphenylhexatriene fluorescence decay. Biophys J. 1985;48:221–234. doi: 10.1016/S0006-3495(85)83775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, Bielawska A. Comprehensive quantitative analysis of bioactive sphingolipids by high-performance liquid chromatography-tandem mass spectrometry. Methods Mol Biol. 2009;579:443–467. doi: 10.1007/978-1-60761-322-0_22. [DOI] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method for total lipid extraction and purification. Can. J. Bioch. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Bogdanov M, Umeda M, Dowhan W. Phospholipid-assisted refolding of an integral membrane protein. Minimum structural features for phosphatidylethanolamine to act as a molecular chaperone. J Biol Chem. 1999;274:12339–12345. doi: 10.1074/jbc.274.18.12339. [DOI] [PubMed] [Google Scholar]
- Brown DA. Lipid rafts, detergent-resistant membranes, and raft targeting signals. Physiology. 2006;21:430–439. doi: 10.1152/physiol.00032.2006. [DOI] [PubMed] [Google Scholar]
- Brown DA, London E. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- Burns AR. Atomic force microscopy of lipid domains in supported model membranes. Methods Mol Biol. 2007;398:263–282. doi: 10.1007/978-1-59745-513-8_18. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR. Cryptococcus neoformans. ASM Press; Washington, DC: 1998. pp. 381–405.pp. 271–324. [Google Scholar]
- Castro BM, de Almeida RF, Silva LC, Fedorov A, Prieto M. Formation of ceramide/sphingomyelin gel domains in the presence of an unsaturated phospholipid: a quantitative multiprobe approach. Biophys J. 2007;93:1639–1650. doi: 10.1529/biophysj.107.107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland JP, Anczykowski B, Schmid AE, Elings VB. Energy dissipation in tapping-mode atomic force microscopy. Appl Phys Lett. 1998;72:2613–2615. [Google Scholar]
- Connell SD, Smith DA. The atomic force microscope as a tool for studying phase separation in lipid membranes. Mol Membr Biol. 2006;23:17–28. doi: 10.1080/09687860500501158. [DOI] [PubMed] [Google Scholar]
- Datta K, Bartlett KH, Baer R, Byrnes E, Galanis E, Heitman J, et al. Spread of Cryptococcus gattii into Pacific Northwest region of the United States. Emerg Infect Dis. 2009;15:1185–1191. doi: 10.3201/eid1508.081384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr., Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–861. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia J, Shea J, Alvarez-Vasquez F, Qureshi A, Luberto C, Voit EO, Del Poeta M. Mathematical modeling of pathogenicity of Cryptococcus neoformans. Mol Syst Biol. 2008;4:183. doi: 10.1038/msb.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giocondi MC, Yamamoto D, Lesniewska E, Milhiet PE, Ando T, Le Grimellec C. Surface topography of membrane domains. Biochim Biophys Acta. 2010;1798:703–718. doi: 10.1016/j.bbamem.2009.09.015. [DOI] [PubMed] [Google Scholar]
- Hakomori S. Glycosphingolipids in cellular interaction, differentiation, and oncogenesis. Annu Rev Biochem. 1981;50:733–764. doi: 10.1146/annurev.bi.50.070181.003505. [DOI] [PubMed] [Google Scholar]
- Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G. Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem. 1994;225:641–649. doi: 10.1111/j.1432-1033.1994.00641.x. [DOI] [PubMed] [Google Scholar]
- Heinz E. In: Advances in lipid methodologiy three. Christie WW, editor. The Oily Press; Dundee, UK: 1996. pp. 211–332. [Google Scholar]
- Heung LJ, Kaiser AE, Luberto C, Del Poeta M. The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J Biol Chem. 2005;280:28547–28555. doi: 10.1074/jbc.M503404200. [DOI] [PubMed] [Google Scholar]
- Heung LJ, Luberto C, Plowden A, Hannun YA, Del Poeta M. The sphingolipid pathway regulates Pkc1 through the formation of diacylglycerol in Cryptococcus neoformans. J Biol Chem. 2004;279:21144–21153. doi: 10.1074/jbc.M312995200. [DOI] [PubMed] [Google Scholar]
- Idnurm A, Bahn YS, Shen WC, Rutherford JC, Muhlschlegel FA. Sensing extracellular signals in Cryptococcus neoformans. In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Cryptococcus: from human pathogen to model yeast. ASM Press; Washington DC: 2011. p. 175. [Google Scholar]
- Kechichian TB, Shea J, Del Poeta M. Depletion of alveolar macrophages decreases the dissemination of a glucosylceramide-deficient mutant of Cryptococcus neoformans in immunodeficient mice. Infect Immun. 2007;75:4792–4798. doi: 10.1128/IAI.00587-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinhans FW, Lees ND, Bard M, Haak RA, Woods RA. ESR determinations of membrane permeability in a yeast sterol mutant. Chem Phys Lipids. 1979;23:143–154. doi: 10.1016/0009-3084(79)90042-2. [DOI] [PubMed] [Google Scholar]
- Lang T, Bruns D, Wenzel D, Riedel D, Holroyd P, Thiele C, Jahn R. SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazcano O, Speights VO, Jr., Strickler JG, Bilbao JE, Becker J, Diaz J. Combined histochemical stains in the differential diagnosis of Cryptococcus neoformans. Mod Pathol. 1993;6:80–84. [PubMed] [Google Scholar]
- Lees ND, Bard M, Kemple MD, Haak RA, Kleinhans FW. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979;553:469–475. doi: 10.1016/0005-2736(79)90302-x. [DOI] [PubMed] [Google Scholar]
- Leipelt M, Warnecke D, Zahringer U, Ott C, Muller F, Hube B, Heinz E. Glucosylceramide synthases, a gene family responsible for the biosynthesis of glucosphingolipids in animals, plants, and fungi. J Biol Chem. 2001;276:33621–33629. doi: 10.1074/jbc.M104952200. [DOI] [PubMed] [Google Scholar]
- Lin WC, Blanchette CD, Ratto TV, Longo ML. Lipid domains in supported lipid bilayer for atomic force microscopy. Methods Mol Biol. 2007;400:503–513. doi: 10.1007/978-1-59745-519-0_34. [DOI] [PubMed] [Google Scholar]
- Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- Litvintseva AP, Thakur R, Reller LB, Mitchell TG. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J Infect Dis. 2005;192:888–892. doi: 10.1086/432486. [DOI] [PubMed] [Google Scholar]
- Luna LG. In Histopathologic methods and color atlas of special stains and tissue artifacts. 1992.
- Lynch DV. In: Lipid metabolism in plants. Moore TSJ, editor. CRC Press, Inc.; Boca Raton, FL: 1993. pp. 285–308. [Google Scholar]
- MacKinnon R. Potassium channels. FEBS Lett. 2003;555:62–65. doi: 10.1016/s0014-5793(03)01104-9. [DOI] [PubMed] [Google Scholar]
- Mandala SM, Thornton RA, Frommer BR, et al. The discovery of australifungin, a novel inhibitor of sphinganine N-acyltransferase from Sporormiella australis. Producing organism, fermentation, isolation, and biological activity. J. Antibiot. (Tokyo) 1995;48:349–356. doi: 10.7164/antibiotics.48.349. [DOI] [PubMed] [Google Scholar]
- Merrill AH, Jr., Sullards MC, Wang E, Voss KA, Riley RT. Sphingolipid metabolism: roles in signal transduction and disruption by fumonisins. Environ Health Perspect. 2001;109(Suppl 2):283–289. doi: 10.1289/ehp.01109s2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen EG, Janbon G, Chaloupka J, Steegborn C, Fu MS, Moyrand F, et al. Cryptococcus neoformans senses CO2 through the carbonic anhydrase Can2 and the adenylyl cyclase Cac1. Eukaryot. Cell. 2006;5:103–111. doi: 10.1128/EC.5.1.103-111.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J, Pirofski LA, Scharff MD, Casadevall A. Antibody-mediated protection in mice with lethal intracerebral Cryptococcus neoformans infection. Proc Natl Acad Sci U S A. 1993;90:3636–3640. doi: 10.1073/pnas.90.8.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DJ. AFM: a nanotool in membrane biology. Biochemistry. 2008;47:7986–7998. doi: 10.1021/bi800753x. [DOI] [PubMed] [Google Scholar]
- Niemeyer MI, Cid LP, Barros LF, Sepulveda FV. Modulation of the two-pore domain acid-sensitive K+ channel TASK-2 (KCNK5) by changes in cell volume. J Biol Chem. 2001;276:43166–43174. doi: 10.1074/jbc.M107192200. [DOI] [PubMed] [Google Scholar]
- Niemeyer MI, Gonzalez-Nilo FD, Zuniga L, Gonzalez W, Cid LP, Sepulveda FV. Neutralization of a single arginine residue gates open a two-pore domain, alkali-activated K+ channel. Proc Natl Acad Sci U S A. 2007;104:666–671. doi: 10.1073/pnas.0606173104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nature genetics. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oura T, Kajiwara S. Candida albicans sphingolipid C9-methyltransferase is involved in hyphal elongation. Microbiology. 2010;156:1234–1243. doi: 10.1099/mic.0.033985-0. [DOI] [PubMed] [Google Scholar]
- Park BJ, Lockhart SR, Brandt ME, Chiller TM. Public health importance of cryptococcal disease: epidemiology, burden and control. In: Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A, editors. Cryptococcus: from human pathogen to model yeast. ASM Press; Washington DC: 2011. pp. 585–593. [Google Scholar]
- Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- Pelling AE, Li Y, Shi W, Gimzewski JK. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc Natl Acad Sci U S A. 2005;102:6484–6489. doi: 10.1073/pnas.0501207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle A, Keller P, Florin EL, Simons K, Horber JK. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett ST, Bushnev A, Hagedorn K, Adiga M, Haynes CA, Sullards MC, et al. Biodiversity of sphingoid bases (“sphingosines”) and related amino alcohols. J Lipid Res. 2008;49:1621–1639. doi: 10.1194/jlr.R800012-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamoorthy V, Cahoon EB, Thokala M, Kaur J, Li J, Shah DM. Sphingolipid C-9 methyltransferases are important for growth and virulence but not for sensitivity to antifungal plant defensins in Fusarium graminearum. Eukaryot Cell. 2009;8:217–229. doi: 10.1128/EC.00255-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes Mateo C, Brochon JC, Lillo M. Pilar, Acuna A. Ulises. Lipid clustering in bilayers detected by the fluorescence kinetics and anisotropy of trans-parinaric acid. Biophys J. 1993;65:2237–2247. doi: 10.1016/S0006-3495(93)81257-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhome R, McQuiston T, Kechichian T, Bielawska A, Hennig M, Drago M, et al. Biosynthesis and immunogenicity of glucosylceramide in Cryptococcus neoformans and other human pathogens. Eukaryot Cell. 2007;6:1715–1726. doi: 10.1128/EC.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhome R, Singh A, Kechichian T, Drago M, Morace G, Luberto C, Del Poeta M. Surface Localization of Glucosylceramide during Cryptococcus neoformans Infection Allows Targeting as a Potential Antifungal. PLoS One. 2011;6:e15572. doi: 10.1371/journal.pone.0015572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittershaus PC, Kechichian TB, Allegood JC, Merrill AH, Jr., Hennig M, Luberto C, Del Poeta M. Glucosylceramide synthase is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest. 2006;116:1651–1659. doi: 10.1172/JCI27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Travassos LR, Miranda KR, Franzen AJ, Rozental S, de Souza W, et al. Human antibodies against a purified glucosylceramide from Cryptococcus neoformans inhibit cell budding and fungal growth. Infect Immun. 2000;68:7049–7060. doi: 10.1128/iai.68.12.7049-7060.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth BL, Poot M, Yue ST, Millard PJ. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl Environ Microbiol. 1997;63:2421–2431. doi: 10.1128/aem.63.6.2421-2431.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Takakuwa N, Ohnishi M, Oda Y. Presence of glucosylceramide in yeast and its relation to alkali tolerance of yeast. Applied microbiology and biotechnology. 2006;71:515–521. doi: 10.1007/s00253-005-0187-3. [DOI] [PubMed] [Google Scholar]
- Sandhoff R. Very long chain sphingolipids: tissue expression, function and synthesis. FEBS Lett. 2010;584:1907–1913. doi: 10.1016/j.febslet.2009.12.032. [DOI] [PubMed] [Google Scholar]
- Scheuring S, Boudier T, Sturgis JN. From high-resolution AFM topographs to atomic models of supramolecular assemblies. J Struct Biol. 2007;159:268–276. doi: 10.1016/j.jsb.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Schlicker C, Hall RA, Vullo D, Middelhaufe S, Gertz M, Supuran CT, et al. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J Mol Biol. 2009;385:1207–1220. doi: 10.1016/j.jmb.2008.11.037. [DOI] [PubMed] [Google Scholar]
- Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. Proc Natl Acad Sci U S A. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea JM, Kechichian TB, Luberto C, Del Poeta M. The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect Immun. 2006;74:5977–5988. doi: 10.1128/IAI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan DC, Hrapchak BB. Connective Tissue and Muscle Fiber Stains London. The C.V. Mosby Company; 1980. pp. 196–197. [Google Scholar]
- Siafakas AR, Wright LC, Sorrell TC, Djordjevic JT. Lipid rafts in Cryptococcus neoformans concentrate the virulence determinants phospholipase B1 and Cu/Zn superoxide dismutase. Eukaryot Cell. 2006;5:488–498. doi: 10.1128/EC.5.3.488-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Qureshi A, Del Poeta M. Quantitation of cellular components if Cryptococcus neoformans for system biology analysis. Methods Mol Biol. 2011 doi: 10.1007/978-1-61779-086-7_16. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar LA, Hudson BS, Petersen M, Diamond J. Conjugated polyene fatty acids on fluorescent probes: spectroscopic characterization. Biochemistry. 1977;16:813–819. doi: 10.1021/bi00624a001. [DOI] [PubMed] [Google Scholar]
- Sonnino S, Aureli M, Loberto N, Chigorno V, Prinetti A. Fine tuning of cell functions through remodeling of glycosphingolipids by plasma membrane-associated glycohydrolases. FEBS Lett. 2010;584:1914–1922. doi: 10.1016/j.febslet.2009.11.020. [DOI] [PubMed] [Google Scholar]
- Ternes P, Sperling P, Albrecht S, Franke S, Cregg JM, Warnecke D, Heinz E. Identification of fungal sphingolipid C9-methyltransferases by phylogenetic profiling. J Biol Chem. 2006;281:5582–5592. doi: 10.1074/jbc.M512864200. [DOI] [PubMed] [Google Scholar]
- Thevissen K, Terras FR, Broekaert WF. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl Environ Microbiol. 1999;65:5451–5458. doi: 10.1128/aem.65.12.5451-5458.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran D, Carpentier JL, Sawano F, Gorden P, Orci L. Ligands internalized through coated or noncoated invaginations follow a common intracellular pathway. Proc Natl Acad Sci U S A. 1987;84:7957–7961. doi: 10.1073/pnas.84.22.7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer-Janssen YP, van Galen J, Batenburg JJ, Helms JB. Lipids in host-pathogen interactions: pathogens exploit the complexity of the host cell lipidome. Prog Lipid Res. 2010;49:1–26. doi: 10.1016/j.plipres.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Hoetzl S. Sphingolipid topology and the dynamic organization and function of membrane proteins. FEBS Lett. 2010;584:1800–1805. doi: 10.1016/j.febslet.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Vanegas JM, Faller R, Longo ML. Influence of ethanol on lipid/sterol membranes: phase diagram construction from AFM imaging. Langmuir. 2010;26:10415–10418. doi: 10.1021/la1012268. [DOI] [PubMed] [Google Scholar]
- Walther TC. Keeping sphingolipid levels nORMal. Proc Natl Acad Sci U S A. 2010;107:5701–5702. doi: 10.1073/pnas.1001503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warth R, Barriere H, Meneton P, Bloch M, Thomas J, Tauc M, et al. Proximal renal tubular acidosis in TASK2 K+ channel-deficient mice reveals a mechanism for stabilizing bicarbonate transport. Proc Natl Acad Sci U S A. 2004;101:8215–8220. doi: 10.1073/pnas.0400081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Bittman R, Duportail G, Heissler D, Vilcheze C, London E. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J Biol Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- Yellen G. The voltage-gated potassium channels and their relatives. Nature. 2002;419:35–42. doi: 10.1038/nature00978. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.