SUMMARY
Rickettsiae are obligate intracellular pathogens that are transmitted to humans by arthropod vectors and cause diseases such as spotted fever and typhus. Although rickettsiae require the host cell actin cytoskeleton for invasion, the cytoskeletal proteins that mediate this process have not been completely described. To identify the host factors important during cell invasion by Rickettsia parkeri, a member of the spotted fever group (SFG), we performed an RNAi screen targeting 105 proteins in Drosophila melanogaster S2R+ cells. The screen identified 21 core proteins important for invasion, including the GTPases Rac1 and Rac2, the WAVE nucleation promoting factor complex and the Arp2/3 complex. In mammalian cells, including endothelial cells, the natural targets of R. parkeri, the Arp2/3 complex was also crucial for invasion, while requirements for WAVE2 as well as Rho GTPases depended on the particular cell type. We propose that R. parkeri invades S2R+ arthropod cells through a primary pathway leading to actin nucleation, whereas invasion of mammalian endothelial cells occurs via redundant pathways that converge on the host Arp2/3 complex. Our results reveal a key role for the WAVE and Arp2/3 complexes, as well as a higher degree of variation than previously appreciated in actin nucleation pathways activated during Rickettsia invasion.
INTRODUCTION
Rickettsiae are Gram-negative, obligate intracellular alpha-proteobacteria that infect both mammalian and arthropod hosts. The spotted fever group (SFG) of Rickettsia includes Rickettsia parkeri, an emerging cause of mild-to-moderate spotted fever disease in North and South America (Paddock et al., 2004). Infection of endothelial cells by Rickettsia parkeri and related SFG species such as Rickettsia rickettsii and Rickettsia conorii results in systemic disease including vascular damage, edema, a characteristic petechial rash, and a necrotic eschar at the inoculation site (Walker et al., 2008).
To gain access to non-phagocytic host cells, rickettsiae must induce their own uptake. For other bacterial pathogens, invasion generally occurs either when bacterial surface proteins bind host cell surface receptors and activate local extension of the host membrane, or when secretion-system mediated injection of bacterial effector proteins induces membrane ruffling and actin polymerization (Cossart et al., 2004). While Rickettsia genomes encode a type IV secretion system (T4SS) as well as a number of proteins with eukaryotic-specific sequence motifs that could function as effectors (Gillespie et al., 2009), no effectors have been demonstrated to date. However, a number of Rickettsia outer membrane proteins contribute to bacterial adherence and invasion. Both the R. rickettsii surface protein rOmpA and the R. conorii surface protein Sca1 contribute to adherence (Li et al., 1998; Riley et al., 2010) while the R. conorii and Rickettsia japonica rOmpB and R. conorii Sca2 proteins are functionally important for bacterial entry and their expression in E. coli is sufficient to allow invasion of host cells (Uchiyama et al., 2006; Cardwell et al., 2009; Chan et al., 2009). Unfortunately, a lack of robust genetic tools for Rickettsia species has hindered analysis of how the type IV secretion system, secreted effectors, and outer membrane proteins might cooperate to mediate host cell invasion.
In addition to bacterial proteins, Rickettsia invasion requires the activation of host signaling pathways upstream of actin polymerization. The only known receptor for Rickettsia entry is the DNA-dependent protein kinase subunit Ku70, which binds to R. conorii rOmpB (Martinez et al., 2005; Chan et al., 2009). Downstream of receptor engagement, R. conorii invasion of mammalian cells requires host protein tyrosine kinases, results in the accumulation of tyrosine-phosphorylated proteins around invading bacteria, and also requires phosphoinositide 3-kinase activity (Martinez et al., 2004). In addition, the Rho family GTPase Cdc42 was implicated in invasion (Martinez et al., 2004).
Actin polymerization during invasion could be initiated by formin-family proteins, tandem monomer-binding nucleators, or nucleation promoting factors (NPFs) that act in conjunction with the Arp2/3 complex (Campellone et al., 2010). Based on experiments implicating Cdc42, it was proposed that the host NPF N-WASP was important for R. conorii invasion (Martinez et al., 2004). Indeed, the Arp3 subunit of the Arp2/3 complex was observed around invading bacteria and expression of an Arp2/3-activating WCA domain from WAVE1 (also known as WASF1 or Scar1) inhibited R. conorii invasion (Martinez et al., 2004). However, a direct role for the Arp2/3 complex has not been demonstrated via depletion or inhibition studies. In addition, it is unclear which NPF proteins act during Rickettsia entry, whether additional bacteria or host actin nucleators may be involved, and whether invasion by diverse Rickettsia species or of physiologically relevant host cell types utilize the same or different pathways.
To achieve a comprehensive understanding of the host cytoskeletal proteins important for Rickettsia invasion, we investigated the invasion of multiple cultured cell lines by the SFG species Rickettsia parkeri, which is genetically similar to R. rickettsii and R. conorii (Ralph et al., 1990), displays a similar ability to invade cells and form actin tails (Serio et al., 2010), but is less pathogenic and is not a select agent (Paddock et al., 2008). We performed a targeted RNAi-based screen in Drosophila S2R+ cells to identify a core group of host cytoskeletal proteins required for this process. We identified 21 proteins including Rho-family GTPases, and the WAVE and Arp2/3 complexes, which played a key role in invasion. During invasion of mammalian cells, including a human endothelial cell line, the requirement for WAVE family proteins and Rho family GTPases was not as stringent as in Drosophila cells, but the Arp2/3 complex was critical. Overall, these results suggest a pathway activating actin nucleation around invading rickettsiae and demonstrate that the molecular requirements for invasion vary depending on host cell type.
RESULTS
R. parkeri invasion of Drosophila and mammalian cells is rapid and depends on viable bacteria and host actin
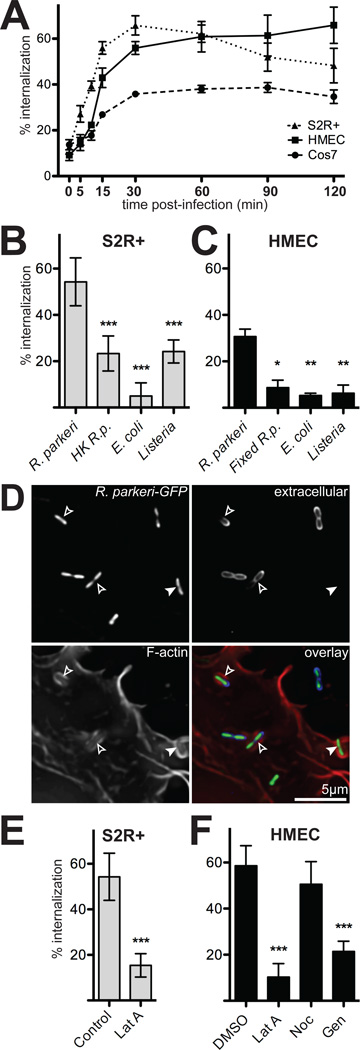
Invasion of host cells by Rickettsia has been reported to occur within 5 min to 2 h post-infection (Walker et al., 1978; Walker, 1984; Teysseire et al., 1995; Martinez et al., 2004). To determine the kinetics of R. parkeri invasion, we infected immortalized human microvascular endothelial cells (HMEC-1), African green monkey kidney-derived cells (COS-7), and adherent Drosophila embryo-derived hemocyte-like cells (S2R+) with R. parkeri and determined the percentage of internalized bacteria at various times post-infection by differential fluorescence staining of internal and external bacteria (Figure 1A). In all cell types examined, internalization plateaued between 30–60 min post-infection and was >50% complete by 15 min. Based on the rapid speed of invasion, subsequent experiments were conducted using 15 min infection times.
Figure 1. R. parkeri invade cells quickly in a process dependent on viable bacteria and host actin.
(A) Percent of cell-associated R. parkeri that were internalized by S2R+ (triangles), HMEC-1 (squares) or COS-7 (circles) cells at various times post-infection (n=3). (B) Percent internalization by S2R+ cells infected for 15 min with live R. parkeri, heat-killed R. parkeri (HK R.p.), E. coli XL-10 Gold (E. coli), or L. monocytogenes 10403S (Listeria). (C) Percent internalization by HMEC-1 infected for 15 min with live R. parkeri, or formaldehyde-fixed R. parkeri (Fixed R.p.), L. monocytogenes, or E. coli. Note: live R.p. in panel C were incubated at RT while samples were fixed (D) Maximum intensity projection of a deconvolved image of COS-7 cells transfected with pLifeact-mCherry (lower left, red in merge) and infected 48 h later with GFP-expressing R. parkeri (GFP, top left, green in merge) for 15 min. Extracellular bacteria were stained with anti-Rickettsia antibody without permeabilization (upper right, blue in merge). Open arrowheads; partially internalized bacteria, closed arrowheads; fully internalized bacterium. (E) Internalization by S2R+ cells infected for 15 min with R. parkeri and untreated (Control) or treated for 30 min prior to infection with 4 µM latrunculin A (Lat A). (F) Internalization by HMEC-1 cells after 15 min infection; cells were either pretreated with 1% DMSO alone, or treated 30 minutes before infection with 4 µM latrunculin A (LatA), 20 µM nocodazole (Noc) or 250 µM genistein (Gen) in 1% DMSO. For all panels: graphs plot the mean percent internalization for at least three independent experiments. Error bars indicate SEM (A), SD (B–D); * p<0.05, ** p<0.01, *** p<0.001 versus mean of control (R. parkeri or DMSO) by unpaired t-test.
We next sought to determine whether rapid invasion of S2R+ and HMEC-1 cells was due to generalized phagocytosis by these cells. We compared internalization of live R. parkeri versus heat-treated or formaldehyde-fixed bacteria, non-invasive E. coli (commercial strain XL-10), and the invasive Listeria monocytogenes strain 10403S (Greiffenberg et al., 1998; Agaisse et al., 2005) (Figure 1B,C). In S2R+ cells, live R. parkeri were internalized more than two-fold more efficiently than nonviable or other bacteria (Figure 1B; Dataset S1). In HMEC-1 cells, live R. parkeri were internalized at least three-fold more efficiently than nonviable or other bacteria (Figure 1C). Therefore, under these experimental conditions, invasion of host cells occurs through an active, R. parkeri-specific process.
We also confirmed that R. parkeri invasion requires host actin polymerization and tyrosine kinase activity, as is the case for R. conorii (Martinez et al., 2004; Chan et al., 2009). Using Lifeact-mCherry as a marker for F-actin, we observed actin accumulation around invading R. parkeri in S2R+, HMEC-1, and COS-7 cells (Figure 1D, Videos S1–S2 and data not shown). The actin depolymerizing agent latrunculin A reduced R. parkeri invasion of both S2R+ and HMEC-1 cells by 3.5 and 5.7-fold, respectively, while the microtubule-disrupting drug nocodazole did not significantly alter invasion (Figure 1E,F; Dataset S1; Figure S1A). In addition, invasion of HMEC-1 cells depended on tyrosine kinase activity, as genistein significantly reduced internalization (Figure 1E). Treatment with latrunculin A and genistein also reduced the total number of bacteria associated with host cells (Figure S1B). Treatment of S2R+, HMEC-1 and COS-7 cells with specific inhibitors of Abl (nilotinib), Src/Abl-family (dasatinib), Src/LCK/FYN-family (PP2), and phosphatidylinositol 3-kinases (wortmannin) had little or no effect on invasion or binding, suggesting that entry does not depend on one specific tyrosine kinase signaling pathway (Figure S1A–B). Overall, these results demonstrate that R. parkeri, like R. conorii, rapidly invade a variety of cell types in a manner dependent on host actin polymerization and tyrosine kinase activity.
RNAi screening identifies 21 proteins important for R. parkeri invasion of S2R+ cells
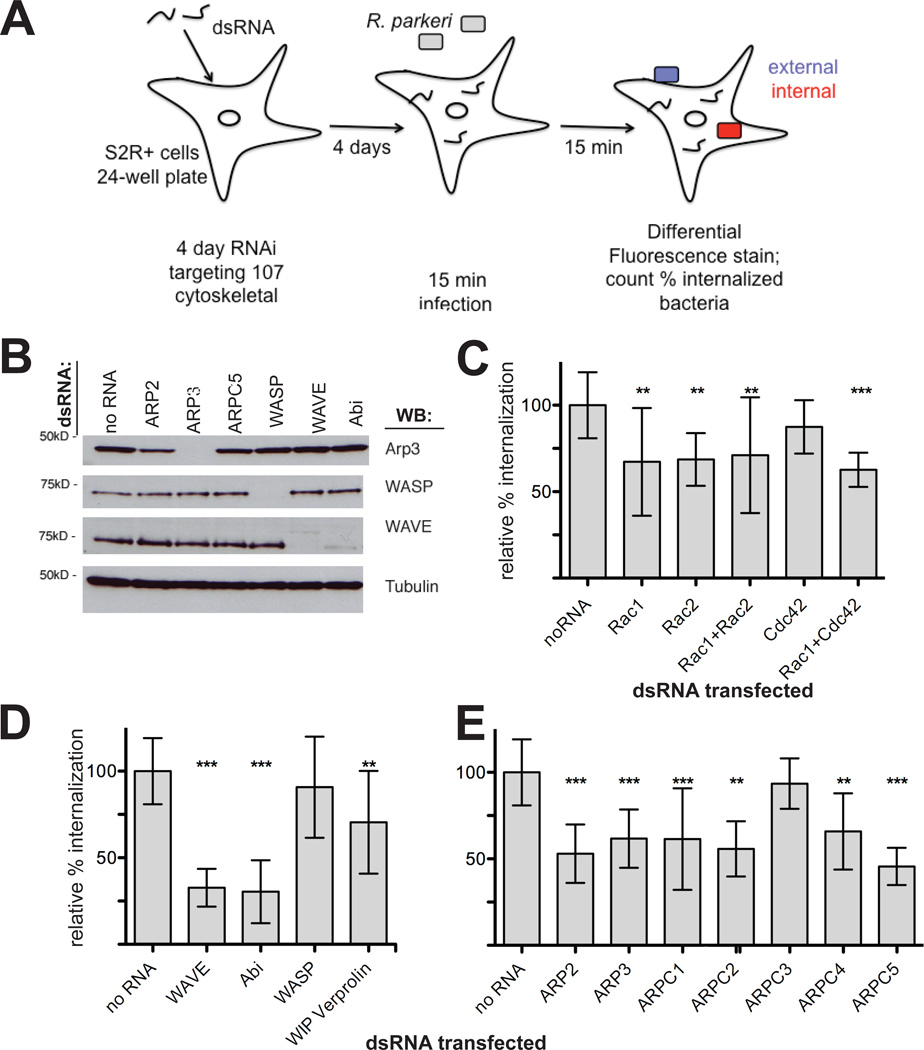
Because of the essential role of actin in R. parkeri invasion, we sought to determine which host proteins direct actin polymerization during this process. To this end, we carried out a screen for invasion using S2R+ cells, which are highly amenable to RNAi-mediated gene silencing and have been used previously to find host factors important for R. parkeri motility as well as L. monocytogenes, Chlamydia trachomatis, Pseudomonas aeruginosa, and Candida albicans host cell invasion (Agaisse et al., 2005; Stroschein-Stevenson et al., 2005; Elwell et al., 2008; Pielage et al., 2008; Serio et al., 2010). We targeted 105 genes encoding proteins that regulate, modify, or comprise the actin cytoskeleton, drawing from a list previously screened for involvement in R. parkeri actin tail formation (Serio et al., 2010) and Drosophila lamellipodia formation (Rogers et al., 2003). S2R+ cells were treated with dsRNA for 4 d, infected with R. parkeri for 15 min, fixed and differentially stained for intracellular and extracellular bacteria (Figure 2A). We confirmed protein depletion for a subset of targets (Figure 2B) and consistently observed changes in cell shape and actin organization that served as a visual confirmation of successful RNA interference during each experiment (Zallen et al., 2002; Rogers et al., 2003).
Figure 2. RNAi screening in S2R+ cells reveals that Rac, the WAVE complex and the Arp2/3 complex are important for R. parkeri invasion.
(A) Schematic representation of the RNAi screening approach. All genes targeted are listed in Dataset S1. (B) Western blot of S2R+ cell lysates after RNAi targeting the indicated proteins for 4 d. (C–E) Subset of the S2R+ RNAi screening results represented as relative % internalization after targeting (C) Rho-family GTPases alone or in combination, (D) the NPFs WAVE/Scar or WASP and their complex subunits, or (E) Arp2/3 complex subunits. Results are normalized to the mean of all untreated control coverslips. Data represent at least three independent experiments. Error bars represent SD; ** p<0.01, *** p<0.001 versus untreated controls by unpaired Student’s t-test.
Twenty-one proteins were identified as having potential functions in R. parkeri invasion of host cells based on a statistically significant (p≤0.01 by students t-test) decrease in the percentage of internalized bacteria compared to untreated control cells (Table 1). When data were normalized to matched control cells for each day’s experiments, differences remained statistically significant (Table 1, Dataset S1). The 21 proteins could be segregated into five functional classes, including: (1) small G proteins; (2) Arp2/3 complex subunits, NPFs and NPF-binding proteins; (3) phagocytosis-associated myosin motors and adapters; (4) membrane-cytoskeleton linking proteins; and (5) actin filament organizing, bundling and severing proteins.
Table 1.
Proteins implicated in Rickettsia invasion of S2R+ cells by RNAi screening
| Functional group | Protein implicated in invasion |
Internalization a (%) |
p-valueb | Normalized internalizationc |
Hit shared with other screensd |
|---|---|---|---|---|---|
| --- | untreated control cells | 54 ± 10 | --- | 1 ± 0.09 | --- |
| Rho family GTPase | Rac1 | 37 ± 17 | 0.0021 | 0.84 ± 0.5 | 4,5,7,10 |
| Rac2 | 37 ± 8 | 0.002 | 0.77 ± 0.09 | 3,4,5,10 | |
| Rac1 + Cdc42 | 34 ± 5 | 0.0003 | 0.68 ± 0.11 | --- | |
| Rac1 + Rac2 | 39 ± 18 | 0.0031 | 0.84 ± 0.46 | --- | |
| Actin filament nucleator | ARP2 | 29 ± 9 | <0.0001 | 0.59 ± 0.15 | 2,3,9 |
| ARP3 | 34 ± 9 | 0.0002 | 0.67 ± 0.07 | 2,3,4,6,9 | |
| ARPC1 (p41) | 33 ± 16 | 0.0013 | 0.66 ± 0.25 | ||
| ARPC2 (p34) | 30 ± 9 | <0.0001 | 0.63 ± 0.21 | 2,3 | |
| ARPC4 (p20) | 36 ± 12 | 0.001 | 0.61 ± 0.22 | 1,2,3,4,5,6,7,8,10 | |
| ARPC5 (p16) | 25 ± 6 | <0.0001 | 0.5 ± 0.04 | 2,3,4,5,10 | |
| Actin nucleation promoting factor | WAVE (SCAR) | 18 ± 6 | <0.0001 | 0.38 ± 0.08 | 1,4,5,6,7,8,9,10 |
| WAVE complex member, regulator | Abi | 17 ± 10 | <0.0001 | 0.34 ± 0.18 | 1,2,3,4,5,7,8 |
| Kette HEM-protein | 31 ± 9 | <0.0001 | 0.57 ± 0.24 | 1,7,8 | |
| Sra-1 | 35 ± 6 | 0.0027 | 0.55 ± 0.07 | 1,4,6,7 | |
| WASP binding, regulator | WIP veroprolin | 38 ± 16 | 0.005 | 0.78 ± 0.34 | |
| Myosin motor protein | Myosin IA | 28 ± 12 | <0.0001 | 0.62 ± 0.37 | 10 |
| Myosin II (zipper) | 33 ± 6 | 0.0002 | 0.72 ± 0.27 | 1 | |
| Endocytic / phagocytic adapter | Hip1R | 35 ± 7 | 0.0005 | 0.74 ± 0.17 | 10 |
| Membrane-cytoskeleton linker | MIM homolog | 33 ± 8 | 0.0001 | 0.69 ± 0.21 | 9 |
| Myoblast city | 38 ± 4 | 0.0089 | 0.87 ± 0.11 | 7 | |
| Actin filament bundling/organizing | α-actinin | 34 ± 14 | 0.0004 | 0.71 ± 0.19 | 7,8,10 |
| Fimbrin | 38 ± 6 | 0.0078 | 0.86 ± 0.05 | 7,10 | |
| Actin monomer dynamics | Cofilin (twinstar) | 40 ± 12 | 0.0088 | 0.8 ± 0.09 | 1,7,10 |
Percent of R. parkeri internalized 15 mins after infection in cells treated with dsRNA targeting the indicated protein(s). The data presented are the mean ± SD of at least three independent experiments.
Determined by pairwise comparison with the untreated control using the Student’s t-test. p<0.01 was considered statistically significant.
Data were adjusted with the mean percent internalization of matched untreated control cells (duplicate samples) for each day’s experiment set as 1 (100%).
RNAi targeting this protein was also found to 1reduce lamella formation (Rogers et al., 2003); 2Listeria monocytogenes and/or 3Mycobacterium fortuitum infection (Agaisse et al., 2005); 4Candida albicans, 5E. coli or 6latex bead phagocytosis (Stroschein-Stevenson et al., 2005); 7Chlamydia trachomatis infection (Elwell et al., 2008); 8Pseudomonas aeruginosa internalization (Pielage et al., 2008); 9Rickettsia parkeri infection, or 10R. parkeri actin tail formation (Serio et al., 2010).
Individual proteins identified in the screen included Rac1 and Rac2, suggesting an involvement of Rho-family GTPases in R. parkeri invasion (silencing Rac1/Rac2 or Rac1/Cdc42 in combination also decreased entry; Table1, Figure 2C). Downstream of Rac1 and Rac2, RNAi targeting of the Rac effector and Arp2/3 complex NPF WAVE (Drosophila SCAR) as well as subunits of the WAVE complex (Abi, Sra-1, Kette), decreased R. parkeri invasion to levels similar to invasion of heat-killed bacteria (Table 1, Figure 1B, Figure 2D). Depletion of subunits of the Arp2/3 complex (ARP2, ARP3, ARPC1, ARPC2, ARPC4, and ARPC5) also significantly reduced invasion (Table 1, Figure 2D). Targeting of WASP, a second Arp2/3 complex NPF, did not significantly reduce internalization (Dataset S1), although targeting of the WASP-interacting protein WIP did inhibit internalization (Table 1, Figure 2D). Other targets for which RNAi reduced internalization included the myosin motors Myosin 1A and Myosin II, the actin organizing proteins α-actinin and fimbrin, the severing protein cofilin, the membrane-cytoskeleton linker proteins MIM homolog and Myoblast city, and the endocytic/phagocytic adapter protein Hip1R (Table 1). In contrast, notable proteins not implicated in internalization included a seventh Arp2/3 complex subunit (ARPC3), three other WAVE-interacting proteins (Drk, HSPC300, Nck), four other Arp2/3 complex regulators (Coronin, Cortactin, Dcarmil, POD-1), six formin actin nucleation and elongation proteins, and the Spire tandem monomer-binding actin nucleator (Dataset S1). Overall, our targeted screen implicated numerous cytoskeletal proteins in actin assembly during R. parkeri invasion of Drosophila S2R+ cells, and identified a specific actin nucleation pathway involving Rac proteins, the WAVE/Abi NPF complex, and the Arp2/3 complex.
Mammalian Rho-family GTPases are recruited to invading R. parkeri and are important for entry
We hypothesized that the proteins implicated in R. parkeri invasion of Drosophila cells might also be utilized during invasion of mammalian cells. To test this, we employed two mammalian cell types, COS-7 and HMEC-1 (Ades et al., 1992), the latter of which are endothelial cells derived from the tissue naturally infected by Rickettsia species (Walker et al., 2008). Given the importance of Rho GTPase signaling for R. parkeri invasion in S2R+ cells, and the previously-implied role for Cdc42 in R. conorii invasion of Vero cells (Martinez et al., 2004), we sought to define the requirement for Rho-family GTPases in mammalian cell invasion.
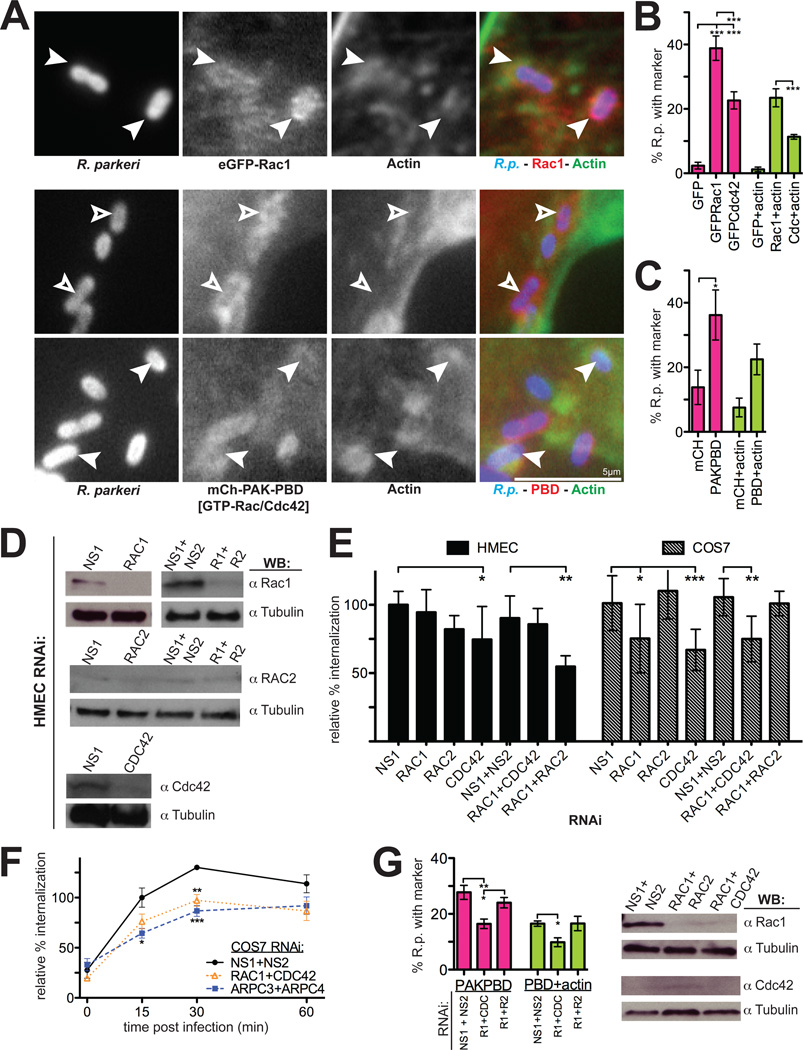
We first tested whether active GTPases were recruited to sites of invasion. HMEC-1 cells were transfected with a plasmid expressing either eGFP-Rac1, eGFP-Cdc42 (Subauste et al., 2000) or the p21-binding domain (PBD) of p21-activated kinase (PAK) fused to mCherry (PAK1-PBD-mCherry), a marker for active (GTP-bound) Rac1 and Cdc42 (Srinivasan et al., 2003; Chenette et al., 2006). Both eGFP-Rac1 and eGFP-Cdc42 proteins, but not eGFP-RhoA, were observed around invading, actin-associated R. parkeri (Figure 3A–B, data not shown). Immunolocalization of Rac1 and Cdc42 was inconclusive due to nonspecific binding of antibodies to bacteria (data not shown). In addition, PAK1-PBD-mCherry was robustly recruited to sites of bacterial invasion 5–15 min after infection (Figure 3A, 3C), and could be imaged by time-lapse microscopy while invasion was progressing (Videos S3, S4). The marker was found around bacteria both with and without associated actin, suggesting that recruitment occurs before actin polymerization begins.
Figure 3. Rho-family GTPases are recruited to invading R. parkeri in mammalian cells and are important for invasion.
(A) R. parkeri (left, blue in merge) are surrounded by eGFP-Rac1 or PAK1-PBD-mCherry (middle, red in merge) and actin (right, green in merge) in HMEC-1 cells at 10 min post-infection. Open arrowheads, Rickettsia associated with PAK1-PBD alone; filled arrowheads, Rickettsia surrounded by actin and eGFP-Rac1 or PAK1-PBD. (B) Percentage of Rickettsia associated with GFP, eGFP-Rac1, eGFP-Cdc42 and actin 10 min post-infection. Pink bars, mean association with marker; light green bars, mean association with both GFP-marker and actin. (C) Percentage of Rickettsia associated with mCherry, PAK1-PBD-mCherry, and actin as in panel (B). (D) Western blots of HMEC-1 cell lysates 48 h post-transfection with the indicated siRNAs, corresponding to one experiment represented in (E). COS-7 cell Western blots are shown in Fig S2A. (E) Relative percent internalization at 15 min post-infection with R. parkeri in either HMEC-1 (solid bars) or COS-7 cells (hashed bars), normalized to nonspecific-RNA treated cells. (F) Relative percentage internalization in COS-7 cells from 0–60 min post-infection with R. parkeri, normalized to 15 min nonspecific-RNA treated cells. (G) Percentage of Rickettsia associated with PAK1-PBD-mCherry and actin after cotransfection with indicated siRNAs, as in (C), with corresponding Western blots. Abbreviations: NS1, nonspecific control siRNA 1; NS2, nonspecific control siRNA 2; R1 + R2, Rac1 and Rac2. Error bars, SEM (B–C, F–G) and SD (E). * p<0.05, ** p<0.01, *** p<0.001 versus indicated control by unpaired Student’s t-test (E–F) or ANOVA with Bonferroni’s post-test (B,C,G). (B–C, F–G) are the mean of two independent experiments performed in duplicate. (E) are the mean of at least three independent experiments performed in duplicate.
To evaluate the function of Rho GTPases during entry, HMEC-1 and COS-7 cells were transfected with siRNAs targeting Rac1, Rac2, or Cdc42 alone or in combination, infected with R. parkeri, and the percentage of internalized bacteria was measured. Gene silencing was confirmed for Rac1 and Cdc42 by immunoblotting (Figure 3D, Figure S2A–B). We were unable to detect significant levels of Rac2 in HMEC-1 cells by either immunoblotting or RT-PCR, suggesting it may not be expressed at appreciable levels in this cell type (Figure 3D, data not shown). In HMEC-1 cells, siRNA targeting Cdc42 alone or Rac1 and Rac2 in combination caused a significant decrease in invasion compared to control-siRNA transfected cells (Figure 3E). Co-transfection of siRNAs with PAK1-PBD-mCherry revealed that recruitment of PAK1-PBD, and PAK1-PBD together with actin, were significantly reduced (p<0.05) after Rac1 and Cdc42 codepletion, to levels similar to mCherry-only controls (Figure 3C, 3G). In COS-7 cells, knockdown of Rac1, Cdc42, or both GTPases resulted in a significant reduction in invasion (Figure 3E) at 15 min, and this decrease persisted through 30–60 min post-infection (Figure 3F). The overall requirement for Rac1 and Cdc42 was generally independent of an effect on bacterial binding to cells, as siRNA treatments had little or no effect on binding (Figure S3A, S3B). Thus, Rho-family GTPases play a role during R. parkeri invasion of mammalian host cells, while the specific requirements for Rac1 and Cdc42 vary between cell types.
WAVE, but not N-WASP, is the NPF important for optimal R. parkeri invasion of mammalian cells
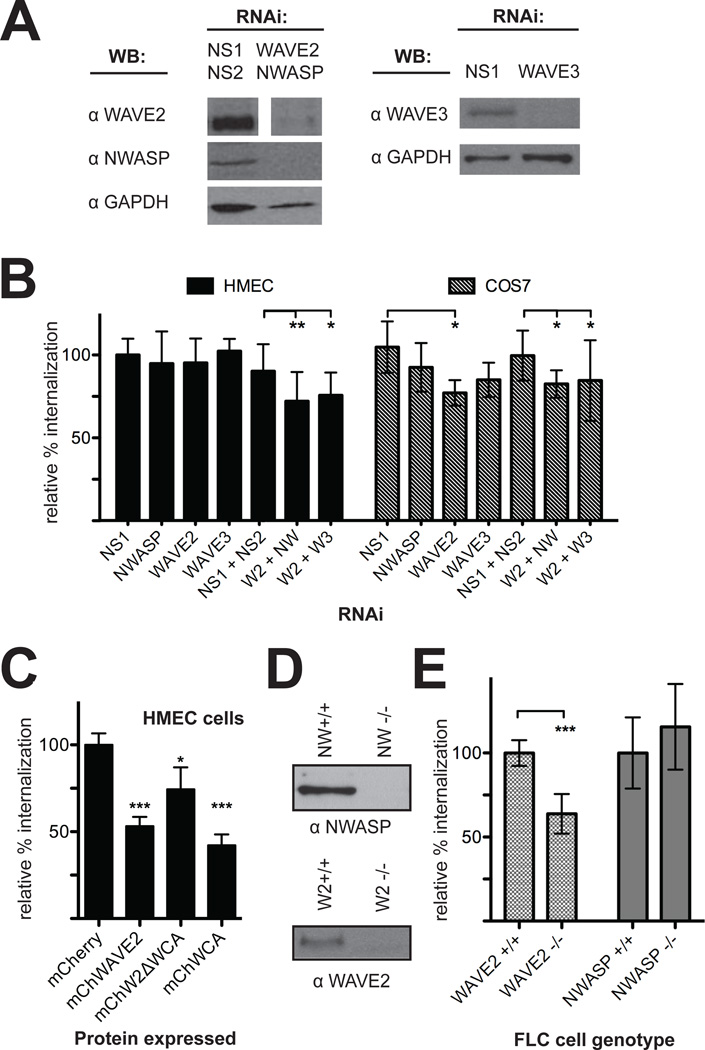
Downstream of Rho-family GTPases, R. parkeri invasion of Drosophila cells required the NPF WAVE and its binding partner Abi, but not the NPF WASP (Figure 2D). There are three mammalian isoforms of WAVE, and two isoforms of WASP: WAVE1 and WAVE3 are expressed in brain and hematopoietic cells, whereas WAVE2 is expressed ubiquitously, while WASP is expressed in hematopoietic cells and N-WASP is ubiquitous (Campellone et al., 2010). We first sought to determine which of these NPF proteins were expressed in the mammalian cell lines used for our experiments. Immunoblotting of extracts from various mammalian cell lines revealed that WAVE2, WAVE3 and N-WASP were abundant in HMEC-1 and COS-7 cells whereas WAVE1 and WASP were not expressed at detectable levels (Figure S4). Neither immunolocalization of N-WASP and WAVE2, nor expression of GFP or mCherry-fused N-WASP, WAVE2, WHAMM, WASH, and JMY, indicated specific colocalization of NPF proteins with invading R. parkeri (data not shown).
To examine the functional importance of NPF proteins during R. parkeri invasion, WAVE2, WAVE3, and N-WASP were targeted by RNAi in both HMEC-1 and COS-7 cells and bacterial internalization and binding were measured. Protein depletion was confirmed by immunoblotting (Figure 4A, Figure S2B). RNAi targeting of WAVEs had little to no effect on bacterial binding, and targeting of N-WASP increased binding in Cos7 cells (Figure S3). Silencing of WAVE2 resulted in an approximately 20% decrease of invasion in COS-7 cells, but had no significant effect in HMEC-1 cells. However, in both HMEC-1 and COS-7 cells, silencing WAVE2 in combination with WAVE3 or N-WASP caused a 20–25% decrease of invasion. Knockdown of N-WASP alone had no discernable affect on entry in either cell type (Figure 4B). We also treated HMEC-1 cells with wiskostatin, a specific inhibitor of N-WASP (Peterson et al., 2001) and saw no significant change in invasion efficiency or bacterial association (Figure S1). Therefore, WAVE2 appears to be consistently important for invasion, and WAVE3 or N-WASP may functionally replace WAVE2 if present, especially in HMEC-1 cells.
Figure 4. Depletion, overexpression or genetic deletion of WAVE2 reduces R. parkeri invasion of mammalian cells.
(A) Western blots of HMEC-1 cell lysates 48 h post-transfection with the indicated siRNAs, corresponding to one experiment represented in (C). COS-7 Western blots are shown in Figure S2B. (B) Relative percent internalization at 15 min post-infection with R. parkeri following RNAi of the indicated targets in HMEC-1 (solid bars) or COS-7 cells (hashed bars). (C) Relative percent internalization at 15 min post-infection with R. parkeri in HMEC-1 cells visibly expressing the indicated proteins. (D) Western blots of cell lysates collected from FLC cells showing a lack of expression of N-WASP (top) or WAVE2 (bottom). (E) Relative percent internalization at 2 h post-infection with R. parkeri in FLC cells with a genetic deletion of WAVE2 or N-WASP. Data represent the mean of at least three independent experiments and were normalized using (B) nonspecific-RNA transfected HMEC-1 cells, (C) mCherry-expressing HMEC-1 cells, or (D) sibling control cells for each deletion line. Abbreviations: NS1, nonspecific control RNA 1; NS2, nonspecific control RNA 2; W2, WAVE2; NW, N-WASP; W3, WAVE3. Error bars indicate SD. * p<0.05, ** p<0.01, *** p<0.001 versus indicated control values by unpaired t-test.
To further examine the role of WAVE2 in R. parkeri invasion of HMEC-1 cells, we tested whether expression of full-length WAVE2 or truncation derivatives might inhibit the process. HMEC-1 cells were transfected with plasmids expressing either the full-length WAVE2 protein (mChWAVE2), or the N-terminal (mChW2ΔWCA) or C-terminal (mChW2WCA) domains fused to mCherry, and invasion efficiency and binding were quantified in cells visibly expressing fusion proteins. Expression of either the full-length WAVE2 protein or the C-terminal WCA domain reduced invasion of R. parkeri by 50–60% compared with mCherry alone (Figure 4C). Interestingly, expression of the WAVE2 N-terminus also reduced invasion by 25% (Figure 4C). Binding was not significantly affected by expression of any WAVE derivatives (Figure S5A). The dominant negative effect of full-length WAVE2 expression on Rickettsia invasion was previously assumed to be due to excess C-terminal WCA domain inhibiting the Arp2/3 complex (Machesky et al., 1998; Magdalena et al., 2003). Our results indicate that inhibition may also involve the N-terminal domain of WAVE binding to and disrupting upstream interacting proteins such as Rac1 (Miki et al., 1998), Abi, or HSPC300 (Shi et al., 2005).
To definitively clarify the roles of WAVE2 or N-WASP during R. parkeri invasion of mammalian cells, we measured invasion in mouse embryonic fibroblast-like cell lines (FLC) genetically deficient in each protein. FLC cells deficient for WAVE2 (Yan et al., 2003) or N-WASP (Snapper et al., 2001), along with matched control cells, were infected with R. parkeri for 2 h and bacterial internalization and binding were quantified. Absence of NPF expression was confirmed by immunoblotting (Figure 4D). WAVE2 knockout cell lines had 40% fewer internalized bacteria than control cells, while there was no significant difference between N-WASP-deleted cell lines and controls (Figure 4E). Genetic deletion of WAVE2 or N-WASP did not significantly affect bacterial binding (Figure S5B). Therefore, similar to HMEC-1 cells, WAVE2 is important during invasion of murine fibroblasts and N-WASP is dispensable. Overall, these results suggest that N-WASP is not crucial for R. parkeri entry, and that WAVE2 is important, with the extent of the requirement depending on the cell type being infected.
The host Arp2/3 complex is recruited by R. parkeri and is required for invasion
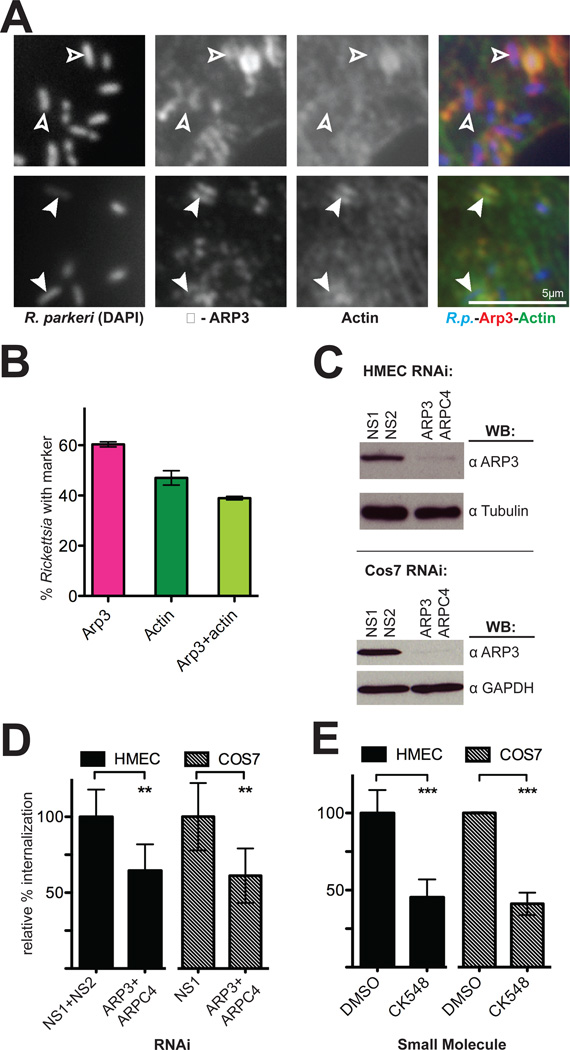
The results of our RNAi screen in S2R+ cells, along with the involvement of Rac1 and the WAVE family proteins in mammalian cells, strongly suggested a role for the Arp2/3 complex during invasion of mammalian cells by R. parkeri. Indeed, we observed robust localization of Arp3 protein around R. parkeri invading HMEC-1 cells (Figure 5A) and COS-7cells (Figure S6) at 5–15 min post-infection. On average, Arp2/3 complex was associated with a higher proportion of invading Rickettsia than was actin (Figure 5B), suggesting that the Arp2/3 complex is recruited before F-actin is polymerized around entering bacteria.
Figure 5. The Arp2/3 complex is recruited to invading R. parkeri and is required for efficient invasion.
(A) Bacteria (left, stained with DAPI, blue in merge) are surrounded by Arp3 (middle, anti-Arp3 immunofluorescence, red in merge) and actin (right, anti-RFP immunofluorescence of Lifeact-mCherry, green in merge). Open arrowheads indicate R. parkeri associated with Arp3 protein only, filled arrowheads indicate R. parkeri surrounded by both actin and Arp3. (B) Percentage of Rickettsia associated with Arp3 and actin. Pink, mean association with Arp3; dark green, mean association with actin; light green, subset of cells associated with both Arp3 and actin. For (A–B), HMEC-1 cells were transfected with pLifeact-mCherry for 48 h before infection and fixed 10 min post-infection. (C) Western blots of HMEC-1 and COS-7 cell lysates at 48 h post-transfection with the indicated siRNAs, corresponding to one experiment represented in (D). (D) Relative percent internalization at 15 min post-infection with R. parkeri in either HMEC-1 (solid bars) or COS-7 (hashed bars) cells. (E) Relative percent internalization at 15 min post-infection with R. parkeri within HMEC-1 or COS-7 cells treated for 30 min prior to infection with either 1% DMSO or 100 µM CK548. (D) and (E) are the mean of at least three independent experiments and were normalized with non-specific RNA-transfected cells (D) or DMSO-treated cells (E) set as 100%. Abbreviations: NS1, nonspecific control RNA 1; NS2, nonspecific control RNA 2; ** p<0.01, *** p<0.001, versus indicated control values by unpaired Student’s t-test.
To investigate the functional importance of the Arp2/3 complex during bacterial invasion, HMEC-1 and COS-7 cells were transfected with two siRNAs targeting the Arp3 and ARPC4 subunits, which were previously shown to deplete the entire complex (Campellone et al., 2008), or with control siRNAs, and silencing was confirmed using immunoblotting (Figure 5C, Figure S2B). Fifteen min after invasion, an average of 45% fewer intracellular bacteria were observed in Arp3/ARPC4-depleted cells (Figure 5D) while bacterial binding was unaffected (Figure S3). This invasion defect persisted through 30–60 min post-infection in COS-7 cells (Figure 3F). To further confirm the requirement for Arp2/3 complex activity, HMEC-1 or COS-7 cells were treated for 30 min with either a specific chemical inhibitor of the Arp2/3 complex, CK-548 (Nolen et al., 2009), or with DMSO as a control, and were infected with R. parkeri in the presence of the inhibitor. Chemical inhibition of the Arp2/3 complex resulted in approximately 55% fewer intracellular bacteria (Figure 5D) and decreased bacterial binding (Figure S1B). The inhibition of entry in cells after Arp2/3 complex inactivation or depletion indicates that the complex is a major nucleator of F-actin around entering Rickettsia in mammalian cells.
DISCUSSION
As an obligate intracellular pathogen, Rickettsia parkeri must invade host cells to survive. Here, we identify core host factors responsible for actin polymerization during invasion of both arthropod and mammalian cells. During invasion, actin is nucleated primarily by the Arp2/3 complex, which is likely activated by a WAVE-dependent and WASP/N-WASP-independent pathway that is in turn controlled by the Rho-family GTPases Rac and Cdc42. Our results reveal differences between actin regulation in arthropod versus mammalian cells due to either different levels of redundancy in the core pathway or to the existence of alternative pathways for polymerizing actin.
We began with an RNAi-based screen in Drosophila S2R+ cells that focused on identifying which of ~100 proteins previously shown to be important for actin polymerization, organization and regulation (Rogers et al., 2003) played a role in R. parkeri invasion. Our results identified 21 proteins, which include an actin nucleating complex and its regulators, as well as actin organizing and binding proteins likely to contribute to actin network stability (Figure 6). A subset of these 21 proteins was also identified in a previous screen for cytoskeletal proteins involved in Rickettsia actin tail formation and infection (Serio et al., 2010), indicating that defects in invasion may perturb later steps in the Rickettsia life cycle. One question that arises from our work is, what upstream signaling pathways regulate actin cytoskeletal proteins during invasion? Multiple Rickettsia outer-membrane autotransporter proteins mediate adherence to host cells and bind to receptors including Ku70 (Martinez et al., 2005; Chan et al., 2009; silencing Ku70 caused a modest but significant defect in invasion of S2R+ cells; Dataset S1). Downstream of receptor engagement, a general inhibitor of tyrosine kinase activity blocks Rickettsia invasion of Drosophila and mammalian cells (Martinez et al., 2004; this study), but our results did not implicate any individual tyrosine kinases. This suggests that multiple tyrosine kinase signaling pathways converge to activate Rho-family GTPases during Rickettsia invasion of host cells.
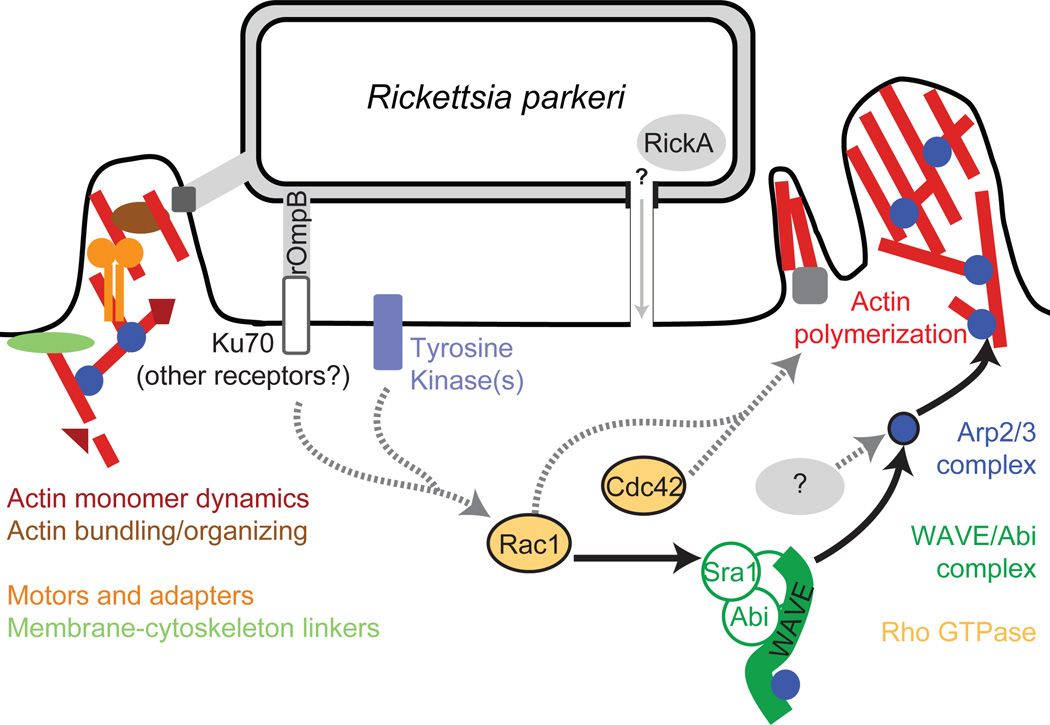
Figure 6. Model of host pathways activated during Rickettsia invasion.
Multiple pathways are likely to be activated during R. parkeri invasion of host cells. Solid arrows and colored proteins were identified in this work, while grey proteins and dashed arrows indicate potential pathways suggested from previous work.
R. parkeri invasion of both Drosophila and mammalian cells was sensitive to depletion of the Rho GTPases Rac or Cdc42, or the combined depletion of both. Cdc42 was initially thought to activate WASP-dependent filopodia extension while Rac1 and Rac2 activate WAVE-dependent lamellipodia formation (Hall, 1998; Miki et al., 1998), but more recent findings indicate that both GTPases can contribute to lamellipodia formation in Drosophila (Rogers et al., 2003) and mammalian cells (Kurokawa et al., 2004). Moreover, Rac1, Rac2 and/or Cdc42 have been previously identified as important for the invasion of S2R+ cells by several pathogens in RNAi screens (Agaisse et al., 2005; Stroschein-Stevenson et al., 2005; Elwell et al., 2008; Serio et al., 2010). Only Cdc42 was previously implicated in a R. conorii invasion of Vero cells (Martinez et al., 2004), in a study making use of dominant-negative Rho GTPase mutants. Because expression of mutant proteins may cause more severe phenotypes than knockdown of the same proteins (Pertz, 2010) and may inhibit GEFs for both GTPases (Rabiet et al., 2002; Ladwein et al., 2008; Pertz, 2010), an RNAi approach provides a clearer picture of specific GTPase requirements for Rickettsia invasion. Interestingly, we observed a varying phenotype after RNAi targeting Rac1, Rac2 and Cdc42, depending on the cell line. Thus, during R. parkeri invasion of host cells, we propose a model where the Rho GTPases cooperatively promote actin polymerization, with Cdc42 and Rac1 playing parallel roles (Figure 6).
We also observed a strong inhibition of R. parkeri invasion after RNAi silencing of WAVE and WAVE-interacting proteins in Drosophila S2R+ cells, whereas depletion of WASP had no discernible effect. WAVE and Abi1 were shown to be required for both lamellipodia and filopodia formation and were identified in all previous RNAi-based screens for pathogen invasion in S2R+ cells (Table 1), while WASP is important for only P. aeruginosa invasion (Pielage et al., 2008) and is not required for the formation of cellular protrusions (Rogers et al., 2003; Biyasheva et al., 2004). We conclude that WAVE is required for actin-dependent cell surface remodeling in S2R+ cells, especially during the invasion of intracellular pathogens including R. parkeri (Figure 6).
We found that R. parkeri invasion of a variety of mammalian cell lines was reduced when WAVE2 was depleted, overexpressed or genetically deleted. On the other hand, our data do not support the previously suggested role for N-WASP (Martinez et al., 2004), nor do they support a role for other host NPFs. Thus, of the many host NPF proteins, only WAVE is important for R. parkeri invasion of cell lines from diverse species (Figure 6). WAVE2 and WAVE3 have been previously implicated in lamellipodia formation, cell migration, and the invasion of mammalian cells by Chlamydia and Salmonella, indicating that their function in membrane remodeling and pathogen invasion is conserved (Yamazaki et al., 2003; Shi et al., 2005; Carabeo et al., 2007). Interestingly, the combined depletion of WAVE2 and WAVE3 or WAVE2 and N-WASP caused a more modest decrease in R. parkeri invasion in HMEC-1 or COS-7 cells than that observed after WAVE depletion in S2R+ cells, suggesting that other host or bacterial proteins may provide an additional degree of redundancy in mammalian cells.
Downstream of the WAVE complex, our results clearly indicate that depletion, inhibition, or sequestration of the Arp2/3 complex strongly inhibits R. parkeri invasion of both Drosophila and mammalian cells, implying that upstream pathways that stimulate actin nucleation converge on the Arp2/3 complex. This is consistent with our observation that invasion of S2R+ cells was not significantly affected by RNAi silencing of other actin nucleators. Although a recent report demonstrated that Salmonella enterica Typhimurium invades cells via both Arp2/3- and Rho/Myosin II-mediated pathways (Hänisch et al., 2011), and we observed reduced invasion of Drosophila cells after Myosin II depletion, treatment of Drosophila and mammalian cells with the Myosin II inhibitor blebbistatin had no effect on invasion, suggesting that R. parkeri invasion can occur independently of Myosin II. Invasion of S2R+ cells by L. monocytogenes, Candida albicans, E. coli, C. trachomatis, and P. aeruginosa is also reduced by knockdown of Arp2/3 complex subunits (Agaisse et al., 2005; Stroschein-Stevenson et al., 2005; Elwell et al., 2008; Pielage et al., 2008). Likewise, the Arp2/3 complex is important for invasion of non-phagocytic mammalian cells by Yersinia pseudotuberculosis, L. monocytogenes, S. enterica Typhimurium and C. trachomatis (Alrutz et al., 2001; Unsworth et al., 2004; Carabeo et al., 2007; Hybiske et al., 2007; Sousa et al., 2007). Thus Rickettsia, like other pathogens, primarily utilize Arp2/3-dependent actin nucleation during invasion of host cells.
The robust nature of invasion in mammalian cells following knockdown or inhibition of individual Arp2/3 complex activators suggests that Arp2/3 is activated via redundant pathways. Depletion of multiple Rho family GTPases or NPFs also caused a more modest inhibition of invasion compared with depletion or inhibition of the Arp2/3 complex. Thus it is likely that Arp2/3 activation occurs independently of WAVE and Rac1. Another host NPF protein could activate Arp2/3, although our results do not support this notion. A second possibility is that a bacterial protein directly activates the Arp2/3 complex during invasion. SFG Rickettsia possess an Arp2/3 activating protein, RickA (Gouin et al., 2004; Jeng et al., 2004), that is a candidate for promoting actin assembly during invasion (Figure 6). However, it remains to be determined whether RickA is secreted into the host cell during this process.
Integrating our data from Drosophila and mammalian cells with previously published research suggests a model for the initiation of actin assembly during Rickettsia invasion (Figure 6). Receptor binding by Rickettsia proteins initiates activation of host protein tyrosine kinases, leading to Cdc42 activation and Rac-dependent activation of WAVE, which may act in parallel with other NPFs to activate Arp2/3. Finally, the Arp2/3 complex acts as the major nucleator of actin filaments during Rickettsia invasion of host cells. Our work provides a framework for further exploration of the host factors required for invasion of host cells by rickettsiae. It will be interesting to determine whether the molecular pathways identified in Drosophila cells are also critical for infection of cells in Rickettsia vectors such as ticks and fleas. The robust nature of Rickettsia invasion is probably due to a combination of redundant host pathways available for actin polymerization and the action of multiple bacterial proteins on these pathways, a common strategy of intracellular pathogens to ensure efficient uptake by a host cell. Future work will define the relative contribution of bacterial and host proteins to actin assembly during invasion.
MATERIALS AND METHODS
Antibodies
Antibodies were obtained from the following sources (in parentheses): mouse anti-Dm-profilin (developed by L. Cooley at Yale School of Medicine, maintained by the Developmental Studies Hybridoma Bank [DSHB] at the University of Iowa); rat anti-Dm-Arp3 (L. Cooley; Hudson et al., 2002); guinea pig anti-Dm-SCAR (J. Zallen, Sloan-Kettering Institute; Zallen et al., 2002); anti-Dm-WASP (G. Borisy, Northwestern University Medical School; Biyasheva et al., 2004), mouse anti-GAPDH (Ambion); mouse anti-tubulin (developed by M. Klymkowsky at the University of Colorado, Boulder, maintained by the DSHB); mouse anti-RFP ab65856 (Abcam); rabbit anti-Arp3 (Welch et al., 1997a); rabbit anti-ARPC2(p34) (Welch et al., 1997b); rabbit anti-WAVE2 (T. Takenawa, University of Tokyo; Yamazaki et al., 2003); goat anti-WAVE1 (sc-10390) and rabbit anti-WAVE2 (sc-33548) (Santa Cruz Biotechnology); rabbit anti-WAVE3 (09–145) (Millipore); guinea pig anti-N-WASP (Duleh et al., 2010); mouse anti-Rac1 (ARC03) and anti-Cdc42 (ACD03) (Cytoskeleton), rabbit anti-Rac2 (sc-96) (Santa Cruz Biotechnology); mouse anti-Rickettsia M14-13 and rabbit anti-Rickettsia R4668 and I7205 (T. Hackstadt, NIH/NIAID Rocky Mountain Laboratories; Anacker et al., 1987; Policastro et al., 1997); rabbit polyclonal anti-Listeria O antibody (BD Biosciences); horseradish peroxidase conjugated anti-mouse, rabbit and guinea pig secondary antibodies for immunoblotting (GE Healthcare); AlexaFluor 488-, 568-, and AMCA-conjugated anti-rabbit and anti-mouse secondary antibodies for immunofluorescence (Invitrogen Molecular Probes). Abbreviation: Dm, Drosophila melanogaster.
Plasmids
To generate the plasmid pLifeact-mCherry, the eGFP-encoding region of a Lifeact-GFP expression plasmid derived from pEGFP-N1 (Clontech) (Serio et al., 2010) was removed and replaced with PCR-amplified DNA encoding mCherry, resulting in a plasmid expressing the Lifeact peptide (Riedl et al., 2008) fused to the N-terminus of mCherry. The plasmid pmCherry was constructed similarly by replacing eGFP in the vector pEGFP-C1 (Clontech). For WAVE2 expression plasmids, DNA encoding full-length Mus musculus WAVE2 (amino acids M1-D497), the WAVE2 WCA region (amino acids T423-D497), or WAVE2ΔWCA (amino acids M1-L429) were excised from GFP-fusion expression plasmids (K. Campellone, University of California, Berkeley) and cloned into the KpnI and XbaI sites of pmCherry. The eGFP-Rac1 plasmid (Subauste et al., 2000; Addgene plasmid 12980) was a gift from G. Bokoch via addgene.org. The PAK1-PBD-mCherry expression plasmid, with Homo sapiens PAK1 amino acids N65-S149 fused to mCherry, and the GFP-Arp3 plasmid, were a gift from O. Weiner (University of California, San Francisco). Rickettsia insertional mutagenesis plasmid PMW1650 was a gift from D. Wood (University of South Alabama; Liu et al., 2007).
Bacterial strains, growth and purification
Rickettsia parkeri Portsmouth strain was a gift from C. Paddock (Centers for Disease Control and Prevention, Atlanta). Listeria monocytogenes strain 10403S was a gift from D. Portnoy (University of California, Berkeley). E. coli strain XL-10 Gold was from Stratagene.
GFP-expressing R. parkeri clonal strain GFPUV-3 was generated by electroporating R. parkeri suspended in 100 µl 250 mM sucrose with 20 µg of plasmid PMW1650 at 2.5 kV, 200 ohms, 25 µF, for 5 ms using a Gene Pulser Xcell (Bio-Rad). Bacteria were immediately suspended in 500 µl Brain Heart Infusion (BHI) media (Difco), and used to infect a 25 cm2 flask of Vero cells. Cells were grown overnight at 34°C, 5% CO2 in DMEM with 2% FBS, and after 24 h media was replaced with media containing 200 ng/ml rifampicin (Sigma). After 6 d, plaques were visible in the cell monolayer. The resulting polyclonal stock was collected, amplified and plaque purified to yield a clonal strain. The insertion site of the transposon cassette was determined essentially as described previously (Liu et al., 2007). Briefly, genomic DNA from R. parkeri was digested with HindIII, the restriction enzyme was heat-inactivated, and the DNA fragments were self-ligated. E. coli were transformed with the resulting plasmids, and selected for resistance to 100 µg/ml rifampicin. The insertion sites were sequenced using primers 5’-CGCCACCTCTGACTTGAGCGTCG and 5’-CCATATGAAAACACTCCAAAAAAC and found to be in a position corresponding to nucleotides 13,843–13,846 of the R. rickettsii Sheila Smith strain (Ellison et al., 2008) in the gene encoding hypothetical protein A1G_00085.
For all other procedures, R. parkeri was propagated in Vero cells grown at 33°C with 5% CO2, purified by Renografin density gradient centrifugation as described previously (Hackstadt et al., 1992), and stored at −80° C. For same-day purification of GFP-expressing R. parkeri, infected monolayers of Vero cells were scraped from culture flasks, pelleted by centrifugation, resuspended in K-36 buffer (0.05 M KH2PO4, 0.05 M K2HPO4, 0.1 M KCl, 0.015 M NaCl, pH 7) and dounced to release intracellular R. parkeri. Cell suspensions were centrifuged to remove nuclei and cell debris, and aliquots from the supernatant were centrifuged at 16000 g for 5 min to pellet R. parkeri, then resuspended in the appropriate culture medium and added to cells for infection and visualization. L. monocytogenes was grown in liquid Brain Heart Infusion (BHI; Difco) at 37°C without agitation. E. coli strain XL-10 Gold was grown in Luria Broth (LB) at 37°C with agitation.
Cell growth and bacterial infection
Drosophila S2R+ cells were a gift from R. Tjian (University of California, Berkeley), and were grown at 28°C in M3 Shields and Sang media (Sigma), supplemented with 0.5 g/L KHCO3, 1 g/L yeast extract (Biotech Sources LLC), 2.5 g/L meat peptone extract (Merck) and 10% FBS (Invitrogen). African green monkey kidney fibroblast cells (COS-7) and epithelial cells (Vero) were from the University of California, Berkeley tissue culture facility, and were grown at 37°C with 5% CO2 in DMEM (Invitrogen) with 2–10% FBS (JR Scientific). Mouse embryonic fibroblast-like cells with genetic deletions of WAVE2 (Yan et al., 2003) or N-WASP (Snapper et al., 2001) and matched control cells were a gift from S. Snapper (Harvard Medical School). The human microvascular endothelial cell line HMEC-1 (Ades et al., 1992) was obtained via material transfer agreement from the Centers for Disease Control, Biological Products Branch, and was grown at 37°C with 5% CO2 in MCDB 131 (Invitrogen) with 10% FBS (Hyclone), 2 mM L-Glutamine (Gibco), 10 ng/mL Epidermal Growth Factor (BD Biosciences), and 1 ug/mL Hydrocortisone (Sigma).
To measure invasion, bacterial infections were carried out as follows: cells were seeded onto glass coverslips in 24-well plates 4 d (S2R+ cells) or 24 h (mammalian cells) prior to infection. For most experiments, culture medium was removed and replaced with 0.5 mL ice-cold growth media, and bacteria were added to each well (R. parkeri at a final MOI of 1–5 or ~5×105 pfu/well, L. monocytogenes and E. coli at an MOI of 5–10). Infected cells were then centrifuged at 200 g for 5 min at 4°C, followed by addition of 0.5 mL of 37°C media per well. Cells were incubated at 28°C (S2R+) or 37°C (HMEC-1, COS-7) for the remainder of infection times. For drug treatments, colocalization and live-cell imaging, infections were carried out in culture medium at 37°C or 28°C (S2R+) without replacement and centrifuge steps were at room temperature. Drugs were diluted in culture medium in 1% DMSO for a final concentration of 250 µM genistein (Sigma), 4 µM latrunculin A, 20 µM nocodazole, 100 µM CK-666 (all three from EMD Chemicals), 100 µM CK-548 (CK-0993548, from Cytokinetics), 200 nM wortmannin, 250 nM dasatinib, 500 nM nilotinib (all three from LC Laboratories), 1 µM PP2, 5 µM wiskostatin, and 1µM AAL-993 (all three from Biomol International) and cells were treated with drugs for 30 min prior to infection.
RNA synthesis and RNAi screening
Primers for PCR amplification of target genes from Drosophila genomic DNA were derived from those published by Rogers et al. (Rogers et al., 2003) or from the Drosophila RNAi Screening Center (http://flyrnai.org), and amplification and reverse transcription were performed as described previously (Serio et al., 2010). For RNAi screening, monolayers of S2R+ cells on 12-mm coverslips were treated with dsRNA at a final concentration of 20 µg/ml for 4 d, and then infected with R. parkeri for 15 min to allow for invasion, as described above. Cells were fixed and processed for the fluorescence internalization assay, as described below. The number of intracellular and extracellular bacteria was counted in at least 5 fields of view, with >200 bacteria counted on each coverslip. At least three replicates of RNAi and infection were performed for each individual target or control. The percent internalization was calculated and the value for each target was compared pairwise with the untreated control by the Student’s t-test using Prism v5.0 (Graphpad Software). For normalized data, the mean of three no-RNA control coverslips for each day’s experiment was set as 1.0 and other data was transformed relative to the mean. Differences were considered to be significant if p ≤0.01.
Immunoblotting and immunofluorescence staining
For immunoblotting, cells were lysed in RIPA buffer (50 mM Tris pH 7.6, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS plus 10 mg/ml of aprotinin, leupeptin, pepstatin, and chymostatin, and 1 mM PMSF) or in Laemmli sample buffer. Equal molar amounts of protein were separated by SDS-PAGE and transferred to nitrocellulose. Membranes were blocked with 5% milk in phosphate buffered saline (PBS), probed with primary followed by secondary antibodies, and visualized with ECL detection reagents (GE Healthcare). For detection of Rac1, Rac2 and Cdc42, proteins were separated using 4–20% TGX polyacrylamide gels and blotted to Immun-blot PVDF membranes (both from Bio-Rad). Membranes were dried completely before blocking and probing with primary and secondary antibodies as described above.
For immunofluorescence staining, cells were fixed with 2.5% formaldehyde in PBS at room temperature for 15 min or using −20°C Cytoskelfix (Cytoskeleton) for 5 min. Antibodies were diluted in PBS with 2% bovine serum albumin (BSA) (anti-Arp3 and anti-RFP, 1:100; anti-Rickettsia 14-13, 1:200) and antibody staining steps were carried out at room temperature using standard procedures. For anti-Arp3 staining, coverslips were blocked in PBS + 2% BSA + 2% dry milk for 30 min prior to staining.
For fluorescence internalization assays, coverslips were incubated with primary antibodies against bacteria followed by AMCA or Alexa 488-conjugated secondary antibodies. Cells were permeabilized in PBS with 0.5% Triton X-100 for 5 min, rinsed, and again incubated with primary antibodies against R. parkeri or Listeria followed by Alexa 568-conjugated secondary antibodies, or E. coli were stained using DAPI. To visualize actin, 4 U/µl of Alexa 488 or 568-conjugated phalloidin (Invitrogen) was included with the secondary antibody. Coverslips were mounted with Prolong Gold anti-fade (Invitrogen) and stored at 4°C.
Transfection and RNAi in mammalian cells
For expression of mCherry/GFP-tagged proteins, 50–250 ng of plasmid DNA was transiently transfected into cells using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. For gene silencing, siRNAs (Ambion) were transfected at a final concentration of 50 nM (COS-7 cells) or 20 nM (HMEC-1 cells) using Lipofectamine-RNAi-MAX (Invitrogen), according to the manufacturer’s instructions. At 24 h post-transfection, cells were reseeded from 6-well culture plates to 24-well plates containing glass coverslips and parallel 6-well plates for protein lysate collection. At 48 h post-transfection, cells were infected with R. parkeri and processed for immunofluorescence microscopy, and uninfected cell lysates were collected for immunoblotting, as described above. For RNA/DNA cotransfections, siRNAs were transfected with RNAiMAX, and siRNA and plasmid DNA were transfected 24h later with Lipofectamine 2000. Cells were reseeded at 48h and infections were carried out at 72h post-transfection. For RNAi-internalization assays, the number of intracellular and extracellular bacteria were counted in at least 5 fields of view, with >200 bacteria counted on each of 2–3 coverslips. An average of ~1000 (301–2562) Rickettsia per experiment were scored for protein recruitment in ≥20 cells from 2–3 coverslips.
Imaging
Images were captured using an Olympus IX71 microscope equipped with a 100X (1.35 NA) PlanApo objective lens and a Photometrics CoolSNAP HQ camera. Fixed and live-cell images were captured in the TIFF 16-bit format using MetaMorph software (Molecular Devices) or Micro-Manager Software (http://www.micro-manager.org). Following image acquisition, Adobe Photoshop was used to convert to 8-bit images, brightness/contrast levels were adjusted, and images were cropped. For Listeria and E. coli internalization assays, WAVE-fusion internalization assays and colocalization quantitation, images were imported into ImageJ and counted using the Cell Counter plugin (http://rsbweb.nih.gov/ij/plugins/cell-counter.html). For live-cell experiments, cells were plated on glass-bottomed 24-well culture dishes (MatTek) and transfected with Lifeact-mCherry or PAK1-PBD-mCherry plasmids 48 h before infection with freshly prepared GFP-expressing R. parkeri at room temperature. Movies were assembled and adjusted using ImageJ and Adobe Photoshop. Deconvolution images were taken using an Applied Precision DeltaVision 4 Spectris microscope with a 100X (1.4 NA) PlanApo objective equipped with a Photometrics CH350 CCD camera. Images were captured using SoftWoRx v3.3.6 software (Applied Precision), deconvolved with Huygens Professional v3.1.0p0 software (Scientific Volume Imaging), and processed using Imaris (Bitplane).
Supplementary Material
Acknowledgements
We thank members of the Welch lab for technical assistance and comments on the manuscript. We also thank T. Clark, S. Dooley, T. Hackstadt and members of the T. Hackstadt lab for assistance with R. parkeri transformations. We are grateful to J. Hartman and Cytokinetics for providing the Arp2/3 complex inhibitor CK-548. We also thank G. Bokoch, K. Campellone, L. Cooley, T. Hackstadt, T. Ohkawa, C. Paddock, D. Portnoy, S. Snapper, T. Takenawa, R. Tjian, O. Weiner, and D. Wood for providing strains or reagents. M.D.W. and S.C.R. were supported by NIH/NIAID grant AI 074760. A.W.S. was supported by NIH/NIGMS Ruth L. Kirschstein National Research Service Award 5F32GM084359.
REFERENCES
- Ades EW, Candal FJ, Swerlick RA, George VG, Summers S, Bosse DC, Lawley TJ. HMEC-1: Establishment of an Immortalized Human Microvascular Endothelial Cell Line. J Investig Dermatol. 1992;99:683–690. doi: 10.1111/1523-1747.ep12613748. [DOI] [PubMed] [Google Scholar]
- Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- Alrutz MA, Srivastava A, Wong K-W, D'Souza-Schorey C, Tang M, Ch'ng L-E, et al. Efficient uptake of Yersinia pseudotuberculosis via integrin receptors involves a Rac1-Arp 2/3 pathway that bypasses N-WASP function. Molecular Microbiology. 2001;42:689–703. doi: 10.1046/j.1365-2958.2001.02676.x. [DOI] [PubMed] [Google Scholar]
- Anacker RL, Mann RE, Gonzales C. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. Journal of Clinical Microbiology. 1987;25:167–171. doi: 10.1128/jcm.25.1.167-171.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biyasheva A, Svitkina T, Kunda P, Baum B, Borisy G. Cascade pathway of filopodia formation downstream of SCAR. Journal of Cell Science. 2004;117:837–848. doi: 10.1242/jcs.00921. [DOI] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD. WHAMM Is an Arp2/3 Complex Activator That Binds Microtubules and Functions in ER to Golgi Transport. Cell. 2008;134:148–161. doi: 10.1016/j.cell.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo RA, Dooley CA, Grieshaber SS, Hackstadt T. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cellular Microbiology. 2007;9:2278–2288. doi: 10.1111/j.1462-5822.2007.00958.x. [DOI] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. The Sca2 Autotransporter Protein from Rickettsia conorii Is Sufficient To Mediate Adherence to and Invasion of Cultured Mammalian Cells. Infection and Immunity. 2009;77:5272–5280. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YGY, Cardwell MM, Hermanas TM, Uchiyama T, Martinez JJ. Rickettsial outer-membrane protein B (rOmpB) mediates bacterial invasion through Ku70 in an actin, c-Cbl, clathrin and caveolin 2-dependent manner. Cellular Microbiology. 2009;11:629–644. doi: 10.1111/j.1462-5822.2008.01279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenette EJ, Mitin NY, Der CJ. Multiple Sequence Elements Facilitate Chp Rho GTPase Subcellular Location, Membrane Association, and Transforming Activity. Mol. Biol. Cell. 2006;17:3108–3121. doi: 10.1091/mbc.E05-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P, Sansonetti PJ. Bacterial Invasion: The Paradigms of Enteroinvasive Pathogens. Science. 2004;304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton. 2010;67:193–206. doi: 10.1002/cm.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. Genomic Comparison of Virulent Rickettsia rickettsii Sheila Smith and Avirulent Rickettsia rickettsiiIowa. Infect. Immun. 2008;76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. RNA Interference Screen Identifies Abl Kinase and PDGFR Signaling in Chlamydia trachomatis Entry. PLoS Pathogens. 2008;4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, et al. An Anomalous Type IV Secretion System in Rickettsia Is Evolutionarily Conserved. PLoS ONE. 2009;4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E, Egile C, Dehoux P, Villiers V, Adams J, Gertler F, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–461. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- Greiffenberg L, Goebel W, Kim KS, Weiglein I, Bubert A, Engelbrecht F, et al. Interaction of Listeria monocytogenes with Human Brain Microvascular Endothelial Cells: InlB-Dependent Invasion, Long-Term Intracellular Growth, and Spread from Macrophages to Endothelial Cells. Infect. Immun. 1998;66:5260–5267. doi: 10.1128/iai.66.11.5260-5267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackstadt T, Messer R, Cieplak W, Peacock MG. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 1992;60:159–165. doi: 10.1128/iai.60.1.159-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. Rho GTPases and the Actin Cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- Hänisch J, Kölm R, Wozniczka M, Bumann D, Rottner K, Stradal TEB. Activation of a RhoA/Myosin II-Dependent but Arp2/3 Complex-Independent Pathway Facilitates Salmonella Invasion. Cell Host & Microbe. 2011;9:273–285. doi: 10.1016/j.chom.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Hudson AM, Cooley L. A subset of dynamic actin rearrangements in Drosophila requires the Arp2/3 complex. J Cell Biol. 2002;156:677–687. doi: 10.1083/jcb.200109065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hybiske K, Stephens RS. Mechanisms of Chlamydia trachomatis Entry into Nonphagocytic Cells. Infection and Immunity. 2007;75:3925–3934. doi: 10.1128/IAI.00106-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng RL, Goley ED, D'Alessio JA, Chaga OY, Svitkina TM, Borisy GG, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–769. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- Kurokawa K, Itoh RE, Yoshizaki H, Nakamura YOT, Matsuda M. Coactivation of Rac1 and Cdc42 at Lamellipodia and Membrane Ruffles Induced by Epidermal Growth Factor. Molecular Biology of the Cell. 2004;15:1003–1010. doi: 10.1091/mbc.E03-08-0609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwein M, Rottner K. On the Rho'd: The regulation of membrane protrusions by Rho-GTPases. FEBS Letters. 2008;582:2066–2074. doi: 10.1016/j.febslet.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Li H, Walker DH. rOmpA is a critical protein for the adhesion of Rickettsia rickettsii to host cells. Microbial Pathogenesis. 1998;24:289–298. doi: 10.1006/mpat.1997.0197. [DOI] [PubMed] [Google Scholar]
- Liu Z-M, Tucker AM, Driskell LO, Wood DO. mariner-Based Transposon Mutagenesis of Rickettsia prowazekii. Appl. Environ. Microbiol. 2007;73:6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Current Biology. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- Magdalena J, Millard TH, Etienne-Manneville S, Launay S, Warwick HK, Machesky LM. Involvement of the Arp2/3 Complex and Scar2 in Golgi Polarity in Scratch Wound Models. Mol. Biol. Cell. 2003;14:670–684. doi: 10.1091/mbc.E02-06-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JJ, Cossart P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J Cell Sci. 2004;117:5097–5106. doi: 10.1242/jcs.01382. [DOI] [PubMed] [Google Scholar]
- Martinez JJ, Seveau S, Veiga E, Matsuyama S, Cossart P. Ku70, a component of DNA-dependent protein kinase, is a mammalian receptor for Rickettsia conorii. Cell. 2005;123:1013–1023. doi: 10.1016/j.cell.2005.08.046. [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. Embo J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock C, Sumner J, Comer J, Zaki S, Goldsmith C, Goddard J, et al. Rickettsia parkeri: A Newly Recognized Cause of Spotted Fever Rickettsiosis in the United States. Clinical Infectious Diseases. 2004;38:805–811. doi: 10.1086/381894. [DOI] [PubMed] [Google Scholar]
- Paddock CD, Finley RW, Wright CS, Robinson HN, Schrodt BJ, Lane CC, et al. Rickettsia parkeri Rickettsiosis and Its Clinical Distinction from Rocky Mountain Spotted Fever. Clinical Infectious Diseases. 2008;47:1188–1196. doi: 10.1086/592254. [DOI] [PubMed] [Google Scholar]
- Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? Journal of Cell Science. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- Peterson JR, Lokey RS, Mitchison TJ, Kirschner MW. A chemical inhibitor of N-WASP reveals a new mechanism for targeting protein interactions. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10624–10629. doi: 10.1073/pnas.201393198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage JF, Powell KR, Kalman D, Engel JN. RNAi Screen Reveals an Abl Kinase-Dependent Host Cell Pathway Involved in Pseudomonas aeruginosaInternalization. PLoS Pathog. 2008;4:e1000031. doi: 10.1371/journal.ppat.1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policastro PF, Munderloh UG, Fischer ER, Hackstadt T. Rickettsia rickettsii growth and temperature-inducible protein expression in embryonic tick cell lines. J Med Microbiol. 1997;46:839–845. doi: 10.1099/00222615-46-10-839. [DOI] [PubMed] [Google Scholar]
- Rabiet M-J, Tardif M, Braun L, Boulay F. Inhibitory effects of a dominant-interfering form of the Rho-GTPase Cdc42 in the chemoattractant-elicited signaling pathways leading to NADPH oxidase activation in differentiated HL-60 cells. Blood. 2002;100:1835–1844. doi: 10.1182/blood-2001-12-0193. [DOI] [PubMed] [Google Scholar]
- Ralph D, Pretzman C, Daugherty N, Poetter K. Genetic Relationships among the Members of the Family Rickettsiaceae As Shown by DNA Restriction Fragment Polymorphism Analysis. Annals of the New York Academy of Sciences. 1990;590:541–552. doi: 10.1111/j.1749-6632.1990.tb42264.x. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, et al. Lifeact: a versatile marker to visualize F-actin. Nat Meth. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YGY, Martinez JJ. The Rickettsia conorii Autotransporter Protein Sca1 Promotes Adherence to Nonphagocytic Mammalian Cells. Infection and Immunity. 2010;78:1895–1904. doi: 10.1128/IAI.01165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. Defining a Core Set of Actin Cytoskeletal Proteins Critical for Actin-Based Motility of Rickettsia. Cell Host & Microbe. 2010;7:388–398. doi: 10.1016/j.chom.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Scita G, Casanova JE. WAVE2 Signaling Mediates Invasion of Polarized Epithelial Cells by Salmonella Typhimurium. J. Biol. Chem. 2005;280:29849–29855. doi: 10.1074/jbc.M500617200. [DOI] [PubMed] [Google Scholar]
- Snapper SB, Takeshima F, Anton I, Liu CH, Thomas SM, Nguyen D, et al. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat Cell Biol. 2001;3:897–904. doi: 10.1038/ncb1001-897. [DOI] [PubMed] [Google Scholar]
- Sousa S, Cabanes D, Bougneres L, Lecuit M, Sansonetti P, Tran-Van-Nhieu G, Cossart P. Src, cortactin and Arp2/3 complex are required for E-cadherin-mediated internalization of Listeriainto cells. Cellular Microbiology. 2007;9:2629–2643. doi: 10.1111/j.1462-5822.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Wang F, Glavas S, Ott A, Hofmann F, Aktories K, et al. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. The Journal of Cell Biology. 2003;160:375–385. doi: 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila Gene Products Required for Phagocytosis of Candida albicans. PLoS Biol. 2005;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subauste MC, Von Herrath M, Benard V, Chamberlain CE, Chuang T-H, Chu K, et al. Rho Family Proteins Modulate Rapid Apoptosis Induced by Cytotoxic T Lymphocytes and Fas. Journal of Biological Chemistry. 2000;275:9725–9733. doi: 10.1074/jbc.275.13.9725. [DOI] [PubMed] [Google Scholar]
- Teysseire N, Boudier JA, Raoult D. Rickettsia conorii entry into Vero cells. Infect Immun. 1995;63:366–374. doi: 10.1128/iai.63.1.366-374.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Kawano H, Kusuhara Y. The major outer membrane protein rOmpB of spotted fever group rickettsiae functions in the rickettsial adherence to and invasion of Vero cells. Microbes and Infection. 2006;8:801–809. doi: 10.1016/j.micinf.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Unsworth KE, Way M, McNiven M, Machesky L, Holden DW. Analysis of the mechanisms of Salmonella-induced actin assembly during invasion of host cells and intracellular replication. Cellular Microbiology. 2004;6:1041–1055. doi: 10.1111/j.1462-5822.2004.00417.x. [DOI] [PubMed] [Google Scholar]
- Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6:375–386. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- Walker TS. Rickettsial interactions with human endothelial cells in vitro: adherence and entry. Infect. Immun. 1984;44:205–210. doi: 10.1128/iai.44.2.205-210.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TS, Winkler HH. Penetration of Cultured Mouse Fibroblasts L Cells by Rickettsia prowazeki. Infect. Immun. 1978;22:200–208. doi: 10.1128/iai.22.1.200-208.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, DePace AH, Verma S, Iwamatsu A, Mitchison TJ. The human Arp2/3 complex is composed of evolutionarily conserved subunits and is localized to cellular regions of dynamic actin filament assembly. J Cell Biol. 1997a;138:375–384. doi: 10.1083/jcb.138.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997b;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S-I, Fujiwara T, et al. WAVE2 is required for directed cell migration and cardiovascular development. Nature. 2003;424:452–456. doi: 10.1038/nature01770. [DOI] [PubMed] [Google Scholar]
- Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. Embo J. 2003;22:3602–3612. doi: 10.1093/emboj/cdg350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen JA, Cohen Y, Hudson AM, Cooley L, Wieschaus E, Schejter ED. SCAR is a primary regulator of Arp2/3-dependent morphological events in Drosophila. J Cell Biol. 2002;156:689–701. doi: 10.1083/jcb.200109057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.