Abstract
DNA detection is commonly used in molecular biology, pathogen analysis, genetic disorder diagnosis and forensic tests. While traditional methods for DNA detection such as polymerase chain reaction (PCR) and DNA microarrays have been well developed, they require sophisticated equipment and operations and thus it is still challenging to develop a portable and quantitative DNA detection method for the public use at home or in the field. Although many other techniques and devices have been reported to make the DNA detection simple and portable, very few of them are currently accessible to the public for quantitative DNA detection because of either the requirement of laboratory-based instrument or lack of quantitative detection.. Herein we report application of personal glucose meters (PGMs), which are widely available, low cost and simple to use, for quantitative detection of DNA, including a hepatitis B virus DNA fragment. The quantification is based on target-dependent binding of complementary DNA-invertase conjugate with the analyte DNA, thereby transforming the concentration of DNA in the sample into glucose through invertase-catalyzed hydrolysis of sucrose. Instead of amplifying DNA strands through PCR, which is vulnerable to contaminations commonly encountered for home and field usage, we demonstrate here signal amplifications based on enzymatic turnovers, making it possible to detect 40 pM DNA using PGM that can detect glucose only at mM level. The method also shows excellent selectivity toward single nucleotide mismatches.
Introduction
Developing portable and quantitative methods for the public to monitor various analytes related to health and environment is one of the greatest challenges in analytical chemistry.1 Although many well developed methods and techniques are being used routinely in medical centres and research labs for the detection of these analytes, few of them are available for the public to use at home or in the field, so that much time and cost is required for the public to take the samples to the professional institutes and wait for the result delivery. In contrast, portable and low-cost methods that is directly usable by the public can facilitate early detections to reduce the hazard of diseases or pollutions, and also are affordable to the people in low income countries and regions.1
DNA detection has attracted increasing attentions owing to its important roles in pathogen analysis, genetic disorder diagnosis and forensic tests.2–13 Traditional methods for the detection of DNA include those based on polymerase chain reaction (PCR) and DNA microarrays.14,15 These methods, although highly sensitive and efficient, are expensive and require sophisticated instrument and operations. The challenge remains in developing a simple, portable and quantitative DNA detection method that can be operated by the public with accessible resources at home or in the field, especially for those under resource limiting conditions or in low income countries.1 Many techniques and devices have been successfully developed by different research groups13,16–20 to achieve simpler and portable DNA detections over the traditional methods, such as nanomaterial-based amplification,7,8,21–26 colorimetry,4,10,13,27,28 fluorescence,3,12,20,29–36 electrochemistry,9,16,25,37–43 surface enhanced Raman scattering (SERS),44–47 and others including surface plasmon, light scattering and force.48–50 However, these methods are based on laboratory-based instrument or customized devices that are not currently easily accessible to the public. Some of the colorimetric or fluorescent methods can monitor DNA by naked eye without instrument, but they can only achieve qualitative or semi-quantitative detections and the color observation may vary between different people or be affected by the light contrast of the surroundings. To achieve portable and quantitative detection, these colorimetric or fluorescent assays still require expensive or customized portable spectrometers that are not readily available to the public.
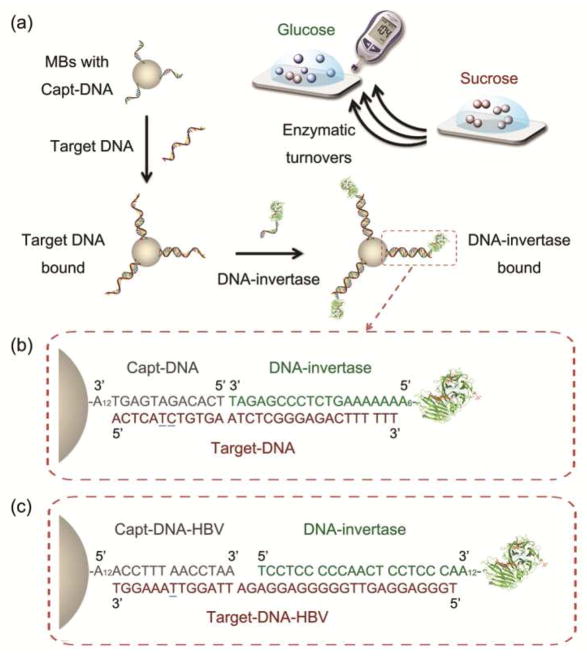
To meet this challenge, we report here the use of a ubiquitous public device, personal glucose meter (PGM), for portable and quantitative DNA detection. The advantages of using PGMs for analytical applications are portability, low cost, simple operation, reliable quantitative results, and most importantly, the wide accessibility to the public.51,52 PGMs are available in stores worldwide with low price (as low as $10 for a meter and $0.5 per test), and have been integrated into cell phones for an even larger number of users.53 However, the current state-of-art PGMs can only be used by diabetes patients to monitor blood glucose. Previously, we reported the successful quantification of a broad range of targets including organic molecules, proteins and metal ions using PGMs linked with functional DNA sensors.54 In this work, by introducing DNA-invertase conjugates54–57 to a DNA sandwich assay, we successfully develop a methodology to quantify DNA using PGMs, thus adding DNA as another analyte in the family of targets that PGMs can detect. The method is based on transforming the concentration of target DNAs in samples into that of PGM-detectable glucose produced from invertase-catalyzed hydrolysis of PGM-inert sucrose. The enzymatic turnovers allow signal amplifications without the need for accurate control of temperature cycles and advanced instrument associated with PCR amplification, and without the risk of possible contamination associated with PCR amplification, such as inhibitors of DNA polymerase and reagents lowering the specificity of polyermase. In addition, since the method doesn’t produce large quantities of DNA copies, there is no DNA to carry over for potential non-specific amplification as in PCR, making it able to quantify target DNA under a variety of non-laboratory conditions using PGMs (Figure 1a).
Figure 1.
(a) The mechanism of target DNA detection by a PGM via the sandwich hybridization assay using Capt-DNA coated magnetic beads (MBs) and DNA-invertase conjugates. (b) DNA sequences used for a proof-of-concept model DNA (Target-DNA) detection. (c) DNA sequences used for a Hepatitis B virus (HBV) DNA fragment (Target-DNA-HBV) detection. The underlined nucleotides are the mismatch sites for controls.
The greatest challenge to realize DNA detection using a personal glucose meter (PGM) is how to establish the link between the target DNA at pM to nM level in the sample and the glucose at mM level needed for a signal readout in PGMs, as the dynamic range of PGMs is typically 0.6~33 mM (10~600 mg/dL) glucose.51,52 Traditionally PCR has been used for amplifying DNA strands to achieve sensitive detection. However, PCR needs well-controlled temperature cycles and laboratory-based device, and has been shown to be subjective to contaminations such as inhibitors of DNA polymerase and reagents lowering the specificity of polyermase, thus can be carried out only in clean and mostly laboratory conditions, making it difficult for home or field use. To achieve the goal of signal amplifications without the complications associated with PCR, we here introduce DNA-invertase conjugates that take advantage of the enzymatic turnovers for signal amplifications without the need to amplify the DNA strands (Figure 1a). First, a biotinylated capture DNA (Capt-DNA) whose sequence is partially complementary to the 5′-half of the target DNA is immobilized onto streptavidin-coated magnetic beads (MBs) via biotin-streptavidin interaction.58 The Capt-DNA coated MBs can capture the target DNA in solution and concentrate the target DNA through DNA hybridization and magnetic separation, respectively. This step is then followed by subsequent addition of a DNA-invertase conjugate, whose DNA sequence is complementary to the 3′ half of the target DNA. Therefore, only in the presence of target DNA can the sandwich assay be successful in making the DNA-invertase bound on the MBs, and the concentration of the target DNA is proportional to the amount of DNA-invertase bound to the MBs. After washing away unbound DNA molecules and invertase, the bound DNA-invertase through the DNA hybridization sandwich can in turn act as a highly efficient signal amplifier to catalyze the hydrolysis of sucrose into glucose with millions of turnovers,59 therefore transforming pM concentration of target DNA into mM levels of glucose for the readout in a PGM (Figure 1a).
Experimental Section
Materials
Streptavidin-coated magnetic beads (1 μm in diameter) and Amicon centrifugal filters (10K and 100K Da molecular weight cut-off) were purchased from Bangs Laboratories Inc. (Fishers, IN) and Millipore Inc. (Billerica, MA), respectively. Grade VII invertase from baker’s yeast (S. cerevisiae), sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC), Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), bovine serum albumin (BSA) and other chemicals for buffers and solvents were purchased from Sigma-Aldrich, Inc. (St. Louis, MO). The following oligonucleotides (left to right: 5′ to 3′) were purchased from Integrated DNA Technologies, Inc. (Coralville, IA):
Capt-DNA: TCACAGATGAGTAAAAAAAAAAAA-Biotin
Thiol-DNA for invertase conjugation: HS-AAAAAAAAAAAAGTCTCCCGAGAT
Target DNA (Target-DNA): ACTCATCTGTGAATCTCGGGAGACTTTTTT
Target DNA with G mismatch (1MS G): ACTCATGTGTGAATCTCGGGAGACTTTTTT
Target DNA with A mismatch (1MS A): ACTCATATGTGAATCTCGGGAGACTTTTTT
Target DNA with T mismatch (1MS T): ACTCATTTGTGAATCTCGGGAGACTTTTTT
Target DNA with 2 mismatches (2MS): ACTCAAGTGTGAATCTCGGGAGACTTTTTT
Capt-DNA for hepatitis B virus (Capt-DNA-HBV): Biotin-AAAAAAAAAAAAACCTTTAACCTAA
Thiol-DNA for invertase conjugation for HBV: TCCTCCCCCAACTCCTCCCAAAAAAAAAAAAA-SH
Target DNA for HBV (Target-DNA-HBV): TGGGAGGAGTTGGGGGAGGAGATTAGGTTAAAGGT
Target DNA for HBV with A mismatch (A-mis): TGGGAGGAGTGGGGGGAGGAGATTAGGTAAAAGGT
Target DNA for HBV with G mismatch (G-mis): TGGGAGGAGTGGGGGGAGGAGATTAGGTGAAAGGT
Target DNA for HBV with C mismatch (C-mis): TGGGAGGAGTGGGGGGAGGAGATTAGGTCAAAGGT
In the model study here, the locations of the mismatch sites were design in the middle of the binding arms with the capture DNA to potentially maximize the ability to recognize the nucleotides at the mismatch sites. Target DNAs are normally much longer than capture DNAs used for the detection, so the capture DNAs can always be designed to make sure the mismatch sites are around the middle of the binding arm.
Buffers used in this study:
Buffer A: 0.1 M NaCl, 0.1 M sodium phosphate buffer, pH 7.3, 0.05% Tween-20
Buffer B: 0.25 M NaCl, 0.15 M sodium phosphate buffer, pH 7.3, 0.05% Tween-20
DNA-invertase conjugation54,56,57
To 30 μL of 1 mM Thiol-DNA or Thiol-DNA-HBV in Millipore water, 2 μL of 1 M sodium phosphate buffer at pH 5.5 and 2 μL of 30 mM TCEP in Millipore water were added and mixed. This mixture was kept at room temperature for 1 hour and then purified by Amicon-10K using Buffer A without Tween-20 by 8 times. For invertase conjugation, 400 μL of 20 mg/mL invertase in Buffer A without Tween-20 was mixed with 1 mg of sulfo-SMCC. After vortexing, the solution was placed on a roller for 1 hour at room temperature. The mixture was then centrifuged and the insoluble excess sulfo-SMCC was removed. The clear solution was then purified by Amicon-100K using Buffer A without Tween-20 by 8 times. The purified solution of sulfo-SMCC-activated invertase was mixed with the above solution of Thiol-DNA or Thiol-DNA-HBV. The resulting solution was kept at room temperature for 48 hours. To remove un-reacted Thiol-DNA or Thiol-DNA-HBV, the solution was purified by Amicon-100K for 8 times using Buffer A without Tween-20.
Procedures for DNA detection using PGM
(1) Sensor preparation
A portion of 2 mL 1 mg/mL streptavidin-coated magnetic beads (MBs) were buffer exchanged to Buffer A twice, and then dispersed in 2 mL Buffer A. Capt-DNA was added to the solution to achieve a final concentration of 5 μM and the mixture was mixed on a roller for 30 min at room temperature. After that, the MBs were separated from the mixture by a magnet. The MBs were further washed by Buffer A for 3 times and then separated as each portion of 50 μL 1 mg/mL MBs in Buffer A.
(2) DNA detection
To each portion of the above MBs residues after removal of Buffer A, 100 μL DNA sample of various concentration of target DNA in Buffer A was added and the mixture was mixed on a roller for 2 h at room temperature. After washing the MBs residue by 3 times using Buffer A containing 2 mg/mL BSA to remove unbound target DNA and block non-specific binding sites by BSA, 100 μL 5 mg/mL DNA-invertase conjugate in Buffer A was added and the mixture was mixed on a roller for 30 min at room temperature. After washing the MBs residue by 5 times using Buffer A, 100 μL 0.5 M sucrose in Buffer A was added to the MBs residue and then mixed on a roller for 16 h at room temperature. A portion of 5 μL of the final solution was tested by a PGM. The time for the enzymatic reaction could be reduced by increasing the pre-concentrating effect of MBs (for example, after washing, disperse the MBs residue in 10 μL instead of 100 μL, so that the time required for producing the same amount of glucose is only 1/10, which is 1.6 h) and using invertase of higher enzyme activity.
Procedures for HBV DNA fragment detection using PGM
(1) Sensor preparation
A portion of 2 mL 1 mg/mL streptavidin-coated MBs were buffer exchanged to Buffer B twice, and then dispersed in 2 mL Buffer B. Capt-DNA-HBV was added to the solution to achieve a final concentration of 5 μM and the mixture was mixed on a roller for 30 min at room temperature. After that, the MBs were separated from the mixture by a magnet. The MBs were further washed by Buffer B for 3 times and then separated as each portion of 50 μL 1 mg/mL MBs in Buffer B.
(2) HBV DNA detection
To each portion of the above MBs residues after removal of Buffer B, 100 μL DNA sample of various concentration of target HBV DNA in Buffer B was added and the mixture was mixed on a roller for 1 h at room temperature. After washing the MBs residue by 3 times using Buffer B containing 2 mg/mL BSA to remove unbound target HBV DNA and block non-specific binding sites by BSA, 100 μL 5 mg/mL DNA-invertase conjugate in Buffer B was added and the mixture was mixed on a roller for 30 min at room temperature. After washing the MBs residue by 5 times using Buffer B, 25 μL 1 M sucrose in Buffer B was added to the MBs residue and then mixed on a roller for 3 h at room temperature. A portion of 5 μL of the final solution was tested by a PGM. The time required for the enzymatic reaction could be reduced by increasing the pre-concentrating effect of MBs and using invertase of higher enzyme activity.
Results and Discussion
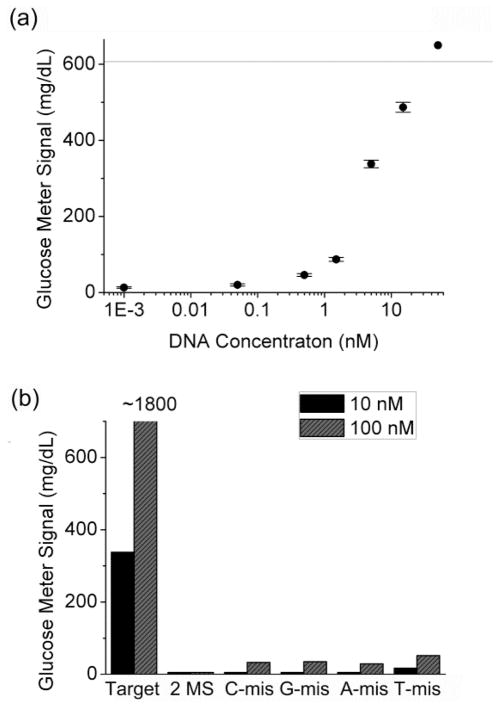
To demonstrate the methodology shown in Figure 1a, we first investigate the detection of a 12-mer model DNA (see DNA sequences in Figure 1b). As shown in Figure 2a, the absence of the model DNA produced negligible readout in a PGM because of the lack of the target-DNA facilitated DNA sandwich hybridization. In the presence of 10 nM target DNA, however, more than 40-fold enhancement of glucose reading in the PGM was observed in comparison with that in the absence of the target DNA, and the signal increases with increasing concentration of the target DNA in the sample. A detection limit of ~50 pM (5 fmol for a 0.1 mL sample) DNA (defined by 3σb/slope) was achieved under the experimental condition (Figure 2a). This detection limit is comparable or better than some other published methods without sophisticated instruments or signal simplification procedures,7,26,60,61 even though the PGMs can detect only mM levels of glucose. Due to the large surface area of the MBs for DNA hybridization, the capacity of DNA binding is large and the dynamic range (from detection limit to the detectable concentration before signal saturated) of the detection was found to be between 0.1 and 10 nM target DNA. Samples containing higher concentrations (>10 nM) of target DNA showed signals beyond the upper limit of PGMs so that they were not included in the calibration curve. To evaluate the selectivity, the same experiment was conducted using DNA molecules with either 1 or 2 mismatches (see the Experimental Section for the details of the mismatches). Under the same condition, little glucose signal was observed in the PGM (Figure 2b) for these control DNAs, demonstrating the excellent selectivity of the method.
Figure 2.
(a) Detection of a 12-mer model DNA using a PGM (the signal plot of a DNA-free sample cannot be shown in the logarithmic chart, which is almost the same as that of the sample with 1 pM target DNA). (b) Selectivity against single or double mismatches. The detection was conducted in 0.1 M sodium phospate buffer, pH 7.3, 0.1 M NaCl, 0.05% Tween-20. The line in the figure indicates the upper limit of the PGM (600 mg/dL glucose). The signal of ~1800 mg/dL is calculated from a test by PGM with 4 times dilution of the sample.
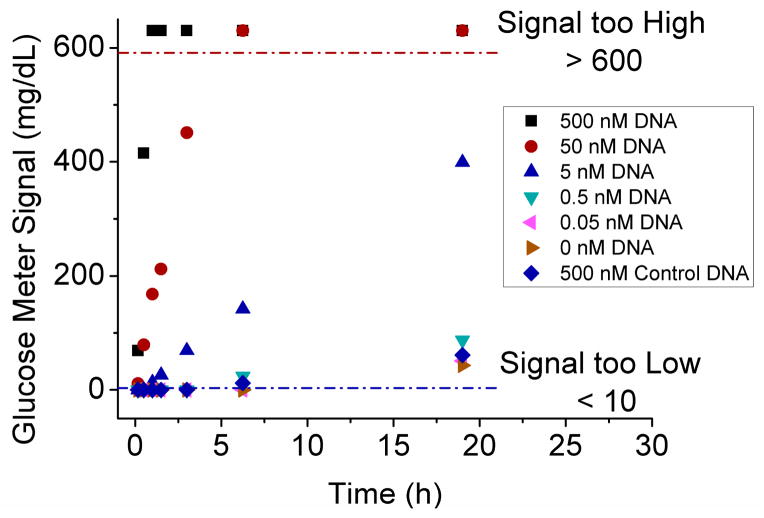
In our method, the glucose was produced via the enzymatic hydrolysis of sucrose by the invertase conjugates. Because excess sucrose (0.5 M) was added to the MBs containing the DNA-invertase conjugates to achieve maximum reaction rate, the kinetics of glucose production should be dependent on the concentration of invertase conjugates bound to the MBs, and thus the concentration of target DNA in the sample. By measuring the glucose signal at different time of the enzymatic reaction, the kinetics of glucose production was studied and shown in Figure 3. For most of the samples (such as 0.5, 5, 50 nM DNA), the glucose concentration increased linearly with time. The signals lower than 10 mg/dL or higher than 600 mg/dL are beyond the detectable range of PGMs so that those data plots do not represent the real glucose concentrations. The more target DNA in the solution, the earlier time for the samples to show a detectable signal over blank. To ensure low concentrations of DNA could be detected, 16 h reaction time was used in the case of the model DNA detection.
Figure 3.
The kinetics of glucose production for the samples containing different concentrations of target DNA or control DNA (with 2 mismatches).
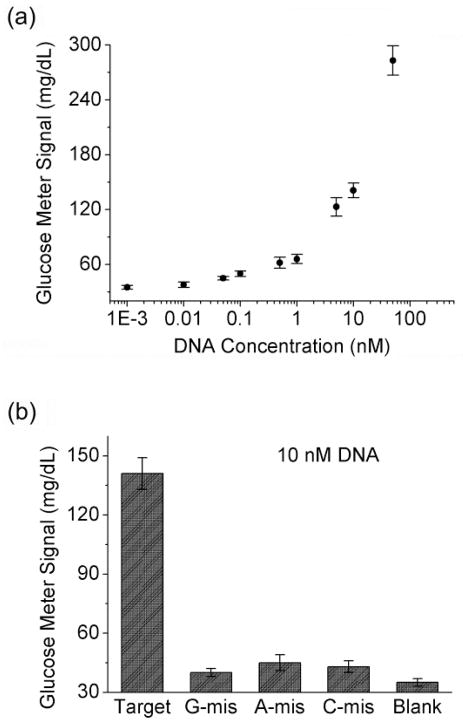
To further demonstrate its versatility in detecting a wide range of target DNA sequences and its potentials for practical applications in disease diagnostics, we further apply the method to detect and quantify a 35-nucleotide hepatitis B virus (HBV) DNA fragment62–64 using a PGM. The fragment can be obtained from the virus DNA by applying suitable endonucleases or other DNA cleaving reagents. As shown in Figure 1c, the number of base pairs for hybridizations between Capt-DNA and target DNA has been increased from 12 bp in the model DNA detection to 13 bp in the HBV-DNA detection in order to enhance the binding affinity between the two DNA strands. Similarly, the number of base pairs between the target DNA and DNA-invertase has also been extended from 19 bp in model DNA detection to 20 bp in the HBV-DNA detection. Buffers of higher ionic strength and MBs of higher concentrations were also used to ensure efficient DNA hybridization and invertase loading. Such a combination of longer binding arms, highly ionic strength and more concentrated MBs can increase the concentrations of target DNA and DNA-invertase conjugates bound to the MBs so that the time required for glucose production by the DNA-invertase can be reduced and the sensitivity can be improved. As illustrated in Figure 4a, about 8-fold enhancement of glucose production could be achieved in the presence of 50 nM HBV DNA fragment. Samples containing higher concentrations (>=100 nM) of HBV DNA fragment produced glucose signals beyond the upper limit of PGMs so that they were not included in the calibration curve. A detection limit of about 40 pM (4 fmol for a 0.1 mL sample) was calculated according to the definition of 3σb/slope. The sensitivity is comparable or better than that of some other reported methods for DNA detection7,26,29–31,42,43,45,60,61 and suitable for the analysis of samples containing high HBV DNA concentrations. The sensitivity can be further improved by increasing the efficiency of pre-concentrating effect of capturing MBs or the time of enzymatic amplification, for potential applications in samples with lower DNA concentrations. Similar to the model DNA detection mentioned above, the HBV DNA fragment detection here was also sequence-specific. In the presence of 1 mismatch (see the Experimental Section for the details of the mismatches), the HBV DNA fragment sample could only produce modest glucose signal enhancement as detected by the glucose meter, in comparison with the fully-matched DNA under the same condition (Figure 4b).
Figure 4.
(a) Detection of a 35-mer HBV DNA fragment using a PGM (the signal plot of a DNA-free sample cannot be shown in the logarithmic chart, which is almost the same as that of the sample with 1 pM target DNA). (b) Single mismatch selectivity of the method. The detection was conducted in 0.15 M sodium phospate buffer, pH 7.3, 0.25 M NaCl, 0.05% Tween-20.
Conclusion
In summary, we have successfully developed a methodology for portable, low-cost and quantitative DNA detection using easily accessible personal glucose meters (PGMs). By a DNA sandwich assay in the presence of Capt-DNA-coated magnetic beads (MBs) and DNA-invertase conjugates, the concentration of target DNA in the sample was rendered to be proportional to the amount of DNA-invertase conjugates immobilized on the MBs via DNA hybridization. Subsequently, the immobilized DNA-invertase conjugates could catalyze the hydrolysis of PGM-inert sucrose to yield PGM-detectable glucose. The catalytic reaction is very efficient in signal amplification, thus pM~nM level of target DNA could produce mM level of glucose required for the readout in a PGM. The method was also applied to the detection of a hepatitis B virus (HBV) DNA fragment. A detection limit as low as 40 pM (4 fmol for a 0.1 mL sample) was achieved. In addition, the method exhibited excellent sequence-selectivity, being able to differentiate a single mismatch in the target DNA. The employment of the MBs allows not only sample capture and concentration, but also washing procedures to remove sample matrix that may interfere in the detection, making it possible to apply the method to complicated biological samples such as body fluids. By integrating with lateral flow or microfluid devices, the method can also be simplified furhter to avoid the washing and transfering steps for the public users. Furthermore, because of the wide accessibility of PGMs to the public and the portability, simple operation and quantitative results of the tests, our method makes it possible by the public for quantitative DNA detection at home and in the field.
Acknowledgments
We wish to thank the U.S. Department of Energy (DE-FG02-08ER64568), National Institutes of Health (ES16865), and National Science Foundation (CTS-0120978) for financial support.
References
- 1.Daar AS, Thorsteinsdottir H, Martin DK, Smith AC, Nast S, Singer PA. Nat Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 2.Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 3.Nakayama S, Yan L, Sintim HO. J Am Chem Soc. 2008;130:12560–12561. doi: 10.1021/ja803146f. [DOI] [PubMed] [Google Scholar]
- 4.Nakayama S, Sintim HO. J Am Chem Soc. 2009;131:10320–10333. doi: 10.1021/ja902951b. [DOI] [PubMed] [Google Scholar]
- 5.Huang Y, Zhang Y, Xu X, Jiang J, Shen G, Yu R. J Am Chem Soc. 2009;131:2478–2480. doi: 10.1021/ja808700d. [DOI] [PubMed] [Google Scholar]
- 6.Xiao Y, Plakos KJI, Lou X, White RJ, Qian J, Plaxco KW, Soh HT. Angew Chem, Int Ed. 2009;48:4354–4358. doi: 10.1002/anie.200900369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, Xue X, Li T, Zeng H, Liu X. Angew Chem, Int Ed. 2009;48:6849–6852. doi: 10.1002/anie.200901772. [DOI] [PubMed] [Google Scholar]
- 8.Xue X, Xu W, Wang F, Liu X. J Am Chem Soc. 2009;131:11668–11669. doi: 10.1021/ja904728v. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang Y, Wang H, Jiang J, Shen G, Yu R, Li J. Anal Chem. 2009;81:1982–1987. doi: 10.1021/ac802512d. [DOI] [PubMed] [Google Scholar]
- 10.Li J, Deng T, Chu X, Yang R, Jiang J, Shen G, Yu R. Anal Chem. 2010;82:2811–2816. doi: 10.1021/ac100336n. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson BS, Buchsbaum SF, Wu T, Hsieh K, Xiao Y, Sun R, Soh HT. J Am Chem Soc. 2011;133:9129–9135. doi: 10.1021/ja203981w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Z, Wang H, Guo M, Tang L, Yu R, Jiang J. Anal Chem. 2011;83:3104–3111. doi: 10.1021/ac1033769. [DOI] [PubMed] [Google Scholar]
- 13.Duan X, Liu LB, Feng F, Wang S. Acc Chem Res. 2010;43:260–270. doi: 10.1021/ar9001813. [DOI] [PubMed] [Google Scholar]
- 14.Niemeyer CM, Blohm D. Angew Chem, Int Ed. 1999;38:2865–2869. doi: 10.1002/(sici)1521-3773(19991004)38:19<2865::aid-anie2865>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 15.Makrigiorgos GM, Chakrabarti S, Zhang Y, Kaur M, Price BD. Nat Biotechnol. 2002;20:936–939. doi: 10.1038/nbt724. [DOI] [PubMed] [Google Scholar]
- 16.Fan C, Plaxco KW, Heeger AJ. Trends Biotechnol. 2005;23:186–192. doi: 10.1016/j.tibtech.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Zhu L, Anslyn EV. Angew Chem, Int Ed. 2006;45:1190–1196. doi: 10.1002/anie.200501476. [DOI] [PubMed] [Google Scholar]
- 18.Graham D, Faulds K. Chem Soc Rev. 2008;37:1042–1051. doi: 10.1039/b707941a. [DOI] [PubMed] [Google Scholar]
- 19.Ho HA, Najari A, Leclerc M. Acc Chem Res. 2008;41:168–178. doi: 10.1021/ar700115t. [DOI] [PubMed] [Google Scholar]
- 20.Feng X, Liu L, Wang S, Zhu D. Chem Soc Rev. 2010;39:2411–2419. doi: 10.1039/b909065g. [DOI] [PubMed] [Google Scholar]
- 21.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 22.Park SJ, Taton TA, Mirkin CA. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 23.Nam JM, Stoeva SI, Mirkin CA. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 24.Stoeva SI, Lee JS, Thaxton CS, Mirkin CA. Angew Chem, Int Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Song S, Zhang L, Wang L, Wu H, Pan D, Fan C. J Am Chem Soc. 2006;128:8575–8580. doi: 10.1021/ja061521a. [DOI] [PubMed] [Google Scholar]
- 26.Hong M, Zhou X, Lu Z, Zhu J. Angew Chem, Int Ed. 2009;48:9503–9506. doi: 10.1002/anie.200905267. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Appella DH. J Am Chem Soc. 2007;129:8424–8425. doi: 10.1021/ja072744j. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H, Liu J, Xu J, Chen H. Chem Commun. 2011;47:8358–8360. doi: 10.1039/c1cc12413g. [DOI] [PubMed] [Google Scholar]
- 29.Pavlov V, Shlyahovsky B, Willner I. J Am Chem Soc. 2005;127:6522–6523. doi: 10.1021/ja050678k. [DOI] [PubMed] [Google Scholar]
- 30.Graf N, Goritz M, Kramer R. Angew Chem, Int Ed. 2006;45:4013–4015. doi: 10.1002/anie.200504319. [DOI] [PubMed] [Google Scholar]
- 31.Weizmann Y, Cheglakov Z, Pavlov V, Willner I. Angew Chem, Int Ed. 2006;45:2238–2242. doi: 10.1002/anie.200503810. [DOI] [PubMed] [Google Scholar]
- 32.Li M, He F, Liao Q, Liu J, Xu L, Jiang L, Song Y, Wang S, Zhu D. Angew Chem, Int Ed. 2008;47:7258–7262. doi: 10.1002/anie.200801998. [DOI] [PubMed] [Google Scholar]
- 33.Zhu S, Liu Z, Zhang W, Han S, Hu L, Xu G. Chem Commun. 2011;47:6099–6101. doi: 10.1039/c1cc10952a. [DOI] [PubMed] [Google Scholar]
- 34.Niu S, Jiang Y, Zhang S. Chem Commun. 2010;46:3089–3091. doi: 10.1039/c000166j. [DOI] [PubMed] [Google Scholar]
- 35.Li H, Zhang Y, Wang L, Tian J, Sun X. Chem Commun. 2011;47:961–963. doi: 10.1039/c0cc04326e. [DOI] [PubMed] [Google Scholar]
- 36.Hunt EA, Deo SK. Chem Commun. 2011;47:9393–9395. doi: 10.1039/c1cc13495g. [DOI] [PubMed] [Google Scholar]
- 37.Patolsky F, Lichtenstein A, Willner I. J Am Chem Soc. 2001;123:5194–5205. doi: 10.1021/ja0036256. [DOI] [PubMed] [Google Scholar]
- 38.Fan C, Plaxco KW, Heeger AJ. Proc Nat Acad Sci USA. 2003;100:9134–9137. doi: 10.1073/pnas.1633515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia F, White RJ, Zuo X, Patterson A, Xiao Y, Kang D, Gong X, Plaxco KW, Heeger AJ. J Am Chem Soc. 2010;132:14346–14348. doi: 10.1021/ja104998m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drummond TG, Hill MG, Barton JK. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 41.Lai RY, Lagally ET, Lee SH, Soh HT, Plaxco KW, Heeger AJ. Proc Nat Acad Sci USA. 2006;103:4017–4021. doi: 10.1073/pnas.0511325103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu Y, Li F, Bai X, Li D, Hua S, Wang K, Niu L. Chem Commun. 2011;47:1743–1745. doi: 10.1039/c0cc04514d. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Sun Y, Zhang S. Chem Commun. 2011;47:1595–1597. doi: 10.1039/c0cc03921g. [DOI] [PubMed] [Google Scholar]
- 44.Bell SEJ, Sirimuthu NMS. J Am Chem Soc. 2006;128:15580–15581. doi: 10.1021/ja066263w. [DOI] [PubMed] [Google Scholar]
- 45.Fabris L, Dante M, Braun G, Lee SJ, Reich NO, Moskovits M, Nguyen TQ, Bazan GC. J Am Chem Soc. 2007;129:6086–6087. doi: 10.1021/ja0705184. [DOI] [PubMed] [Google Scholar]
- 46.Barhoumi A, Halas NJ. J Am Chem Soc. 2010;132:12792–12793. doi: 10.1021/ja105678z. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Z, Wen Y, Ma Y, Luo J, Jiang L, Song Y. Chem Commun. 2011;47:7407–7409. doi: 10.1039/c1cc11062d. [DOI] [PubMed] [Google Scholar]
- 48.Du B, Li Z, Liu C. Angew Chem, Int Ed. 2006;45:8022–8025. doi: 10.1002/anie.200603331. [DOI] [PubMed] [Google Scholar]
- 49.Chu L, Forch R, Knoll W. Angew Chem, Int Ed. 2007;46:4944–4947. doi: 10.1002/anie.200605247. [DOI] [PubMed] [Google Scholar]
- 50.Jung Y, Hong B, Zhang W, Tendler SJB, Williams PM, Allen S, Park JW. J Am Chem Soc. 2007;129:9349–9355. doi: 10.1021/ja0676105. [DOI] [PubMed] [Google Scholar]
- 51.Clark LC, Lyons C. Ann NY Acad Sci. 1962;102:29–45. doi: 10.1111/j.1749-6632.1962.tb13623.x. [DOI] [PubMed] [Google Scholar]
- 52.Montagnana M, Caputo M, Giavarina D, Lippi G. Clin Chim Acta. 2009;402:7–13. doi: 10.1016/j.cca.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 53.Carroll AE, Marrero DG, Downs SM. Diabetes Technol Ther. 2007;9:158–164. doi: 10.1089/dia.2006.0002. [DOI] [PubMed] [Google Scholar]
- 54.Xiang Y, Lu Y. Nat Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niemeyer CM. Trends Biotechnol. 2002;20:395–401. doi: 10.1016/s0167-7799(02)02022-x. [DOI] [PubMed] [Google Scholar]
- 56.Hermanson GT. Bioconjugate Techniques. Elsevier; London: 2008. [Google Scholar]
- 57.Niemeyer CM. Angew Chem, Int Ed. 2010;49:1200–1216. doi: 10.1002/anie.200904930. [DOI] [PubMed] [Google Scholar]
- 58.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 59.Reddy A, Maley F. J Biol Chem. 1996;271:13953–13958. doi: 10.1074/jbc.271.24.13953. [DOI] [PubMed] [Google Scholar]
- 60.Xiao Y, Qu X, Plaxco KW, Heeger AJ. J Am Chem Soc. 2007;129:11896–11897. doi: 10.1021/ja074218y. [DOI] [PubMed] [Google Scholar]
- 61.Connolly AR, Trau M. Angew Chem, Int Ed. 2010;49:2720–2723. doi: 10.1002/anie.200906992. [DOI] [PubMed] [Google Scholar]
- 62.Takemura F, Ishii T, Fujii N, Uchida T. Nucleic Acids Res. 1990;18:4587–4587. doi: 10.1093/nar/18.15.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Preisleradams S, Schlayer HJ, Peters T, Korp R, Rasenack J. Nucleic Acids Res. 1993;21:2258–2258. doi: 10.1093/nar/21.9.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cai S, Xin L, Lau C, Lu J, Zhang X. Chem Commun. 2011;47:6120–6122. doi: 10.1039/c1cc10914f. [DOI] [PubMed] [Google Scholar]