Abstract
Autism spectrum disorder (ASD) is a highly heritable disorder of complex and heterogeneous aetiology. It is primarily characterized by altered cognitive ability including impaired language and communication skills and fundamental deficits in social reciprocity. Despite some notable successes in neuropsychiatric genetics, overall, the high heritability of ASD (~90%) remains poorly explained by common genetic risk variants. However, recent studies suggest that rare genomic variation, in particular copy number variation, may account for a significant proportion of the genetic basis of ASD. We present a large scale analysis to identify candidate genes which may contain low-frequency recessive variation contributing to ASD while taking into account the potential contribution of population differences to the genetic heterogeneity of ASD. Our strategy, homozygous haplotype (HH) mapping, aims to detect homozygous segments of identical haplotype structure that are shared at a higher frequency amongst ASD patients compared to parental controls. The analysis was performed on 1,402 Autism Genome Project trios genotyped for 1 million single nucleotide polymorphisms (SNPs). We identified 25 known and 1,218 novel ASD candidate genes in the discovery analysis including CADM2, ABHD14A, CHRFAM7A, GRIK2, GRM3, EPHA3, FGF10, KCND2, PDZK1, IMMP2L and FOXP2. Furthermore, 10 of the previously reported ASD genes and 300 of the novel candidates identified in the discovery analysis were replicated in an independent sample of 1,182 trios. Our results demonstrate that regions of HH are significantly enriched for previously reported ASD candidate genes and the observed association is independent of gene size (odds ratio 2.10). Our findings highlight the applicability of HH mapping in complex disorders such as ASD and offer an alternative approach to the analysis of genome-wide association data.
Electronic supplementary material
The online version of this article (doi:10.1007/s00439-011-1094-6) contains supplementary material, which is available to authorized users.
Introduction
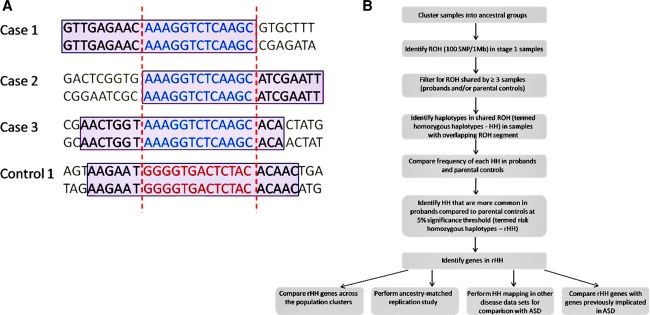
Extended runs of homozygosity (ROH) have recently been highlighted as a genomic feature that may be useful to map recessive disease genes in outbred populations (Hildebrandt et al. 2009; Lencz et al. 2007; Yang et al. 2010). Furthermore, even in complex disorders, we expect to find an unusually high number of affected individuals to have the same haplotype in the region surrounding a disease mutation (Durand et al. 2007; Lesch et al. 2008; Wong et al. 2002). Therefore, a rare pathogenic variant and surrounding haplotype is often enriched in frequency in a group of affected individuals compared with the haplotype frequency in a cohort of unaffected controls (The International HapMap Consortium 2003). We propose that homozygous haplotypes (HH) that are shared by multiple affected individuals may be important for the discovery of recessive disease genes in complex disorders. We have extended the traditional homozygosity mapping method by analysing the haplotype within shared ROH regions to identify homozygous segments of identical haplotype that are present uniquely or at a higher frequency in ASD probands compared to parental controls (Fig. 1). Such regions are termed risk homozygous haplotypes (rHH). We postulate that rHH may contain low-frequency recessive variants that contribute to ASD risk in a subset of ASD patients.
Fig. 1.
The principles and analytical approach of homozygous haplotype mapping. a The schematic outlines the principle of homozygous haplotype (HH) mapping. SNP genotype data is collected on each case and control. Homozygous and heterozygous SNPs are shown in black and grey respectively. Firstly, runs of homozygosity (ROH) are identified in the samples (outlined in purple boxes). The overlapping ROH region shared by a minimum of three individuals (shown between red dashed lines) is considered for the HH analysis. The haplotypes within the overlapping ROH region are identified and a Fisher’s exact test applied to determine if a particular HH is significantly more common in ASD cases compared to parental controls. Only the haplotypes of those individuals who have an ROH in the region in question are considered. In the above example all four individuals (3 ASD cases and 1 parental control) have an overlapping ROH. However, the haplotype in the overlapping ROH may differ. The 3 ASD cases have haplotype A (blue) while the parental control has haplotype B (red). Haplotype A is shared at a higher frequency in ASD cases compared to parental controls (apply Fisher’s test) and is termed a risk homozygous haplotype (rHH). This is an example of a rHH that is specific to ASD probands; b Flowchart of homozygous haplotype analysis of ASD cohort. The discovery analysis was performed on 1,402 AGP trios from the AGP stage 1 collection. The replication study involved an additional 1,182 AGP trios from the stage 2 collection. The stage 1 and 2 samples were clustered together to (1) separate stage 1 and 2 individuals into population clusters of similar ancestry and (2) classify stage 2 individuals into the joint ancestry-matched population clusters for the stage 2 replication study. The same rHH mapping strategy was applied to the discovery (stage 1) and replication (stage 2) data sets independently. The genes located in homozygous haplotypes significantly more common in ASD cases compared to parental controls were identified in each analysis. The rHH candidate genes were then compared for the ancestry-matched groups that had at least 50 probands in both the discovery and replication sample sets. To assess the contribution of genomic architecture to the rHH findings in ASD, the same strategy was applied to two additional disease data sets; bipolar disorder (BD) and coronary artery disease (CAD). The location of the rHH in ASD, BD and CAD were compared
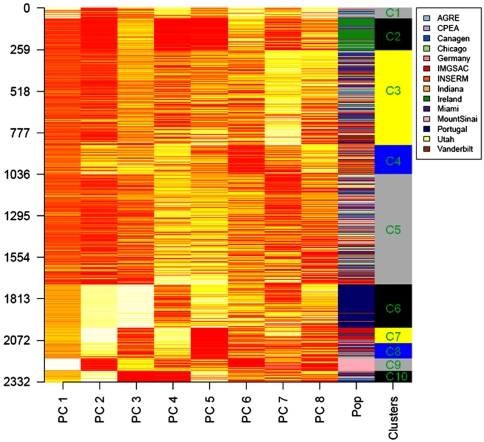
Allelic and locus heterogeneity are major challenges in the identification of ASD risk loci (Lamb et al. 2000). In cases where distinct populations share the same risk allele, differences in allele frequency and LD structure between populations may result in the risk allele segregating on different haplotype backgrounds. Correction for population substructure in large scale studies minimises the rate of false positives and dilution of a population-specific signal. Furthermore, Nothnagel et al. (2010) recently reported that the distribution of SNP-defined ROHs is highly structured across European populations and highlighted the importance of accounting for ancestry when undertaking ROH-based analyses. Our analysis accounts for population effects by separating the samples into groups of common ancestry and applying the HH mapping to each population group independently. It also involves the use of parental controls to address variation in low-frequency alleles across populations (Cardon and Palmer 2003). The genetic ancestry of the sample set was examined by principal component analysis (PCA), Hopach clustering (van der Laan and Pollard 2002) and genetic distance F st calculations (Supplementary Material 1, Supplementary Fig. 1 and 2, Supplementary Tables 2 and 3). Ten distinct population clusters were identified ranging in size from 27 to 289 probands (Fig. 2). Population clusters with a minimum of 50 probands were selected for the discovery analysis (n = 5). Each cluster was analysed independently by HH mapping and the genes identified in each population cluster were then compared. In this manner (taking a gene-centric approach) it is possible to identify genes that may confer risk across different populations regardless of whether the causal haplotype is population-specific or not.
Fig. 2.
Genetic ancestry of AGP sample set. Principal component analysis (PCA) of 2,584 ASD proband samples (discovery stage 1 = 1,402 samples, replication stage 2 = 1,182 samples) was performed in EIGENSOFT. Tracy–Widom statistics indicated that the first eight principal components (PCs) were significantly contributing to the genetic variation of the sample set (Supplementary Table 2). The Hopach hierarchical clustering algorithm was applied to eigenvalues (y-axis) from the first eight PCs (x-axis) (van der Laan and Pollard 2002). In the ‘Pop’ column each sample is coloured according to the AGP site at which it was collected (see legend). Hopach clustering, non-parametric bootstrapping and genetic distance calculations (Supplementary Table 3) identified ten ancestral population clusters labelled C1 to C10. The five population clusters with a minimum of 50 probands (C2–C6) were used in the discovery analysis (n = 1,019 trios)
Subjects and methods
Cohort description
The samples used in the HH analysis were collected as part of an international consortium, the Autism Genome Project (AGP). Informed consent was obtained from all participants. The AGP sample set is a trio based collection, comprising an affected proband and two parents, grouped into the three distinct diagnostic classes of autism; strict, broad and spectrum. Affected individuals were diagnosed using the Autism Diagnostic Interview-Revised (ADI-R) and/or the Autism Diagnostic Observation Schedule (ADOS). A detailed description of the AGP sample set is provided in Supplementary Material 1 and by Pinto et al. (2010). Raw genotype data for the ASD trios is deposited at NCBI dbGAP (accession phs000267.v1.p1). The HH analysis was performed on trios in the autism spectrum diagnostic category (n = 2,584 trios). The ASD spectrum trios were further subdivided into stage 1 and stage 2 collections. In the current study, the 1,402 stage 1 trios were used for the initial discovery analysis and the 1,182 stage 2 trios were used for the independent replication study.
Genotyping and quality control
Stage 1 samples were genotyped using the Illumina 1M-single array while the stage 2 samples were genotyped on a combination of 1M and 1M-duo chips. The 1M SNP array contains 1,072,820 SNP markers at an average inter-marker distance of 2.7 kb while the 1M-duo chip contains 1,199,187 SNPs with a mean marker spacing of 1.5 kb. The 1,003,736 SNPs that were genotyped on both the 1M and 1M-duo platforms were considered for the analysis. Possible gender miscalls were assessed through analysis of chromosome X genotypes in PLINK (version 1.04) (http://pngu.mgh.harvard.edu/~purcell/plink/) (Purcell et al. 2007). Duplicates were identified by calculating identity by state (IBS) values using PLINK and one sample from each duplicate pair was removed. After quality control and filtering for autosomal markers, 887,716 SNPs and 7,719 individuals were retained for analysis (Supplementary Material 1).
Ancestry analysis
The population structure of the sample set was assessed through principal component analysis (PCA), Hopach hierarchical clustering and F st calculations. PCA was performed with EIGENSTRAT from EIGENSOFT (version 3.0) (Price et al. 2006) using 70,175 independent autosomal SNPs with a minor allele frequency >5% and a call rate of 100%. Tracy–Widom statistics indicated that the first eight principal components (PCs) were significantly contributing to the population structure of the sample set. Individuals were assigned to clusters based on similarity in eigenvalues for the first eight PCs using the Hopach hybrid hierarchical clustering algorithm available in the R package (van der Laan and Pollard 2002). Hierfstat was used to calculate pair-wise F st metrics for the clusters using 5,000 SNPs randomly chosen from the panel of 70,175 SNPs used for PCA (Goudet 2005). Clusters displaying high similarity (F st value < 1 × 10−4) were merged, resulting in 10 population clusters for the HH analysis (Supplementary Material 1 and Supplementary Table 3).
Homozygous haplotype analysis
Long series of consecutive homozygous SNPS, referred to as runs of homozygosity (ROHs), were identified in each sample using the ‘homozyg’ function in PLINK. A threshold of 100 consecutive homozygous SNPs spanning at least 1 Mb at a minimum density of 1 SNP/50 kb was implemented. Using the homozyg-group function in PLINK, samples (min 3) with overlapping ROHs were pooled and subdivided into haplotype groups (min 95% allelic identity). Each haplotype within the overlapping ROH region is referred to as a homozygous haplotype (HH). For each overlapping ROH, the frequency of each HH within cases and controls was evaluated using the Fisher’s exact test (R script). HH that were significantly more common in ASD probands compared to parental controls (p value < 0.05) were considered rHH (Supplementary Tables 1a–m). All subsequent analysis was limited to these regions. The rHH regions were annotated using the Illumina 1M annotation file (Human Genome build 36.1 RefSeq). In cases where rHH were located in intergenic regions, the nearest centromeric and telomeric gene was noted.
Inspection of LD structure and copy number variants
Patterns of linkage disequilibrium (LD) within the rHH were visualised and analysed in Haploview using HapMap CEU as a reference (Barrett et al. 2005). LD was measured as r 2 values and calculated between each pair of SNPs. The Tagger algorithm (Haploview) was used to determine the number of tagging SNPs within each rHH at an r 2 threshold of 0.8. Genes located in rHH comprising <10 tagging SNPs were noted. For each stage 1 population group, the samples contributing to significant rHH were inspected for CNV content as detected by Pinto et al. (2010). If present, rHH identified as CNVs were removed prior to application of the Fisher’s exact test.
Replication study
A replication study was undertaken using the AGP follow-up stage 2 data set (freezes 5–8) which comprises 1,182 ASD trios genotyped on a combination of the Illumina 1M and 1M-duo platforms. The stage 2 data was cleaned with the stage 1 samples to ensure that the same markers would be used in both analyses. The stage 1 and stage 2 probands were clustered to identify ancestry-matched replication groups. The four population clusters (C3–C6) with ≥50 probands in both stage 1 and 2 analyses were considered for the replication study. The same HH mapping method was applied to the four stage 2 population clusters. The rHH genes identified in the discovery (stage 1) and replication (stage 2) analyses were then compared to identify overlap.
Statistical analyses
Comparison of ROH burden
For each population group, the number and length of autosomal ROH in ASD probands and parental controls was compared using a paired t test. ASD probands were compared to their mothers and fathers separately.
Identification of HHs that are more prevalent in ASD
A Fisher’s exact test was used to identify HH that were more prevalent in ASD probands compared to parental controls at a 5% Fisher’s significance level. As we are assuming a recessive model a one-sided test was used.
Comparison of results across the five stage 1 population groups
Each of the five population clusters were analysed independently and the rHH genes subsequently compared to identify overlaps. A gene was considered a candidate in multiple population clusters regardless of whether the position of the rHH in each population differed (to allow for population-specific effects). In this manner, it is possible to identify genes that may confer susceptibility to ASD across multiple populations even though the underlying risk allele may be population-specific. Genes located in/near rHH over-represented in ASD probands in at least two population groups were noted.
Enrichment for previously reported ASD genes
A χ2 test with Yates’ correction factor was used to determine if the rHH genes displayed enrichment for previously reported ASD candidate genes (n = 202). To account for a potential bias towards large genes, a logistic multiple regression was performed in STATA including both ASD gene status and gene size (as defined in UCSC hg18) as covariates. Thus, if rHH genes were primarily associated with larger genes, and this was merely a confounding factor arising simply because of a preponderance of larger genes in the ASD list of genes (mean length of 185 kb compared to 57 kb for other genes), then the multiple regression model would identify gene size as the significant predictor, and fail to identify ASD genes as being significantly over-represented amongst the rHH genes.
Results
Identification of risk homozygous haplotypes
Runs of homozygosity (ROH) of at least 1 Mb and 100 SNPs were identified in samples from each of the five population clusters using PLINK (“Subjects and methods” and Supplementary Material 1). A paired t test demonstrated that ASD probands did not have a higher genome-wide burden of ROH compared to parental controls (Supplementary Fig. 3). The findings from the current ASD study reflect those of a related neuropsychiatric condition, bipolar disorder, where it was recently reported that individuals with bipolar disorder do not have an excess of runs of homozygosity (Vine et al. 2009). A one-sided Fisher’s exact test was applied to identify HH that were more prevalent in ASD probands compared to parental controls at a 5% significance threshold. Such HH are considered regions of interest and are referred to as rHH. Since low-frequency variants will rarely be present at a high enough frequency to survive multiple testing, they will not be detected with the correction methods currently available. Therefore unless otherwise stated, p values have not been corrected for multiple testing. Genes located within or overlapping the rHH were noted. In cases where the rHH was intergenic, the neighbouring centromeric and telomeric genes were used. We found that, on average, 76% of the rHH were genic (Supplementary Fig. 4).
Haplotype sharing in homozygous segments
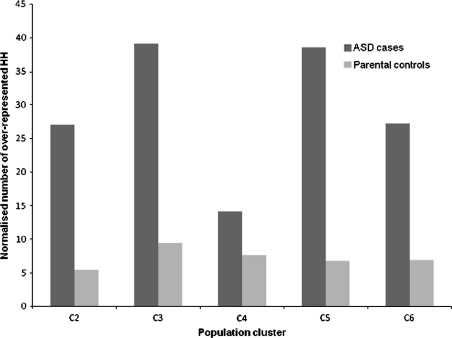
To determine if excess HH sharing is also likely to occur in a non-disease cohort we compared the number of HHs that are more common in ASD probands compared to parental controls and the number of HHs that are more common in parental controls compared to ASD cases. We observed that ASD probands shared a significantly higher number of homozygous segments of identical haplotype compared to the parental control group (paired t test p value = 0.008) (Fig. 3). This finding suggests that, although ASD probands may not have a higher burden of homozygous segments compared to parental controls, the probands display a much higher degree of haplotype sharing within overlapping homozygous regions. Excess haplotype sharing often indicates the presence of a disease locus (Bahlo et al. 2006), an observation that forms the basis of the current study. Two distinct types of rHH were identified in the affected cohort; (1) homozygous segments with a haplotype that is present in ASD probands but absent in parental controls and (2) homozygous segments with a haplotype that is present at a higher frequency in ASD cases compared to parental controls. A summary of the rHH results is shown in Table 1. The average rHH size across the five population clusters was 541.27 kb, 24-fold larger than the average haplotype block (Gabriel et al. 2002). Visual inspection of LD structure showed that the rHH extended across multiple LD blocks and 90% of rHH contained at least 10 tagging SNPs, indicating that it is unlikely that the observed HH sharing is a consequence of long-range LD.
Fig. 3.
Comparison of HH sharing in ASD cases and parental controls. The normalised number of rHH (HH that are more common in one group compared to the other) in five population clusters with a minimum of 50 probands. The dark grey bars represent the number of HH that are more common (Fisher’s exact test right p value <0.05) in ASD probands compared to parental controls. Such regions are referred to as rHH throughout the paper. The light grey bars denote the number of HH that are more common (Fisher’s exact test left p value <0.05) in parental controls compared to ASD probands. To account for differences in sample size, counts have been normalised to a group of 100 samples (Supplementary Material 1). The number of rHH identified in ASD probands is significantly greater than the number of rHH identified in parental controls across the five population clusters (paired t test p value = 0.008)
Table 1.
Summary of rHH results for each population cluster
| Cluster | No. of ASD probands | No. of parental controls | Significant rHH | Total no. of rHH genes | ||
|---|---|---|---|---|---|---|
| Total | ASD-specific | Enriched | ||||
| C2 | 148 | 294 | 40 | 23 | 17 | 243 |
| C3 | 289 | 584 | 99 | 44 | 55 | 341 |
| C4 | 85 | 170 | 11 | 5 | 6 | 79 |
| C5 | 280 | 560 | 100 | 48 | 52 | 417 |
| C6 | 217 | 434 | 57 | 18 | 39 | 372 |
| Total | 1,019 | 2,024 | 307 | 138 | 169 | 1,452a |
A summary of results for each of the ancestry-matched population clusters in the discovery analysis. The number of homozygous haplotypes over-represented [5% significance level, risk homozygous haplotypes (rHH)] in the ASD cohort is further subdivided into ASD-specific (only present in probands) and enriched (more common in probands than controls). The number of genes implicated by the rHH in each population cluster is shown in the final column. A total of 307 rHH were identified across the 5 population clusters. These regions contained or were adjacent to 1,452 genes
aWhen genes that are found in more than one population cluster are considered only once, the final number of genes is 1,243
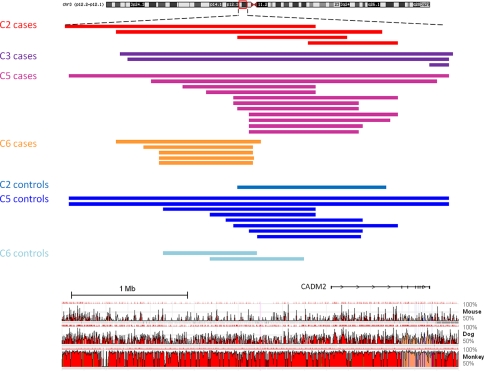
Genes located in rHH across multiple population clusters
We identified 192 genes (1 gene in 4 populations, 15 in 3 populations and 176 in 2 populations) that are in or near rHH regions significantly more prevalent among the ASD probands in two or more population clusters (Supplementary Table 4). In four of the five population clusters, an rHH was found in an evolutionary-conserved intergenic region on 3p12.1 in the vicinity of CADM2, a novel ASD candidate gene (Fig. 4). The rHH adjacent to CADM2 were identified in 23/1,019 ASD cases and 11/2,031 parental controls [Yates’ corrected χ2 p = 1.9 × 10−5, OR = 4.26 (2.1, 8.6)]. Recent studies have highlighted the presence of long-range regulatory mutations in several diseases and suggest that cis-regulatory domains of developmental genes may extend over megabases of DNA flanking its coding sequences (Benko et al. 2009; Kleinjan and van Heyningen 2005). CADM2 is a member of the synaptic cell adhesion molecule (SynCAM) immunoglobulin superfamily which has potential roles in early postnatal development of the central nervous system (specifically synapse formation) and is predominantly expressed in neurons of the developing and adult brain (Thomas et al. 2008). The encoded protein localises to both excitatory and inhibitory neurons which is intriguing given that there is compelling evidence for dysfunction in neuronal communication and excitatory/inhibitory imbalance in autism (Persico and Bourgeron 2006; Sudhof 2008). Of interest, CADM2 contains a neurexin domain. Rare variation contributing to autism has previously been identified in neurexin and neurexin-binding genes (Arking et al. 2008; Betancur et al. 2009; Kim et al. 2008). In our study, the location of the rHH in relation to CADM2 differed between each of the four population clusters. This may indicate population-specific risk loci arising from different founder events involving the gene or a control element proximal to the gene. Given the recent implication of synaptic genes in ASD susceptibility (Durand et al. 2008; Zoghbi 2003) and the strong functional candidacy of CADM2, its involvement in ASD warrants further investigation.
Fig. 4.
rHH identified in four population clusters in the vicinity of CADM2. An rHH located in a non-coding evolutionary-conserved region on 3p12.1 was identified in four of the five population clusters. The coloured bars represent the run of homozygosity in each patient/parental control carrying the rHH. For each population cluster, the rHH is the shared ROH segment. The ROH profile is presented with a conservation plot (ECR browser; conservation throughout mouse, dog and rhesus monkey of fragments >350 bp at 75% identity indicated in red). rHH adjacent to CADM2 were identified in 23/1019 ASD cases and 11/2031 parental controls [Yates’ corrected χ2 p value = 1.9 × 10−5, OR = 4.26 (2.1, 8.6)]
Other novel ASD candidate genes located in rHH across multiple population clusters include GRIK2; an ionotropic glutamate receptor associated with autosomal recessive mental retardation and autism (Jamain et al. 2002; Motazacker et al. 2007), GRM3; a metabotropic glutamate receptor that impacts on aspects of cognition dependent on hippocampal and prefrontal cortical function (Egan et al. 2004), EPHA3; an ephrin receptor involved in axon guidance and synaptic plasticity, FGF10; a fibroblast growth factor involved in regulating the onset of neurogenesis and cortical brain size (Sahara and O’Leary 2009), ABHD14A; a cerebellar abhydrolase protein with a role in granule neuron development (Hoshino et al. 2003), NTS; a brain and gastrointestinal peptide involved in passive avoidance behaviour and the modulation of dopaminergic neurotransmission and serotonin levels (Shugalev et al. 2008), SCAMP5; a brain-enriched membrane trafficking protein located 10 kb centromeric to the breakpoint of a 15q de novo balanced translocation in a patient with autism (Castermans et al. 2010), CHRFAM7A; a hybrid gene involving a partial duplication of the alpha7 nicotinic acetylcholine receptor in the postsynaptic membrane and that has previously been associated with schizophrenia, bipolar disorder, dementia, Alzheimer’s disease and attention-deficit hyperactivity disorder and KCND2; a brain-specific voltage-gated ion channel component that regulates neurotransmitter release and neuronal excitability at the glutamatergic synapse where SHANK3 and NLGN products are formed.
ASD-specific rHH across multiple populations
Of particular interest in this study are the rHH that are ASD-specific; shared by multiple ASD patients but not present in any of the parental controls. We identified 8 ASD-specific rHH, implicating 12 genes (POLR3C, CD160, ZNF364, PDZK1, NUDT17, GBE1, HTR1E, C10orf95, CUECDC2, CHORDC1, MGAT4C and C12orf50) and a cluster of defensins (8p23.1), which are significant in at least two population clusters. The ASD-specific rHH at 1q21.1 is shared by three population clusters and the maximal overlapping region contains a single gene; PDZK1. PDZK1 encodes a scaffold protein that connects plasma membrane proteins and regulatory components. The PDZK1 protein is part of a complex that includes SYNGAP1, KLHL17 and NMDA receptors and this complex is important for maintaining the integrity of actin cytoskeleton structures in neurons (Chen and Li 2005). In addition, the ASD-specific rHH at 1q21.1 overlaps with a rare deletion associated with schizophrenia in two independent studies (Tam et al. 2009). Furthermore, we observed that 8 of 996 ASD cases from the AGP sample set analysed for rare copy number variants in the study by Pinto and colleagues have a rare duplication (n = 6; 145–1,470 kb) or deletion (n = 2; 189 and 528 kb) involving the PDZK1 gene.
Enrichment for genes previously implicated in ASD
The genes located in rHH regions were compared to autosomal genes that have previously been implicated in ASD in association studies, expression analyses and chromosomal anomaly studies, as reviewed by Yang and Gill (2007). We extended the literature search to 2009 using the same search criteria provided by Yang and Gill (Supplementary Table 5). Of the 1,243 genes identified in the study, 25 are published ASD candidate genes (Table 2). However, considerable caution is required in interpreting such findings, since both rHH genes and previously reported ASD candidate genes tend to be larger (p < 10−3). Accordingly, we performed a logistic multiple regression including both ASD gene status and gene size as covariates. While the odds ratio decreased slightly, it remained highly significant, indicating that there is an association between rHH status and ASD status that is independent of gene size [size adjusted p = 0.001, OR = 2.10, (1.36, 3.25)]. In addition, 9 ASD candidates (BTN2A1, FOXP2, GBE1, GRIK2, IMMP2L, LRFN5, LRRN3, ROBO1 and SLC4A10) are amongst the 192 genes located in rHH in two or more population clusters [size adjusted p = 6.9 × 10−5, OR = 4.76 (2.34, 9.64)].
Table 2.
Previously identified ASD candidate genes located in rHH
| Pop. | No. genes identified in rHH regions | No. rHH genes previously implicated in ASD | ASD candidate genes located in rHH regions |
|---|---|---|---|
| Discovery stage 1 | |||
| C2 | 243 | 7 | BTN2A1, CSMD3, FNTA, FOXP2, GABRA2, GABRG1, GBE1 |
| C3 | 341 | 12 | ALAS1, FBXO33, GBE1, IMMP2L, LRFN5, LRRN3, NRXN1, PRCP, PTGS2, REEP3, ROBO1, SLC4A10 |
| C4 | 79 | – | – |
| C5 | 417 | 5 | ACO2, FOXP2, GRIK2, HERC2, TSPAN12 |
| C6 | 372 | 11 | BTN2A1, DOCK4, GBE1, GRIK2, GRM8, IMMP2L, LRFN5, LRRN3, ROBO1, SLC4A10, WDR75 |
| Replication stage 2 | |||
| C3 | 529 | 8 | ALAS1, FOXP2, GBE1, GRIK2, GRM8, IMMP2L, SLC4A10, STOM |
| C4 | 70 | 1 | CSMD3 |
| C5 | 731 | 9 | ACO2, CSMD3, FBXO33, GABRG1, GRM8, NAGLU, PCDH10, SEPHS2, SLC6A4 |
| C6 | 46 | – | – |
A number of genes that have previously been implicated in ASD were found to be located in rHH regions in both the discovery and replication HH mapping studies. In the discovery analysis, 25 previously reported ASD candidate genes occurred in rHH regions. Nine ASD candidate genes were located in rHH in more than one population group and are shown in bold. Another 16 ASD candidates were located in rHH in a single population group and may represent population-specific susceptibility genes. We also found that 16 previously implicated ASD genes were located in rHH regions in the replication study. Two ASD candidate genes were located in rHH in more than one population group and 14 ASD genes were population-specific. Ten ASD candidate genes (ALAS1, CSMD3, FOXP2, GABRG1, GBE1, GRM8, FBXO33, IMMP2L, SLC4A10 and ACO2) occurred in rHH regions in both the discovery and replication HH mapping analyses
Replication study
To investigate our findings further, we performed a replication study on an additional 1,182 independent AGP trios. The replication cohort (stage 2) was clustered with the discovery sample set (stage 1) to identify ancestry-matched replication groups (Supplementary Material 1). The four population clusters (C3–C6) with at least 50 probands in both the discovery and replication sample sets were considered for the replication study (Supplementary Table 6). The same analytical strategy was applied to the replication sample set and identified 1,190 genes in rHH regions. The rHH genes identified in the discovery (number of genes = 1,086) and replication (number of genes = 1,190) analyses of population clusters C3–C6 were subsequently compared. We found that 28.5% (310/1086 genes) of the rHH genes identified in the discovery analysis occurred in a rHH in at least one of the replication population clusters (Supplementary Table 7). In particular, the replication study provided further evidence for the possible involvement of the novel candidate genes ABHD14A, CADM2, EPHA3, FGF10, GRIK2, GRM3, and KCND2. SCAMP5 and CHRFAM7A (found in rHH in multiple population clusters in the discovery analysis) did not replicate in their corresponding replication clusters while NTS was significant in one of two ancestry-matched replication groups. Furthermore, 10 previously identified ASD candidate genes (ALAS1, CSMD3, FOXP2, GABRG1, GBE1, GRM8, FBXO33, IMMP2L, SLC4A10 and ACO2) were located in rHH regions in both the discovery and replication HH mapping analyses [size adjusted p = 0.001, OR = 2.99 (1.50, 5.90)]. The ASD-specific rHH at 1p21.1 was also significant in two of four population clusters in the replication study providing further evidence of an ASD risk gene at this locus.
rHH located in significant ASD linkage peaks
The rHH regions identified in the discovery (stage 1) and replication (stage 2) analyses were compared to the genomic regions that have previously displayed significant linkage (LOD score >3.3) with ASD. We found that 31 rHH identified in the discovery and replication analyses are located under significant ASD linkage peaks (Supplementary Table 8). Of interest, three of the ASD linkage peaks harbour rHH from multiple population groups.
Firstly, three population groups in the replication analysis (stage 2: C4, C5, C6) have an overlapping rHH within the 7q22.1 ASD linkage peak (International Molecular Genetic Study of Autism Consortium (IMGSAC) 2001). The rHH at 7q22.1 range in size from 545 to 808 kb. The maximal shared region is 545.3 kb and includes 19 genes. Three of the genes at this locus (CYP3A4, CYP3A5 and CYP3A7) are members of the cytochrome P450 superfamily that plays important roles in hormone synthesis and breakdown, cholesterol synthesis, vitamin D metabolism and the metabolism of drugs and toxic compounds. Data from the KEGG databases suggests that members of the cytochrome P450 family such as CYP3A4 and CYP3A5 are involved in 5-HT (serotonin) catabolism (Jia et al. 2004). Moreover, in the brain, mRNA expression appears to be specific to neurons in the cerebral cortex and basal ganglia, regions of the brain that are consistently implicated in ASD (Dutheil et al. 2008). In addition, genetic polymorphisms of cytochrome P450 enzymes have been linked to ASD and schizophrenia (Currenti 2010).
Secondly, a rHH shared by two populations in the discovery analysis is located within the significant ASD linkage peak at 15q13.1–q14 (Lauritsen et al. 2006; Liu et al. 2008; Philippe et al. 1999). This particular locus has been implicated in several neuropsychiatric disorders. The overlapping rHH region identified in the current study is 579 kb and contains five genes, only two of which are coding (CHRFAM7A and ARHGAP11B). Of most relevance to ASD is CHRFAM7A which represents a fusion product involving a partial duplication of the alpha7 nicotinic acetylcholine receptor in the postsynaptic membrane and a gene from the family with sequence similarity 7. CHRFAM7A has previously been associated with schizophrenia, bipolar disorder, dementia, Alzheimer’s and attention-deficit hyperactivity disorder (Feher et al. 2009; Flomen et al. 2006; Manchia et al. 2010; Martin et al. 2007; Sinkus et al. 2009). This is the first implication of CHRFAM7A in ASD.
Thirdly, we noted a rHH at 20q11.21–q13.12 that was significantly more common in ASD patients compared to parental controls in two population groups in the replication study. The shared rHH region includes 11 genes. The strongest functional candidate at this locus is MYH7B which encodes a myosin heavy chain protein that maintains excitatory synaptic function. Reduction of MyH7B in rats causes a profound alteration in dendritic spine structure and excitatory synaptic strength (Rubio et al. 2011). The resulting abnormal spine phenotype was strikingly similar to the morphological changes induced by over-expression of SynGAP1, a gene linked to mental retardation and ASD (Hamdan et al. 2009; Pinto et al. 2010). The advantage of the rHH analysis is that the ASD-specific rHH regions are much smaller (~0.53 Mb) than ASD linkage regions which range from 2 to 242 Mb and may facilitate identification of the causative gene within these loci.
Discussion
Despite some notable successes in neuropsychiatric genetics, overall the high heritability of ASD (~90%) remains poorly explained by common genetic risk variants. Instead, early studies of rare variation, in particular copy number variation, have suggested that rare variants may account for a significant proportion of the genetic basis of ASD. The study of excess haplotype sharing within homozygous regions offers a complimentary approach to the analysis of GWAS data in the study of complex disease and particularly focuses on the contribution of low-frequency variation to disease risk. We report the first genome-wide rHH mapping study to identify novel recessive candidate genes and loci involved in ASD susceptibility. The strategy was applied to one of the largest ASD trio collections, subdivided into 10 distinct population clusters. We identified 307 rHH regions containing 1,243 genes for further investigation. In cases where ancestry-matched replication groups were available, almost one-third of the discovery phase rHH genes (310/1,086) were located in homozygous haplotypes that were significantly more common in ASD patients compared to parental controls in the replication analysis. Importantly, we identified novel ASD genes that merit further study including ABHD14A, CADM2, CHRFAM7A, EPHA3, FGF10, GRIK2, GRM3, KCND2 and PDZK1. The current study also provides further support for 25 previously reported ASD genes. Interestingly 192 of the rHH genes were significant in multiple population clusters. The variation in haplotype of the rHH across the different populations may suggest the existence of common risk genes but population-specific risk alleles. The findings of the current study serve as a starting point to screen for causal variants and elucidate the underlying pathogenesis of ASD. In particular, sequence analysis will be more economically feasible since the search for causal variants is limited to 1,243 genes and specifically identifies the patients carrying the rHH of interest, allowing a more targeted follow-up approach.
Sample size as a limiting factor
Sample size is an important feature of the HH mapping approach and is a limiting factor in the current study. A number of the population clusters in the ASD study are of modest sample size (n < 100) and five population clusters could not be included because of insufficient sample numbers. Larger collections of genetically homogenous samples would increase the power to detect low-frequency events. Similarly, due to the small sample sizes, we have not corrected for multiple testing. The HH sharing strategy aims to identify genes that may contain low-frequency disease variants. Given the small sample sizes, such events are highly unlikely to survive correction for multiple testing. Although this may be considered a weakness it is important to note that correcting for multiple testing will not change the relative order of the rHH results (Zaykin and Zhivotovsky 2005). The distributions of ranks will remain the same with HHs that show the greatest difference in frequency between ASD cases and parental controls remaining as the most important findings. We acknowledge that by not correcting for multiple testing, there is an increased risk of false positive findings. To address this we have focused on the genes that occur in rHH in multiple population clusters and replicate in the stage 2 sample set, as they are less likely to be false positives.
Genomic structure underlying rHH regions
Analysis of Log R ratios and B allele frequencies confirmed that the rHH regions are not attributed to copy number deletions. To ascertain whether some unknown genomic architecture contributed to the observed rHH findings and also to reduce the risk of false positives, we undertook rHH mapping in two additional disease data sets from the Wellcome-Trust Case–Control Consortium; bipolar disorder (BD) (cases = 1,875 and controls = 2,954) and coronary artery disease (CAD) (cases = 1,963 and controls = 2,978). We hypothesised that genome architecture could possibly be contributing to the results of the ASD rHH analysis if (1) the rHH in the BD and CAD studies showed a significant overlap with the rHH identified in the current ASD study and (2) the BD and CAD rHH genes showed a significant enrichment for previously identified ASD genes. There was a 2 and 2.6% overlap between BD-ASD and CAD-ASD rHH regions, respectively, neither of which are greater than expected by chance (J.P.C., unpublished data). Furthermore the BD and CAD rHH genes did not display an enrichment for known ASD candidate genes (BD Yates’ corrected χ2 p = 0.815, CAD Yates’ corrected χ2 p = 1.000) suggesting that the rHH identified in the current study are more likely to be related to the ASD phenotype than to the genomic architecture in the rHH regions.
Homozygous haplotype sharing in complex disorders
There are very few analytical strategies designed to identify rare recessive disease genes for complex traits. The homozygous haplotype sharing strategy addresses this issue and proposes a novel concept for searching for genes that may contain low-frequency disease variants. The most important feature of the HH mapping approach presented in this study is the concept; analysis of the haplotype within shared homozygous segments provides an additional level of information that has been overlooked in ROH-based analyses and excess sharing of a homozygous haplotype amongst patients may support the presence of a rare recessive disease mutation in the region. In the future, the strategy itself will undoubtedly benefit from further modifications and improvements, particularly in the areas of modelling, simulation and statistics for rare genetic events.
We applied the HH mapping approach to one of the largest international ASD cohorts (4,206 samples) and identified novel and known ASD candidate genes which were replicated in an independent sample set. We also found that homozygous haplotypes over-represented in ASD patients were significantly enriched for previously identified ASD candidate genes, further validating our approach. Although HH mapping does not identify causative alleles, the regions reported in the current study provide narrow genomic intervals containing highly plausible candidate genes for further investigation. The findings reported in this study suggest that the analysis of homozygous haplotype sharing may be an important tool in uncovering some of the missing heritability in a variety of complex disorders.
Ethical standards The experiments comply with the current laws of the country in which they were performed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors acknowledge the families participating in the study and the main funders of the Autism Genome Project Consortium (AGP): Autism Speaks (USA), the Health Research Board (HRB; Ireland), The Medical Research Council (MRC; UK), Genome Canada/Ontario Genomics Institute, and the Hilibrand Foundation (USA). Additional support for individual groups was provided by the US National Institutes of Health (NIH grants HD055751, HD055782, HD055784, HD35465, MH52708, MH55284, MH57881, MH061009, MH06359, MH066673, MH080647, MH081754, MH66766, NS026630, NS042165, NS049261), the Canadian Institute for Advanced Research (CIFAR), the Canadian Institutes for Health Research (CIHR), Assistance Publique–Hôpitaux de Paris (France), Autistica, Canada Foundation for Innovation/Ontario Innovation Trust, Deutsche Forschungsgemeinschaft (grant Po 255/17-4) (Germany), EC Sixth FP AUTISM MOLGEN, Fundação Calouste Gulbenkian (Portugal), Fondation de France, Fondation FondaMental (France), Fondation Orange (France), Fondation pour la Recherche Médicale (France), Fundação para a Ciência e Tecnologia (Portugal), the Hospital for Sick Children Foundation and University of Toronto (Canada), INSERM (France), Institut Pasteur (France), the Italian Ministry of Health (convention 181 of 19.10.2001), the John P. Hussman Foundation (USA), McLaughlin Centre (Canada), Ontario Ministry of Research and Innovation (Canada), the Seaver Foundation (USA), the Swedish Science Council, The Centre for Applied Genomics (Canada), the Utah Autism Foundation (USA) and the Wellcome Trust core award 075491/Z/04 (UK). We acknowledge support from the Autism Genetic Resource Exchange (AGRE) and Autism Speaks. We gratefully acknowledge the resources provided by the AGRE consortium and the participating AGRE families. AGRE is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). We wish to acknowledge the National Children’s Research Centre Our Lady’s Children’s Hospital Crumlin Ireland for providing additional support and the Wellcome Trust Case–Control Consortium for providing data sets that were used as part of this study. J.P.C is supported by an EMBARK postgraduate award from the Irish Research Council for Science, Engineering and Technology (IRCSET).
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Appendix: The AGRE Consortium
Dan Geschwind, M.D., Ph.D., UCLA, Los Angeles, CA; Maja Bucan, Ph.D., University of Pennsylvania, Philadelphia, PA; W.Ted Brown, M.D., Ph.D., F.A.C.M.G., N.Y.S. Institute for Basic Research in Developmental Disabilities, Staten Island, NY; Rita M. Cantor, Ph.D., UCLA School of Medicine, Los Angeles, CA; John N. Constantino, M.D., Washington University School of Medicine, St. Louis, MO; T.Conrad Gilliam, Ph.D., University of Chicago, Chicago, IL; Joachim Hallmayer, Ph.D., Stanford University, Stanford, CA; Martha Herbert, M.D., Ph.D., Harvard Medical School, Boston, MA Clara Lajonchere, Ph.D, Autism Speaks, Los Angeles, CA; David H. Ledbetter, Ph.D., Emory University, Atlanta, GA; Christa Lese-Martin, Ph.D., Emory University, Atlanta, GA; Janet Miller, J.D., Ph.D., Autism Speaks, Los Angeles, CA; Stanley F. Nelson, M.D., UCLA School of Medicine, Los Angeles, CA; Gerard D. Schellenberg, Ph.D., University of Washington, Seattle, WA; Carol A. Samango-Sprouse, Ed.D., George Washington University, Washington, D.C.; Jonathan Shestack, Autism Speaks, NY, NY; Sarah Spence, M.D., Ph.D., UCLA, Los Angeles, CA; Matthew State, M.D., Ph.D., Yale University, New Haven, CT.Rudolph E. Tanzi, Ph.D., Massachusetts General Hospital, Boston, MA.
Footnotes
Annemarie Poustka: deceased.
References
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82:160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahlo M, Stankovich J, Speed TP, Rubio JP, Burfoot RK, Foote SJ. Detecting genome wide haplotype sharing using SNP or microsatellite haplotype data. Hum Genet. 2006;119:38–50. doi: 10.1007/s00439-005-0114-9. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, Kleinjan DJ, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, McBride D, Golzio C, Fisher M, Perry P, Abadie V, Ayuso C, Holder-Espinasse M, Kilpatrick N, Lees MM, Picard A, Temple IK, Thomas P, Vazquez MP, Vekemans M, Roest Crollius H, Hastie ND, Munnich A, Etchevers HC, Pelet A, Farlie PG, Fitzpatrick DR, Lyonnet S. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 2009;41:359–364. doi: 10.1038/ng.329. [DOI] [PubMed] [Google Scholar]
- Betancur C, Sakurai T, Buxbaum JD. The emerging role of synaptic cell-adhesion pathways in the pathogenesis of autism spectrum disorders. Trends Neurosci. 2009;32:402–412. doi: 10.1016/j.tins.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- Castermans D, Volders K, Crepel A, Backx L, De Vos R, Freson K, Meulemans S, Vermeesch JR, Schrander-Stumpel CT, De Rijk P, Del-Favero J, Van Geet C, Van De Ven WJ, Steyaert JG, Devriendt K, Creemers JW. SCAMP5, NBEA and AMISYN: three candidate genes for autism involved in secretion of large dense-core vesicles. Hum Mol Genet. 2010;19:1368–1378. doi: 10.1093/hmg/ddq013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Li M. Interactions between CAP70 and actinfilin are important for integrity of actin cytoskeleton structures in neurons. Neuropharmacology. 2005;49:1026–1041. doi: 10.1016/j.neuropharm.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Currenti SA. Understanding and determining the etiology of autism. Cell Mol Neurobiol. 2010;30:161–171. doi: 10.1007/s10571-009-9453-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Chaste P, Fauchereau F, Betancur C, Leboyer M, Bourgeron T. Alterations in synapsis formation and function in autism disorders. Med Sci (Paris) 2008;24:25–28. doi: 10.1051/medsci/200824125. [DOI] [PubMed] [Google Scholar]
- Dutheil F, Beaune P, Loriot MA. Xenobiotic metabolizing enzymes in the central nervous system: Contribution of cytochrome P450 enzymes in normal and pathological human brain. Biochimie. 2008;90:426–436. doi: 10.1016/j.biochi.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Egan MF, Straub RE, Goldberg TE, Yakub I, Callicott JH, Hariri AR, Mattay VS, Bertolino A, Hyde TM, Shannon-Weickert C, Akil M, Crook J, Vakkalanka RK, Balkissoon R, Gibbs RA, Kleinman JE, Weinberger DR. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proc Natl Acad Sci USA. 2004;101:12604–12609. doi: 10.1073/pnas.0405077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feher A, Juhasz A, Rimanoczy A, Csibri E, Kalman J, Janka Z. Association between a genetic variant of the alpha-7 nicotinic acetylcholine receptor subunit and four types of dementia. Dement Geriatr Cogn Disord. 2009;28:56–62. doi: 10.1159/000230036. [DOI] [PubMed] [Google Scholar]
- Flomen RH, Collier DA, Osborne S, Munro J, Breen G, St Clair D, Makoff AJ. Association study of CHRFAM7A copy number and 2 bp deletion polymorphisms with schizophrenia and bipolar affective disorder. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:571–575. doi: 10.1002/ajmg.b.30306. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Goudet J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes. 2005;5:184–186. doi: 10.1111/j.1471-8286.2004.00828.x. [DOI] [Google Scholar]
- Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, Dobrzeniecka S, Cote M, Perreau-Linck E, Carmant L, D’Anjou G, Fombonne E, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Mouaffak F, Joober R, Mottron L, Drapeau P, Marineau C, Lafreniere RG, Lacaille JC, Rouleau GA, Michaud JL. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360:599–605. doi: 10.1056/NEJMoa0805392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt F, Heeringa SF, Ruschendorf F, Attanasio M, Nurnberg G, Becker C, Seelow D, Huebner N, Chernin G, Vlangos CN, Zhou W, O’Toole JF, Hoskins BE, Wolf MT, Hinkes BG, Chaib H, Ashraf S, Schoeb DS, Ovunc B, Allen SJ, Vega-Warner V, Wise E, Harville HM, Lyons RH, Washburn J, Macdonald J, Nurnberg P, Otto EA. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino J, Aruga J, Ishiguro A, Mikoshiba K. Dorz1, a novel gene expressed in differentiating cerebellar granule neurons, is down-regulated in Zic1-deficient mouse. Brain Res Mol Brain Res. 2003;120:57–64. doi: 10.1016/j.molbrainres.2003.10.004. [DOI] [PubMed] [Google Scholar]
- International Molecular Genetic Study of Autism Consortium (IMGSAC) A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Betancur C, Quach H, Philippe A, Fellous M, Giros B, Gillberg C, Leboyer M, Bourgeron T. Linkage and association of the glutamate receptor 6 gene with autism. Mol Psychiatry. 2002;7:302–310. doi: 10.1038/sj.mp.4000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, Yu X, Zhang B, Yuan Y, Xu Q, Shen Y. No association between polymorphisms in three genes of cytochrome p450 family and paranoid schizophrenia in northern Chinese Han population. Eur Psychiatry. 2004;19:374–376. doi: 10.1016/j.eurpsy.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, Descartes M, Holt L, Braddock S, Troxell R, Kaplan L, Volkmar F, Klin A, Tsatsanis K, Harris DJ, Noens I, Pauls DL, Daly MJ, MacDonald ME, Morton CC, Quade BJ, Gusella JF. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Moore J, Bailey A, Monaco AP. Autism: recent molecular genetic advances. Hum Mol Genet. 2000;9:861–868. doi: 10.1093/hmg/9.6.861. [DOI] [PubMed] [Google Scholar]
- Lauritsen MB, Als TD, Dahl HA, Flint TJ, Wang AG, Vang M, Kruse TA, Ewald H, Mors O. A genome-wide search for alleles and haplotypes associated with autism and related pervasive developmental disorders on the Faroe Islands. Mol Psychiatry. 2006;11:37–46. doi: 10.1038/sj.mp.4001754. [DOI] [PubMed] [Google Scholar]
- Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, Kucherlapati R, Malhotra AK. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci USA. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Timmesfeld N, Renner TJ, Halperin R, Roser C, Nguyen TT, Craig DW, Romanos J, Heine M, Meyer J, Freitag C, Warnke A, Romanos M, Schafer H, Walitza S, Reif A, Stephan DA, Jacob C. Molecular genetics of adult ADHD: converging evidence from genome-wide association and extended pedigree linkage studies. J Neural Transm. 2008;115:1573–1585. doi: 10.1007/s00702-008-0119-3. [DOI] [PubMed] [Google Scholar]
- Liu XQ, Paterson AD, Szatmari P. Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biol Psychiatry. 2008;64:561–570. doi: 10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchia M, Viggiano E, Tiwari AK, Renou J, Jain U, De Luca V, Kennedy JL. Smoking in adult attention-deficit/hyperactivity disorder: interaction between 15q13 nicotinic genes and Temperament Character Inventory scores. World J Biol Psychiatry. 2010;11:506–510. doi: 10.3109/15622970902980788. [DOI] [PubMed] [Google Scholar]
- Martin LF, Leonard S, Hall MH, Tregellas JR, Freedman R, Olincy A. Sensory gating and alpha-7 nicotinic receptor gene allelic variants in schizoaffective disorder, bipolar type. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:611–614. doi: 10.1002/ajmg.b.30470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motazacker MM, Rost BR, Hucho T, Garshasbi M, Kahrizi K, Ullmann R, Abedini SS, Nieh SE, Amini SH, Goswami C, Tzschach A, Jensen LR, Schmitz D, Ropers HH, Najmabadi H, Kuss AW. A defect in the ionotropic glutamate receptor 6 gene (GRIK2) is associated with autosomal recessive mental retardation. Am J Hum Genet. 2007;81:792–798. doi: 10.1086/521275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothnagel M, Lu TT, Kayser M, Krawczak M. Genomic and geographic distribution of SNP-defined runs of homozygosity in Europeans. Hum Mol Genet. 2010;19:2927–2935. doi: 10.1093/hmg/ddq198. [DOI] [PubMed] [Google Scholar]
- Persico AM, Bourgeron T. Searching for ways out of the autism maze: genetic, epigenetic and environmental clues. Trends Neurosci. 2006;29:349–358. doi: 10.1016/j.tins.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, Gillberg C, Rastam M, Sponheim E, Coleman M, Zappella M, Aschauer H, Van Maldergem L, Penet C, Feingold J, Brice A, Leboyer M. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bolte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio MD, Johnson R, Miller CA, Huganir RL, Rumbaugh G. Regulation of synapse structure and function by distinct myosin II motors. J Neurosci. 2011;31:1448–1460. doi: 10.1523/JNEUROSCI.3294-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, O’Leary DD. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugalev NP, Stavrovskaya AV, Ol’shanskii AS, Hartmann G, Lenard L. Serotoninergic mechanisms of the effects of neurotensin on passive avoidance behavior in rats. Neurosci Behav Physiol. 2008;38:517–521. doi: 10.1007/s11055-008-9011-8. [DOI] [PubMed] [Google Scholar]
- Sinkus ML, Lee MJ, Gault J, Logel J, Short M, Freedman R, Christian SL, Lyon J, Leonard S. A 2-base pair deletion polymorphism in the partial duplication of the alpha7 nicotinic acetylcholine gene (CHRFAM7A) on chromosome 15q14 is associated with schizophrenia. Brain Res. 2009;1291:1–11. doi: 10.1016/j.brainres.2009.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam GW, Redon R, Carter NP, Grant SG. The role of DNA copy number variation in schizophrenia. Biol Psychiatry. 2009;66:1005–1012. doi: 10.1016/j.biopsych.2009.07.027. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Thomas LA, Akins MR, Biederer T. Expression and adhesion profiles of SynCAM molecules indicate distinct neuronal functions. J Comp Neurol. 2008;510:47–67. doi: 10.1002/cne.21773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan MJ, Pollard KS. A new algorithm for hybrid hierarchical clustering with visualisation and the bootstrap. J Stat Plan Inference. 2002;117:275–303. doi: 10.1016/S0378-3758(02)00388-9. [DOI] [Google Scholar]
- Vine AE, McQuillin A, Bass NJ, Pereira A, Kandaswamy R, Robinson M, Lawrence J, Anjorin A, Sklar P, Gurling HM, Curtis D. No evidence for excess runs of homozygosity in bipolar disorder. Psychiatr Genet. 2009;19:165–170. doi: 10.1097/YPG.0b013e32832a4faa. [DOI] [PubMed] [Google Scholar]
- Wong W, Newell EW, Jugloff DG, Jones OT, Schlichter LC. Cell surface targeting and clustering interactions between heterologously expressed PSD-95 and the Shal voltage-gated potassium channel, Kv4.2. J Biol Chem. 2002;277:20423–20430. doi: 10.1074/jbc.M109412200. [DOI] [PubMed] [Google Scholar]
- Yang MS, Gill M. A review of gene linkage, association and expression studies in autism and an assessment of convergent evidence. Int J Dev Neurosci. 2007;25:69–85. doi: 10.1016/j.ijdevneu.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Yang TL, Guo Y, Zhang LS, Tian Q, Yan H, Papasian CJ, Recker RR, Deng HW. Runs of homozygosity identify a recessive locus 12q21.31 for human adult height. J Clin Endocrinol Metab. 2010;95:3777–3782. doi: 10.1210/jc.2009-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Zhivotovsky LA. Ranks of genuine associations in whole-genome scans. Genetics. 2005;171(2):813–823. doi: 10.1534/genetics.105.044206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.