Abstract
Human cytomegalovirus (HCMV)-encoded NK cell evasion functions include an MHC-I homologue (UL18) with high affinity for the leukocyte inhibitory receptor LIR-1 (CD85j, ILT2 or LILRB1) and a signal peptide (SPUL40) that acts by upregulating cell surface expression of HLA-E. Detailed characterization of SPUL40 revealed that the N-terminal 14 amino acid residues bestowed TAP-independent upregulation of HLA-E, while c-region sequences delayed processing of SPUL40 by a signal peptide peptidase-type intramembrane protease. Most significantly, the consensus HLA-E-binding epitope within SPUL40 was shown to promote cell surface expression of both HLA-E and gpUL18. UL40 was found to possess two transcription start sites, with utilization of the downstream site resulting in translational being initiated within the HLA-E-binding epitope (P2). Remarkably, this truncated SPUL40 was functional and retained the capacity to upregulate gpUL18, but not HLA-E. Our findings thus identify an elegant mechanism by which an HCMV signal peptide differentially regulates two distinct NK cell evasion pathways. Moreover, we describe a natural SPUL40 mutant that provides the first example of an HCMV clinical virus with a defect in an NK cell evasion function and exemplifies issues that confront the virus when adapting to immunogenetic diversity in the host.
Keywords: Human, Natural Killer Cells, Viral, MHC, Cell Surface Molecules, Cytotoxicity, Gene Regulation
Introduction
Human cytomegalovirus (HCMV; species Human herpesvirus 5) is the prototype member of subfamily Betaherpesvirinae in the family Herpesviridae, and an important opportunistic pathogen. In most individuals, primary infection is subclinical and is followed by a life-long, persistent infection that is controlled by host immune surveillance. Nevertheless, HCMV is a major cause of morbidity and mortality in immunocompromised transplant recipients, and late stage HIV AIDS patients. Congenital transmission can result in spontaneous abortion or overt cytomegalic inclusion disease on birth. A proportion of asymptomatically infected neonates go on to exhibit sensorineural hearing loss. Disease can be manifest in many different forms, including infectious mononucleosis, hepatitis, post-transplant arteriosclerosis, pneumonia, colitis, immune senescence and alteration to the immune repertoire, including expansion in LIR-1+ and CD94/NKG2C+ natural killer (NK) cell subsets (1). During persistent infections the virus is a significant and sustained burden on the host’s immune system, which in turn exerts strong selective pressures on the virus.
HCMV has amassed an impressive collection of genes dedicated to subjugating host immune systems (2, 3). At least four genes act in concert efficiently to downregulate cell surface expression of MHC class I (MHC-I): US6 inhibits the transporter associated with antigen processing (TAP), US3 promotes retention of MHC-I molecules in the endoplasmic reticulum (ER) while US2 and US11 induce rapid degradation of MHC-I heavy chains by the ER-associated degradation pathway. In vivo studies with rhesus CMV mutants imply that evasion of CD8+ cytotoxic T lymphocytes is not critical during primary infection, but is required for superinfection of seropositive animals (4). Evasion of CD8+ T cell recognition may thus promote virus transmission, yet it also has the potential to sensitize infected cells to NK cell attack. Endogenous MHC-I molecules are key ligands for a wide range of NK cell inhibitory receptors (5). In vitro, HCMV-infected cells are remarkably resistant to NK cell attack, and this is absolutely dependent on HCMV-encoded NK cell evasion functions. To date, six HCMV encoded proteins (UL16, UL18, UL40, UL83, UL141 and UL142) and one miRNA have been shown to suppress NK cell activation (reviewed in (1)). Among them, UL18 and UL40 counteract down-regulation of endogenous classical MHC-I. HCMV encodes its own MHC-I homologue, gpUL18, which forms a trimeric complex with β2-microglobulin (6-11). The inhibitory leukocyte receptor LIR-1 (also known as CD85j, ILT2, LILRB1) binds the mature gpUL18 complex with >1000-fold higher affinity than MHC-I, and hence even low levels of gpUL18 efficiently compete for LIR-1 binding (12, 13). LIR-1 is expressed on most monocytes, dendritic and B cells and subsets of T and NK cells. Although UL18 has a strong net inhibitory effect on LIR-1+ NK cells, it also stimulates LIR− NK cells. The outcome of a polyclonal NK cell response to UL18 thus correlates with the size of the LIR-1+ NK cell subset (14).
HCMV UL40 promotes efficient cell surface expression of the non-classical MHC-I molecule HLA-E (15). HLA-E binds the NK cell inhibitory receptor CD94/NKG2A (16-18). Cell surface expression of HLA-E depends on binding of a conserved nonameric peptide normally derived from the N-terminal signal peptide (SP) of classical MHC-I molecules (19). The SP is cleaved from the nascent peptide chain by signal peptidase (SPase), and this is followed by further processing of the SP (SPHLA) by the intramembrane protease SP peptidase (SPP) (20, 21). Cleavage in the centre of the membrane-spanning hydrophobic h-region releases a fragment containing the HLA-E-binding peptide into the cytosol (20). This peptide is further trimmed by the proteasome before being transported to the ER in a TAP-dependent mechanism (18, 22). HLA-E thus acts as a cellular indicator/reporter of normal MHC-I synthesis and TAP function. By inhibiting TAP, US6 would also be expected to inhibit HLA-E maturation. However, the UL40 SP (SPUL40) contains a nonameric sequence with exact sequence identity to an endogenous HLA-E-binding peptide. HCMV UL40 is able to upregulate HLA-E independently of TAP function (15, 23), and thereby promotes efficient protection against lysis by CD94/NKG2A+ NK cells (15, 23, 24).
In addition to CD94/NKG2A, HLA-E also interacts with CD94/NKG2C, albeit with lower affinity. CD94/NKG2C is an activating receptor expressed on a relatively small population of NK cells. Interestingly, the frequency of this receptor increases in HCMV-infected individuals (25). Neither the biological significance of this subset nor the mechanism driving their expansion in HCMV infections is clear (26). HLA-E-restricted CD8+ cytotoxic T cell responses can also be induced. The HLA-E-binding peptide encoded by UL40 does not always have an exact match to a host MHC-I derived peptide, and consequently T cell tolerance is not acquired. In such individuals, UL40 has the potential to provide a T cell target and thereby potentially provides a mean for the immune system to eradicate HCMV-infected targets (27).
In this study, we sought better to define the roles played by sequence elements within SPUL40 in promoting maturation of HLA-E. To this end, we undertook the characterization of natural HCMV SPUL40 sequence variation in clinical isolates, the construction of a panel of UL40:HLA-A2 chimeric signal peptides and targeted mutations in SPUL40, as well as performing a detailed transcriptional analysis of UL40. These studies confirm that the UL40 nonamer peptide plays a critical role in promoting HLA-E maturation. In addition, we observed that in 721.221 cells, SPUL40 was processed by an SPP-type protease activity, in contrast to previous studies in isolated ER-derived rough microsomes (28). Remarkably, the n-region of SPUL40 was required to mediate TAP-independent HLA-E loading. Moreover, SPUL40 was found to be capable of upregulating both HLA-E and gpUL18, and this property mapped within the UL40 nonameric peptide (VMAPRTLIL). Although the peptide-binding specificities of HLA-E and UL18 appeared to overlap, differential regulation of HLA-E and UL18 could be achieved by UL40 sequence variants. Genetic selection in SPUL40 has the potential to influence NK and T cell recognition via both HLA-E and UL18.
Materials and Methods
Vector constructs
Recombinant adenoviruses (RAds) were generated as described previously (29). Briefly, PCR products generated as follows were inserted into an adenovirus (Ad) transfer vector under the control of the HCMV major immediate-early promoter, sequenced, then co-transfected with pJM17 plasmid into 293 cells (Effectene, Qiagen). Resulting RAds were propagated and titrated in 293 cells. RAd-HLA-A2.His was generated by attaching a RGS(6xHis) tag to the C-terminus of HLA-A2 by PCR using the plasmid pA2-RSV5neo as a template (a gift from P. Lehner, University of Cambridge, UK). RAd-UL40(Δ2-14) construct was generated by replacing the 14 N-terminal amino acid residues (aa) of SPUL40 (MNKFSNTRIGFTCA) with 2 N-terminal aa from HLA-C (MR) by PCR using HCMV strain AD169 DNA as a template. The RAd-UL40(1-14)HLA-A2.His construct was made by replacing the 2 N-terminal aa from HLA-A2 (MA) with 14 N-terminal aa from AD169 UL40 (MNKFSNTRIGFTCA), and attaching a RGS(6xHis) tag to the C-terminus of HLA-A2 by PCR using pA2-RSV5neo as a template. RAd-UL40.His was made by replacing the UL40 C-terminal YALKKA sequence with HHHHHH sequence, thus generating a 5xHis tag, by PCR using RAd-UL40 as a template. RAd-UL40(M16T) was generated by replacing M at position 16 with T by PCR using RAd-UL40 as a template. RAd-UL40(M16T).His was generated by replacing the UL40 C-terminal YALKKA sequence with HHH sequence by PCR using RAd-UL40(M16T) as a template. RAd-ToledoUL40 and RAd-3157UL40, which express gpUL40 from HCMV strains Toledo and 3157, were generated by PCR from respective genomic DNAs. RAd-UL40(1-41)RFP and RAd-HLA-A2(1-28)RFP were generated by attaching complete SPUL40 (aa 1-41) and SPHLA-A2 (aa 1-28) to the N-terminus of the red fluorescent protein (RFP) (Clontech). RAd-UL40, RAd-UL18 and RAd-HLA-E, which encode AD169 UL40, AD169 UL18 and HLA-E*0101, have been described previously (15). RAd-CTRL is an equivalent adenovirus lacking a transgene insertion. The following RAds were generated by recombineering as described elsewhere (30). RAd-UL40(GAG) and RAd-UL40(MelanA) were made by replacing the UL40 HLA-E-binding peptide (VMAPRTLIL) with an HIV-1 GAG peptide (SLYNTIATL) or a MelanA peptide (ELAGIGILTV). RAd-HLA-A2(12-24)UL40 was generated by replacing the C-terminal end of SPUL40 (aa 24-37 TVGLLCMRIRSLLC) with the C-terminal end of SPHLA-A2 (aa 12-24 LLSGALALTQTWA). RAd-UL40(GAG), RAd-UL40(MelanA) and HLA-A2(12-24)UL40 were cloned in-frame with a C-terminal V5 tag.
HCMV strains
Strains Merlin (Genbank AY446894), Toledo (Genbank AC146905), 3157 (Genbank GQ221974), AD169varUK (Genbank BK000394) and the AD169varUK ΔUL16, ΔUL18 and ΔUL40 deletion mutants have all been described (24, 31-35). Strain AD169varUK deletion mutants ΔUL16 and ΔUL18 were kindly provided by Helena Browne (Cambridge University) and strain Toledo by E. Mocarski (Emory University School of Medicine).
Cells and infections
Human fetal foreskin fibroblasts (HFFFs) were immortalized with human telomerase reverse transcriptase (hTERT) (36) and transduced with a retrovirus expressing the human Coxsackie-adenovirus receptor (hCAR) (1) (HFFF-hTERT-hCAR). 293 cells were derived from human embryonal kidney cells transformed by sheared human adenovirus type 5 DNA. TAP-deficient skin fibroblasts (Npi) cells (37) (kindly provided by V. Cerundolo, University of Oxford) were immortalized with hTERT (36). Fibroblasts and 293 cells were grown in DMEM supplemented with 10% fetal calf serum and antibiotics (Invitrogen). For HLA-E experiments, cells were infected on day 1 with RAd-HLA-E (50 PFU/cell), then on day 2 with the RAd construct under test (50 PFU/cell), and incubated for a further 48 h prior to analysis. For co-expression of UL18 with UL40, cells were infected with both RAds at the same time, and analyzed 72 h post-infection. For HCMV infections, fibroblasts were infected for 72-96 h at 10 PFU/cell.
Antibodies for flow cytometry
HLA-E expression was measured with the DT9 (anti HLA-E/HLA-C) or 3D12 (anti HLA-E) mAbs (eBioscience). Polyhistidine-tagged proteins were detected with Tetra.His mAb (Qiagen). HLA-A2 expression was detected with the MA2.1 mAb (anti HLA-A2/B17, ATCC) and MHC-I expression was detected with the W6/32 mAb (anti HLA-A/B/C, ECACC). The expression of HCMV glycoproteins (gB, gH and gN) was assessed with specific mAbs (a gift from W. Britt, University of Alabama at Birmingham, USA). gpUL18 expression was detected with the 10C7 mAb (ATCC). Primary Abs were visualized with anti-mouse IgG-FITC conjugated goat F(ab’)2 fragments (Sigma) or anti-mouse IgG-Alexa Fluor® 647 conjugated goat F(ab’)2 fragments (Invitrogen). In immunoblot experiments HLA-E expression was detected with the mAb MEM-E/02 (Serotek), gpUL18 with M71 (Amgen), HLA-HC with HC-10 (ATCC), CD155 with 5D1 (A. Nomoto, Microbial Chemistry Research Foundation, Tokyo) and actin with A-2066 (Sigma). Polyclonal mouse antibody to gpUL40 was prepared as described previously (15).
SDS-PAGE and western blotting
Cells were lysed for 30 min in NuPAGE® LDS Sample Buffer (Invitrogen) supplemented with protease inhibitor cocktail (P8340, Sigma), and supernatants were digested with endoglycosidase H (Endo H) or peptide-N-glycosidase F (PNGase F) according to the manufacturer’s instructions (New England Biolabs). Enzyme was omitted from mock-digested samples. Proteins were then separated on NuPAGE® Novex 10% Bis-Tris Midi Gels (Invitrogen) and transferred to nitrocellulose membranes (Hybond™-C, GE Healthcare). Membranes were blocked for 16 h at 4°C in SuperBlock® Blocking Buffer (Pierce), incubated with mAbs specific for gpUL18 (M71, Amgen) or actin (Sigma) for 16 h in Tris-buffered saline Tween-20 (TBST) 10% SuperBlock® Blocking Buffer, washed three times in TBST 5% dried milk, incubated with Goat Anti-Mouse IgG (H+L)-horseradish peroxidase (HRP) Conjugate or Goat Anti-Rabbit IgG (H+L)-HRP Conjugate (Bio-Rad) in TBST for 1 h and washed three times in TBST. SuperSignal® West Pico Chemiluminescent Substrate (Pierce) was used to detect the HRP signal. Western blots were imaged on an AutoChemi System (UVP) with Labworks 4.6 software (UVP).
Preparation of digitonin-permeabilized cells
Cells were permeabilized as described previously (38), except that digitonin was used instead of saponin. Briefly, 721.221 cells were washed with buffer H (50 mM HEPES-KOH, pH 7.4, 165 mM KOAc, 2 mM Mg(OAc)2) and centrifuged at 250 g for 5 min. The cell pellet was resuspended (107 cell/ml) in ice-cold buffer H supplemented with 6 μg/ml digitonin, 1 mM DTT and 10 μg/ml each of phenylmethylsulfonyl fluoride, chymostatin, leupeptin, antipain and pepstatin. After 5 min incubation on ice, the cell suspension was diluted with a volume of buffer H. Cells were then pelleted and washed once with buffer H. The extent of permeabilization was controlled by trypan blue exclusion, with 5% living cells remaining on average. Subsequently, digitonin-permeabilized cells were resuspended in buffer H supplemented with protease inhibitors as outlined above, 1 mM CaCl2 and 20 U/ml micrococcal nuclease and incubated 10 min at 22°C. The reaction was stopped by adding 2 mM EGTA. The cells were washed and finally resuspended in buffer H (105 cells/μl).
In vitro transcription, translation and SP processing
To generate templates for in vitro transcription, UL40 with a hybrid HLA-C/UL40 signal peptide (MNKFSNTRIGFTCAVMAPRTLVLLLSGALALTQTWA, to generate pAL303) and HLA-A2 with a hybrid HCMV AD169 UL40/HLA-A2 signal peptide (MNKFSNTRIGFTCAVMAPRTLVLLLSGALALTQTWA, to generate pAL307) were amplified by PCR using suitable plasmid templates. The forward primer introduced the SP6 promoter and a Kozak initiation sequence, while the reverse primer inserted a stop codon at the desired position of the open reading frame. The PCR products were transcribed in vitro with SP6 RNA polymerase at 42 °C in the presence of 500 μM m7G(5′)ppp(5′)G CAP analogue (New England Biolabs). mRNAs were translated in 25 μl reticulocyte lysate (Promega) containing [35]S-methionine and [35]S-cysteine, and two equivalents of nuclease-treated rough microsomes prepared from dog pancreas (39)or digitonin-permeabilized 721.221 cells (105 cells) as indicated. 10 μM (Z-LL)2-ketone or 30 μM N-nenzoyl-Asn-Leu-Thr-methylamide was added to inhibit SPP or N-glycosylation respectively(40). Microsomes and digitonin-permeabilized cells were extracted with 500 mM KOAc and isolated by centrifugation through a sucrose cushion as described previously (40). For extraction with alkali, KOAc-extracted microsomes were treated with 100 mM NaCO3, pH 11.3 (39). Proteins were analyzed by SDS-PAGE using Tris-Bicine gels with radiolabeled proteins being visualized by phosphorimaging (40, 41).
RNA preparation
HFFF cell monolayers were infected with 5 PFU/cell of HCMV strain Merlin in the presence (for early RNA) or absence (for late RNA) of 300 μg/ml phosphonoacetic acid. Total cellular RNA was isolated at 48 h (early RNA) or 72 h (late RNA) post infection using Tri Reagent (Sigma).
Northern blotting
Northern blotting was carried out using standard techniques. Briefly, aliquots of early and late RNA (3 μg) were electrophoresed alongside DIG-labelled I RNA markers (Roche) in a formaldehyde-agarose gel. The RNA was transferred to a nylon membrane (Roche) and fixed by ultraviolet irradiation. A 318 bp PCR product was generated from the UL40 coding region by using primers 5′-AATGCCCACAGTGTACAT-3′ and 5′-CGCCAGACCTCCAGCAACACCGTC-3′, and cloned into pGEM-T Easy (Promega). A strand-specific, biotin-labelled UL40 RNA probe was generated from this plasmid by using a DIG northern starter kit (Roche). The probe was hybridized to the blot and rendered light-emitting by using the same kit, and detected by exposure to X-ray film.
Mapping of transcript ends
Rapid amplification of cDNA ends (RACE) was carried out by using a SMARTer RACE kit (Clontech). Briefly, two UL40-specific primers (5′-CAACGTGTTATCG GCAGGATGATG-3′ and 5′-CAGTACACGTCGAGCGTCATGAGG-3′) were employed separately with the kit UPM-A primer to generate 5′-RACE products by PCR, and a single UL40-specific primer (5′-CGCCAGACCTCCAGCAACACCG TC-3′) was used to generate 3′-RACE products. The products were purified by agarose gel electrophoresis and cloned into pGEM-T Easy (Promega). Several clones were generated for each product and sequenced using universal primers.
Cell surface biotinylation
Carbohydrates exposed on cell surface of live cells were oxidated with 3mM sodium periodate (Sigma), then labeled with 0.2mM aminooxybiotin (Biotium) in presence of aniline (Sigma) catalyst. Cells were harvested from the dish, extracted in lysis buffer (10mM Tris pH8, 1% TritonX100, 150mM NaCl), biotinylated proteins were purified using Streptavidin agarose (Thermo Scientific) and analyzed by immunoblot.
Results
Induction of HLA-E cell surface expression by SPUL40
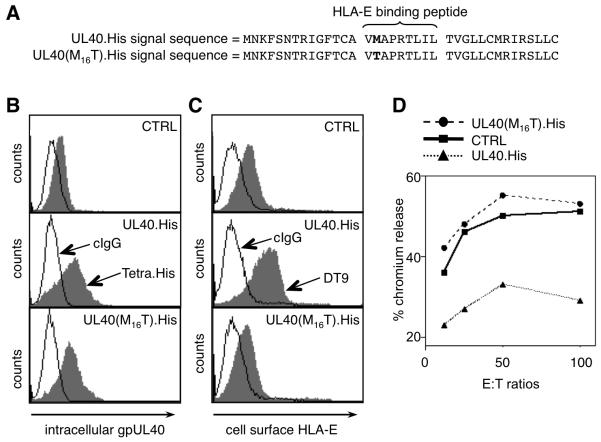
UL40-mediated evasion of NK cell recognition is thought to be induced by the conserved HLA-E-binding peptide (VMAPRTLIL in strain AD169) being excised from SPUL40 and delivered into the ER by a TAP-independent mechanism (15). To interrogate this model experimentally, we initially investigated whether the SPUL40-derived HLA-E-binding peptide fulfills the same criteria as epitopes derived from classical MHC-I signal peptides. A natural sequence variant present in two thirds of all HLA-B alleles has a threonine at the anchor position P2 (VTAPRTLIL); this variant neither binds HLA-E*0101 nor upregulates its surface expression (16, 42). UL40 was therefore modified to insert a threonine at P2 (M16T with respect to the UL40 ORF; Fig. 1A). The amino acid substitution did not affect gpUL40 expression levels (Fig. 1B), but it ablated the capacity of UL40 both to upregulate cell surface expression of HLA-E in HFFF and to elicit protection against NK cell lysis (Fig. 1C and D). The result is consistent with the UL40-derived epitope binding HLA-E directly to promote its maturation and thereby avoid NK cell activation.
Figure 1. Consensus VMAPRTLIL sequence implicated in UL40 function.
RAd-UL40.His has a wild type HLA-E-binding epitope (VMAPRTLIL), whereas RAd-UL40(M16T).His has a mutated form (VTAPRTLIL). RAd-CTRL is the same vector without a transgene. (A) The signal peptide present in each construct showing the M/T mutation in bold. (B) Intracellular expression of gpUL40 and (C) cell surface expression of HLA-E*0101 in HFFF infected with RAd-CTRL, RAd-UL40.His and RAd-UL40(M16T).His measured using antibodies to the C-terminal epitope tag and to HLA-E/-C (DT9), respectively. (D) An NK cell cytolysis assay was performed using NKL cells incubated with HFFF cells infected with the RAds as indicated. Results are representative of three independent experiments.
Several regions of the HCMV genome exhibit an extraordinary degree of sequence variation (33) Polymorphisms extend into UL40 to include the consensus HLA-E-binding peptide (15, 24, 43, 44). A substitution at P8 in strain Toledo (VMAPRTLVL) mimics a naturally occurring sequence variant found in HLA-A2 alleles, but nonetheless the VMAPRTLVL peptide has been shown experimentally to bind to HLA-E (17, 19). In the context of UL40, this substitution also induced cell surface expression of HLA-E during HCMV strain Toledo productive infection and when strain Toledo UL40 was expressed in isolation using an adenovirus vector (Fig. 2) (15). The clinical isolate strain 3157 (GenBank accession GQ221974) was of particular interest because it has a base substitution in the translation initiation codon. This is predicted to lead to initiation of translation at methionine P2, which is actually located within the HLA-E-binding peptide. Both HCMV strain 3157 and an Ad vector encoding strain 3157 UL40 (RAd3157 UL40) lacked the capacity to upregulate HLA-E (Fig. 2). RAd3157 was still capable of efficient gpUL40 expression (data not shown), and thus the truncated signal peptide retains its function in ER targeting and translocation. The DNA sequence encoding SPUL40 in the original clinical sample matched that of the sequenced strain 3157 passage 3 virus (result not shown, (34)). Consequently, strain 3157 is the first natural HCMV isolate shown to have a defect in an NK cell evasion function.
Figure 2. Modulation of HLA-E expression by natural UL40 sequence variants.
Cell surface expression of HLA-E as measured by the mAb DT9 following (A) infection of HFFF with HCMV strains AD169, Toledo and 3157 (72 h post infection). Downregulation of MHC-I (mAb W6/32) was used as a marker of HCMV infection. (B) Infection of HFFF with RAds expressing UL40 from different strains. The sequences of SPUL40 from the different virus strains are shown.
SPP-mediated processing of the non-canonical substrate SPUL40
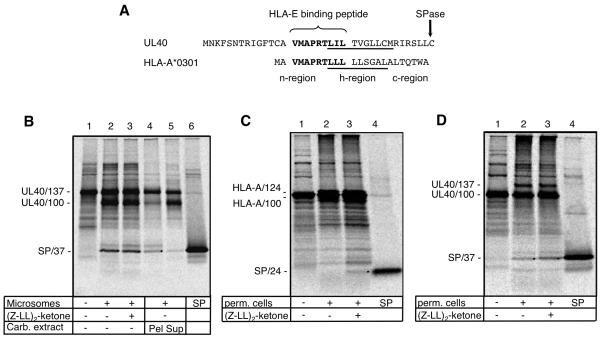
To analyze the molecular events leading to generation of the HLA-E-binding epitopes, the processing of HLA and UL40 signal peptides was examined in two distinct in vitro translocation assays using either purified canine ER membranes (rough microsomes), or digitonin-permeabilized 721.221 cells. To this end, radiolabeled polypeptides were synthesized in reticulocyte lysates in the presence of these different ER membranes, then recovered by high salt extraction and centrifugation. Immediately after cleavage by SPase, SPs are normally anchored in the membrane with the N-terminus exposed to the cytosol and the C-terminus facing the ER lumen (20). For both HLA and UL40, the HLA-E-binding epitopes are predicted to be located at the junction of the cytosolic n-regions and the membrane spanning h-region (Fig. 3A) (39). In view of their differential requirement for TAP, the HLA-E-binding peptide donated by HLA and UL40 must reach the ER by distinct routes. A difference in the processing of SPUL40 and SPHLA was observed previously using ER-derived rough microsomes. Although SPHLA was rapidly cleaved by SPP, SPUL40 was not processed further, irrespective of whether the SPP inhibitor (Z-LL)2-ketone was added (20) (28). Indeed, carbonate extraction showed SPUL40 remained anchored in the membrane like a bona fide membrane protein (Fig. 3B). However, to generate a product capable of binding HLA-E, SPUL40 must presumably be processed further. The fate of membrane anchored SPUL40 was therefore investigated using a more physiological translocation assay performed in digitonin-permeabilized 721.221 cells (38). As observed previously using isolated rough microsomes, SPHLA was rapidly processed and could only be recovered in the membrane fraction when (Z-LL)2-ketone was added (Fig. 3C). In contrast to this efficient clearance from the membrane, significant amounts of SPUL40 were detected in the isolated membrane fraction of digitonin-permeabilized 721.221 cells, consistent with it having a prolonged close association with the ER membrane (Fig. 3D). A striking and reproducible difference to the result in isolated rough microsomes was that (Z-LL)2-ketone stabilized SPUL40. This result implies that in 721.221 cells an SPP-type protease activity is able to cleave more stable SPs such as SPUL40. While SPP is a likely candidate for this ‘SPP-type’ activity, this needs to be formally investigated.
Figure 3. Processing of HLA-A*0301 and UL40 signal peptides.
(A) Alignment of the UL40 and HLA-A*0301 SPs illustrating the SPase cleavage sites plus the n, h (hydrophobic; underlined) and c sequences in HLA-A*0301 (B) In vitro translation of mRNA coding for the signal peptide plus 100 aa of UL40 (UL40/137; lane 1) in the presence of ER-derived microsomes (lanes 2-5) and (Z-LL)2-ketone (lane 3). One aliquot of microsomes was extracted with sodium carbonate and separated into pellet (Pel, lane 4) and supernatant (Sup, lane 5). Dots indicate the SPs; lane 6 shows in vitro-translated UL40 SP. (C) and (D) In vitro translation of mRNA coding for the signal peptide plus 100 aa of HLA-A*0301 (HLA-A/124; lane 1) or UL40 (UL40/137; lane 1) in the presence of selectively permeabilized 721.221 cells (lanes 2-3) and (Z-LL)2-ketone (lane 3). Dots indicate the SPs; lane 4 shows in vitro-translated reference SP.
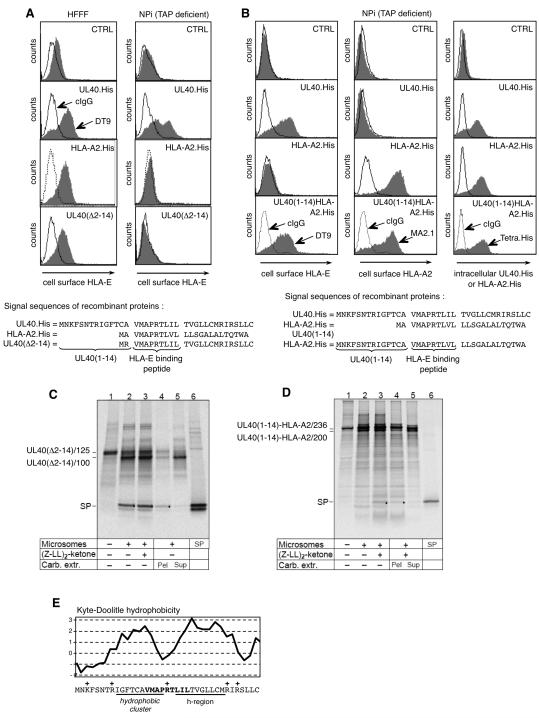
The n-region of SPUL40 mediates TAP-independent HLA-E loading
The delay in SPP-catalyzed turnover leads to prolonged membrane attachment of SPUL40. We sought to test whether this property of SPUL40 was instrumental in facilitating TAP-independent cell surface expression of HLA-E. A series of UL40/HLA-A chimera and mutant constructs were cloned into an Ad vector and analyzed in both normal (HFFF) and TAP-deficient (NPi) fibroblasts. HLA-A2 induced TAP-dependent up-regulation of HLA-E cell surface expression, whereas UL40 induced expression in both normal and TAP-deficient cells (Fig. 4A). SPUL40 was modified to encode the two N-terminal residues (MR) present in HLA-C in place of the 14 residues of the UL40 n-region (Fig. 4A). To retain the charge distribution of the UL40 signal peptide, which is predicted to be important for correct membrane insertion of the nascent peptide chain (45), arginine at P2 (as in HLA-C) was preferred to alanine (as in HLA-A2). In contrast to UL40, UL40(Δ2-14) upregulated cell surface expression of HLA-E in a TAP-dependent manner (Fig. 4A). Equal amounts of SPUL40(Δ2-14) were observed in canine microsomes irrespective of whether SPP-inhibitor was added or not (Fig. 4C), and prolonged membrane attachment was also observed in digitonin-permeabilized 721.221 cells (result not shown). Consequently, the sequence responsible for delayed SPP-catalyzed cleavage maps to the c-region, and the c-region is not responsible for TAP-independent processing of SPUL40. In order to test whether the n-region of SPUL40 was able by itself to mediate TAP-independent HLA-E loading, the two N-terminal residues (MA) of SPHLA-A2 were replaced with the 14 N-terminal residues from SPUL40 (RAd-UL40(1-14)HLA-A2.His). Similarly to RAd-HLA-A2.His, HLA-A2 from RAd-UL40(1-14)HLA-A2.His was expressed on the surface of both normal (data not shown) and TAP-deficient cells (Fig. 4B), and consistent with HLA-A2 binding to hydrophobic peptides as has been previously reported (46). In contrast, TAP-deficient cells infected with RAd-UL40(1-14)HLA-A2.His, but not RAd-HLA-A2.His, exhibited elevated levels of HLA-E on the surface. In vitro translocation assays revealed that SPUL40(1-14)HLA-A2.His was an efficient substrate for SPP in rough microsomes (Fig. 4D) and digitonin-permeabilized 721.221 cells (data not shown). Taken together, these results show that TAP-independent presentation is determined by the ‘n-region’, irrespective of how efficiently SPUL40 is cleaved by SPP. The n-region of common signal peptides contains only polar and charged residues, which are followed by a stretch of 5-15 hydrophobic residues of the h-region (47). A striking feature of the SPUL40 n-region is a short hydrophobic cluster (Fig. 4E), a property of UL40 that is distinctive and provides insight into the potential mechanism. The HLA-E-binding peptide thus effectively lies across a charged valley flanked by two hydrophobic elements (Fig. 4E).
Figure 4. The SPUL40 ‘n-region’ mediates TAP-independent HLA-E cell surface expression.
(A) HFFF (TAP+) or NPi (TAP−) cells infected with RAd-CTRL, RAd-UL40.His, RAd-HLA-A2.His or RAd-UL40(Δ2-14). Cell surface expression of HLA-E was assessed using the mAb DT9. (B) NPi cells were infected with RAd-CTRL, RAd-UL40.His, RAd-HLA-A2.His or RAd-UL40(1-14)HLA-A2.His and analyzed for cell surface expression of HLA-E, HLA-A2 or intracellular expression of His-tagged proteins using the indicated mAbs. The signal peptide structure of each construct is shown. Results are representative of three independent experiments. In vitro translation of mRNA coding for (C) the signal peptide plus 100 aa of UL40(Δ2-14) (UL40(Δ2-14)/125; lane 1) or (D) the signal peptide plus 200 aa of UL40(1-14)HLA-A2 (UL40(1-14)HLA-A2/236, lane 1) in the presence of ER-derived microsomes (lanes 2-5) and (Z-LL)2-ketone as indicated. One aliquot of microsomes was extracted with sodium carbonate and separated into pellet (Pel, lane 4) and supernatant (Sup, lane 5). Dots indicate the SPs; lanes 6 show in vitro-translated SPs. (E) Hydrophobicity plot of the SPUL40 (strain AD169) illustrating in addition to the canonical h-region, which is the prime determinant for ER targeting and insertion of the nascent chain, an additional hydrophobic cluster within the n-region. The HLA-E/gpUL18-binding peptide is indicated in bold; position of positive charged amino acid residues with a +.
UL40 enhances cell surface expression of UL18 and HLA-E
During the characterization of an HCMV UL40 deletion mutant (AD169ΔUL40), flow cytometry experiments consistently revealed higher cell surface expression of the HCMV MHC-I homologue gpUL18 in the presence of UL40 (Fig. 5A). This capacity of UL40 to enhance expression of a glycoprotein was restricted to gpUL18; there were no comparable consistent changes to HCMV gB, gH or gN expression (Fig. 5A). During HCMV productive infection, cell surface expression of gpUL18 can be clearly detected by flow cytometry, but only when optimized high-sensitivity detection systems are used. The specificity of the gpUL18 signal was therefore validated by including an HCMV UL18 deletion mutant (AD169ΔUL18) in the experiment; gpUL18 was both detectable on the cell surface and enhanced by UL40 (Fig. 5B). More robust gpUL18 expression can be achieved by using Ad vectors (48), thus this technology was adopted to investigate the mechanism by which UL40 promotes gpUL18 cell surface expression. UL40 reliably upregulated gpUL18 surface expression to levels proportionate to the amount of RAd-UL18 introduced into the assay (Fig. 5C and D). Moreover, expression of HLA-E on the surface of HCMV infected human fibroblast was clearly detected with an HLA-E-specific mAb and shown to be dependent on UL40 but independent of UL18 (Fig 5E). The downregulation of HLA-E observed with AD169ΔUL40, relative to mock-infected control, was presumably due to the effects of US6. Likewise, the more efficient downregulation of total HLA-I observed with AD169ΔUL40 may reflect the contribution HLA-E makes to the total HLA-I signal. UL16 does not impact on UL18 or HLA-E expression (not shown). AD169ΔUL16 was used as an alternative to the parental strain AD169 because it also retains expression of a selectable marker (LacZ) used in the construction of AD169ΔUL18 (31, 32), and a potential cause of a background on western blots (48). Western blot analyses also demonstrated gpUL18 expression was upregulated by co-expression of UL40 both in the context of a productive HCMV infection and when the genes were expressed in isolation by an adenovirus vector (Fig. 5F and G). Most significantly, the proportion of the fully mature (Endo H-resistant form) of gpUL18 (>100 kDa glycoform) was enhanced in the presence of UL40 relative to the 67 kDa Endo H-sensitive form (Fig. 5F) both in the context of a productive HCMV infection and when expressed by using an adenovirus vector (Fig. 5G) UL40 thus promotes the maturation of gpUL18 to its fully glycosylated form. CD155 is included as a positive infection control as a cellular protein whose expression is activated by strain AD169. Previously, biotinylation studies had indicated that the EndoH-sensitive form of gpUL18 may be expressed on the cell surface (48, 49). However, this finding was counterintuitive as cell surface proteins normally acquire resistance during transit through the Golgi apparatus. We therefore performed a surface labeling experiment in HCMV-infected cells utilizing an improved technique with reduced potential for cell permeabilization. We now show that only the fully glycosylated form of gpUL18 reaches the surface of HCMV infected cells. This finding is important as it eliminates the possibility that the EndoH-sensitive form could provide a differential stimulus to NK cells during productive HCMV infection. Most significantly, UL40 was clearly shown to promote surface expression of both gpUL18 and HLA-E (Fig. 5H).
Figure 5. UL40 promotes cell surface expression of gpUL18 and HLA-E.
(A) Cell surface expression of gpUL18, MHC-I, gB, gH and gN compared on HFFF cells infected with HCMV strain AD169 or AD169ΔUL40 for 72h (30 PFU/cell). (B) Cell surface expression of gpUL18 on cells infected with HCMV strain AD169, AD169ΔUL40, AD169ΔUL18 (30 PFU/cell) for 72h. (C) gpUL18 surface expression on cells co-infected with RAd-UL18 (100 PFU/cell) and either RAd-UL40 or the vector control (RAd-CTRL) (30 PFU/cell) for 72h. (D) HFFF infected with RAd-UL18 at the multiplicities of infection (m.o.i) indicated and RAd-UL40 or RAd-CTRL (30 PFU/cell) for 72h. Results are means ± SEM of duplicate samples (Two-Way ANOVA test with Bonferroni post-tests). (E) Cell surface expression of HLA-E (antibody 3D12) and MHC-I on HFFF infected with strain AD169ΔUL16, AD169ΔUL18 or AD169ΔUL40 (15 PFU/cell) for 96h (F) Western blot of HFFF infected with AD169ΔUL16, AD169ΔUL18 or AD169ΔUL40 (15 PFU/cell) for 96h. (G) gpUL18 detected by western blot in HFFF infected with RAd-UL18 (100 PFU/cell) and either RAd-UL40 or RAd-CTRL (30 PFU/cell) for 72h. Cell extracts were digested with Endo H [E] or PNGase F [P] or no enzyme [Mock; M]. Endo H-resistant gpUL18 glycoform ( ), Endo H-sensitive gpUL18 glycoform before (
), Endo H-sensitive gpUL18 glycoform before ( ) and after (
) and after ( ) digestion, PNGase-digested gpUL18 forms (
) digestion, PNGase-digested gpUL18 forms ( ) are indicated. (H) Following cell surface biotinylation and immunoprecipitation, gpUL18, HLA-E, HLA-HC and CD155 cell surface expression were compared on western blot in cells infected with AD169ΔUL16, AD169ΔUL18 or AD169ΔUL40 (15 PFU/cell) for 96h.
) are indicated. (H) Following cell surface biotinylation and immunoprecipitation, gpUL18, HLA-E, HLA-HC and CD155 cell surface expression were compared on western blot in cells infected with AD169ΔUL16, AD169ΔUL18 or AD169ΔUL40 (15 PFU/cell) for 96h.
An analysis of a panel of UL40 deletion mutants demonstrated that the gpUL18-enhancing function mapped specifically to SPUL40 (Fig. 6 and data not shown). Cell surface expression of gpUL18 could be upregulated by using a construct comprising SPUL40 fused upstream of the red fluorescent protein (RFP), but not by the HLA-A2 signal peptide fused to RFP (HLA-A2(1-28)RFP) (Fig. 6A and B). In addition, replacing SPUL40 with the signal peptide of HCMV UL141 did not stimulate gpUL18 maturation (data not shown). To define more precisely the sequence element responsible for modulating UL18, we analyzed chimeric and/or truncated versions of SPUL40. When the C-terminal element in SPUL40 (aa 24-37) was replaced with an equivalent domain of HLA-A2 (TVGLLCMRIRSLLC to LLSGALALTQTWA), or the 14 residues of the SPUL40 n-region was substituted with the dipeptide MR, then the ability to upregulate surface gpUL18 was retained (Fig. 6C and D respectively). However, substitution of the HLA-E-binding epitope with either an HIV-1 GAG or MelanA epitope destroyed the capacity to upregulate cell surface expression of either gpUL18 or HLA-E (Fig. 6E and data not shown). This study thus implicates the VMAPRTLIL sequence within SPUL40 in modulating cell surface expression of both HLA-E and UL18.
Figure 6. SPUL40 signal enhances cell surface expression of gpUL18.
Cell surface expression of gpUL18 was monitored by flow cytometry on HFFF cells infected for 72 h with the RAds indicated. RAd-UL18 (100 PFU/cell) plus RAd-CTRL (30 PFU/cell) and RAd-UL18 plus RAd-UL40 (30 PFU/cell) were used as standards in all panels. RAd-UL18 plus a second RAd (30 PFU/cell) encoding: (A) SPUL40 cloned in-frame with RFP, (B) SPHLA-A2 in-frame with RFP, (C) UL40 with residues 24-37 replaced with residues 12-24 from HLA-A2, (D) UL40 with residues 1-14 replaced with first 2 residues from HLA-C, (E) UL40 with HLA-E-binding peptide replaced with an HIV-1 GAG epitope, (F) strain Toledo UL40, (G) UL40 with the M16T mutation, or (H) strain 3157 UL40. Results are representative of three independent experiments.
Differential regulation of HLA-E and gpUL18
Single amino acid sequence differences in SPUL40 present naturally in HCMV clinical strains have the capacity to impact on HLA-E and/or gpUL18 cell surface expression. Significantly, while the VMAPRTLVL peptide variant (I/V at P8) in strain Toledo induced both HLA-E and gpUL18 expression (Fig. 2 and 6F respectively), two other UL40 constructs (one artificial and one natural) could stimulate expression of gpUL18 but not HLA-E. This property was shared by the designed mutant RAd-UL40(M16T) (Fig. 1C and 6G) and the natural variant encoded by strain 3157 (Fig. 2 and 6H). As discussed above, strain 3157 is unusual in being predicted to initiate UL40 translation at P2, and thus a peptide derived from it must lack the valine at P1, as defined in the HLA-E-binding peptide. This implies that the 3157-derived octamer (which also has a I/L substitution; MAPRTLLL) can bind and stabilize gpUL18 while leaving the first position in the gpUL18 peptide-binding groove vacant.
Fine mapping of the UL40 transcriptional start site was undertaken using the NCBI reference strain Merlin. The most abundant RNA species detected in early and late RNA by northern blotting using a UL40 probe was estimated at 600-1000 nt in size (Fig. 7A). This is consistent with transcripts containing the UL40 coding region (666 nt) flanked by relatively short 5′- and 3′-untranslated regions. RACE experiments demonstrated the existence of two 5′-ends for UL40 mRNAs located approximately 110 bp apart, each lacking a TATA box (Fig. 7B and C). Both mRNAs were shown to terminate close downstream from a consensus AATAAA polyadenylation signal, and thus have measured sizes of approximately 800 and 700 nt (not including the polyA tail). The longer mRNA would be translated from the first methionine codon in the UL40 coding region to generate the 37-residue full-length SP capable of upregulating both HLA-E and gpUL18 (Fig. 7C). The shorter mRNA, which is relatively less abundant in late RNA, would be translated from the second methionine codon (P2 in the consensus HLA-E-binding peptide) to generate a 22-residue SP (Fig. 7C). This peptide is equivalent to that produced by strain 3157 (except for the I/L substitution described above), and is predicted to be capable of upregulating gpUL18 only. During productive infection, HCMV would thus be predicted to express UL40 gene products with two distinct SPs: a 37-residue full length version capable of upregulating both HLA-E and gpUL18, and a second encoding a 22-residue truncated version that is predicted to be capable of upregulating gpUL18 only.
Figure 7. UL40 transcriptional analysis.
(A) Northern blot of a strand-specific UL40 probe hybridized to strain Merlin early (E) and late (L) RNAs. The size range (in nt) of the UL40 mRNAs is indicated on the right. Larger RNAs that are 3′-coterminal with UL40 were also detected: major species at approximately 1400 nt (UL41A) and 4500 nt (UL44), plus less abundant species. (B) Agarose gel of RACE products generated from the 5′- and 3′-ends of strain Merlin E and L RNAs using UL40-specific primers. Approximate sizes of the UL40 products from 3′-RACE (560 bp) and 5′-RACE (420 and 310 bp, each with an artefactual, more slowly migrating band above) are shown on the left. DNA markers (M) are shown on the right (sizes in bp). The larger products at approximately 900 bp in the 5′-L lane represent the 5′-end of an upstream, 3′-coterminal gene (UL41A), which maps to residue 54804 in the Merlin sequence, downstream from a potential TATA box (TATATT). (C) Locations of the 5′- and 3′-ends of UL40 mRNAs aligned with the relevant DNA and amino acid sequences (predicted SPs). Dots indicate the central portion of the UL40 coding region, which is not shown. The major 5′-ends are in bold, underlined type and marked with broad arrows. The upstream and downstream occurrences were identified from 21/25 and 23/27 clones, respectively. The ATG codons that initiate the long and short forms of the UL40 protein are in bold, underlined type; an upstream ATG in another reading frame is also underlined. The major 3′-ends are in bold, underlined type and marked by narrow arrows. Their occurrences, as read from left to right, were identified from 3, 7 and 4 clones (from a total of 18 clones), and are located downstream from the bold, underlined canonical polyadenylation signal (AATAAA).
Discussion
The upregulation of HLA-E cell surface expression by UL40 involves a conserved nonameric peptide being excised from SPUL40 and delivered to HLA-E in a TAP-independent manner (15, 23, 24). This study sought both to test the validity of this model and gain insight into the molecular processes involved. Using a series of UL40 mutants and chimeric molecules, the function responsible for upregulating HLA-E was formally mapped to the VMAPRTLIL sequence in SPUL40. This result is consistent with a recent study that showed the VMAPRTLIL peptide could be eluted from cells co-transfected with HLA-E and UL40 (50) and our finding that mutation of the key methionine anchor residue at P2 (to VTAPRTLIL) abrogated the capacity of UL40 to both upregulate HLA-E and protect against NK cell recognition. Previously, our investigations into the processing of SPUL40 in purified ER-derived rough microsomes found the peptide to be resistant to SPP (28). This result was recapitulated here, yet SPUL40 was clearly cleaved by an SPP-type protease when crude cellular membranes (digitonin-permeabilized 721.221 cells) were used. The difference between the systems implies that the processing of more stable SPP substrates may be facilitated by accessory factors, that may be depleted, or inactivated, in the isolated microsomal fraction; e.g. components of the ER-associated degradation pathway. SPP has recently been shown to form three distinct oligomeric complexes in the ER membrane, each exhibiting different binding specificity for SPs and misfolded membrane proteins (51). It will be important to delineate the role played by these distinct SPP complexes in processing SPUL40 and the process by which the peptide is trimmed to enable the epitope to be loaded onto either HLA-E or gpUL18.
UL40 upregulates HLA-E independently of TAP, and this function was mapped to 14-residues in the n-region of SPUL40 (MNKFSNTRIGFTCA). TAP-independent presentation has been reported for at least two signal peptide-derived epitopes (39, 46, 52): For IP-30 the peptide epitope is located in the c-region, which upon SPP-catalyzed processing is released directly into the ER-lumen. For calreticulin, the epitope comprising the N-terminal signal peptide fragment is small and hydrophobic probably allowing it to transit the ER membrane unassisted by TAP. For SPUL40, the signal peptide fragment is unlikely to be able to transit the membrane unassisted as it has an extended size and carries a positive net charge (Fig. 4C and Fig. 8). However, how the UL40 nascent chain inserts into the translocation channel and how the hydrophobic cluster in the n-region associates with the membrane is uncertain. In analogy to bona-fide signal peptides, we predict that the nascent polypeptide initially enters the ER membrane with the n-region extruding into the cytosol. SPase-catalyzed cleavage after the SPUL40 c-region is likely to take place only after the h-region is in the plane of the membrane. To deliver the HLA-E binding epitope to the ER lumen, we predict SPUL40 must undergo the topological rearrangement depicted in Fig. 8. A comparable sequential topogenesis and inversion of a signal peptide with two hydrophobic clusters binding a positive charged amino acid had been reported for the arenaviral glycoprotein C precursor (for review see (53)). In the final stages of processing, SPP is postulated to cleave N-terminal residues of the HLA-E epitope while SPase may remove residual C-terminal residues.
Figure 8. Postulated sequential topogenesis and processing of the SP UL40.
Upon targeting and translocation, the gpUL40 precursor (pre-gpUL40) is cleaved by SPase. The liberated SP (SPUL40) remains anchored in the membrane. Proposed association of the hydrophobic N-terminal segment (yellow box labeled *) with the membrane, and subsequent flipping of the hydrophilic spacer (green) and the c-region (blue) are indicated. The canonical n- and h-region are indicted in yellow (h) and red (n), respectively. In semi-intact cells, SPUL40 is further processed by an SPP-type intramembrane protease. The consensus HLA-E-binding peptide is delivered to the ER in a TAP-independent manner, where it binds and stabilizes both endogenous HLA-E and the HCMV-encoded MHC-I homologue gpUL18.
HLA-E is nearly monomorphic, yet flexibility exists with respect to the pool of peptides it can bind (15, 16, 19). Indeed, HIV and hepatitis C virus encode HLA-E-binding peptides that exhibit minimal sequence identity with the UL40-encoded epitope (54, 55). Although the VMAPRTLIL peptide is conserved in most HCMV clinical isolates, some exhibit sequence variation at this locus (15, 27, 44). The strength of the interaction with CD94/NKG2A is influenced by the sequence of peptides bound to HLA-E (40, 56, 57). Although the single amino acid polymorphism detected in strain Toledo (I/V at P8) sustains NK cell evasion (15, 44), it reduces the affinity of the interaction with CD94/NKG2A (58). Importantly, a single amino acid polymorphism within the UL40-encoded HLA-E-binding peptide can lead to the induction of robust HLA-E-restricted CD8+ T cell responses. Individuals whose HLA genotype is such that they do not encode an HLA-E-binding peptide that matches that of the infecting HCMV strain, may produce HLA-E-restricted T cells. Such ‘NK-CTL’ are particularly sensitive to the amino acid at P8 in the nonamer peptide, and are capable of differentiating the single amino acid substitution at that position, and thus between strains AD169 and Toledo (27). In the presence of an HLA-E specific T cell response, selective pressure may be exerted to mutate SPUL40 to adapt to the HLA genotype of its host. Strain 3157 is remarkable in this context as it is disabled in its capacity to upregulate HLA-E. Interestingly, a substitution (M/V) in the P2 anchor residue of the HLA-E-binding epitope has recently been reported for strain Toledo SPUL40 (59). However, since this is a high-passage strain it is possible this mutation arose in vitro. Strain 3157 is a low-passage clinical isolate cultured from the urine of a congenitally-infected neonate, and the first natural isolate to be reported with a defect in an NK cell evasion function.
Our study adds yet another dimension to the role of the short SPUL40 in modulating the immune response during HCMV infection. UL40 stimulates cell surface expression of gpUL18 both in the context of HCMV productive infection and when the genes are expressed in isolation. This function also mapped within the VMAPRTLIL peptide in SPUL40, thus the same peptide appears capable of promoting both HLA-E and gpUL18 cell surface expression (Fig. 8). The capacity of gpUL18 to bind peptide is well-established, and indeed required to provide thermal stability to the protein (11, 60). Although our evidence that VMAPRTLIL binds gpUL18 directly is circumstantial, the model is encouraged by a prediction that the VMAPRTLIL peptide fitted with the UL18 consensus motif (7). Moreover, in an independent study Reyburn and co-workers have also eluted the endogenous VMAPRTLIL peptide from a soluble form of gpUL18 expressed in isolation by DNA transfection (H. Reyburn, personal communication). Interestingly, during productive HCMV infection UL18 interferes with the ability of US6 to inhibit TAP (61), and thus peptide-loading on UL18 does not need to involve a TAP-independent mechanism. Additional putative UL18 binding peptides have been proposed, and it is likely that the UL18 peptide-binding domain can accommodate additional viral and cellular epitopes. Nevertheless, the fact the phenotype was identified in the context of a productive infection with an HCMV UL40 mutant implies that the UL40-derived peptide is a chief source. Interestingly, co-expression of the HLA-A2 signal peptide did not stimulate UL18 surface expression even though it contains a suitable ‘HLA-E-binding peptide’ (Fig. 4B). Thus, it is possible that the context in which the epitope is presented is important and therefore host cell epitopes may not be able to upregulate UL18 cell surface expression.
Although the pools of peptides capable of stabilizing HLA-E and UL18 overlap, differential regulation of HLA-E and UL18 can be achieved by UL40 sequence variants. Strain 3157 UL40 initiates translation at amino acid 16 (relative to the prototype sequence, or P2 in the HLA-E-binding peptide), and thereby lacks the capacity to upregulate cell surface HLA-E while retaining the capacity to stabilize gpUL18 expression (Fig. 2 and 6H). This result also implies that the truncated UL40 signal peptide can bind and stabilize UL18 expression while leaving position 1 in the UL18-peptide binding groove vacant. Although unconventional, this situation has been described for epitopes that bind classical MHC-I molecules (62, 63). In certain individuals, the selection pressure exerted by UL40-specific HLA-E restricted T cells may be biased against the UL40-specific NK cell evasion function. In this context, it is important to note that we observed in strain Merlin UL40 that transcription was initiated efficiently at two distinct sites. Selection of the downstream site results in expression of a protein initiating at amino acid 16, producing a product capable of selectively upregulating gpUL18. Differential transcription from the UL40 promoter has an in-built capacity to produce products capable of upregulating both HLA-E and gpUL18, or gpUL18 alone.
Although the function of the glycoprotein encoded by HCMV UL40 remains unknown, multiple functions have now been assigned to its signal peptide. The VMAPRTLIL peptide is responsible for upregulating cell surface expression of HLA-E and gpUL18 to inhibit recognition by CD94/NKG2A+ and LIR-1+ NK cells, respectively, yet HLA-E can also be recognized by the NK cell activating receptor CD94/NKG2C and the peptide may play a critical role in the expansion of CD94/NKG2C+ NK cells in HCMV seropositive individuals. Moreover, gpUL18 can also stimulate an as yet undefined LIR-1− NK cell subset. In this context, it is interesting to note that CD94/NKG2C can bind efficiently to gpUL18 in vitro (64). It will be interesting to investigate whether this interaction is functional and whether the UL40 signal peptide plays a role. The capacity for SPUL40 to promote NK cell recognition may develop over time during persistent infection, potentially akin to an adaptive response. Whether the virus adapts by mutation to evade HLA-E-restricted T cell responses, as may have happened for strain 3157, needs to be evaluated. A version of UL40 capable of upregulating HLA-E and UL18 is retained in most virus strains, and thus this function/sequence must be selected to promote virus transmission and/or persistence. The immunogenetic diversity that exists within human populations presents special challenges for HCMV, a virus that appears to have evolved a complex strategy directed at suppressing NK cell activation (3). The high level of sequence variation that exists amongst HCMV clinical isolates may, in part, reflect adaptation to immunological pressures in vivo.
Acknowledgements
We are grateful to Siân Llewellyn-Lacey for kindly providing technical support and to Amgen for providing an anti-UL18 mAb (M71). The authors thank Hugh Reyburn for kindly allowing reference to unpublished results.
This research was supported was supported by grant funding from the Wellcome Trust (WT090323MA), UK Medical Research Council (G1000236) and the BBSRC (BBF0098361).
References
- 1.McSharry BP, Burgert HG, Owen DP, Stanton RJ, Prod’homme V, Sester M, Koebernick K, Groh V, Spies T, Cox S, Little AM, Wang EC, Tomasec P, Wilkinson GW. Adenovirus E3/19K promotes evasion of NK cell recognition by intracellular sequestration of the NKG2D ligands major histocompatibility complex class I chain-related proteins A and B. J Virol. 2008;82:4585–4594. doi: 10.1128/JVI.02251-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mocarski ES., Jr. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 2002;10:332–339. doi: 10.1016/s0966-842x(02)02393-4. [DOI] [PubMed] [Google Scholar]
- 3.Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod’homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, Wang EC, Griffin CA, Davison AJ. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, Axthelm MK, Nelson JA, Jarvis MA, Picker LJ, Fruh K. Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science. 2010;328:102–106. doi: 10.1126/science.1185350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 6.Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913–919. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- 7.Yang Z, Bjorkman PJ. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne H, Smith G, Beck S, Minson T. A complex between the MHC class I homologue encoded by human cytomegalovirus and beta 2 microglobulin. Nature. 1990;347:770–772. doi: 10.1038/347770a0. [DOI] [PubMed] [Google Scholar]
- 9.Fahnestock ML, Johnson JL, Feldman RM, Neveu JM, Lane WS, Bjorkman PJ. The MHC class I homolog encoded by human cytomegalovirus binds endogenous peptides. Immunity. 1995;3:583–590. doi: 10.1016/1074-7613(95)90129-9. [DOI] [PubMed] [Google Scholar]
- 10.Wagner CS, Ljunggren HG, Achour A. Immune modulation by the human cytomegalovirus-encoded molecule UL18, a mystery yet to be solved. J Immunol. 2008;180:19–24. doi: 10.4049/jimmunol.180.1.19. [DOI] [PubMed] [Google Scholar]
- 11.Wagner CS, Rolle A, Cosman D, Ljunggren HG, Berndt KD, Achour A. Structural elements underlying the high binding affinity of human cytomegalovirus UL18 to leukocyte immunoglobulin-like receptor-1. J Mol Biol. 2007;373:695–705. doi: 10.1016/j.jmb.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu ML. A novel immunoglobulin superfamily receptor for cellular and viral MHC class I molecules. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 13.Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–613. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 14.Prod’homme V, Griffin C, Aicheler RJ, Wang EC, McSharry BP, Rickards CR, Stanton RJ, Borysiewicz LK, Lopez-Botet M, Wilkinson GW, Tomasec P. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1-NK cells. J Immunol. 2007;178:4473–4481. doi: 10.4049/jimmunol.178.7.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S, Cerundolo V, Borysiewicz LK, McMichael AJ, Wilkinson GW. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 16.Braud VM, Allan DS, O’Callaghan CA, Soderstrom K, D’Andrea A, Ogg GS, Lazetic S, Young NT, Bell JI, Phillips JH, Lanier LL, McMichael AJ. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 17.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee N, Goodlett DR, Ishitani A, Marquardt H, Geraghty DE. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160:4951–4960. [PubMed] [Google Scholar]
- 19.Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- 20.Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol. 2001;167:6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 21.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–2218. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 22.Braud VM, Allan DS, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1998;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
- 23.Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M, Weiss EH. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol. 2000;164:5019–5022. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- 24.Wang EC, McSharry B, Retiere C, Tomasec P, Williams S, Borysiewicz LK, Braud VM, Wilkinson GW. UL40-mediated NK evasion during productive infection with human cytomegalovirus. Proc Natl Acad Sci U S A. 2002;99:7570–7575. doi: 10.1073/pnas.112680099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guma M, Angulo A, Vilches C, Gomez-Lozano N, Malats N, Lopez-Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 2004;104:3664–3671. doi: 10.1182/blood-2004-05-2058. [DOI] [PubMed] [Google Scholar]
- 26.Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, Lopez-Botet M. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood. 2006;107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 27.Pietra G, Romagnani C, Mazzarino P, Falco M, Millo E, Moretta A, Moretta L, Mingari MC. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc Natl Acad Sci U S A. 2003;100:10896–10901. doi: 10.1073/pnas.1834449100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10:735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson GW, Akrigg A. Constitutive and enhanced expression from the CMV major IE promoter in a defective adenovirus vector. Nucleic Acids Res. 1992;20:2233–2239. doi: 10.1093/nar/20.9.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stanton RJ, McSharry BP, Armstrong M, Tomasec P, Wilkinson GW. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function BioTechniques. 2008;45:659–668. doi: 10.2144/000112993. [DOI] [PubMed] [Google Scholar]
- 31.Kaye J, Browne H, Stoffel M, Minson T. The UL16 gene of human cytomegalovirus encodes a glycoprotein that is dispensable for growth in vitro. J Virol. 1992;66:6609–6615. doi: 10.1128/jvi.66.11.6609-6615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browne H, Churcher M, Minson T. Construction and characterization of a human cytomegalovirus mutant with the UL18 (class I homolog) gene deleted. J Virol. 1992;66:6784–6787. doi: 10.1128/jvi.66.11.6784-6787.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. Genetic content of wild-type human cytomegalovirus. J Gen Virol. 2004;85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 34.Cunningham C, Gatherer D, Hilfrich B, Baluchova K, Dargan DJ, Thomson M, Griffiths PD, Wilkinson GW, Schulz TF, Davison AJ. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J Gen Virol. 2010;91:605–615. doi: 10.1099/vir.0.015891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanton RJ, Baluchova K, Dargan DJ, Cunningham C, Sheehy O, Seirafian S, McSharry BP, Neale ML, Davies JA, Tomasec P, Davison AJ, Wilkinson GW. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Invest. 2010;120:3191–3208. doi: 10.1172/JCI42955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McSharry BP, Jones CJ, Skinner JW, Kipling D, Wilkinson GW. Human telomerase reverse transcriptase-immortalized MRC-5 and HCA2 human fibroblasts are fully permissive for human cytomegalovirus. J Gen Virol. 2001;82:855–863. doi: 10.1099/0022-1317-82-4-855. [DOI] [PubMed] [Google Scholar]
- 37.Moins-Teisserenc HT, Gadola SD, Cella M, Dunbar PR, Exley A, Blake N, Baykal C, Lambert J, Bigliardi P, Willemsen M, Jones M, Buechner S, Colonna M, Gross WL, Cerundolo V. Association of a syndrome resembling Wegener’s granulomatosis with low surface expression of HLA class-I molecules. Lancet. 1999;354:1598–1603. doi: 10.1016/s0140-6736(99)04206-3. [DOI] [PubMed] [Google Scholar]
- 38.Jadot M, Hofmann MW, Graf R, Quader H, Martoglio B. Protein insertion into the endoplasmic reticulum of permeabilized cells. FEBS Lett. 1995;371:145–148. doi: 10.1016/0014-5793(95)00881-9. [DOI] [PubMed] [Google Scholar]
- 39.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 40.Weihofen A, Lemberg MK, Ploegh HL, Bogyo M, Martoglio B. Release of signal peptide fragments into the cytosol requires cleavage in the transmembrane region by a protease activity that is specifically blocked by a novel cysteine protease inhibitor. J Biol Chem. 2000;275:30951–30956. doi: 10.1074/jbc.M005980200. [DOI] [PubMed] [Google Scholar]
- 41.Lemberg MK, Martoglio B. Analysis of polypeptides by sodium dodecyl sulfate-polyacrylamide gel electrophoresis alongside in vitro-generated reference peptides. Anal Biochem. 2003;319:327–331. doi: 10.1016/s0003-2697(03)00298-7. [DOI] [PubMed] [Google Scholar]
- 42.Strong RK, Holmes MA, Li P, Braun L, Lee N, Geraghty DE. HLA-E allelic variants. Correlating differential expression, peptide affinities, crystal structures, and thermal stabilities. J Biol Chem. 2003;278:5082–5090. doi: 10.1074/jbc.M208268200. [DOI] [PubMed] [Google Scholar]
- 43.Garrigue I, Faure-Della Corte M, Magnin N, Recordon-Pinson P, Couzi L, Lebrette ME, Schrive MH, Roncin L, Taupin JL, Dechanet-Merville J, Fleury H, Lafon ME. UL40 human cytomegalovirus variability evolution patterns over time in renal transplant recipients. Transplantation. 2008;86:826–835. doi: 10.1097/TP.0b013e3181859edd. [DOI] [PubMed] [Google Scholar]
- 44.Cerboni C, Mousavi-Jazi M, Wakiguchi H, Carbone E, Karre K, Soderstrom K. Synergistic effect of IFN-gamma and human cytomegalovirus protein UL40 in the HLA-E-dependent protection from NK cell-mediated cytotoxicity. Eur J Immunol. 2001;31:2926–2935. doi: 10.1002/1521-4141(2001010)31:10<2926::aid-immu2926>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci U S A. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei ML, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. 1992;356:443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 47.von Heijne G. Ribosome-SRP-signal sequence interactions. The relay helix hypothesis. FEBS Lett. 1985;190:1–5. doi: 10.1016/0014-5793(85)80414-2. [DOI] [PubMed] [Google Scholar]
- 48.Griffin C, Wang EC, McSharry BP, Rickards C, Browne H, Wilkinson GW, Tomasec P. Characterization of a highly glycosylated form of the human cytomegalovirus HLA class I homologue gpUL18. J Gen Virol. 2005;86:2999–3008. doi: 10.1099/vir.0.81126-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Park B, Oh H, Lee S, Song Y, Shin J, Sung YC, Hwang SY, Ahn K. The MHC class I homolog of human cytomegalovirus is resistant to down-regulation mediated by the unique short region protein (US)2, US3, US6, and US11 gene products. J Immunol. 2002;168:3464–3469. doi: 10.4049/jimmunol.168.7.3464. [DOI] [PubMed] [Google Scholar]
- 50.Millo E, Pietra G, Armirotti A, Vacca P, Mingari MC, Moretta L, Damonte G. Purification and HPLC-MS analysis of a naturally processed HCMV-derived peptide isolated from the HEK-293T/HLA-E+/Ul40+ cell transfectants and presented at the cell surface in the context of HLA-E. J Immunol Methods. 2007;322:128–136. doi: 10.1016/j.jim.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 51.Schrul B, Kapp K, Sinning I, Dobberstein B. Signal peptide peptidase (SPP) assembles with substrates and misfolded membrane proteins into distinct oligomeric complexes. Biochem J. 427:523–534. doi: 10.1042/BJ20091005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henderson RA, Michel H, Sakaguchi K, Shabanowitz J, Appella E, Hunt DF, Engelhard VH. HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science. 1992;255:1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 53.Kapp K, Schrempf S, Lemberg MK, Dobberstein B. Post-Targeting Functions of Signal Peptides. I. In: Zimmermann R, editor. Protein Transport into the Endoplasmic Reticulum. Landes Bioscience; Austin: 2009. [Google Scholar]
- 54.Nattermann J, Nischalke HD, Hofmeister V, Ahlenstiel G, Zimmermann H, Leifeld L, Weiss EH, Sauerbruch T, Spengler U. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am J Pathol. 2005;166:443–453. doi: 10.1016/S0002-9440(10)62267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nattermann J, Nischalke HD, Hofmeister V, Kupfer B, Ahlenstiel G, Feldmann G, Rockstroh J, Weiss EH, Sauerbruch T, Spengler U. HIV-1 infection leads to increased HLA-E expression resulting in impaired function of natural killer cells. Antivir Ther. 2005;10:95–107. doi: 10.1177/135965350501000107. [DOI] [PubMed] [Google Scholar]
- 56.Hoare HL, Sullivan LC, Clements CS, Ely LK, Beddoe T, Henderson KN, Lin J, Reid HH, Brooks AG, Rossjohn J. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. J Mol Biol. 2008;377:1297–1303. doi: 10.1016/j.jmb.2008.01.098. [DOI] [PubMed] [Google Scholar]
- 57.Kaiser BK, Barahmand-Pour F, Paulsene W, Medley S, Geraghty DE, Strong RK. Interactions between NKG2x immunoreceptors and HLA-E ligands display overlapping affinities and thermodynamics. J Immunol. 2005;174:2878–2884. doi: 10.4049/jimmunol.174.5.2878. [DOI] [PubMed] [Google Scholar]
- 58.Sullivan LC, Clements CS, Beddoe T, Johnson D, Hoare HL, Lin J, Huyton T, Hopkins EJ, Reid HH, Wilce MC, Kabat J, Borrego F, Coligan JE, Rossjohn J, Brooks AG. The heterodimeric assembly of the CD94-NKG2 receptor family and implications for human leukocyte antigen-E recognition. Immunity. 2007;27:900–911. doi: 10.1016/j.immuni.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 59.Magri G, Muntasell A, Romo N, Saez-Borderias A, Pende D, Geraghty DE, Hengel H, Angulo A, Moretta A, Lopez-Botet M. NKp46 and DNAM-1 NK-cell receptors drive the response to human cytomegalovirus-infected myeloid dendritic cells overcoming viral immune evasion strategies. Blood. 2010;117:848–856. doi: 10.1182/blood-2010-08-301374. [DOI] [PubMed] [Google Scholar]
- 60.Chapman TL, Bjorkman PJ. Characterization of a murine cytomegalovirus class I major histocompatibility complex (MHC) homolog: comparison to MHC molecules and to the human cytomegalovirus MHC homolog. J Virol. 1998;72:460–466. doi: 10.1128/jvi.72.1.460-466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim Y, Park B, Cho S, Shin J, Cho K, Jun Y, Ahn K. Human cytomegalovirus UL18 utilizes US6 for evading the NK and T-cell responses. PLoS Pathog. 2008;4:e1000123. doi: 10.1371/journal.ppat.1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khan AR, Baker BM, Ghosh P, Biddison WE, Wiley DC. The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J Immunol. 2000;164:6398–6405. doi: 10.4049/jimmunol.164.12.6398. [DOI] [PubMed] [Google Scholar]
- 63.Glithero A, Tormo J, Doering K, Kojima M, Jones EY, Elliott T. The crystal structure of H-2D(b) complexed with a partial peptide epitope suggests a major histocompatibility complex class I assembly intermediate. J Biol Chem. 2006;281:12699–12704. doi: 10.1074/jbc.M511683200. [DOI] [PubMed] [Google Scholar]
- 64.Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci U S A. 2008;105:6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]