Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that play crucial regulatory roles by targeting mRNAs for silencing. To identify miRNAs in Jatropha curcas L, a bioenergy crop, cDNA clones from two small RNA libraries of leaves and seeds were sequenced and analyzed using bioinformatic tools. Fifty-two putative miRNAs were found from the two libraries, among them six were identical to known miRNAs and 46 were novel. Differential expression patterns of 15 miRNAs in root, stem, leave, fruit and seed were detected using quantitative real-time PCR. Ten miRNAs were highly expressed in fruit or seed, implying that they may be involved in seed development or fatty acids synthesis in seed. Moreover, 28 targets of the isolated miRNAs were predicted from a jatropha cDNA library database. The miRNA target genes were predicted to encode a broad range of proteins. Sixteen targets had clear BLASTX hits to the Uniprot database and were associated with genes belonging to the three major gene ontology categories of biological process, cellular component, and molecular function. Four targets were identified for JcumiR004. By silencing JcumiR004 primary miRNA, expressions of the four target genes were up-regulated and oil composition were modulated significantly, indicating diverse functions of JcumiR004.
Keywords: Jatropha, Biofuel, miRNA, fatty acid synthesis.
Introduction
There are several classes of 19-24 nt short RNAs that regulate gene expression. The most conserved class is the microRNAs (miRNAs), which are small, noncoding RNAs that can play crucial regulatory roles in eukaryotes by targeting mRNAs for silencing 1. Many miRNAs are conserved between species, others are only conserved between more closely related species such as C. elegans and C. briggsae 2, 3. So far, identification of miRNAs has been limited to a few model species with their genomes sequenced. The main approach to discover miRNAs and their targets in plants is based on prediction programs that scan genomic or cDNA sequence 4. The weakness of this approach resides in the use of genome sequence or cDNA sequence, the substantial effort and time spent trying to validate false in silico candidates before a real target is identified. Studies were undertaken to identify miRNAs that are difficult to predict in silico or not conserved in model plants. Such studies have been hampered, however, by the lack of sensitive cloning methods for miRNAs. Because the available computational approaches can only identify miRNAs that are conserved, a cloning approach was employed to identify non model plants' miRNAs that may not be conserved or may have atypical features 5.
Jatropha curcas L is a perennial poisonous shrub belonging to the Euphorbiaceae family 6. Its seeds contain about 30% oil that is usable in a standard diesel engine 7, therefore jatropha is regarded as a promising candidate for producing biodiesel and becoming one of the world's key energy crops 8, 9. We found that the variation in DNA level within Jatropha curcas was very limited despite of large phenotypic variation 10, and we conducted marker-traits analysis by using interspecies crossing populations between Jatropha curcas and Jatropha integerrima 11. Herein we are searching for epigenetic factors including miRNAs in jatropha, which could be an important contributor to the large phenotypic variations within Jatropha curcas. However, it is generally difficult to identify miRNAs in non model plant species due to its poor genome information.
As for predicting miRNA targets, of the predicted targets of the first 14 Arabidopsis miRNA families, 70% are transcription factor genes 12. As more nonconserved miRNAs are being identified, the diversity of target genes expands beyond the dominating transcription factor genes to include classes associated with other metabolic and cellular processes 13. miRNA networks may also modulate species specific processes such as wood formation in trees 14. To gain an understanding of these jatropha-specific processes that likely require the regulation of many genes, we endeavor to investigate the miRNA networks in leaf and seed tissues of jatropha, which is a promising key energy crop in the near future.
The identification of miRNA targets is essential for understanding miRNA functions. Identification of miRNAs and their targets is challenging, which was often done computationally. The main criterion for target prediction is the sequence complementarities between a miRNA and its target genes. Even though computational target prediction has been successful, but false positive rates exist 15. Therefore, experimental validation must be performed. The biological significance of silencing miRNAs was described previously 16, briefly, the action on anti-miRNA oligonucleotides is to silence miRNAs but to upregulate gene expression by relieving the repressive effect of miRNAs on their target protein-coding genes. In some cases, upregulation of gene expression is desirable. Anti-miRNA oligonucleotides were proved to be useful in inhibiting individual miRNAs, thereby helping to unravel the function of miRNAs and their targets in human 17 and mice 18. An experimental approach to target identification was presented where the cartilage-specific miR-140 was silenced in mouse cells. The mRNAs derepressed by silencing of miR-140 was identified 19. Significantly reduced accumulation of miR163 and miR171a was achieved using hairpin RNAi constructs that were designed to target both the primary miRNA transcripts. Reduction of miRNA accumulation resulted in an increase in accumulation of the mRNA targets of these miRNAs 20. In this study, we validated the putative targets predicted based on complementarities through examining their expression variation by directly silencing the corresponding primary miRNA through virus-induced gene silencing (VIGS). VIGS offers an attractive alternative as it allows the investigation of gene functions without plant transformation. For plants with long life cycle such as jatropha, however, transgenic technology is unsuitable for routine testing of gene function because it is time consuming and laborious 21.
In this study, we isolated a collection of miRNAs and their target genes in jatropha, and examined the expression patterns of miRNAs in different tissues. We further analyzed the functions of JcumiR004, as an example of the miRNA directly isolated from jatropha, and showed JcumiR004 may play important roles in a broad range of biological functions related to agronomic traits including oil composition.
Results
Cloning and identification of miRNAs in jatropha
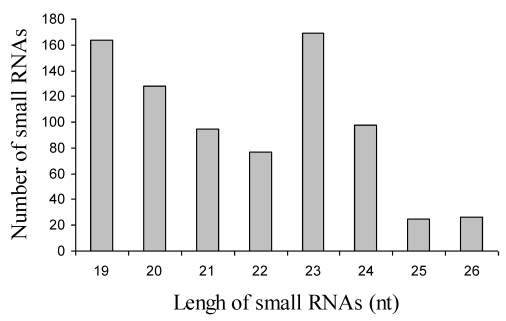
In order to identify miRNAs in jatropha, we generated two small RNA libraries ranging in size from 18-26 nt using RNA isolated from leaves and developing seeds. The isolated small RNAs were separated by 15% denaturing PAGE and small RNAs of 19-26 nt were gel-purified, ligated with 3' and 5'-adaptors and RT-PCR-amplified as described 22. PCR products were isolated by electrophoresis, cloned into the pGEMT vector and sequenced. A total of 426 and 356 sequences were collected from leaf and seed libraries respectively. Analysis of these sequences resulted in identification of 233 and 114 unique sequences ranging in size from 19-26 nt in length, which is of the typical size range for endogenous small RNAs. The number of small RNA of 23 nt was highest compare to those of other size (Fig. 1).
Figure 1.
Size of distribution of 882 small RNAs in Jatropha curcas L.
One important feature of miRNAs is the hairpin stem-looped structure in the precursors23. The RNA sequences were subjected to BLAST analysis against the genomes of Arabidopsis thaliana, Oryza sativa, Vitis vinifera, Populus trichocarpa, Euphorbia genistoides and Jatropha curcas. From these plants, 52 sequences were screened which harboring putative miRNAs from jatropha leaf or seed library (Table 1), and four of which were found both in the leaf and seed small RNA libraries, i.e. JcumiR001, JcumiR002, JcumiR005 and JcumiR022 (Table 1). These sequences were capable of forming stem-loop structures characteristic of miRNA precursors (Additional file 1: Supplementary Figure 1). Among these putative miRNAs, six were homologous to known miRNAs, JcumiR012-osa mir166e; JcumiR015-Rattus norvegicus mir-3596a; JcumiR017-Osa MIR457 precursor; JcumiR018-Osa microRNA 169c gene; JcumiRNA027-Osa microRNA 167d gene; JcumiRNA042-Capsella rubella miR319a; while the other 46 were novel.
Table 1.
miRNAs isolated from jatropha.
| miRNA | Sequence | Length | small RNA library (leaf/seed) |
No of clones |
Precursor in other plants GenBank Access No |
ΔG | known miRNAs in other species |
|---|---|---|---|---|---|---|---|
| JcumiR001 | UGUCGCGAUGGUAAUUCAACC | 21 | leaf and seed | 6 and 2 | Hevea brasiliensis DQ306824.1 | -23.2 | |
| JcumiR002 | GAUCGCGUGGCCUAAUGGA | 19 | leaf and seed | 26 and 2 | Vitis vinifera AM463399 | -30.4 | |
| JcumiR003 | UCCUCUGAGCUAGGCAAUG | 19 | leaf | 1 | Jatropha curcas EZ413062.1 | -31.1 | |
| JcumiR004 | UGAUUGAGCCGUGCCAAUAUC | 21 | leaf | 1 | Oryza sative AY551250.1 | -54.5 | |
| JcumiR005 | GUCGUUGUAGUAUAGUGGU | 19 | leaf and seed | 12 and 5 | Populus trichocarpapa AC209103 | -27.6 | |
| JcumiR006 | GGCAUGGGCGAUAUGGGCAAG | 21 | leaf | 2 | Strongylocentrotus purpuratus XM_001189429 | -37.8 | |
| JcumiR007 | AUCAAAAGGGUUGGUAUUGCUCCU | 24 | leaf | 1 | Euphorbia genistoidesAM040819 | -23.3 | |
| JcumiR008 | GAUGCUGGUGUUCGGGCUA | 19 | leaf | 2 | Medicago truncatu AC156629 | -18.1 | |
| JcumiR009 | AUUCGGCUCAAUCCUUUUAG | 20 | leaf | 1 | Jatropha curcas FJ695500.1 | -21.1 | |
| JcumiR010 | GGAAACAGAAUUGGCGGCUU | 20 | leaf | 1 | Arabidopsis thaliana AL161537 | -30.11 | |
| JcumiR011 | UGGUUCAAGGCGUAGCAUUGG | 21 | leaf | 3 | Vitis vinifera AM477478 | -28.57 | |
| JcumiR012 | GAAUGUUGUCUGGUUCAAGG | 20 | leaf | 1 | Oryza sativa HM139377.1 | -58.7 | Precursor of Osa-miR166e |
| JcumiR013 | GAAUUAUAGGUGUUGAAUAUGGU | 23 | leaf | 1 | Populus trichocarpapa AC213088 | -18.6 | |
| JcumiR014 | UGGUAGAGCGGUCGGCUGU | 19 | leaf | 1 | Phaseolusvulgari EU196765 | -44.1 | |
| JcumiR015 | UGAGGUAGUAGGUUGUAUAGU | 21 | leaf | 2 | Rattus norvegicus NR_037385.1 | -34.2 | Rattus norvegicus mir-3596a |
| JcumiR016 | UCGCACACAUGUCAGACUCCCC | 22 | leaf | 2 | Mimulus guttatusAC182572 | -38.2 | |
| JcumiR017 | UCCACUGCGCUAUGCGGUC | 19 | leaf | 2 | Oryza sativa AY946832 | -40.05 | Osa mir457 precursor |
| JcumiR018 | CCGGCAAGUCAUCCUUGGCUG | 21 | leaf | 1 | Oryza sativa AY551241 | -69.06 | Osa mirRNA 169c gene |
| JcumiR019 | GGGGAUGUAGCUCAAAUGGUAGAG | 24 | leaf | 5 | Oryza sativa AK288811.1 | -48.7 | |
| JcumiR020 | UCGAAAAUUCAACGCAAGC | 19 | leaf | 1 | Oryza sativa AL607000.3 | -49.8 | |
| JcumiR021 | UUCCCGUUUCCGCCCAACUCA | 21 | leaf | 1 | Vitis vinifera AM436815 | -38.08 | |
| JcumiR022 | GGGUGCGAUCAUACCAGCAC | 20 | leaf and seed | 2 and 7 | Vitis vinifera AM48227 | -44.84 | |
| JcumiR023 | GGGGGGUCCCAGUCCCGAAC | 20 | leaf | 3 | Arachis duranensis HP035294.1 | -66.9 | |
| JcumiR024 | GGGCACGUCUGCCUGGGUGUCA | 22 | leaf | 4 | Jatropha curcas EZ408819.1 | -55.6 | |
| JcumiR025 | GGAAUAGCUCAGUUGGUAGAG | 21 | leaf | 1 | Solanum lycopersi AC215427 | -27.4 | |
| JcumiR026 | UGCUUUUAGGUAGGUUUGGUGAAC | 24 | leaf | 1 | Vitis vinifera AM444108 | -27.9 | |
| JcumiR027 | UGAAGCUGCCAGCAUGAUCUGG | 22 | leaf | 2 | Populus trichocarpa AC213492 | -26.2 | Osa mir167d gene |
| JcumiR028 | GACCGAGUGCCUCCCACGC | 19 | leaf | 2 | Jatropha curcas EZ414173.1 | -33.1 | |
| JcumiR029 | GCCGCUAUGAUGAAAUCGGU | 20 | seed | 1 | Vitis vinifera AM451225 | -38.12 | |
| JcumiR030 | ACAGACCGGUAGACUUGAAC | 20 | seed | 1 | Vitis vinifera AM45651 | -24.2 | |
| JcumiR031 | GUCCCAGUCCCGAACCCGUCGG | 22 | seed | 1 | Vitis vinifera AM456511, Populus EST CU231567 |
-67.3 | |
| JcumiR032 | UGGUGCCCGAUAGCGUGGUC | 20 | seed | 1 | Oryza sativa AP008211 | -18.9 | |
| JcumiR033 | UCCAUUAGGCUACGCGAUC | 19 | seed | 1 | Oryza sativa CT832877 | -19.8 | |
| JcumiR034 | CGAGUUCUAUCGGGUAAAGCCAA | 23 | seed | 1 | Populus trichocarpa AC216416 | -32.7 | |
| JcumiR035 | UGAGGUAGUUGUUUGUAUGG | 20 | seed | 8 | Vitis vinifera AM436351 | -40.6 | |
| JcumiR036 | UCAAAAUAUAACCUUCCCA | 19 | seed | 4 | Vitis vinifera AM454629 | -74.43 | |
| JcumiR037 | GGGUUCGUUUCCCACAGACGGCGC | 24 | seed | 4 | Vitis vinifera AM478487 | -25.05 | |
| JcumiR038 | GGGAGGAGGUGGUGGUGGGC | 20 | seed | 7 | Oryza minuta AC229917 | -147.25 | |
| JcumiR039 | UACCAUUUGAGCUAAUCCCCC | 21 | seed | 2 | Vitis vinifera AM486140 | -79 | |
| JcumiR040 | UGGAAUUCGCGGUUAAAUU | 20 | seed | 3 | Oryza sativa AK241705 | -19.9 | |
| JcumiR041 | CGGUGUGCACCUGUCGGCUCGUCCC | 25 | seed | 7 | Vitis vinifera AM423427 | -26.7 | |
| JcumiR042 | UGAGGUAGUAGGUUGUGUGGUU | 22 | seed | 10 | Vitis vinifera AM463593 | -90.5 | Capsella rubella miR319a |
| JcumiR043 | UGAGGUAGUUGGUUGUAUGGU | 21 | seed | 5 | Vitis vinifera AM458795 | -65.22 | |
| JcumiR044 | UUCUGAAAUGUGCGUGUGU | 19 | seed | 3 | Vitis vinifera AM458673 | -60.04 | |
| JcumiR045 | GAGUCCGGAGACGUCGGCGGGGGC | 24 | seed | 7 | Arabidopsis thali AK230153 | -75.9 | |
| JcumiR046 | UGCUGUGAUCAGUGUCGCU | 19 | seed | 2 | Populus trichocarpa AC185361 | -19.4 | |
| JcumiR047 | UCACAGUGUCAUUUAGCUGA | 20 | seed | 7 | Oryza sativa AP008209 | -36.9 | |
| JcumiR048 | UGUGGGAUGAUGAUCUUGCG | 20 | seed | 10 | Oryza sativa AP008214 | -28.7 | |
| JcumiR049 | CGCCACACCUCCCUCGCCC | 19 | seed | 2 | Oryza sativa CR855194 | -30.7 | |
| JcumiR050 | UGCCUUAGACUAAAUUCGUGUC | 22 | seed | 2 | Vitis vinifera AM456883 | -17.7 | |
| JcumiR051 | UGGUUCCCGAUAACGCGUGGUC | 22 | seed | 2 | Arabidopsis thali NM_106553 | -36 | |
| JcumiR052 | UGAAAAGGUGUUGGUUGAUA | 20 | seed | 2 | Vitis vinifera AM483931 | -54.1 |
Expression profiling of miRNAs
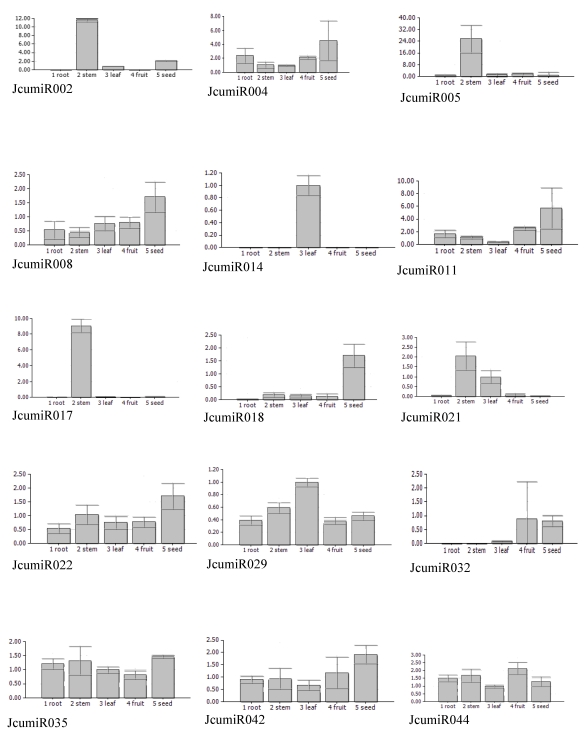
Knowledge about the expression patterns of miRNAs can provide clues about their functions. To get an insight into possible tissue-dependent roles of miRNAs in Jatropha curcas, the expression patterns of miRNAs in different tissues, including root, stem, leave, fruit and seed were examined using quantitative real time PCR. Expression patterns of some miRNAs could not be well evaluated through real time PCR due to non-specific amplicons. Fifteen miRNAs, which have been well detected, revealed different expression patterns. Half of miRNAs isolated from leaf (JcumiR014, 017, 021) did not expressed in fruit or seed, while all miRNAs isolated from seed expressed abundantly in seed (Fig. 2). Besides the miRNA from seed library, JcumiR004, 008 and 018 from leaf library also showed to be highly expressed in seed. JcumiR014 and JcumiR029 were found to be strongly expressed in leaf. Expression of JcumiR002, 005 and 007 was higher in stem, but lower in other tissues tested. JcumiR004, 029, 035, 042 and 044 showed moderate expression in roots. JcumiR004, 008 and 022 from leaf library, and JcumiR029, 035, 042 and 044 from seed library showed ubiquitous expression in all tissues, although the expression of JcumiR008, 022 and 042 were relatively higher in seeds (Fig. 2). These observations suggest that these miRNAs display differential tissue-specific expression patterns.
Figure 2.
Expressions of 15 primary JcumiRNAs in different tissues. The 18S rRNA was selected as a control. 1: root, 2: stem, 3: leaf, 4: fruit and 5: seed.
Predicted targets of jatropha miRNAs
In plants, the miRNA target sites were found predominantly in the coding regions 24. We searched putative miRNA targets with a cDNA library constructed by Dr Yin Zhongchao's group in our institute. Twenty eight predicted target genes have target sites in the coding region (Additional file 2: Supplementary Table 1). More than two targets were screened for JcumiR004, 008, 014, 021 and 036. We were unable to predict targets for half of the miRNAs by applying the above rules, due to the limited number of jatropha EST sequences available in the databases.
Gene ontology of the targets
The 28 ESTs of putative targets were used to search for similar protein sequences in the Uniprot database (BLASTX). The functional information is presented in Additional file 2: Supplementary Table 1. Using the best hits found by BLASTX, an inferred gene ontology annotation was found for 16 of the sequences through QuickGO. Using GO Slimmer in AmiGO database, of the 16 known functional ESTs, one was associated with genes belonging to biological process, 7 with cellular component, and 8 with molecular function. The result showed that the miRNA target genes encoded a broad range of proteins, with majority (15/16) of the predicted protein products involved in molecular function and cellular component ontologies. The predicted protein description of the target EST sequences were listed in Additional file 2: Supplementary Table 1, with some interesting proteins such as DNA cross-link repair protein, vacuolar ATP synthase proteolipid, elongation factor 1-alpha, Cytochrome c-type biogenesis protein, 14-3-3 protein, heat stress transcription factor A-5, etc. Thus, the miRNA target genes encode a broad range of proteins.
Functional classification of JcumiR004 targets
The highest number of ESTs were found to be putative targets of JcumiR004 in the jatropha cDNA library, which are homologues to four genes or mRNA in Arabidopsis, i.e. Ubiquitin-conjugating enzyme E22 (UBC22), Protein BUD31 homolog 1(Os01g0857700), Proteasome subunit alpha type-3 (PAG1), Auxin response factor 7 (ARF7). These genes play important roles in a broad range of biological functions. The products of UBC22 and ARF7 were classified into molecular function ontology, involved in the elemental activities of a gene product at the molecular level, such as binding or catalysis; while Os01g0857700 and PAG1, cellular component, involved in the parts of a cell or its extracellular environment.
Function analysis of JcumiR004
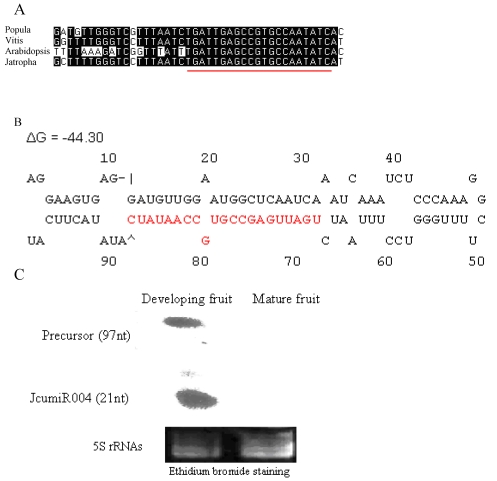
Mature miRNA of JcumiR004 is conserved in populus, vitis, Arabidopsis and jatopha (Fig. 3A).
Figure 3.
JcumiR004 miRNA and precursor. (A): JcumiR004 is conserved in populus, vitis and Arabidopsis. (B): Precursor of JcumiR004 in jatropha with free energies ΔG -44.30. (C): RNA gel blots: total RNA from fruit was probed with labeled oligonucleotides. The 5S RNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls. miRCURY locked nucleic acid (LNA:+) probe (Exiqon, Denmark): 5DigN/G+A+T +ATT GG+C +ACGGC+TCA+ATCA
Expressions of primary JcumiR004 were relatively high in developing fruit while not in mature seed (Fig. 2). RNA gel blot of JcumiR004 was done in fruit, the most important organ for jatropha oil yield and quality traits. The size of the RNAs were estimated to be around 20 and 100nt corresponding to mature (21nt) and precursor (~97nt) of JcumiR004. The result showed that both mature miRNA and precursor of JcumiR004 were abundant in developing fruits while not in matured (Fig. 3C). It was therefore suggested that JcumiR004 could be actively involved in fruit formation or development. Precursor of JcumiR004 was isolated by 3' and 5' RACE for further analysis. A good loop stem structure of the precursor was isolated with free energies ΔG=-44.30 as shown in Fig. 3B. The primary miRNA harboring this precursor is used for further function analysis by VIGS.
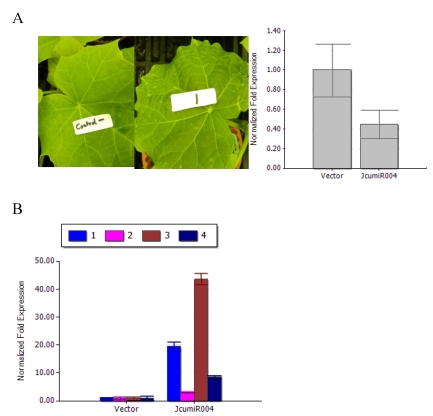
When examining the effect of TRV VIGS to silence JcumiR004 primary miRNA gene, we observed that TRV:JcumiR004 was able to induce a photo-bleaching and different spots on the leaf. After twenty days post-infiltration (dpi), the photo bleaching phenotype of JcumiR004 was seen (Fig. 4A).
Figure 4.
Variation of phenotypes of jatropha leaf and JcumiRNA target gene expression. (A) Left: Phenotypes of jatropha plants at 18 days post-infiltration (dpi) with TRV vector (left) and TRV: JcumiR004 (right) plants; Right: Primary miRNA of JcuLmiR004 was reduced in plants infiltrated with TRV: JcuLmiR004 compared to plants with TRV vector. (B) JcumiR004 target gene expression levels in plants infiltrated with TRV vectors and TRV: JcumiR004 plants. Numbers represent mean relative values from three independent experiments with standard deviation 1: UBC22 gene, 2: Os01g0857700, 3: PAG1 and 4: ARF7.
By silencing JcumiR004 primary miRNA in TRV:JcumiR004 plants, the expression level of JcumiR004 was decreased after the primary miRNA knockdown (Fig. 4A). The expression levels of JcumiR004 target genes in plants infiltrated with TRV: JcumiR004 plants were significantly increased as compared to the plants with vector control (Fig. 4B). The results revealed that expression levels of these target genes could be obviously increased by silencing primary miRNA of JcumiR004. The transcripts of UBC22, Os01g0857700, PAG1 and ARF7 were significantly increased by 41.5, 6.5, 93.4 and 18.5 folds.
Gas chromatographic (GC) analysis showed significantly lower linoleic acid (C18:2) levels in plants with JcumiR004 primay miRNA silenced as compared to vector control (Table 2). The other compositions of fatty acid remained unchanged. It would be interesting to further investigate which gene(s) involved in jatropha fatty acid metabolism pathway was modulated by JcumiR004.
Table 2.
Fatty acid (FA) composition of systemic leaves from plants infiltrated with empty vector and TRV:JcumiR004.
| C16:0 | C18:0 | C18:1 | C18:2 | C18:3 | |
|---|---|---|---|---|---|
| Vector | 22.0±2.0 | 22.1±1.5 | 0±0 | 21.2±2.3 | 34.7±4.1 |
| JcumiR004 | 21.1±2.1 | 22.2±1.8 | 0±0 | 0±0 | 40.1±4.6 |
Numbers in each column refer to the relative molar ratios of the different FA with the total being 100%. Means and standard deviation are calculated with three replications.
Discussion
JcumiRNA cloning
The identification of miRNAs and targets will lay the foundation to unravel the complex miRNA-mediated regulatory networks controlling development and other physiological processes. In this study, using an experimental approach, we provide evidence for the existence of 52 miRNA in jatropha. Up to now, miRNA identification in plants using a cloning approach has been limited to Arabidopsis, rice, Populus, wheat 25 and castor bean 26.
IZeng et al. reported that a substantial number of miRNAs previously identified and characterized in model plants were conserved in four agrieconomically important Euphorbiaceous plants, Castor bean (Ricinus communis), Cassava (Manihot esculenta), Rubber tree (Hevea brasiliensis) and jatropha 26. They predicted 85 conserved miRNAs in Castor bean, and experimentally verified and characterized 58 (68.2%) of the 85 miRNAs in at least one of four Euphorbiaceous species, including jatropha during normal seedling development. In this study, cloning directly from jatropha leaves and seeds led to identification of 52 jatropha miRNAs with 45 novel ones. Among the 52 jatropha miRNAs, primary miRNAs of 48 miRNAs were identified in other species than jatropha, revealing most of miRNAs are novel and conserved across plants. Future large scale experimental approaches and jatropha EST data will be likely to identify additional miRNAs from jatropha.
JcumiRNAs exhibit tissue-specific expression patterns
The JcumiRNAs in this study were identified to be homologous miRNAs in other plant species. Despite the existence of miRNA sequence conservation between jatropha and other plants, these homologous miRNAs exhibit contrasting tissue-specific expression patterns. Fruit and seed are most important for jatropha oil productivity. In this study, half of JcumiRNAs isolated from leaf did not expressed in fruit or seed, while all miRNAs isolated from seed expressed abundantly in fruit and seed. There could be certain JcumiRNAs related to seed development or seed oil metabolism pathway.
Predicted targets of JcumiRNAs might play roles in a broad range of biological functions
The identification of miRNA targets is essential for understanding miRNA functions. Although our EST data base is relatively small, we still identified quite many putative targets to the isolated miRNAs, which showing that the miRNA target genes encoded a broad range of proteins. Sixteen targets had clear BLASTX hits to the Uniprot database and were associated with genes belonging to the three major gene ontology categories of biological process, cellular component, and molecular function. The majority of the predicted protein products in this study were involved in molecular function and cellular component ontologies. Four targets of JcumiR004 were screened, which are homologous to four genes. Predicted protein products of the four targets involved in molecular function and cellular component ontologies.
Identification of miRNAs and their targets is challenging, which was often done computationally. Even though computational target prediction has been successful, but false positive rates exist (11). After we predicted four putative targets of JcumiR004 based on complementarities, we have further validated the putative targets through examining their expression variation by silencing the corresponding primary miRNA through VIGS. Further reporter assay is still needed to see how mutation of the JcumiR004 seed match is going to affect this target expression. The biological significance of silencing primary JcumiR004 is to silence or downregulate mature Jcumi004 but to upregulate target gene expression by relieving the repressive effect of mature Jcumi004 on their target protein-coding genes.
In this case, upregulations of the target genes' expressions are desirable. Among these targets, UBC plays important functions in many aspects of plant growth and development, including phytohormone and light signalling, embryogenesis, organogenesis, leaf senescence and plant defense (for review, see 27. In Arabidopsis thaliana, 37 proteins with a UBC domain and active-site cysteine are predicted 28. However, the biological functions of these genes remain largely uncharacterized 29. Here we found that UBC22 gene is a target of JcumiRNA, which will be beneficial for further study on UBC gene family. BUD31 (G10 family) protein gene was reported to be involved in pre-mRNA splicing 30. PAG1, a 20S proteasome subunit in Arabidopsis development, is essential for growth and development 31. Auxin Response Factor (ARF) gene family products regulate auxin-mediated transcriptional activation/repression 32.
Despite of the limited sequences of the cDNA library, we predicted 28 targets which could be expected to play roles in a broad range of biological functions.
JcumiR004 affects jatropha oil traits
Oil traits are among the most important traits in jatropha breeding. Plant oils are mostly composed of five common FA, namely palmitic acid (16:0), stearic acid (18:0), oleic acid (18:1), linoleic acid (18:2) and linolenic acid (18:3). Normal jatropha seed oil is mainly composed of oleate (35%-50%), linoleate (30%-45%) and palmitate (10%) 6. Here we found that JcumiR004 can affect oil composition of jatropha leaf by changing linoleic acid. Although the TRV system applied in this study worked in jatropha leaf, the results provide a useful reference for research on jatropha fruit and seed development 21. It would be intriguing to investigate what gene regulated by JcumiR004 is involved in oil composition. It is necessary to search for more target genes of JcumiR004. Relationships among JcumiR004 and its target genes involved in oil pathway will be investigated to study its metabolism pathway. More targets of Jcumi004 could be indentified as the genome information getting richer.
In this study, real time PCR showed that primary JcumiR004 expressed ubiquitously in all the tissues of root, stem, leaf, developing fruit and mature seed. Silencing primary JcumiR004 led to not only upregulation of the 4 targets involved in various aspects of plant growth and development, but also modulation of C18:2 composition in fatty acid. Taken together, it has been revealed that JcumiR004, as an example of the miRNA directly isolated from jatropha, play important roles in a broad range of biological functions related to agronomic traits including oil composition. We could expect that the miRNAs in jatropha may be an indispensible factor to the rich phenotypic variation in Jatropha curcas.
In conclusion, we produced a collection of 52 miRNAs and 28 targets in jatropha. Different expression patterns of miRNAs in root, stem, leave, fruit and seed were detected. As an example of the miRNA directly isolated from jatropha, we analyzed JcumiR004 on its expression profiles of primary and mature miRNA, its functions on regulation of four targets, and its modulation of C18:2 composition in fatty acid. The data in this study revealed jatropha miRNAs' diverse roles in broad range of biological functions.
Experimental procedures
Plant materials
For small RNA libraries construction and real time PCR, tissues of root, stem and leaf were collected from Jatropha curcas seedlings with 2-3 true leaves while developing fruit and mature seed were from fully grown plants. Jatropha seeds were germinated in a greenhouse and seedlings with 2-3 true leaves were used for VIGS assays.
Small RNA Isolation
Enrichment of small RNAs from total RNA was performed with mirVana™ miRNA isolation kit (Ambion, CA, USA) and then was separated on a denaturing 15% polyacrylamide gel. The nucleotides from positions 19-26 bp were size fractionated. RNA was eluted overnight with 0.4 M NaCl at 4°C and recovered by ethanol precipitation with glycogen. The purified small RNAs were then ligated to (5'-(Pu)uuAACCGCGAATTCCAG(idT)-3'; (where lowercase letters indicate RNA, uppercase letters indicate DNA, Pu denotes 5'-phosphorylated uridine, and idT represents 3'-inverted deoxythymidine.) and 5' adaptor (5'-GACCACGCGTATCGGGCACCACGTATGCTATCGATCGTGAGATGGG-3'), and miRNA libraries were constructed as described 22. Reverse transcription was performed with PowerScript reverse transcriptase (Clontech, CA, USA) and RT primer to get the first strand cDNA for further analysis. The oligonucleotides used for the procedures were described in 22.
Prediction of stem-loop structures
The RNA sequences were subjected to BLAST analysis against other genomes: Arabidopsis thaliana, Oryza sativa and Euphorbia genistoides. We followed the following criteria 23, 33 for selecting the candidates of potential miRNAs or pre-miRNAs: (1) predicted mature miRNAs had only 0-3 nucleotide mismatches in sequence with all previously known plant mature miRNAs; (2) Sequences of miRNA precursors can fold into a hairpin secondary structure that contains the ~22 nt mature miRNA sequence within one arm of the hairpin; (3) miRNA had 30-70% contents of A+U; (4) miRNA had less than six mismatches with the opposite miRNA* sequence in the other arm; and (5) no loop or break in miRNA sequences was allowed.
Secondary structures were predicted by using the mfold program (www. bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) with the default parameters 34. In each case, only the lowest energy structure was selected for manual inspection, as described by 12. Small RNA sequences were folded with flanking sequences in five contexts: (1) 300 bp upstream and 20 bp downstream, (2) 150 bp upstream and 20 bp downstream; (3) 150 bp upstream and 150 downstream, (4) 20 bp upstream and 150 downstream, and (5) 20 bp upstream and 300 bp downstream.
Examining miRNA expressions
Total RNAs of jatropha root, stem, leaf, fruit and seed were isolated using plant RNA purification reagent (Invitrogen). Root, stem and leaf were harvest from the seedlings 2 weeks post seeding. Fruit was harvest 1 to 2 weeks after pollination, which is at its formation and developing stage. Seed are mature ones ready for oil extraction.
For examining expressions of miRNAs in different tissues, quantitative real time PCR was applied. Briefly, poly(A) tails were then added to the 3' end of the RNAs by poly(A) polymerase (Ambion), and the polyadenylated RNAs were reverse transcribed by SuperScript II reverse transcriptase (Invitrogen) with the oligo(dT) 3'-RACE adaptor (Ambion). To amplify the miRNA from the reverse transcribed cDNAs, we used the miRNA sequence as the forward primer and the 3'-RACE outer primer (Ambion) as the reverse primer, as described in 14. Real-time RT-PCR was conducted with Real-Time PCR machine (I-Cycle, BioRad). The Jatropha 18S rRNA was selected as the endogenous reference. After PCR was finished, the PCR specificity is examined by 3% agarose gel using 5 μl from each reaction to check the right product length and make sure no primer dimer or non-specific amplicons.
For examining expressions of miRNAs in fruit harvested 1 to 2 weeks after pollination and mature fruit using northern blot, RNA gel blots of mature miRNA were probed with labeled oligonucleotides. Briefly, fifteen micrograms of total RNA and RNA marker were electrophoresed on a 15% urea/polyacrylamide/Tris-borate-EDTA gel, and transferred to nylon membrane Hybond N+ (Amersham Biotech, Uppsala, Sweden) by overnight capillary transfer. After electrophoresis, we stained the RNA marker lane with SYBR Green II (Takara Bio) for 5 to 10 min and then photographed the gel aligned with a ruler to estimate the size of RNAs in Northern blot. Membranes were UV-cross-linked using the strataLinker (Stratagene, CA, USA), rinsed in 2×SSC, and probed with 5' Digoxigenin-labeled miRCURY locked nucleic acid (LNA) probe (Exiqon, Vedbaek, Danmark) at 42°C. The tRNA and 5S RNA bands were visualized by ethidium bromide staining of polyacrylamide gels and served as loading controls.
Prediction of miRNA targets
For target prediction, we followed a set of rules proposed in earlier reports for predicting miRNA targets 24, 25. These criteria include allowing one mismatch in the region complementary to nucleotide positions 2 to 12 of the miRNA, but not at position 10/11, which is a predicted cleavage site, and three additional mismatches between positions 12 and 22 but with no more than two continuous mismatches. To identify potential targets for jatropha miRNAs, we searched for antisense hits in a jatropha cDNA library constructed by Dr Yin Zhongchao's group in our institute. Target predictions were performed by searching the jatropha cDNA library database for miRNA complementary sequences, with the “contig assembly” algorithm in Sequencher 4.10.1 (GeneCodes, Ann Arbor, MI).
Target gene ontology and functional classification
The targets were categorized on the basis of their homologous gene function. The Blast2GO annotation tool was used to annotate EST sequences of target genes against the SwissProt database using BLASTX with default parameters 35. The annotation information for target genes was functionally classified through the European Bioinformatics Institute QuickGO interface 36. The gene ontology numbers for the best homologous hits were used to find molecular function, cellular component, and biological process ontology for these sequences using GO Slimmer in AmiGO database 37.
Analyzing functions of miRNA using VIGS
Generation of recombinant vectors and agrobacterium infiltration: VIGS was conducted following 21. Briefly, primary miRNA of JcumiR004 were cloned into pTRV2 to generate pTRV2 derivatives. pTRV1, and pTRV2, or pTRV2 derivatives were introduced into Agrobacterium strain GV3101 by electroporation. Agrobacterial cells were grown, collected and resuspended in MMA (10 mM MES, 10 mM MgCl2, 200μM acetylsyringone) solution to a final OD600 of 1.5. Jatropha plants were infiltrated with cultures either with a syringe or by vacuum. For syringe infiltration, agrobacterial-inocula were delivered into the underside of two or three fully expanded leaf using a 1-mL needleless syringe. For vacuum infiltration, whole plants were submerged into agrobacterial-inocula and subjected to 80-90 kPa vacuum for 5 min, and then quickly releasing the vacuum to allow the inoculums to rapidly enter plant tissues. Infiltrated plants were grown in a growth chamber at 25°C with 16h light ⁄ 8h dark photoperiod cycle.
Determining fatty acid composition using gas chromatography: Total lipid, extracted from 100 mg fresh jatropha leaves, was transmethylated with 3N methanolic-HCl (Sigma, St. Louis, MO, USA) plus 400 µL 2,2,-dimethoxypropane (Sigma, St. Louis, MO, USA). Each sample were grinded with liquid nitrogen, divided into 3 copies. The fatty acid methyl esters (FAME) was analyzed by GC using GC Agilent 6890 (Palo Alto, CA, USA) employing helium as the carrier gas and DB-23 columns for components separation. The GC analytical method was performed at 140°C for 50 s and a 30°C min-1 ramp to 240°C, and the final temperature was maintained for 50 s for a total run time of 32 min. FA composition value included in the analyses was calculated based on peak area.
Acknowledgments
The work is part of the project “Genetic Improvement of Jatropha” initiated and coordinated by Professor Nam-Hai Chua. We thank Dr Yin Zhongchao in our institute providing the cDNA library database. We also thank our sequencing facility for helping DNA sequencing and genotyping.
Funding
This project is financially supported by JOIL Pte Limited and the internal fund of the Temasek Life Sciences Laboratory, Singapore.
Supplementary Materials
Supplementary Figure 1. Stem-loop structures of 52 putative miRNA precursors.
Supplementary Table 1. The predicted targets of conserved and newly identified jatropha miRNAs.
References
- 1.Moxon S, Jing R, Szittya G, Schwach F, Rusholme Pilcher RL, Moulton V. et al. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol. 2003;13:807–18. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of Parallel Analysis of RNA Ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nature protocols. 2009;4:356–62. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- 5.Dugas DV, Bartel B. MicroRNA regulation of gene expression in plants. Curr Opin Plant Biol. 2004;7:512–20. doi: 10.1016/j.pbi.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Heller J. Physic nut, Jatropha curcas L. Bioversity International. 1996.
- 7.Shah S, Sharma A, Gupta M. Extraction of oil from Jatropha curcas L. seed kernels by combination of ultrasonication and aqueous enzymatic oil extraction. BioRes Technol. 2005;96:121–3. doi: 10.1016/j.biortech.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 8.Achten WMJ, Maes W, Aerts R, Verchot L, Trabucco A, Mathijs E. et al. Jatropha: from global hype to local opportunity. J Arid Biol. 2010;74:164–5. [Google Scholar]
- 9.Jain S, Sharma M. Prospects of biodiesel from Jatropha in India: A review. Renew Energ Stor. 2010;14:763–71. [Google Scholar]
- 10.Wang CM, Liu P, Yi C, Gu K, Sun F, Li L. et al. A First Generation Microsatellite-and SNP-Based Linkage Map of Jatropha. PloS one. 2011;6:e23632. doi: 10.1371/journal.pone.0023632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu P, Chun Ming Wang, Lei Li, Fei Sun, Peng Liu, Gen Hua Yue. Mapping QTLs for oil traits and eQTLs for oleosin genes in jatropha. BMC Plant J. 2011 doi: 10.1186/1471-2229-11-132. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–20. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 13.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM. et al. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell. 2005;17:2186. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammell M, Long D, Zhang L, Lee A, Carmack CS, Han M. et al. mirWIP: microRNA target prediction based on microRNA-containing ribonucleoprotein-enriched transcripts. Nature methods. 2008;5:813–9. doi: 10.1038/nmeth.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z. MicroRNA interference technologies. Springer Verlag. 2009.
- 17.Weiler J, Hunziker J, Hall J. Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther. 2005;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 18.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M. et al. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas FE, Pais H, Schwach F, Lindow M, Kauppinen S, Moulton V. et al. Experimental identification of microRNA-140 targets by silencing and overexpressing miR-140. RNA. 2008;14:2513. doi: 10.1261/rna.1221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaistij FE, Elias L, George GL, Jones L. Suppression of microRNA accumulation via RNA interference in Arabidopsis thaliana. Plant Mol Biol. 2010:1–7. doi: 10.1007/s11103-010-9625-4. [DOI] [PubMed] [Google Scholar]
- 21.Ye J, Qu J, Bui HTN, Chua NH. Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing. Plant Biotech J. 2009;7:964–76. doi: 10.1111/j.1467-7652.2009.00457.x. [DOI] [PubMed] [Google Scholar]
- 22.Takada S, Berezikov E, Yamashita Y, Lagos-Quintana M, Kloosterman WP, Enomoto M. et al. Mouse microRNA profiles determined with a new and sensitive cloning method. Nucl Acids Res. 2006;34:e115. doi: 10.1093/nar/gkl653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X. et al. A uniform system for microRNA annotation. RNA. 2003;9:277. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunkar R, Girke T, Zhu JK. Identification and characterization of endogenous small interfering RNAs from rice. Nucl Acids Res. 2005;33:4443. doi: 10.1093/nar/gki758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK. et al. Cloning and characterization of microRNAs from wheat (Triticum aestivum L.) Genome Biol. 2007;8:R96. doi: 10.1186/gb-2007-8-6-r96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeng C, Wang W, Zheng Y, Chen X, Bo W, Song S. et al. Conservation and divergence of microRNAs and their functions in Euphorbiaceous plants. Nucl Acids Res. 2010;38:981. doi: 10.1093/nar/gkp1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Botany. 2007;99:787. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS. et al. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant physiology. 2005;139:1597. doi: 10.1104/pp.105.067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu L, Ménard R, Berr A, Fuchs J, Cognat V, Meyer D. et al. The E2 ubiquitin conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009;57:279–88. doi: 10.1111/j.1365-313X.2008.03684.x. [DOI] [PubMed] [Google Scholar]
- 30.Wang BB, Brendel V. The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004;5:R102. doi: 10.1186/gb-2004-5-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egelund J. Blast-, alignment-and phylogenetic analysis of PAG1 [PhD] The Royal Veterinary and Agricultural University. 2001.
- 32.Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C. et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA. Conservation and divergence of plant microRNA genes. Plant J. 2006;46:243–59. doi: 10.1111/j.1365-313X.2006.02697.x. [DOI] [PubMed] [Google Scholar]
- 34.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl Acids Res. 2003;31:3406. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 36.Binns D, Dimmer E, Huntley R, Barrell D, O'Donovan C, Apweiler R. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics. 2009;25:3045. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Camon E, Magrane M, Barrell D, Lee V, Dimmer E, Maslen J. et al. The Gene Ontology annotation (GOA) database: sharing knowledge in Uniprot with Gene Ontology. Nucl Acids Res. 2004;32:D262. doi: 10.1093/nar/gkh021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Stem-loop structures of 52 putative miRNA precursors.
Supplementary Table 1. The predicted targets of conserved and newly identified jatropha miRNAs.