Abstract
The proteasome is a multicatalytic protease complex that degrades most endogenous proteins including misfolded or damaged proteins to ensure normal cellular function. The ubiquitin-proteasome degradation pathway plays an essential role in multiuple cellular processes, including cell cycle progression, proliferation, apoptosis and angiogenesis. It has been shown that human cancer cells are more sensitive to proteasome inhibition than normal cells, indicating that a proteasome inhibitor could be used as a novel anticancer drug. Indeed, this idea has been supported by the encouraging results of the clinical trials using the proteasome inhibitor Bortezomib (Velcade, PS-341), a drug approved by the US Food and Drug Administration (FDA). Several natural compounds, including the microbial metabolite lactacystin, green tea polyphenols, and traditional medicinal triterpenes, have been shown to be potent proteasome inhibitors. These findings suggest the potential use of natural proteasome inhibitors as not only chemopreventive and chemotherapeutic agents, but also tumor sensitizers to conventional radiotherapy and chemotherapy. In this review, we will summarize the structure and biological activities of the proteasome and several natural compounds with proteasome inhibitory activity, and will discuss the potential use of these compounds for the prevention and treatment of human cancers.
Keywords: Proteasome inhibitors, natural products, cancer prevention, cancer treatment
1. THE UBIQUITIN-PROTEASOME PATHWAY
The discovery of the ubiquitin-proteasome pathway opened a new field for cancer therapy. Accumulating evidence from laboratory studies, preclinical and clinical trails supports the concept that targeting the proteasome is a promising strategy for cancer treatment. Since the discovery of first generation of proteasome inhibitor, lactacystin, in the 1990s, more and more proteasome inhibitors were developed by chemical synthesis and isolated from natural resources. Now proteasome inhibitors are widely studied for the treatment of a variety of solid and hematological malignancies as a monotherapy or combination therapy to potentially overcome chemo- or radio-resistance. Proteasome inhibitors can be divided into several groups including, peptide aldehyde, peptide boronate, peptide vinyl sulfone, peptide epoxyketone, and those derived from natural sources [1]. Peptide aldehyde proteasome inhibitors, including MG132, MG115, and LLnL, are the most studied and best understood. However, these tripeptidyl proteasome inhibitors could not be used in vivo because of stability and toxicity issues [2]. Therefore, natural proteasome inhibitors, such as dietary polyphenols with the cancer-preventive and anticancer effects, have been increasingly studied. In this review we will summarize the proteasome inhibitory activities of several proteasome inhibitors with an emphasis on natural compounds and their potential use as anti-cancer therapies.
The proteasome is a multicatalytic enzyme complex with a molecular weight of 2.5 MDa. Usually, the cellular proteasome is referred to as the 26S proteasome in accordance with its sedimentation coefficient. The 26S proteasome complex contains a catalytic core, the 20S proteasome, which contains of four stacked rings that form a barrel with a central cavity. These stacked rings include two non-catalytic α rings (each with seven nonidentical subunits) outside of two catalytic β rings (each with seven nonidentical subunits), and form a special αββα arrangement (Fig. 1). The catalytic activity of the β rings are found within the β1, β2, and β5 subunits, which confer the 20S proteasome with caspase or peptidyl-glutamyl peptide-hydrolyzing-like (PGPH), trypsin-like, and chmotrypsin-like proteolytic activities, respectively. In accordance with general enzymes with similar hydrolytic activities, the β1 subunit cuts peptides after acidic residues, the β2 subunit prefers to cleave after basic amino acids, and the β5 subunit hydrolyzes after hydrophobic residues. In all the three β-subunits, a threonine residue at the amino terminal (Thr1) is considered to be the catalytically active amino acid [3]. Thus the catalytic 20S proteasome itself is capable of hydrolyzing most cellular proteins. However all the three hydrolysis activities are confined to the inner cavity of the 20S proteasome [3, 4], which is so narrow that only unfolded proteins are allowed to get access for degradation [4].
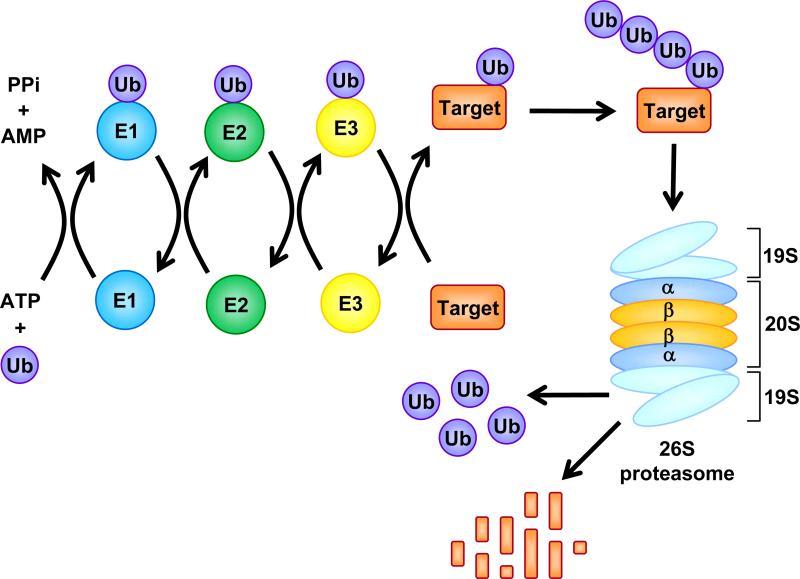
Fig. (1). The Ubiquitin-Proteasome Pathway.
Ubiquitin (Ub) is activated by Ub-activating (E1) enzymes through adenylation and formation of high-energy thiol ester bond and then transferred to Ub-conjugating (E2) enzymes. With the help of Ub-ligating (E3) enzymes, Ub is finally tranferred to a reactive lysine residue of a target protein. Ubiquitinated proteins are recognized by the 19S cap of the 26S proteasome and fed into its 20S catalytic core for degradation into oligopeptides. The Ub is then released and recycled.
Besides being the central processing unit, the 20S proteasome in eukaryotic cells also serves as a proteolytic core of 2 larger proteasomal assemblies: the 19S regulator (PA700) and the 11S activator (PA28). These molecules bind to both ends of the proteasome and modify its proteolytic properties. As a regulatory subcomplex of the 26S proteasome, the 19S particle (700 kDa) controls the access of substrates into the 20S proteasome (Fig. 1). It serves to control which proteins will be degraded, in cooperation with the ubiquitin conjugating system, through recognition, binding, and de-ubiquitination of ubiquitin tagged proteins. Furthermore, the 19S particle is responsible for the unfolding and feeding of the ubiquitinated proteins into the catalytic site of the 20S proteasome [5]. These diverse actions are fulfilled by multiple subcomplex of the 19S regulatory particle. For example, in S. cerevisae, the base of 19S subcomplex usually contains 8 subunits and 6 of them are ATPases that provide ATP to unfold protein substrates [4]. At least nine subunits composing the lid of 19S subcomplex are non ATPases that are important for 19S assembly and deubiquitination of ubiquitinated proteasome target proteins (Fig. 1) [4, 6].
The 11S activator is a 28 kD proteasome activator, associated with one or both ends of the 20S proteasome alone or in combination with a 19S regulatory complex [7]. It is a heteroheptamer or homoheptamer that stimulates the peptidase activity of the 20S proteasome and modulates the proteasome-catalyzed production of antigenic peptides presented to the immune system on MHC class I molecules [8, 9]. The 11S activator cannot recognize and degrade ubiquitinated substrates and it has been hypothesized that the 11S activator acts as an adapter molecule between the 20S proteasome and cytosolic chaperones [10].
Protein degradation by the proteasome is regulated by the ubiquitin system. Ubiquitin is a highly, evolutionarily conserved, 76-amino acid protein that serves as a tag for destruction of targeted proteins through covalent binding. Three different enzymes, Ub-activating (E1), Ub-conjugating (E2), and Ub-ligating (E3), are involved in protein ubiquitination [10]. Initially, the E1 enzyme activates ubiquitin in an ATP-requiring manner by forming a thiol ester bond at its C-terminus (Fig. 1). Activated ubiquitin is transferred from E1, to one of several distinct ubiquitin-conjugating enzymes, E2s, through an additional thiol-ester bond. With the help of E3, the activated ubiquitin is then transferred to the specific substrate at the lysine residues of a target protein (Fig. 1) [11]. One common E1, 20-30 E2s, and at least several hundreds of E3s exist in the cell [3, 7]. Combination of E2 and E3, together with E3s specificity will provide selective ubiquitination of proteasome target proteins for degradation [10].
It is well established that the ubiquitin conjugating system is necessary for proteasome degradation and it acts as a cascade to control the precise degradation of cellular proteins. On the contrary, several substrates such as ornithine decarboxylase, p21, IκBα, retinoblastoma protein (RB), and hypoxia-inducible factor-1alpha (HIF-1α) may be degraded by the proteasome via ubiquitin-tagging or in a ubiquitin-free manner [12, 13]. In fact, it is well established that HIF-1α is degraded under normoxic conditions by the ubiquitin-proteasome system. However, very recently, Kong et al reported that under hypoxic conditions HIF-1α is constitutively degraded by the proteasome, without the requirement of prior ubiquitylation [12]. These conflicting reports challenge the conventional concept that ubiquitination is necessary for protein degradation via the proteasome, but do not diminish the great importance for ubiquitin-dependent proteasome degradation.
2. BIOLOGICAL EFFECTS OF PROTEASOME INHIBITION
2.1. Cell cycle arrest
The primary, immediate consequence of proteasome inhibition is a decrease of overall rates of protein breakdown in cells [14, 15]. Various proteins involved in the processes of carcinogenesis and cancer survival have been identified as targets of the proteasome, including cyclins A, B, D and E [16-19], tumor suppressor protein p53 [20], pro-apoptotic protein Bax [21], cyclin-dependent kinase inhibitor (CKI) p27 [22, 23], and the inhibitor of NF-κB, IκB-α [24].
The cell cycle is driven by the activation/induction of cyclin-dependent kinases (Cdks), while CKIs lead to growth arrest [25, 26]. Progression through the cell cycle requires tightly regulated proteolysis of both cyclins and CKIs and numerous studies support the roles for the CKIs, p16, p21, and p27, as tumor suppressor proteins [27-29]. In fact, cell cycle arrest, due to an accumulation of the CKIs p21 and p27, occurs following treatment with proteasome inhibitors [30-33] and their accumulation is furthermore linked to mechanisms of apoptosis [22].
The CKI p27 is directed to the ubiquitin-proteasome pathway for degradation by the specific ubiquitin-conjugating enzymes Ubc2 and Ubc3 [23] and SCFSkp2 ubiquitin ligase complex consisting of Cul1, Rbx1, Skp1 and F-box Skp2 [34]. p27 must be phosphorylated by Cdk2 at threonine 187 (T187) for recognition by the SCFSkp2 complex [35]. Cdk1 acts as an adaptor that binds to Skp2 and to phosphorylated p27 at phosphorylated T187 [36]. Furthermore, upregulation of p21 has been shown to be induced by the specific proteasome inhibitors lactacystin and MG132 in p53-deficient colon cancer DLD1 cells [37]. Accumulation of either p27 or p21 stops the cell cycle at G1/S by binding cyclin A or E [38].
In addition to CKIs, degradation of cyclins is also regulated by the ubiquitin/proteasome pathway. Cyclin D1 degradation is mediated by phosphorylation-triggered, ubiquitin-dependent proteolysis and its degradation allows the cell passage out of G1 [19]. Similarly, cyclin E protein must be degraded to allow the cell entry into S phase [18]. Cyclin A must be degraded to allow the cell to pass through S into G2, and cyclin B degradation is required for mitosis completion and return to G1 phase [17]. Accumulation of these proteins following proteasome inhibition leads to a halt in cell cycle progression.
2.2. Apoptosis
Apoptosis is an evolutionally conserved cellular suicide program that occurs through the intrinsic, mitochondria pathway or the extrinsic, death receptor pathway and the ratio of pro-apoptotic to anti-apoptotic proteins is critically involved in cellular survival. The central event of intrinsic apoptosis is the release of cytochrome c into the cytosol from mitochondria which is regulated by members of the pro-apoptotic Bcl-2 family of proteins, such as Bax, a monomeric protein distributed in the cytosol. Upon apoptosis stimulation by proteasome inhibitors, for example lactacystin or MG132, Bax undergoes a conformational change to form a functional Bax-Bax dimer. This dimer translocates to the mitochondria resulting in the loss of the mitochondrial membrane potential and cytochrome c release [39]. Furthermore, the tripeptidyl proteasome inhibitor N-carbobenzoxy-l-leucyl-l-leucyl-norvalinal (LLnV) accumulates Bax to mitochondria, where it interacts with the anti-apoptotic Bcl-2 protein to overcome Bcl-2-mediated protection from apoptosis [21, 40]. Therefore, Bax degradation via the proteasome pathway is critical to maintain cancer cell survival and its accumulation via proteasome inhibition, is important for reducing the advancement of some cancers [21]. Conversely, degradation of the anti-apoptotic counterparts to Bax, such as Bcl-2 and Bcl-XL is essential to limit the growth of various tumors [41].
Extrinsic apoptosis is triggered by external stimuli through death receptors, members of the TNF (tumor necrosis factor) receptor gene superfamily [42]. Death receptors contain a cytoplasmic tail of about 80 amino acids called the death domain, which is crucial for transmision of the death signal from the cells surface to intracellular signaling pathways. Death receptors include CD95 (APO-1/Fas), TNF receptor 1 (TNFRI), TNF-related apoptosis-inducing ligand-receptor 1 (TRAIL-R1) and TRAIL-R2 and their corresponding ligands include CD95 ligand (CD95L), TNFα, TRAIL (tumor necrosis factor–related apoptosis-inducing ligand), etc. [42].
Although TNFα is a potent mediator of apoptosis, activation of the transcription factor NF-κB can block the cell death in response to TNFα and other signals and ultimately prevent apoptosis [43]. NF-κB plays an important role in tumorigenesis via transactivation of genes involved in cell proliferation, apoptosis, tumor cell invasion and metastasis, and angiogenesis [44]. NF-κB is normally sequestered in the cytoplasm through association with its endogenous inhibitor IκB-α. SCFβ-TrCP ubiquitin ligase recognizes phosphorylated 19-amino-acid destruction motif of IκB-α at the residues 21-41 and makes it ubiquitinated for proteasome degradation [45], while NF-κB is allowed to be translocated to the nucleus and mediate transcription of anti-apoptotic genes. However, accumulation of the IκB-α protein via proteasome inhibition prevents the activation of anti-apoptotic NF-κB [46], resulting in tumor cell apoptosis.
3. DIFFERENTIAL EFFECTS OF PROTEASOME INHIBITION IN NORMAL CELLS VS. TUMOR CELLS
Accumulating evidence indicates that tumor and normal cells behave differently in response to proteasome inhibition although a well-defined mechanism to elucidate tumor cell susceptibility and normal cell resistance to proteasome inhibitors has not been established. The greater sensitivity of proliferating cells to apoptosis following proteasome inhibition is likely a consequence of the accumulation of the proteins described in Section 2 and their effects on cellular activities.
Several studies have demonstrated that proliferating tumor cells are more sensitive to proteasome inhibition than non-proliferating cells [40, 47-50] and upon proteasome inhibition, these tumor cells undergo apoptosis at early time points, in as early as 15 min [51-54]. In fact, we have shown that SV40-transformed fibroblasts treated with the proteasome inhibitor CEP1612 exhibited an accumulation of the proteasome target protein p27, but this was not the case in the parent, normal fibroblast cell line [40]. Furthermore, IκBα, p27, and Bax are accumulated by a variety of proteasome inhibitors in numerous tumor cell lines but not in non-transformed or normal cell lines [51, 55-57].
Recently, we also reported that a newly-developed proteasome inhibitor, a disulfiram-copper complex, potently inhibits the proteasomal activity in cultured breast cancer MDA-MB-231 and MCF10DCIS.com cells before induction of apoptotic cell death [58]. In sharp contrast, the disulfiram-copper complex neither inhibited the proteasome nor induced apoptosis in normal, immortalized MCF-10A cells [58]. Furthermore, MDA-MB-231 cells, containing copper concentrations similar to those found in patients, treated with disulfiram, demonstrated proteasome-inhibitory and apoptosis inducing effects. Likewise, in mice bearing MDA-MB-231 tumor xenografts, disulfiram significantly inhibited the tumor growth associated with in vivo proteasome inhibition and apoptosis induction. In this system, it appears that inhibition of the proteasomal activity can be achieved by targeting tumor cellular copper with disulfiram to selectively induce apoptosis in tumor cells [58].
Others have found similar, potent proteasomal inhibitory effects in cancer cells while normal cells appear unaffected. Accumulation of NOXA (a pro-apoptotic member of the Bcl-2 family of proteins) can negate the multiple Bcl-2 pro-survival family members and facilitate mitochondrial cytochrome c, SMAC, and apoptosis-inducing factor (AIF) release with subsequent DNA degradation and apoptosis. It has been shown that melanoma and myeloma cells accumulate NOXA upon treatment with MG-132, lactacystin, and bortezomib while NOXA induction was not triggered in untransformed melanocytes [59]. Furthermore, the proteasome inhibitor Z-Ile-Glu(OtBu)-Ala-Leucinal (PSI) has been found to induce apoptosis in multiple human myeloid leukemia cell lines at low doses (IC50 ranging from 5 to 25 nmol/l) indicated by nuclear DNA fragmentation, cleavage of poly (ADP-ribose) polymerase (PARP) and of beta-catenin. Importantly, PSI was found to be cytotoxic to leukemia, but not normal hematopoietic progenitor cells [50]. These studies indicate that the untransformed cell line is much less sensitive to proteasome inhibitor-induced apoptosis.
Finally, B-cell chronic lymphocytic leukemia (B-CLL) cells have been shown to exhibit a hypersensitivity to apoptotic cell death via proteasome inhibition when treated with lactacystin [60]. Furthermore, these leukemia cells were found to have a 3-fold increased level of the chymotrypsin-like activity than normal lymphocytes [61]. These findings suggest that one possible explanation for the difference of tumor vs. normal cells in response to proteasome inhibition is the basal proteasome activity level. At least in the case of B-CLL, these tumor cells may require higher proteasomal chymotrypsin-like activity for rapid proliferation compared to normal cells.
4. BORTEZOMIB
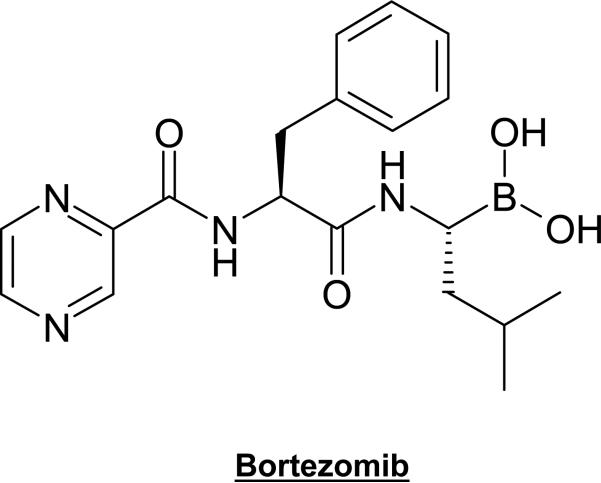
Bortezomib (PS-341, Velcade), the most-described and best known proteasome inhibitor, is a dipeptide boronic acid analogue (Fig. 2) with cell death-inducing activity found in several tumor cell lines and animal models [62-65]. Its inhibitory activity is directed mostly against the β5-(chymotrypsin-like) and β1-(PGPH-like) subunits, with the β5-subunit being the predominant cell-death inducing target [65]. A major mode of action of bortezomib is its ability to block nuclear translocation of NF-κB through the stabilization of its inhibitor, IκB. This leads to decreased expression of myeloma cell adherence factors, and interference with adherence-mediated induction of interleukin-6 production by bone marrow stromal cells. Bortezomib was also shown to suppress p44/42 mitogen-activated protein kinase (MAPK) signaling and to induce accumulation of CDKs p21 and p27 [66].
Fig. (2).
Chemical structure of Bortezomib.
Bortezomib is the first proteasome inhibitor to enter clinical trials and to receive regular approval from the US Food and Drug Administration for the treatment of relapsed and refractory multiple myeloma [67]. After it exhibited considerable success against multiple myeloma and non-Hodgkin lymphoma [66], bortezomib was also tested against other hematological aberrations and solid tumors [68, 69], both as a single agent and in combination with conventional therapies [70, 71].
4.1. Phase I Bortezomib Clinical Trials
The efficacy of bortezomib in combination with melphalan, doxorubicin, and dexamethasone was investigated in phase I clinical trials with relapsed or refractory hematological malignancies. A phase I/II trial showed that the combination of bortezomib and melphalan was significantly superior to melphalan treatment alone in patients not eligible for autologous stem cell transplantation and in those with poor prognostic features [72]. Favorable responses were observed in non-Hodgkin lymphoma, acute myeloid leukemia, and multiple myeloma patients with a combination of bortezomib and doxorubicin [73]. Finally, the improved response without prohibitive toxicity was found in patients with relapsed and/or refractory myeloma after combination treatment with bortezomib and dexamethasone [70].
Phase I clinical studies with solid tumors also showed some promising data when bortezomib was used either as a single agent or as a part of combination therapy. Bortezomib alone showed anti-tumor activity in patients with advanced androgen-independent prostate cancer [69], and the combination of bortezomib and carboplatin elicited an overall response rate of 47% in recurrent ovarian or primary peritoneal cancer, including one complete response in a patient with platinum-resistant disease [74].
4.2. Phase II
Recently, phase II clinical trials indicated beneficial activity of bortezomib against refractory multiple myeloma [75] and relapsed or refractory mantle cell lymphoma [76]. Moreover, bortezomib in combination with dexamethasone was shown to be an effective therapy in untreated symptomatic multiple myeloma patients [70]. In phase II trials with relapsed non-Hodgkin's lymphoma, results with bortezomib treatment were also encouraging, with several partial and complete responses [77, 78].
4.3. Phase III
The APEX (Assessment of Proteasome Inhibition for Extending Remissions), a phase III trial, was a large international study which compared bortezomib with high-dose dexamethasone in patients with multiple myeloma who had relapsed after 1–3 prior therapies [79]. This critical trial exemplified that bortezomib had superior clinical efficacy over dexamethasone in terms of response rate, time to progression, and survival.
4.4. Toxicity of bortezomib
The most frequent toxic side effects associated with bortezomib treatment are nausea, fatigue, and diarrhea [75, 80], and more adverse events include thrombocytopenia, peripheral neuropathy, neutropenia, lymphopenia, and hyponatremia. Patients who started bortezomib treatment with low baseline platelet counts were at the greatest risk of developing clinically significant thrombocytopenia [75]. While baseline symptoms of peripheral neuropathy were often present and were attributed to prior treatment with neurotoxic agents, neuropathy was improved or was resolved in the majority of patients upon discontinuation of bortezomib [75].
5. NATURAL COMPOUNDS WITH PROTEASOME INHIBITORY ACTIVITY
To reduce the recovery period and potentially promote the efficaciousness of chemotherapeutics, it is critical to eliminate noxious substances in the treatment of cancer. The cancer-preventive and anticancer effects of natural products are widely supported by results from epidemiological, cell culture, and animal studies and a number of microbial metabolites and plant polyphenols have been implicated as natural proteasome inhibitors (reviewed in [81] and [82]). We reviewed several natural compounds here based on their common structure, the carbonyl group. The carbonyl carbons such as the ester carbon in lactacystin and EGCG or the ketone carbon in other natural mentioned compounds are electrophilic since the oxygen pulls electron density away from it. Electrophilic functional groups of inhibitors are susceptible to be attacked by the proteolytically active Thr 1 Oγ that is nucleophilic [83], which might be responsible for the observed proteasome inhibition.
5.1. Lactacystin
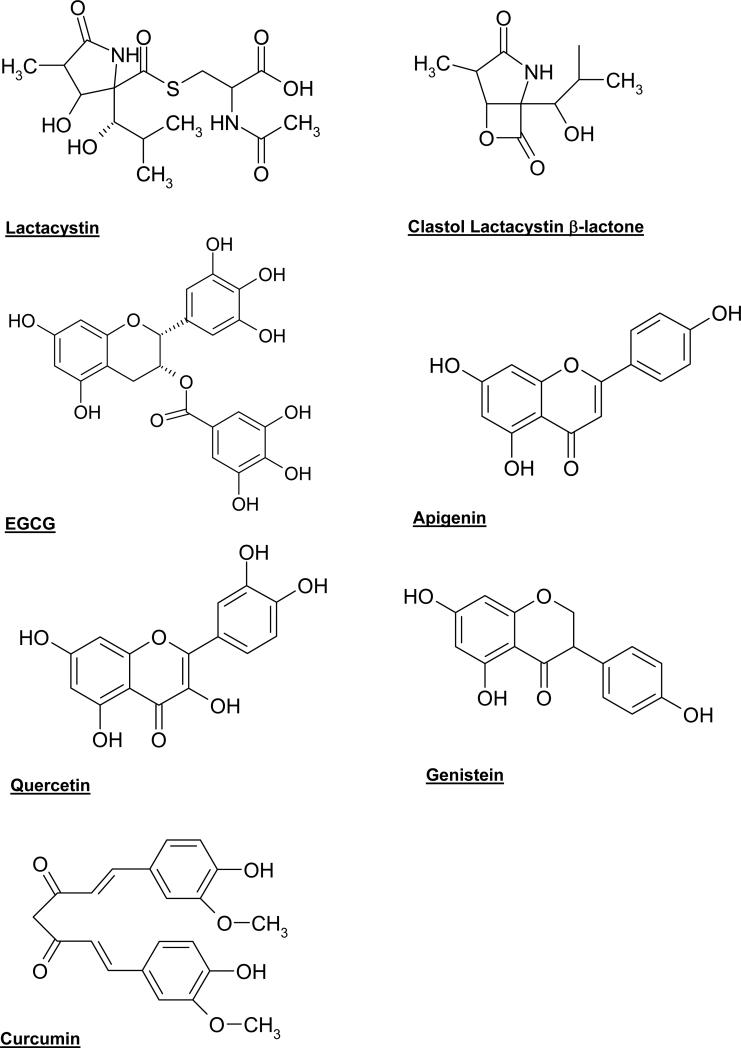
Lactacystin is a microbial natural product found in Streptomyces metabolites that inhibits cell proliferation and induces neurite outgrowth in the murine neuroblastoma cell line Neuro 2A [84, 85]. Additionally, lactacystin inhibits cell cycle progression in human osteosarcoma MG-63 cells [86]. Structure-activity relationship studies revealed that the proteasome was the cellular target of lactacystin. The highly conserved amino-terminal threonine of the mammalian proteasome was found to be covalently modified by lactacystin's electrophilic carbonyl carbon (Fig. 3), a critical site for nucleophilic attack and its biological activity [87]. Lactacystin inhibits all three proteolytic activities of the proteasome: trypsin-like, chymotrypsin-like and peptidylglutamyl-peptide hydrolyzing activities, but prefers to inhibit chymotrypsin-like activity [87]. Under pH 8.0, it gets hydrolyzed to a potent intermediate clasto-lactacystin β-lactone (Fig. 3) [88].
Fig. (3).
Chemical structures of lactacystin, clasto-lactacystin β-lactone and several natural polyphenols.
Specific inhibition of chymotrypsin-like activity is an efficient biological effect for the induction of neurite outgrowth [89], and apoptosis in cancer cells. This property entitles the use of lactacystin as a monotherapy in a variety of cancers [90-92]. However high doses of lactacystin are required to overcome the blood/brain barrier to reach to brain tumors and the accompanied cytotoxicity limits its use in these cancers [93]. However, very recently, one report proposed the use of lactacystin in the treatment of malignant gliomas using a local drug-delivery system for administration. It can be efficiently incorporated into controlled-release polymers that are, at the proposed concentration, safe for CNS delivery, yet effective against the tumor in rat 9L gliosarcoma model [93].
In addition to monotherapy, synergistic effects of lactacystin for cancer treatment have also been extensively investigated in a number of cancer cell lines. A combination of lactacystin with the peptide aldehyde proteasome inhibitor MG132 induced additive apoptosis in prostate cancer cells [94]. In ovarian carcinoma, treatment with 20 μM lactacystin was found to overcome the acquired resistance of cisplatin by preventing the loss of copper transporters that modulate the cellular accumulation of cisplatin [95]. Additionally, lactacystin significantly decreased repair of cisplatin-DNA adducts and reduced expression of the DNA repair gene ERCC-1 (excision repair cross complementation group 1) in cisplatin-resistant ovarian cancer cells, ultimately enhancing tumor cell sensitivity [96]. Furthermore, lactacystin sensitizes refractory colon cancer HT-29 cells to etoposide and doxorubicin through restoration of topo IIα expression in vitro and in vivo [97].
Lactacystin may not only act as sensitizer, but may also enhance the effects of chemotherapeutic agents. One report showed that although lactacystin alone triggered a moderate level of apoptosis in lymphoma Raji B cells, its apoptosis effect was potentiated when combined with cell-permeable, N-terminal Smac peptides [98].
In addition to primary tumor growth inhibition, lactacystin is also capable of inhibiting tumor invasion. One group reported that treatment with 10 μM lactacystin inhibited Matrigel invasion of highly invasive PC-3 cells by 25% via reducing NF-κB activity [99]. Likewise, lactacystin appears to inhibit cell migration and tumor metastasis by reducing expression of L-isoaspartyl methyltransferase (PIMT), the enzyme responsible for repairing damaged proteins that have accumulated abnormal aspartyl residues during cell aging [100].
5.2. Green tea polyphenols
Tea (Camellia sinensis) is the most popular beverage, besides water, in the world. Tea consumption is suggested to bring about cancer-preventive and anti-oxidant effects from tea polyphenols including (–)-epicatechin (EC), (–)-epigallocatechin (EGC), (–)-epicatechin-3-gallate (ECG), and (–)-epigallocatechin-3-gallate (EGCG) [101]. Among them, EGCG is the most abundant and potent constituent that has been shown to be a potent and specific proteasome inhibitor (Fig. 3) [57]. It inhibits the chymotrypsin-like activity of the proteasome in vitro with an IC50 value around 86 to194 nM and in vivo at 1-10 μM. These in vivo EGCG concentrations are comparable with the concentrations found in the serum of green tea drinkers [57].
Green tea polyphenols could activate or stabilize tumor suppressor genes and affect several signal transduction and ubiquitin/proteasome degradation pathways. In a human liver cancer cell system, (-)-EGCG could inhibit cell proliferation, block cell cycle progression in the G1 phase and induce apoptosis in hepatoma (HepG2) cells [102]. Regarding the molecular targets, the results showed that EGCG significantly increased the expression of p53 and p21 protein and an enhancement in Fas/APO-1 and Bax protein was observed as well [102]. Similar results were observed in human prostate cancer cells.
Hastak et al [103] used human prostate carcinoma LNCaP cells with wild-type p53 as a model to study the role of p53 and NF-κB in EGCG-induced growth arrest and apoptosis. They found that EGCG could stabilize p53 protein via phosphorylation of critical serine residues on p53 and modulation of MDM2-p14ARF pathway. Furthermore the results demonstrated that activation of p53-dependent downstream targets p21 and Bax and down-regulation of NF-κB-dependent Bcl-2 resulted in growth arrest and apoptosis [103].
EGCG also exhibits antiangiogenic activities in various experimental studies [104, 105]. Zhang et al tested the effects of EGCG and green tea extracts on HIF-1α and its downstream target, vascular endothelial growth factor (VEGF), which play a critical role in tumor angiogenesis. Contrary to the normal consequences of proteasome inhibition, EGCG significantly inhibited the protein level of transcription factor HIF-1α in human cervical carcinoma (HeLa) and HepG2 cells. Treatment of EGCG also resulted in a drastic decrease in VEGF expression at both mRNA and protein levels in HeLa cells [105]. Similar effects have been found in other proteasome inhibitors. Inoue et al reported that proteasome inhibitor MG115 downregulated X-linked inhibitor of apoptosis protein (XIAP) and MG132 downregulated AKT in human hepatocellular carcinoma cells [106]. We also reported that proteasome inhibitor celastrol downregulated androgen receptor while it upregulated Bax, IkB-a and p27 [53]. HIF-1α is a transcription factor of oxygen-regulated genes that are involved in cell proliferation, cell survival and angiogenesis such as VEGF [107]. It seems that proteasome inhibitors induce apoptosis by selectively upregulating pro-apoptotic proteins and/or downregulating anti-apoptotic proteins. Further study by Zhang et al showed that EGCG inhibited the translational machine of HIF-1α [105], suggesting that EGCG downregulated HIF-1α at a step before protein synthesis. It has been reported that administration of EGCG and green tea extract could inhibit matrix metalloprotease-2 (MMP-2) in human umbilical vein endothelial cells (HUVECs), inhibit angiogenesis in tumors and restrain tumor growth in mice bearing highly vascular Kaposi's sarcoma tumor model xenografts [108]. It was also found that EGCG inhibited the MMP9 or MMP2 upstream regulator, membrane-type 1 matrix metalloproteinase (MT1-MMP), and suppressed ephrin-A1-induced vascular endothelial migration [109-111].
EGCG remains to be the most potent natural polyphenol in green tea. However it is unstable in neutral or alkaline conditions. In order to discover more stable and more potent tea polyphenol as proteasome inhibitors, we synthesized several novel EGCG analogs with -OH groups eliminated from the B- and/or D-rings [51]. In addition, we also synthesized their putative prodrugs with -OH groups protected by acetate, named Pro-EGCG [51]. We found that, compared to EGCG, protected analogs exhibited greater potency to inhibit proliferation and induce apoptosis in human leukemic, prostate and breast cancer cells [56]. Most recently, we compared Pro-EGCG with natural EGCG for their anti-tumor activity in vivo. The results showed that treatment with EGCG for 31 days in human breast MDA-MB-231 tumors induced in nude mice resulted in an inhibition of tumor growth (23%). Interestingly, Pro-EGCG significantly inhibited tumor growth by 56% [112].
Furthermore, the OH groups of EGCG can be modified through major biotransformation reactions including methylation, glucuronidation, and sulfonation, resulting in reduced biological activities of EGCG in vivo [113-115]. A recent report found that women who carried at least one low activity catechol-O-methyltransferase (COMT) allele and were tea drinkers had a significantly reduced risk of breast cancer compared with non-tea drinkers [116]. In contrast, risk of breast cancer did not differ between tea drinkers and non-tea drinkers among those who were homozygous for the high activity COMT allele [116]. Using synthesized methylated tea polyphenols, we found that they were poor proteasome inhibitors and therefore less cancer-protective. These results suggested that high COMT activity could methylate and inactivate EGCG, resulting non-benefit of tea drinking in high COMT patients [117].
5.3. Grape Polyphenols
Grape extracts contain several flavonoids including resveratrol, apigenin and quercetin, all of which have been shown to possess anti-proliferative activities in tumor cells [118-121] in vitro. Resveratrol (trans-3, 4, 5-trihydroxystilbene, Fig. 3), found in wine, grape skins, and other nuts, fruits, and vegetables, is a potential chemopreventive and antitumor compound, which inhibits multiple steps of carcinogenesis in a variety of cancers [122]. Although resveratrol has been shown to have promising activities as a sensitizer for chemotherapeutic combination strategies [81], it is unclear whether resveratrol has direct proteasome-inhibitory effects. However, inhibition of NF-κB [123] suggests proteasome inhibition as a potential mechanism of action of resveratrol.
Apigenin (5,7,4-trihydroxyflavone, Fig. 3) present in grapes, parsley, chamomile tea, etc. [124-126], is a nontoxic, antimutagen, with antioxidant potential [124, 127, 128]. As a proteasome inhibitor, apigenin potently inhibits chymotrypsin-like activity of the proteasome [55]. In fact, an in silico docking method, revealed that the carbonyl carbon of apigenin (Fig. 3) was a site of nucleophilic susceptibility and could bind to the chymotrypsin site in an orientation and conformation that is suitable for a nucleophilic attack by Thr 1 [55]. Furthermore, biological analysis of apigenin indicates strong inhibitory activity against a purified 20S proteasome (IC50 = 1.8-2.3 μM), and 26S proteasome in intact leukemia Jurkat cells (1-10 μM). Accumulation of the proteasome substrates Bax and IκB-α as well as caspase-3 activation and apoptotic PARP cleavage were validated in leukemia Jurkat cells treated with apigenin. A final indicator of apigenin's potential value as a therapeutic proteasome inhibitor was the lack of toxicity in normal, non-transformed cells [55].
Quercetin (3,3',4',5,7-Pentahydroxyflavone, Fig. 3) is one of the most abundant flavonoids found in fruits and vegetables with the highest concentrations found in onions, apples, and red wine [129]. The potential for quercetin to act as a proteasome inhibitor is strongly suggested by its inhibitory activity against a purified 20S proteasome (IC50 = 3.5 μM) and 26S proteasome in intact tumor cells (2 μM) [55]. Again, in silico docking analysis, demonstrated that the carbonyl carbon acted as a site of nucleophilic susceptibility and could bind to the chymotrypsin site in an orientation and conformation suitable for a nucleophilic attack by Thr 1 [55]. The proteasome substrates Bax and IκB-α were found to be accumulated and apoptotic PARP cleavage and caspase-3 activation in a dose- and time-dependent manner were observed. While leukemia Jurkat cells were subject to proteasome inhibition and apoptosis induction, normal, non-transformed cells appeared to be unaffected [55].
5.4. Soy (Genistein)
The soybean is a natural product that is uniquely rich in polyphenols called isoflavones, and numerous studies have evaluated its anticancer activities. As a result, epidemiological studies associate soy consumption with reduced cancer occurrence and genistein (Fig. 3), one of the predominant soy isoflavones, inhibits the growth of numerous human tumor cell lines [130-132].
To examine the possibility that genistein acts as a proteasome inhibitor, we have utilized in silico computational docking studies using genistein as the ligand and the proteasome as the molecular target to show that genistein interacts with the proteasomal β5 subunit in a manner that suggests proteasome inhibition [133]. Consistently, ~30% of the chymotrypsin-like activity of a purified 20S proteasome is inhibited by 1 μM of genistein [133]. It should be noted that plasma levels of genistein are in a range of 0.5–2.5 μM and the concentrations of genistein vary in different tissues and organs [134, 135]. Additionally, numerous studies have observed down-regulation of NF-κB activity, a downstream event of proteasome inhibition, in tumor cells in the presence of genistein (reviewed in [136]). These findings indicate the possibility that a partial inhibition of proteasome activity by genistein at physiological concentrations might contribute to its reported cancer-preventative and anticancer effects.
To fully inhibit cellular proteasome chymotrypsin-like activity in intact prostate and breast cancer (but not normal, untransformed) cells, higher concentrations (as much as 50 μM) of genistein may be required [133], indicating that either genistein is unstable in vivo or other targets may exist. In fact, it has been reported that genistein at 10-50 μM concentrations could inhibit tyrosine kinase activities in hepatic [137] and prostate cells [138]. Therefore, genistein could have multiple cellular targets and simultaneous inhibition of these cancer-specific targets by genistein may be responsible for its cancer-preventive properties.
A phase II clinical trial investigating combined therapy of genistein (optimal dosing to be determined) and gemcitabine hydrochloride for the treatment of patients with stage IV breast cancer (NCI identification: NCT00244933; Karmanos Cancer Institute) is underway to determine if multiple pathways could be targeted using multiple agents against drug resistant cells. Similarly, a phase II study will evaluate the use of genistein (optimal dosing to be determined) together with gemcitabine and erlotinib to treat patients with locally advanced or metastatic pancreatic cancer (NCI identification: NCT00376948; Karmanos Cancer Institute). The dietary cancer-preventive agent genistein is expected to potentiate antitumor activities of common chemotherapeutics in these studies. However, the effective dose of genistein remains to be determined in these in-progress trials.
5.5. Turmeric (Curcumin)
Curcumin (diferuloylmethane, Fig. 3) is a main component of the spice turmeric, commonly used in Indian and Southeast Asian foods. Curcumin suppresses all three stages of carcinogenesis, initiation, promotion and progression, probably due to inhibition of NF-κB, which is controlled by the proteasome-mediated proteolytic degradation pathway [139], and subsequent inhibition of pro-inflammatory pathways [140].
A recent report showed that curcumin enhanced the antitumor activity of celecoxib, indicating their synergistic inhibition of the growth of colorectal cancer cells [141]. Furthermore, curcumin has been shown to enhance the antitumor activities of cisplatin, doxorubicin, and taxol in HA22T/VGH hepatic cancer cells, HeLa cells, CAOV3 and SKOV3 ovarian cancer cells [142-145]. In addition, the combined treatment of malignant glioma cells with curcumin and TRAIL induced enhanced cytotoxicity, as measured by hypo-diploid cells in sub-G1 phase, cleavage of procaspases-3, -8, and -9, and release of cytochrome c from mitochondria [146]. Furthermore, curcumin also acts as a radio-sensitizing agent. Pretreatment of a squamous cell carcinoma cell line with curcumin before exposure to 2.5 Gy of radiation caused a significant decrease in cell counts and colonies, compared to either treatment alone [147]
To determine if the potent in vitro anti-proliferative effects of curcumin could improve current standard chemotherapeutics, human phase I and II trials were conducted and curcumin was found to be safe with no dose-limiting toxicity when taken by mouth at doses up to 10 g/day [148]. Trials currently underway include an evaluation of colon cancer patients treated with gemcitabine in combination with curcumin and celecoxib (NCT00295035; Tel-Aviv Sourasky Medical Center) and pancreatic cancer patients treated with curcumin combined with gemcitabine (NCT00192842; Rambam Health Care Campus). Observations indicate that chemotherapeutics in combination with curcumin may provide a superior therapeutic index and advantage in the clinical setting for treatment of refractory tumors.
5.6. Traditional medicinal triterpenes
5.6.1. Celastrol
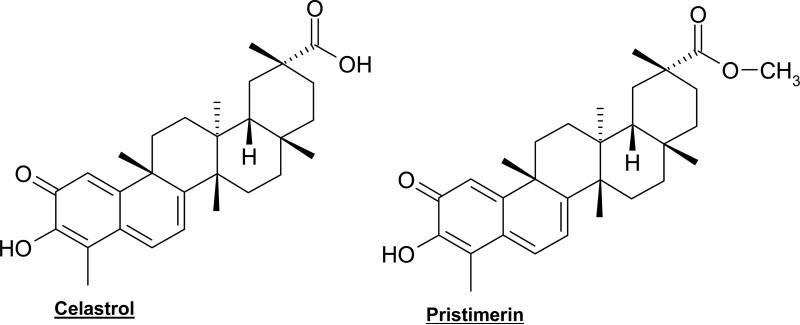
Celastrol (quione methide tripterene, Fig. 4) is a natural compound extracted from Chinese medicinal plant Tripterygium wilfordii Hook F. Due to its antioxidant and anti-inflammatory effects, celastrol has been effectively used in the treatment of autoimmune diseases (rheumatoid arthritis, systemic lupus erythematosus), asthma, chronic inflammation, and neurodegenerative disease [149, 150]. Within physiological conditions, celastrol inhibits cancer cell proliferation and induces cell death in a broad range of cancer cell lines including A549 lung, MCF-7 breast, HCT-8 ileocecal KB epidermoid nasopharyngeal, U-87 glioblastoma, PC-3 prostate, and PTX10 ovarian cell lines [151]. In fact, the antitumor activity of celastrol was shown in MDA-MB-435 breast cancer-bearing nude mice, in which celastrol inhibited ~60% tumor growth [152]. In androgen receptor (AR)-positive LNCaP cells, proteasome inhibition was accompanied by suppression of the AR protein, an event that is likely associated with Hsp90 inhibition since the ATP-binding activity of Hsp90 responsible for AR folding was inhibited [153].
Fig. (4).
Chemical structures of natural triterpenes.
We reported, for the first time, that celastrol induced apoptosis in human prostate cancer in vivo by targeting the tumor proteasome [53]. Celastrol was found to be a potent inhibitor of the proteasomal chymotrypsin-like activity in vitro, in cultured prostate tumor cells, and in animals. Consistent with previous reports [40, 154], inhibition of the proteasomal chymotrypsin-like activity by celastrol in prostate cancer cells and xenografts was associated with its apoptosis-inducing and/or antitumor activities.
Our recent studies suggested that a ketone carbon in dietary flavonoids is responsible for interacting in and inhibiting the proteasome [55] and the conjugated ketone carbons of celastrol (Fig. 4) most likely contribute to its proteasome-inhibitory potency. To provide direct evidence for proteasome inhibition by celastrol, we performed a cell-free proteasome activity assay using a purified rabbit 20S proteasome in the presence of celastrol at various concentrations. Celastrol potently and preferentially inhibited the chymotrypsin-like activity of a purified 20S proteasome with an IC50 value of 2.5 μM.
In intact human prostate cancer cells, the cellular 26S proteasome was inhibited by celastrol at 1-5 μΜ. The inhibition of proteasome activity by celastrol resulted in accumulation of ubiquitinated proteins and three natural proteasome substrates (IκB-α, Bax, and p27), and induction of apoptosis in AR-negative PC-3 cells. Additionally, treatment of PC-3 tumor-bearing nude mice with celastrol (1-3 mg/kg/day, i.p., for 1 to 31 days) resulted in significant inhibition (65-93%) of tumor growth. Multiple assays using the animal tumor tissue samples from both early and end time-points demonstrated in vivo inhibition of the proteasomal chymotrypsin-like activity and induction of apoptosis after celastrol treatment [53].
To determine if the conjugated ketone carbons found in celastrol were required for proteasome inhibition, the proteasome inhibitory activities of celastrol were compared with its reduced form, dihydrocelastrol. The IC50 value of dihydrocelastrol in the presence of a purified proteasome was determined to be 10 μΜ, suggesting that the reduced ketone decreased the proteasome-inhibitory activity by 4-fold compared to celastrol (IC50 value of 2.5 μM). Similarly, LNCaP cells treated with 5 μΜ of celastrol for 16 hours resulted in greater proteasome inhibition than did treatment with dihydrocelastrol under the same conditions (48% vs 4%) and kinetic experiments revealed that celastrol, but not dihydrocelastrol, caused a significant reduction of AR protein expression. Additionally, celastrol was found to have greater apoptosis-inducing ability than dihydrocelastrol, as shown by caspase activation and PARP cleavage at both earlier time points and higher levels. Thus the comparison of the highly oxidized celastrol with a reduced form supports that the conjugated ketone structure is critical for proteasome inhibition and apoptosis induction [155].
Additional mechanisms of celastrol action have also been described. It has been reported that dietary celastrol administration to G93A SOD1 transgenic mice, a mouse model of amyotrophic lateral sclerosis (ALS), significantly improved weight loss, motor performance and delayed the onset of ALS. It potently increased expression of heat shock proteins including HSP70, while reducing TNF-α and iNOS expression in G93A mice tissues compared to the control [156]. As an inhibitor of the NF-κB pathway, celastrol inhibits IκB-α kinase activation, IκB-α phosphorylation, IκB-α degradation, p65 nuclear translocation and phosphorylation, DNA-binding activity and NF-κB-mediated reporter gene expression induced by TNF-α or phorbol myristyl acetate (PMA) [152, 157]. Furthermore, is has been shown that celastrol combined with other chemotherapeutic agents inhibits tumor invasion in human lung adenocarcinoma H1299 cells [157]. Celastrol has been shown to be active as a topoisomerase II inhibitor, exhibiting ~5-fold greater apoptosis inducing potency than etoposide in HL-60 leukemia cells [158]. All these reports demonstrate that celastrol has a great potential to be used for cancer treatment.
As a potential therapy in the clinical setting, celastrol may be useful in the treatement of prostate cancer. Since 2004, docetaxel-based chemotherapy has become the standard chemotherapy used to treat advanced hormone-resistant prostate cancer [159]. However, docetaxel can't overcome two disadvantages that preclude its use as a monotherapy for cancer treatment, one is the constitutive activation of NF-κB in prostate cancer cells and another is that the high-dose required for treatment is accompanied with significant toxicity such as neurotoxicity. Celastrol is capable of compensating these two points, suggesting it might be a good candidate for prostate cancer treatment as a monotherapy or in combination with a current standard chemotherapy.
5.6.2. Pristimerin
Pristimerin is a natural methyl ester of celastrol (Fig. 4) found in the plant sources of Celastraceae or Hippocrateaceae families [151, 160, 161]. It has been known that pristimerin possesses anti-inflammatory, antioxidant, antimalarial, and insecticidal activities [162-165]. Most recently, it has been shown that pristimerin induces apoptosis in breast cancer MDA-MB-231 cells in a caspase-dependent manner [167]. The proteasome was suggested as a potential target of pristimerin after computational modeling was applied. A possible interaction between the conjugated ketone carbon of pristimerin (Fig. 4) and Thr1 of the proteasome β5 subunit was observed. More importantly, pristimerin inhibited the chymotrypsin-like activity of a purified 20S proteasome with an IC50 value of 2.2 μM [168]. Accumulation of proteasome target proteins Bax, p27 and IκB-α demonstrated that the proteasome inhibition by pristimerin is functional. The events of caspase-3 activation and PARP cleavage further supports that proteasome inhibition is responsible for apoptosis induction by pristimerin [166].
CONCLUSIONS
The proteasome inhibitors have attracted an increasingly attention in cancer prevention and treatment. As a single agent for multiple myeloma treatment, the antitumor activity of the proteasome inhibitor bortezomib is well established. Furthermore, the chemo-sensitizing properties of bortezomib have been demonstrated in tumor types other than myeloma. However, bortezomib causes some toxicity, generating attention for the identification of non-toxic natural proteasome inhibitors such as natural products and plant polyphenols.
Natural products such as green tea polyphenols, grape polyphenols, soy isoflavones, etc., are potent bioactive molecules that possess anti-carcinogenic activities. They interfere with the initiation, development and progression of cancer by the modulation of various mechanisms including cellular proliferation, differentiation, apoptosis, angiogenesis, and metastasis. Moreover, cancer cell lines and tumors appear to be more susceptible to the bioactivities of these natural compounds than normal cells. Although the natural compounds examined in this review exhibit various cellular effects, they all share a common carbonyl group and have the ability to inhibit cellular proteasomal activity which is associated with tumor growth suppression. Furthermore, examples of natural compounds combined with conventional synthetic drugs used in cancer treatment have been described, providing an interesting potential for combination therapy.
ACKNOWLEDGEMENT
The research was partially supported by grants from National Cancer Institute (CA112625 and CA120009 to Q.P. Dou.).
ABBREVIATIONS
- PGPH
peptidyl-glutamyl peptide-hydrolyzing-like
- Thr1
threonine residue at the amino terminal of β5 subunit of the proteasome
- HIF-1α
hypoxia-inducible factor-1alpha
- CKI
cyclin-dependent kinase inhibitor
- Cdks
cyclin-dependent kinases
- LLnV
N-carbobenzoxy-l-leucyl-l-leucyl-norvalinal
- PSI
Z-Ile-Glu(OtBu)-Ala-Leucinal
- B-CLL
B-cell chronic lymphocytic leukemia
- PARP
poly(ADP-ribose) polymerase
- MAPK
mitogen-activated protein kinase
- APEX
Assessment of Proteasome Inhibition for Extending Remissions
- PIMT
-isoaspartyl methyltransferase
- EGCG
epigallocatechin-3-gallate
- MT1-MMP
membrane-type 1 matrix metalloproteinase
- HUVECs
human umbilical vein endothelial cells
- COMT
catechol-O-methyltransferase
- TRAIL
tumor necrosis factor–related apoptosis-inducing ligand
- AIF
apoptosis-inducing factor
- PMA
phorbol myristyl acetate
- ERCC-1
excision repair cross complementation group 1
- VEGF
vascular endothelial growth factor
- TNF
tumor necrosis factor
- XIAP
X-linked inhibitor of apoptosis protein
REFERENCES
- 1.Lu M, Dou QP, Kitson RP, Smith DM, Goldfarb RH. J Cell Biochem. 2006;97:122–134. doi: 10.1002/jcb.20543. [DOI] [PubMed] [Google Scholar]
- 2.Adams J, behnke Mark., Chen s., Cruickshank AA, Dick LR, Grenier L, Klunder JM, Ma YT, Plamondon L, Stein RL. Bioorg Med Chem Lett. 1998;8:333–338. doi: 10.1016/s0960-894x(98)00029-8. [DOI] [PubMed] [Google Scholar]
- 3.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM, Cohen RE. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 5.Nandi D, Tahiliani P, Kumar A, Chandu D. J Biosci. 2006;31:137–155. doi: 10.1007/BF02705243. [DOI] [PubMed] [Google Scholar]
- 6.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. PLoS Biol. 2006;4:1314–1323. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hendil KB, Khan S, Tanaka K. Biochem J. 1998;332(Pt 3):749–754. doi: 10.1042/bj3320749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knowlton JR, Johnston SC, Whitby FG, Realini C, Zhang Z, Rechsteiner M, Hill CP. Nature. 1997;390:639–643. doi: 10.1038/37670. [DOI] [PubMed] [Google Scholar]
- 9.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 10.Nalepa G, Rolfe M, Harper JW. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover A, Orian A, Schwartz AL. Bioessays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 12.Kong X, Alvarez-Castelao B, Lin Z, Castano JG, Caro J. J Biol Chem. 2007;282:15498–15505. doi: 10.1074/jbc.M700704200. [DOI] [PubMed] [Google Scholar]
- 13.Hoyt MA, Coffino P. Cell Mol Life Sci. 2004;61:1596–1600. doi: 10.1007/s00018-004-4133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Cell. 1994;78:761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 15.Craiu A, Gaczynska M, Akopian T, Gramm CF, Fenteany G, Goldberg AL, Rock KL. J Biol Chem. 1997;272:13437–13445. doi: 10.1074/jbc.272.20.13437. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Lee J, Cho SY, Fine HA. Cancer Res. 2004;64:3949–3957. doi: 10.1158/0008-5472.CAN-03-3906. [DOI] [PubMed] [Google Scholar]
- 17.Glotzer M, Murray AW, Kirschner MW. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 18.Won KA, Reed SI. Embo J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 19.Diehl JA, Zindy F, Sherr CJ. Genes Dev. 1997;11:957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 20.Blagosklonny MV. Int J Cancer. 2002;98:161–166. doi: 10.1002/ijc.10158. [DOI] [PubMed] [Google Scholar]
- 21.Li B, Dou QP. Proc Natl Acad Sci U S A. 2000;97:3850–3855. doi: 10.1073/pnas.070047997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun J, Nam S, Lee CS, Li B, Coppola D, Hamilton AD, Dou QP, Sebti SM. Cancer Res. 2001;61:1280–1284. [PubMed] [Google Scholar]
- 23.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 24.Perkins ND. Trends Biochem Sci. 2000;25:434–440. doi: 10.1016/s0968-0004(00)01617-0. [DOI] [PubMed] [Google Scholar]
- 25.Sherr CJ, Roberts JM. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 26.Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massague J, Roberts JM, Koff A. Genes Dev. 1994;8:9–22. doi: 10.1101/gad.8.1.9. [DOI] [PubMed] [Google Scholar]
- 27.Serrano M, Gomez-Lahoz E, DePinho RA, Beach D, Bar-Sagi D. Sience. 1995;267:249–252. doi: 10.1126/science.7809631. [DOI] [PubMed] [Google Scholar]
- 28.Sherr CJ, Roberts JM. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 29.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 30.Orlowski RZ. Cell Death Differ. 1999;6:303–313. doi: 10.1038/sj.cdd.4400505. [DOI] [PubMed] [Google Scholar]
- 31.Katayose Y, Kim M, Rakkar AN, Li Z, Cowan KH, Seth P. Cancer Res. 1997;57:5441–5445. [PubMed] [Google Scholar]
- 32.Milacic V, Chen D, Ronconi L, Landis-Piwowar KR, Fregona D, Dou QP. Cancer Res. 2006;66:10478–10486. doi: 10.1158/0008-5472.CAN-06-3017. [DOI] [PubMed] [Google Scholar]
- 33.Momose I, Iijima M, Kawada M, Ikeda D. Biosci Biotechnol Biochem. 2007;71:1036–1043. doi: 10.1271/bbb.60697. [DOI] [PubMed] [Google Scholar]
- 34.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 35.Carrano AC, Eytan E, Hershko A, Pagano M. Nat Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 36.Ganoth D, Bornstein G, Ko TK, Larsen B, Tyers M, Pagano M, Hershko A. Nat Cell Biol. 2001;3:321–324. doi: 10.1038/35060126. [DOI] [PubMed] [Google Scholar]
- 37.Cayrol C, Ducommun B. Oncogene. 1998;17:2437–2444. doi: 10.1038/sj.onc.1202189. [DOI] [PubMed] [Google Scholar]
- 38.Li L, Li Z, Howley PM, Sacks DB. J Biol Chem. 2006;281:1978–1985. doi: 10.1074/jbc.M508545200. [DOI] [PubMed] [Google Scholar]
- 39.Dewson G, Snowden RT, Almond JB, Dyer MJ, Cohen GM. Oncogene. 2003;22:2643–2654. doi: 10.1038/sj.onc.1206326. [DOI] [PubMed] [Google Scholar]
- 40.An B, Goldfarb RH, Siman R, Dou QP. Cell Death Differ. 1998;5:1062–1075. doi: 10.1038/sj.cdd.4400436. [DOI] [PubMed] [Google Scholar]
- 41.Green DR, Reed JC. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 42.Walczak H, Krammer PH. Exp Cell Res. 2000;256:58–66. doi: 10.1006/excr.2000.4840. [DOI] [PubMed] [Google Scholar]
- 43.Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW. Proc Natl Acad Sci U S A. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orlowski RZ, Baldwin AS., Jr. Trends Mol Med. 2002;8:385–389. doi: 10.1016/s1471-4914(02)02375-4. [DOI] [PubMed] [Google Scholar]
- 45.Winston JT, Strack P, Beer-Romero P, Chu CY, Elledge SJ, Harper JW. Genes Dev. 1999;13:270–283. doi: 10.1101/gad.13.3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas DK, Iglehart JD. J Cell Physiol. 2006;209:645–652. doi: 10.1002/jcp.20785. [DOI] [PubMed] [Google Scholar]
- 47.Drexler HC. Proc Natl Acad Sci U S A. 1997;94:855–860. doi: 10.1073/pnas.94.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drexler HC, Risau W, Konerding MA. Faseb J. 2000;14:65–77. doi: 10.1096/fasebj.14.1.65. [DOI] [PubMed] [Google Scholar]
- 49.Orlowski RZ, Eswara JR, Lafond-Walker A, Grever MR, Orlowski M, Dang CV. Cancer Res. 1998;58:4342–4348. [PubMed] [Google Scholar]
- 50.Soligo D, Servida F, Delia D, Fontanella E, Lamorte G, Caneva L, Fumiatti R, Lambertenghi Deliliers G. Br J Haematol. 2001;113:126–135. doi: 10.1046/j.1365-2141.2001.02683.x. [DOI] [PubMed] [Google Scholar]
- 51.Landis-Piwowar KR, Kuhn DJ, Wan SB, Chen D, Chan TH, Dou QP. Int J Mol Med. 2005;15:735–742. [PubMed] [Google Scholar]
- 52.Landis-Piwowar KR, Wan SB, Wiegand RA, Kuhn DJ, Chan TH, Dou QP. J Cell Physiol. 2007 doi: 10.1002/jcp.21124. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Chen D, Cui QC, Yuan X, Dou QP. Cancer Res. 2006;66:4758–4765. doi: 10.1158/0008-5472.CAN-05-4529. [DOI] [PubMed] [Google Scholar]
- 54.Daniel KG, Gupta P, Harbach RH, Guida WC, Dou QP. Biochem Pharmacol. 2004;67:1139–1151. doi: 10.1016/j.bcp.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 55.Chen D, Daniel KG, Chen MS, Kuhn DJ, Landis-Piwowar KR, Dou QP. Biochem Pharmacol. 2005;69:1421–1432. doi: 10.1016/j.bcp.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 56.Kuhn D, Lam WH, Kazi A, Daniel KG, Song S, Chow LM, Chan TH, Dou QP. Front Biosci. 2005;10:1010–1023. doi: 10.2741/1595. [DOI] [PubMed] [Google Scholar]
- 57.Nam S, Smith DM, Dou QP. J Biol Chem. 2001;276:13322–13330. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- 58.Chen D, Cui QC, Yang H, Dou QP. Cancer Res. 2006;66:10425–10433. doi: 10.1158/0008-5472.CAN-06-2126. [DOI] [PubMed] [Google Scholar]
- 59.Qin JZ, Ziffra J, Stennett L, Bodner B, Bonish BK, Chaturvedi V, Bennett F, Pollock PM, Trent JM, Hendrix MJ, Rizzo P, Miele L, Nickoloff BJ. Cancer Res. 2005;65:6282–6293. doi: 10.1158/0008-5472.CAN-05-0676. [DOI] [PubMed] [Google Scholar]
- 60.Masdehors P, Merle-Beral H, Magdelenat H, Delic J. Leuk Lymphoma. 2000;38:499–504. doi: 10.3109/10428190009059268. [DOI] [PubMed] [Google Scholar]
- 61.Masdehors P, Merle-Beral H, Maloum K, Omura S, Magdelenat H, Delic J. Blood. 2000;96:269–274. [PubMed] [Google Scholar]
- 62.Adams J, Palombella VJ, Sausville EA, Johnson J, Destree A, Lazarus DD, Maas J, Pien CS, Prakash S, Elliott PJ. Cancer Res. 1999;59:2615–2622. [PubMed] [Google Scholar]
- 63.Frankel A, Man S, Elliott P, Adams J, Kerbel RS. Clin Cancer Res. 2000;6:3719–3728. [PubMed] [Google Scholar]
- 64.Hideshima T, Richardson P, Chauhan D, Palombella VJ, Elliott PJ, Adams J, Anderson KC. Cancer Res. 2001;61:3071–3076. [PubMed] [Google Scholar]
- 65.Crawford LJ, Walker B, Ovaa H, Chauhan D, Anderson KC, Morris TC, Irvine AE. Cancer Res. 2006;66:6379–6386. doi: 10.1158/0008-5472.CAN-06-0605. [DOI] [PubMed] [Google Scholar]
- 66.Orlowski RZ. Hematology (Am Soc Hematol Educ Program) 2005:220–225. doi: 10.1182/asheducation-2005.1.220. [DOI] [PubMed] [Google Scholar]
- 67.Kane RC, Farrell AT, Sridhara R, Pazdur R. Clin Cancer Res. 2006;12:2955–2960. doi: 10.1158/1078-0432.CCR-06-0170. [DOI] [PubMed] [Google Scholar]
- 68.Dou QP, Goldfarb RH. IDrugs. 2002;5:828–834. [PubMed] [Google Scholar]
- 69.Papandreou CN, Daliani DD, Nix D, Yang H, Madden T, Wang X, Pien CS, Millikan RE, Tu SM, Pagliaro L, Kim J, Adams J, Elliott P, Esseltine D, Petrusich A, Dieringer P, Perez C, Logothetis CJ. J Clin Oncol. 2004;22:2108–2121. doi: 10.1200/JCO.2004.02.106. [DOI] [PubMed] [Google Scholar]
- 70.Jagannath S, Durie BG, Wolf J, Camacho E, Irwin D, Lutzky J, McKinley M, Gabayan E, Mazumder A, Schenkein D, Crowley J. Br J Haematol. 2005;129:776–783. doi: 10.1111/j.1365-2141.2005.05540.x. [DOI] [PubMed] [Google Scholar]
- 71.Oakervee HE, Popat R, Curry N, Smith P, Morris C, Drake M, Agrawal S, Stec J, Schenkein D, Esseltine DL, Cavenagh JD. Br J Haematol. 2005;129:755–762. doi: 10.1111/j.1365-2141.2005.05519.x. [DOI] [PubMed] [Google Scholar]
- 72.Mateos MV, Hernandez JM, Hernandez MT, Gutierrez NC, Palomera L, Fuertes M, Diaz-Mediavilla J, Lahuerta JJ, de la Rubia J, Terol MJ, Sureda A, Bargay J, Ribas P, de Arriba F, Alegre A, Oriol A, Carrera D, Garcia-Larana J, Garcia-Sanz R, Blade J, Prosper F, Mateo G, Esseltine DL, van de Velde H, San Miguel JF. Blood. 2006;108:2165–2172. doi: 10.1182/blood-2006-04-019778. [DOI] [PubMed] [Google Scholar]
- 73.Orlowski RZ, Voorhees PM, Garcia RA, Hall MD, Kudrik FJ, Allred T, Johri AR, Jones PE, Ivanova A, Van Deventer HW, Gabriel DA, Shea TC, Mitchell BS, Adams J, Esseltine DL, Trehu EG, Green M, Lehman MJ, Natoli S, Collins JM, Lindley CM, Dees EC. Blood. 2005;105:3058–3065. doi: 10.1182/blood-2004-07-2911. [DOI] [PubMed] [Google Scholar]
- 74.Aghajanian C, Dizon DS, Sabbatini P, Raizer JJ, Dupont J, Spriggs DR. J Clin Oncol. 2005;23:5943–5949. doi: 10.1200/JCO.2005.16.006. [DOI] [PubMed] [Google Scholar]
- 75.Richardson PG, Barlogie B, Berenson J, Singhal S, Jagannath S, Irwin D, Rajkumar SV, Srkalovic G, Alsina M, Alexanian R, Siegel D, Orlowski RZ, Kuter D, Limentani SA, Lee S, Hideshima T, Esseltine DL, Kauffman M, Adams J, Schenkein DP, Anderson KC. N Engl J Med. 2003;348:2609–2617. doi: 10.1056/NEJMoa030288. [DOI] [PubMed] [Google Scholar]
- 76.Fisher RI, Bernstein SH, Kahl BS, Djulbegovic B, Robertson MJ, de Vos S, Epner E, Krishnan A, Leonard JP, Lonial S, Stadtmauer EA, O'Connor O A, Shi H, Boral AL, Goy A. J Clin Oncol. 2006;24:4867–4874. doi: 10.1200/JCO.2006.07.9665. [DOI] [PubMed] [Google Scholar]
- 77.Goy A, Younes A, McLaughlin P, Pro B, Romaguera JE, Hagemeister F, Fayad L, Dang NH, Samaniego F, Wang M, Broglio K, Samuels B, Gilles F, Sarris AH, Hart S, Trehu E, Schenkein D, Cabanillas F, Rodriguez AM. J Clin Oncol. 2005;23:667–675. doi: 10.1200/JCO.2005.03.108. [DOI] [PubMed] [Google Scholar]
- 78.O'Connor OA, Wright J, Moskowitz C, Muzzy J, MacGregor-Cortelli B, Stubblefield M, Straus D, Portlock C, Hamlin P, Choi E, Dumetrescu O, Esseltine D, Trehu E, Adams J, Schenkein D, Zelenetz AD. J Clin Oncol. 2005;23:676–684. doi: 10.1200/JCO.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 79.Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, Reece D, San-Miguel JF, Blade J, Boccadoro M, Cavenagh J, Dalton WS, Boral AL, Esseltine DL, Porter JB, Schenkein D, Anderson KC. N Engl J Med. 2005;352:2487–2498. doi: 10.1056/NEJMoa043445. [DOI] [PubMed] [Google Scholar]
- 80.Jagannath S, Barlogie B, Berenson J, Siegel D, Irwin D, Richardson PG, Niesvizky R, Alexanian R, Limentani SA, Alsina M, Adams J, Kauffman M, Esseltine DL, Schenkein DP, Anderson KC. Br J Haematol. 2004;127:165–172. doi: 10.1111/j.1365-2141.2004.05188.x. [DOI] [PubMed] [Google Scholar]
- 81.Landis-Piwowar KR, Milacic V, Chen D, Yang H, Zhao Y, Chan TH, Y Yan B, Dou QP. Drug Resist Updat. 2006;9:263–273. doi: 10.1016/j.drup.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 82.Garg AK, Buchholz TA, Aggarwal BB. Antioxid Redox Signal. 2005;7:1630–1647. doi: 10.1089/ars.2005.7.1630. [DOI] [PubMed] [Google Scholar]
- 83.Groll M, Huber R. Biochim Biophys Acta. 2004;1695:33–44. doi: 10.1016/j.bbamcr.2004.09.025. [DOI] [PubMed] [Google Scholar]
- 84.Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. J Antibiot (Tokyo) 1991;44:113–116. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- 85.Omura S, Matsuzaki K, Fujimoto T, Kosuge K, Furuya T, Fujita S, Nakagawa A. J Antibiot (Tokyo) 1991;44:117–118. doi: 10.7164/antibiotics.44.117. [DOI] [PubMed] [Google Scholar]
- 86.Fenteany G, Standaert RF, Reichard GA, Corey EJ, Schreiber SL. Proc Natl Acad Sci U S A. 1994;91:3358–3362. doi: 10.1073/pnas.91.8.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fenteany G, Standaert RF, Lane WS, Choi S, Corey EJ, Schreiber SL. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 88.Dick LR, Cruikshank AA, Grenier L, Melandri FD, Nunes SL, Stein RL. J Biol Chem. 1996;271:7273–7276. doi: 10.1074/jbc.271.13.7273. [DOI] [PubMed] [Google Scholar]
- 89.Fenteany G, Schreiber SL. Chem Biol. 1996;3:905–912. doi: 10.1016/s1074-5521(96)90179-9. [DOI] [PubMed] [Google Scholar]
- 90.Soldatenkov VA, Dritschilo A. Cancer Res. 1997;57:3881–3885. [PubMed] [Google Scholar]
- 91.Kudo Y, Takata T, Ogawa I, Kaneda T, Sato S, Takekoshi T, Zhao M, Miyauchi M, Nikai H. Clin Cancer Res. 2000;6:916–923. [PubMed] [Google Scholar]
- 92.Chen F, Chang D, Goh M, Klibanov SA, Ljungman M. Cell Growth Differ. 2000;11:239–246. [PubMed] [Google Scholar]
- 93.Legnani FG, Pradilla G, Thai QA, Fiorindi A, Recinos PF, Tyler BM, Gaini SM, DiMeco F, Brem H, Olivi A. J Neurooncol. 2006;77:225–232. doi: 10.1007/s11060-005-6937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shirley RB, Kaddour-Djebbar I, Patel DM, Lakshmikanthan V, Lewis RW, Kumar MV. Neoplasia. 2005;7:1104–1111. doi: 10.1593/neo.05520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Holzer AK, Howell SB. Cancer Res. 2006;66:10944–10952. doi: 10.1158/0008-5472.CAN-06-1710. [DOI] [PubMed] [Google Scholar]
- 96.Li QQ, Yunmbam MK, Zhong X, Yu JJ, Mimnaugh EG, Neckers L, Reed E. Cell Mol Biol (Noisy-le-grand), 47 Online Pub. 2001:OL61–72. [PubMed] [Google Scholar]
- 97.Ogiso Y, Tomida A, Lei S, Omura S, Tsuruo T. Cancer Res. 2000;60:2429–2434. [PubMed] [Google Scholar]
- 98.Sun Y, Ottosson A, Pervaiz S, Fadeel B. Leukemia. 2007;21:1035–1043. doi: 10.1038/sj.leu.2404660. [DOI] [PubMed] [Google Scholar]
- 99.Hwang YS, Hodge JC, Sivapurapu N, Lindholm PF. Mol Carcinog. 2006;45:518–529. doi: 10.1002/mc.20183. [DOI] [PubMed] [Google Scholar]
- 100.Lanthier J, Desrosiers RR. Biochem Cell Biol. 2006;84:684–694. doi: 10.1139/o06-055. [DOI] [PubMed] [Google Scholar]
- 101.Yang CS, Wang ZY. J Natl Cancer Inst. 1993;85:1038–1049. doi: 10.1093/jnci/85.13.1038. [DOI] [PubMed] [Google Scholar]
- 102.Kuo PL, Lin CC. J Biomed Sci. 2003;10:219–227. doi: 10.1007/BF02256057. [DOI] [PubMed] [Google Scholar]
- 103.Hastak K, Gupta S, Ahmad N, Agarwal MK, Agarwal ML, Mukhtar H. Oncogene. 2003;22:4851–4859. doi: 10.1038/sj.onc.1206708. [DOI] [PubMed] [Google Scholar]
- 104.Kavantzas N, Chatziioannou A, Yanni AE, Tsakayannis D, Balafoutas D, Agrogiannis G, Perrea D. Vascul Pharmacol. 2006;44:461–463. doi: 10.1016/j.vph.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 105.Zhang Q, Tang X, Lu Q, Zhang Z, Rao J, Le AD. Mol Cancer Ther. 2006;5:1227–1238. doi: 10.1158/1535-7163.MCT-05-0490. [DOI] [PubMed] [Google Scholar]
- 106.Inoue T, Shiraki K, Fuke H, Yamanaka Y, Miyashita K, Yamaquchi Y, Yamamoto N, Ito K, Suqimoto K, Nakano T. Anticancer Drugs. 2006;17:261–268. doi: 10.1097/00001813-200603000-00004. [DOI] [PubMed] [Google Scholar]
- 107.Lee JW, Bae SH, Jeong JW, Kim SH, Kim KW. Exp Mol Med. 2004;36:1–12. doi: 10.1038/emm.2004.1. [DOI] [PubMed] [Google Scholar]
- 108.Fassina G, Vene R, Morini M, Minghelli S, Benelli R, Noonan DM, Albini A. Clin Cancer Res. 2004;10:4865–4873. doi: 10.1158/1078-0432.CCR-03-0672. [DOI] [PubMed] [Google Scholar]
- 109.Tang FY, Chiang EP, Shih CJ. J Nutr Biochem. 2007;18:391–399. doi: 10.1016/j.jnutbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 110.Yamakawa S, Asai T, Uchida T, Matsukawa M, Akizawa T, Oku N. Cancer Lett. 2004;210:47–55. doi: 10.1016/j.canlet.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 111.Yang J, Wei D, Liu J. Biomed Pharmacother. 2005;59:98–103. doi: 10.1016/j.biopha.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 112.Landis-Piwowar KR, Huo C, Chen D, Milacic V, Shi G, Chan TH, Dou QP. Cancer Res. 2007;67:4303–4310. doi: 10.1158/0008-5472.CAN-06-4699. [DOI] [PubMed] [Google Scholar]
- 113.Lu H, Meng X, Yang CS. Drug Metab Dispos. 2003;31:572–579. doi: 10.1124/dmd.31.5.572. [DOI] [PubMed] [Google Scholar]
- 114.Okushio K, Suzuki M, Matsumoto N, Nanjo F, Hara Y. Biosci Biotechnol Biochem. 1999;63:430–432. doi: 10.1271/bbb.63.430. [DOI] [PubMed] [Google Scholar]
- 115.Zhu Q, Zhang A, Tsang D, Huang Y, Chen Z. J Agric Food Chem. 1997:4624–4628. [Google Scholar]
- 116.Wu AH, Tseng CC, Van Den Berg D, Yu MC. Cancer Res. 2003;63:7526–7529. [PubMed] [Google Scholar]
- 117.Landis-Piwowar KR, Wan SB, Wiegand RA, Kuhn DJ, Chan TH, Dou QP. J Cell Physiol. 2007;213:252–260. doi: 10.1002/jcp.21124. [DOI] [PubMed] [Google Scholar]
- 118.Caltagirone S, Rossi C, Poggi A, Ranelletti FO, Natali PG, Brunetti M, Aiello FB, Piantelli M. Int J Cancer. 2000;87:595–600. doi: 10.1002/1097-0215(20000815)87:4<595::aid-ijc21>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 119.Kampa M, Hatzoglou A, Notas G, Damianaki A, Bakogeorgou E, Gemetzi C, Kouroumalis E, Martin PM, Castanas E. Nutr Cancer. 2000;37:223–233. doi: 10.1207/S15327914NC372_16. [DOI] [PubMed] [Google Scholar]
- 120.Soleas GJ, Grass L, Josephy PD, Goldberg DM, Diamandis EP. Clin Biochem. 2006;39:492–497. [Google Scholar]
- 121.Mouria M, Gukovskaya AS, Jung Y, Buechler P, Hines OJ, Reber HA, Pandol SJ. Int J Cancer. 2002;98:761–769. doi: 10.1002/ijc.10202. [DOI] [PubMed] [Google Scholar]
- 122.Delmas D, Lancon A, Colin D, Jannin B, Latruffe N. Curr Drug Targets. 2006;7:423–442. doi: 10.2174/138945006776359331. [DOI] [PubMed] [Google Scholar]
- 123.Liao HF, Kuo CD, Yang YC, Lin CP, Tai HC, Chen YY, Chen YJ. J Radiat Res (Tokyo) 2005;46:387–393. doi: 10.1269/jrr.46.387. [DOI] [PubMed] [Google Scholar]
- 124.Lepley DM, Li B, Birt DF, Pelling JC. Carcinogenesis. 1996;17:2367–2375. doi: 10.1093/carcin/17.11.2367. [DOI] [PubMed] [Google Scholar]
- 125.Janssen K, Mensink RP, Cox FJ, Harryvan JL, Hovenier R, Hollman PC, Katan MB. Am J Clin Nutr. 1998;67:255–262. doi: 10.1093/ajcn/67.2.255. [DOI] [PubMed] [Google Scholar]
- 126.Manach C, Scalbert A, Morand C, Remesy C, Jimenez L. Am J Clin Nutr. 2004;79:727–747. doi: 10.1093/ajcn/79.5.727. [DOI] [PubMed] [Google Scholar]
- 127.Romanova D, Vachalkova A, Cipak L, Ovesna Z, Rauko P. Neoplasma. 2001;48:104–107. [PubMed] [Google Scholar]
- 128.Birt DF, Mitchell D, Gold B, Pour P, Pinch HC. Anticancer Res. 1997;17:85–91. [PubMed] [Google Scholar]
- 129.Hertog MG, Hollman PC, Katan MB, Kromhout D. Nutr Cancer. 1993;20:21–29. doi: 10.1080/01635589309514267. [DOI] [PubMed] [Google Scholar]
- 130.Singletary K, Ellington A. Anticancer Res. 2006;26:1039–1048. [PubMed] [Google Scholar]
- 131.Russo A, Cardile V, Lombardo L, Vanella L, Acquaviva R. J Nutr Biochem. 2006;17:103–108. doi: 10.1016/j.jnutbio.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 132.Yu Z, Li W, Liu F. Cancer Lett. 2004;215:159–166. doi: 10.1016/j.canlet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 133.Kazi A, Daniel KG, Smith DM, Kumar NB, Dou QP. Biochem Pharmacol. 2003;66:965–976. doi: 10.1016/s0006-2952(03)00414-3. [DOI] [PubMed] [Google Scholar]
- 134.Watanabe S, Yamaguchi M, Sobue T, Takahashi T, Miura T, Arai Y, Mazur W, Wahala K, Adlercreutz H. J Nutr. 1998;128:1710–1715. doi: 10.1093/jn/128.10.1710. [DOI] [PubMed] [Google Scholar]
- 135.Uehar M, Arai Y, Watanabe S, Adlercreutz H. Biofactors. 2000;12:217–225. doi: 10.1002/biof.5520120134. [DOI] [PubMed] [Google Scholar]
- 136.Dorai T, Aggarwal BB. Cancer Lett. 2004;215:129–140. doi: 10.1016/j.canlet.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 137.Gu Y, Zhu CF, Iwamoto H, Chen JS. World J Gastroenterol. 2005;11:6512–6517. doi: 10.3748/wjg.v11.i41.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li Y, Sarkar FH. Clin Cancer Res. 2002;8:2369–2377. [PubMed] [Google Scholar]
- 139.Cohen S, Lahav-Baratz S, Ciechanover A. Biochem Biophys Res Commun. 2006;345:7–13. doi: 10.1016/j.bbrc.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 140.Singh S, Khar A. Anticancer Agents Med Chem. 2006;6:259–270. doi: 10.2174/187152006776930918. [DOI] [PubMed] [Google Scholar]
- 141.Lev-Ari S, Strier L, Kazanov D, Madar-Shapiro L, Dvory-Sobol H, Pinchuk I, Marian B, Lichtenberg D, Arber N. Clin Cancer Res. 2005;11:6738–6744. doi: 10.1158/1078-0432.CCR-05-0171. [DOI] [PubMed] [Google Scholar]
- 142.Notarbartolo M, Poma P, Perri D, Dusonchet L, Cervello M, D'Alessandro N. Cancer Lett. 2005;224:53–65. doi: 10.1016/j.canlet.2004.10.051. [DOI] [PubMed] [Google Scholar]
- 143.Bava SV, Puliappadamba VT, Deepti A, Nair A, Karunagaran D, Anto RJ. J Biol Chem. 2005;280:6301–6308. doi: 10.1074/jbc.M410647200. [DOI] [PubMed] [Google Scholar]
- 144.Chan MM, Fong D, Soprano KJ, Holmes WF, Heverling H. J Cell Physiol. 2003;194:63–70. doi: 10.1002/jcp.10186. [DOI] [PubMed] [Google Scholar]
- 145.Chirnomas D, Taniguchi T, de la Vega M, Vaidya AP, Vasserman M, Hartman AR, Kennedy R, Foster R, Mahoney J, Seiden MV, D'Andrea AD. Mol Cancer Ther. 2006;5:952–961. doi: 10.1158/1535-7163.MCT-05-0493. [DOI] [PubMed] [Google Scholar]
- 146.Gao X, Deeb D, Jiang H, Liu YB, Dulchavsky SA, Gautam SC. J Exp Ther Oncol. 2005;5:39–48. [PubMed] [Google Scholar]
- 147.Khafif A, Hurst R, Kyker K, Fliss DM, Gil Z, Medina JE. Otolaryngol Head Neck Surg. 2005;132:317–321. doi: 10.1016/j.otohns.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 148.Aggarwal BB, Kumar A, Bharti AC. Anticancer Res. 2003;23:363–398. [PubMed] [Google Scholar]
- 149.Cleren C, Calingasan NY, Chen J, Beal MF. J Neurochem. 2005;94:995–1004. doi: 10.1111/j.1471-4159.2005.03253.x. [DOI] [PubMed] [Google Scholar]
- 150.Allison AC, Cacabelos R, Lombardi VR, Alvarez XA, Vigo C. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2001;25:1341–1357. doi: 10.1016/s0278-5846(01)00192-0. [DOI] [PubMed] [Google Scholar]
- 151.Chang FR, Hayashi K, Chen IH, Liaw CC, Bastow KF, Nakanishi Y, Nozaki H, Cragg GM, Wu YC, Lee KH. J Nat Prod. 2003;66:1416–1420. doi: 10.1021/np030241v. [DOI] [PubMed] [Google Scholar]
- 152.Lee JH, Koo TH, Yoon H, Jung HS, Jin HZ, Lee K, Hong YS, Lee JJ. Biochem Pharmacol. 2006;72:1311–1321. doi: 10.1016/j.bcp.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 153.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, Maloney KN, Clardy J, Hahn WC, Chiosis G, Golub TR. Cancer Cell. 2006;10:321–330. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 154.Lopes UG, Erhardt P, Yao R, Cooper GM. J Biol Chem. 1997;272:12893–12896. doi: 10.1074/jbc.272.20.12893. [DOI] [PubMed] [Google Scholar]
- 155.Yang H, Shi G, Dou QP. Mol Pharmacol. 2007;71:426–437. doi: 10.1124/mol.106.030015. [DOI] [PubMed] [Google Scholar]
- 156.Kiaei M, Kipiani K, Petri S, Chen J, Calingasan NY, Beal MF. Neurodegener Dis. 2005;2:246–254. doi: 10.1159/000090364. [DOI] [PubMed] [Google Scholar]
- 157.Sethi G, Ahn KS, Pandey MK, Aggarwal BB. Blood. 2007;109:2727–2735. doi: 10.1182/blood-2006-10-050807. [DOI] [PubMed] [Google Scholar]
- 158.Nagase M, Oto J, Sugiyama S, Yube K, Takaishi Y, Sakato N. Biosci Biotechnol Biochem. 2003;67:1883–1887. doi: 10.1271/bbb.67.1883. [DOI] [PubMed] [Google Scholar]
- 159.Sowery RD, So AI, Gleave ME. Curr Urol Rep. 2007;8:53–59. doi: 10.1007/s11934-007-0021-9. [DOI] [PubMed] [Google Scholar]
- 160.Niampoka C, Suttisri R, Bavovada R, Takayama H, Aimi N. Arch Pharm Res. 2005;28:546–549. doi: 10.1007/BF02977756. [DOI] [PubMed] [Google Scholar]
- 161.Buffa Filho W, Corsino J, Bolzani da SV, Furlan M, Pereira AM, Franca SC. Phytochem Anal. 2002;13:75–78. doi: 10.1002/pca.626. [DOI] [PubMed] [Google Scholar]
- 162.Sassa H, Kogure K, Takaishi Y, Terada H. Free Radic Biol Med. 1994;17:201–207. doi: 10.1016/0891-5849(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 163.Dirsch VM, Kiemer AK, Wagner H, Vollmar AM. Eur J Pharmacol. 1997;336:211–217. doi: 10.1016/s0014-2999(97)01245-4. [DOI] [PubMed] [Google Scholar]
- 164.Avilla J, Teixido A, Velazquez C, Alvarenga N, Ferro E, Canela R. J Agric Food Chem. 2000;48:88–92. doi: 10.1021/jf990008w. [DOI] [PubMed] [Google Scholar]
- 165.Luo DQ, Wang H, Tian X, Shao HJ, Liu JK. Pest Manag Sci. 2005;61:85–90. doi: 10.1002/ps.953. [DOI] [PubMed] [Google Scholar]
- 166.Figueiredo JN, Raz B, Sequin U. J Nat Prod. 1998;61:718–723. doi: 10.1021/np9704157. [DOI] [PubMed] [Google Scholar]
- 167.Wu CC, Chan ML, Chen WY, Tsai CY, Chang FR, Wu YC. Mol Cancer Ther. 2005;4:1277–1285. doi: 10.1158/1535-7163.MCT-05-0027. [DOI] [PubMed] [Google Scholar]
- 168.Yang H, Landis-Piwowar KR, Lu D, Yuan P, Li L, Reddy GP, Yuan X, Dou QP. J Cell Biochem. 2008;103:234–244. doi: 10.1002/jcb.21399. [DOI] [PubMed] [Google Scholar]