Abstract
MicroRNAs (miRNAs) play an important role in development and regulate the expression of many animal genes by post-transcriptional gene silencing. Here we describe the cloning and expression of new miRNAs from zebrafish. By high-throughput sequencing of small-RNA cDNA libraries from 5-day-old zebrafish larvae and adult zebrafish brain we found 139 known miRNAs and 66 new miRNAs. For 65 known miRNAs and for 11 new miRNAs we also cloned the miRNA star sequence. We analyzed the temporal and spatial expression patterns for 35 new miRNAs and for 32 known miRNAs in the zebrafish by whole mount in situ hybridization and northern blotting. Overall, 23 of the 35 new miRNAs and 30 of the 32 known miRNAs could be detected. We found that most miRNAs were expressed during later stages of development. Some were expressed ubiquitously, but many of the miRNAs were expressed in a tissue-specific manner. Most newly discovered miRNAs have low expression levels and are less conserved in other vertebrate species. Our cloning and expression analysis indicates that most abundant and conserved miRNAs in zebrafish are now known.
INTRODUCTION
Over the past few years it has become clear that the expression of many genes is extensively controlled at the post-transcriptional level by microRNAs (miRNAs) (1,2). Although miRNAs were initially recognized as an oddity specific to developmental switches in Caenorhabditis elegans (3,4), the cloning and computational prediction of hundreds of miRNAs in both animals and plants uncovered a whole new layer of gene regulation (5). It appears now that a mammalian genome may contain >500 genes encoding miRNAs (6,7).
miRNAs are transcribed as long RNA polymerase II transcripts (8,9) which fold into characteristic stem–loop structures. Once cleaved by the nuclear enzyme Drosha (10), a smaller precursor miRNA is transported to the cytoplasm (11,12), where the Dicer protein mediates maturation of the miRNA into a 20–23 nt species (13–16), a process which is coupled to loading into a miRNP complex. These small-RNA molecules bind to the 3′-untranslated region of mRNAs by partial basepairing, which primarily results in inhibition of mRNA translation (17). mRNAs that are repressed by miRNAs are localized in cytoplasmic foci called P-bodies (18–20).
While plant miRNAs usually bind with perfect complementarity to their target mRNA and induce mRNA degradation (21), animal miRNAs in most cases regulate a mRNA containing a sequence complementary to the 7 nt seed of the miRNA (nt 1–7 or 2–8) (22–24). Computational predictions indicate that thousands of genes might be regulated by miRNAs and that the average number of genes that is targeted by a miRNA is ∼200 (25,26). Many miRNA target sites are evolutionarily conserved and the mRNAs that bear conserved target sites are expressed at lower levels in the tissue where the miRNA is expressed compared with the tissues where the miRNA is not expressed (27,28). In addition, the mRNAs with conserved target sites are often expressed in developmental stages prior to miRNA expression (27).
The important role of miRNAs in animal development has been shown by several approaches. Removal of all miRNAs results in developmental arrest in mouse and fish (29–31). Several other studies have revealed the details of processes where miRNAs act. For example, the miR-1 knockout in Drosophila results in aberrant muscle growth (32), while in mouse, miR-1 regulates the transcription factor Hand2 during heart development (33). In addition, miRNAs may regulate major signaling pathways like the notch signaling pathway in Drosophila (34). In mouse and chick, miR-196 expression regulates the expression of Sonic hedgehog through targeting the transcription factor Hoxb8 (35).
In zebrafish there are currently 369 miRNA genes expressing 168 different miRNAs (5). Many of the conserved miRNAs have a striking organ-specific expression pattern in zebrafish and are mostly expressed at later stages of development (36). Also in Drosophila, miRNAs exhibit diverse spatial expression patterns during embryonic development as indicated by the analysis of the expression of primary miRNA transcripts (37). Strikingly, the expression patterns of some highly conserved miRNAs like miR-1 and miR-124 are similar in flies, fish and mouse, suggesting ancient roles in tissue development (37,38).
Encouraged by recent studies which indicate that there are many more miRNAs than currently known (6,7), we attempted to find new miRNAs in the zebrafish by sequencing small-RNA cDNA libraries made from 5-day-old zebrafish larvae and adult zebrafish brain. We found 139 known miRNAs and 66 new miRNAs in the zebrafish. In addition, we studied the temporal and spatial expression of miRNAs with unknown expression from three sources: 32 miRNAs predicted or cloned previously from zebrafish (7,39), 34 miRNAs cloned in this study and one miRNA cloned from human (E. Berezikov, R. H. A. Plasterk and E. Cuppen, unpublished data). We used locked nucleic acid (LNA) probes to detect these 67 miRNAs in situ in the zebrafish embryo and on northern blots with total RNA from different developmental stages and adult tissues. In contrast to our previous in situ hybridization screen for conserved vertebrate miRNAs (36), we could only detect miRNA expression in situ for a subset of 28 miRNAs. For 53 miRNAs we could detect a ∼22 nt species on northern blots. The remainder of 14 miRNAs could not be detected by in situ hybridization or northern blotting, although 13 of these were cloned in this study and passed our computational analysis. These might thus represent low abundant miRNAs or miRNAs expressed only in a few cells.
Our data show that there are many more miRNAs in zebrafish than so far described. They also demonstrate that next to the highly abundant and tissue-specific miRNAs, most of which are conserved (36), there is a large set of miRNAs expressed at much lower levels, many of which are less conserved.
MATERIALS AND METHODS
Small-RNA library construction
Two small-RNA cDNA libraries were prepared by Vertis Biotechnologie AG (Freising-Weihenstephan, Germany). RNAs smaller than 200 bases were isolated from 5-day-old zebrafish larvae and dissected adult zebrafish brain using the mirVana miRNA isolation kit (Ambion). Subsequently, the population of small-RNAs ranging in size from 15 to 30 bp were purified from 12.5% polyacrylamide gel. This small-RNA fraction was poly(A)-tailed followed by ligation of an RNA linker to the 5′ end of the RNA. First strand cDNA synthesis was performed using oligo(dT)-linker primers and M-MLV-RNase H− reverse transcriptase. The resulting cDNA was then PCR-amplified in 15 (larvae library) or 23 (brain library) cycles. After limited exonuclease treatment to generate 5′ overhangs, the gel purified fraction of the cDNA in the range of 95–110 bp was directionally ligated in the EcoRI and BamHI sites of pBSII SK+. Ligations were electroporated into T1 Phage resistant TransforMax™ EC100™ electrocompetent cells (Epicentre) resulting in 1.25 (larvae library) and 1.3 (brain library) × 106recombinant clones.
Sequencing of small-RNA cDNA libraries
Both libraries were plated on Luria–Bertani (LB) amp plates and 12 288 individual colonies were automatically picked and put into 384-well plates (Genetix QPix2; New Milton Hampshire, UK) containing 75 µl LB-Amp and grown overnight at 37°C with continuous shaking. All following pipetting steps were performed using liquid handling robots (Tecan Genesis RSP200 with integrated TeMo96 and Velocity11 Vprep with BenchCell 4×). 5 µl of culture were transferred to a 384-well PCR plate (Greiner) containing 20 µl water. Cells were lysed by heating for 15 min at 95°C in a PCR machine. Lysate (1 µl) was transferred to a fresh 384-well plate containing 4 µl PCR mix (final concentrations: 0.2 µM M13forward, TGTAAAACGACGGCCAGT; 0.2 µM M13reverse, AGGAAACAGCTATGACCAT, 400 µM of each dNTP, 25 mM Tricine, 7.0% Glycerol (w/v), 1.6% dimethyl sulfoxide (w/v), 2 mM MgCl2, 85 mM Ammonium acetate, pH 8.7 and 0.2 U Taq Polymerase in a total volume of 10 µl) and the insert was amplified by 35 cycles of 20 min at 94°C, 10 min at 58°C, 30 min at 72°C. After adding 30 µl water, 1 µl of PCR product was directly used for dideoxy sequencing by transferring to a new 384-well PCR plate containing 4 µl sequencing mix (0.027 µl BigDye terminator mix v3.1 (Applied Biosystems, Foster City, CA), 1.96 µl of 2.5× dilution buffer (Applied Biosystems), 0.01 µl sequencing oligo (100 µM stock T7, GTAATACGACTCACTATAGGGC) and 2 µl water). Thermocycling was performed for 35 cycles of 10 min at 94°C, 10 min at 50°C, 20 min at 60°C and final products were purified by ethanol precipitation in 384-well plates as recommended by the manufacturer (Applied Biosystems) and analyzed on ABI3730XL sequencers with a modified protocol for generating ∼100 nt sequencing reads.
Sequence analysis
Base calling and quality trimming of sequence chromatograms was done by phred software (40). After masking of vector and adapter sequences and removing redundancy, inserts of length 18 bases and longer were mapped to the zebrafish genome using megablast software (ftp://ftp.ncbi.nlm.nih.gov/blast/). Not all inserts matched perfectly to a genome, and detailed analysis of non-matching sequences indicated that many of them represented known miRNAs with several additional nucleotides added to one of the ends. These non-genomic sequences may be artifacts of the cloning procedure or a result of non-templated modification of mature miRNAs (41). Such sequences were corrected according to the best blast hit to a genome. Next, for every genomic locus matching to an insert, repeat annotations were retrieved from the Ensembl database (http://www.ensembl.org) and repetitive regions were discarded from further analysis, with the exception of the following repeats: MIR, MER, L2, MARNA, MON, Arthur and trf, since these repeat annotations overlap with some known miRNAs. Genomic regions containing inserts with 100 nt flanks were retrieved from Ensembl and a sliding window of 100 nt was used to calculate RNA secondary structures by RNAfold (42). Only regions that folded into hairpins and contained an insert in one of the hairpin arms were used in further analysis. Since every non-redundant insert produced independent hits at this stage, hairpins with overlapping genomic coordinates were merged into one region, tracing locations of matching inserts. In cases when several inserts overlapped, the whole region covered by overlapping inserts was used in downstream calculations as a mature sequence. Next, gene and repeat annotations for hairpin genomic regions were retrieved from Ensembl, and repetitive regions (with above mentioned exceptions) as well as ribosomal RNAs, tRNAs and snoRNAs were discarded.
To find homologous hairpins in other genomes, mature regions were blasted against human, macaca, chimpanzee, mouse, rat, dog, cow, opossum, chicken, tetraodon, zebrafish and fugu genomes. Hits with length of at least 20 nt and identity of at least 70% were extracted from genomes along with flanking sequences of length similar to that observed in original hairpins to which a certain mature query sequence belonged. Extracted sequences were checked for hairpin structures using RNAfold, and positive hairpins were aligned with the original hairpin using clustalw (43). Only homologs with at least 70% overall identity and 90% identity within the mature sequence were considered. In cases where several homologous hairpins in a species were identified, the best clustalw-scoring hairpin was retained. Next, homologs from different organisms were aligned with the original hairpin by clustalw to produce a final multiple alignment of the hairpin region. Chromosomal location of homologous sequences were used to retrieve gene and repeat annotations from respective species Ensembl databases. Hairpins that contained repeat/RNA annotations in one of the species, as well as hairpins containing mature regions longer that 25 nt or with GC-content higher than 85% were discarded. For remaining hairpins, randfold values were calculated for every sequence in an alignment using mononucleotide shuffling and 1000 iterations. The cut-off of 0.01 was used for randfold and only regions that contained a hairpin below this cut-off for at least one species in an alignment, were considered as miRNA genes. Finally, positive hairpins were split into known and novel miRNAs according to annotations. To facilitate these annotations and also to track performance of the pipeline, mature sequences of known miRNAs from miRBase (5) were included into the analysis from the very beginning.
In situ hybridization
Albino zebrafish embryos and larvae of 12, 16, 24, 48, 72 and 120 hpf were fixed in 4% PFA in phosphate-buffered saline (PBS) overnight at 4°C. Proteinase K treatment was done for 2, 5, 10, 30, 45 and 90 min, respectively. In situ hybridization was performed as previously described (36,38). LNA-modified DNA probes (LNA probes) were designed and synthesized by Exiqon (Denmark). The sequences of the LNA probes complementary to the mature miRNAs are listed in Supplementary Table S1. The LNA probes were labeled with digoxigenin (DIG) using the DIG 3′ end labeling kit (Roche) and purified using Sephadex G25 MicroSpin columns (Amersham).
Plastic sectioning
Embryos and larvae stained by whole-mount in situ hybridization were transferred from benzyl benzoate/benzyl alcohol to 100% methanol and incubated for 10 min. Specimens were washed twice with 100% ethanol for 10 min and incubated overnight in 100% Technovit 8100 infiltration solution (Kulzer) at 4°C. Next, embryos were transferred to a mold and embedded overnight in Technovit 8100 embedding medium (Kulzer) deprived of air at 4°C. Sections of 7 µm thickness were cut with a microtome (Reichert-Jung 2050), stretched on water and mounted on glass slides. Sections were dried overnight. Counterstaining was done with 0.05% neutral red for 12 s, followed by extensive washing with water. Sections were preserved with Pertex and mounted under a coverslip.
Image acquisition
Embryos, larvae and sections were analyzed with Zeiss Axioplan and Leica MZFLIII microscopes and subsequently photographed with digital cameras. Images were adjusted with Adobe Photoshop 7.0 software.
Northern blotting
Total RNA was isolated using trizol (Invitrogen). For each sample ∼15 µg RNA was separated on 15% denaturing polyacrylamide gels and blotted according to standard procedures. Blots were prehybridized for 30 min at 60°C in hybridization buffer (0.36 M Na2HPO4, 0.14 M NaH2PO4, 1 mM EDTA and 7% SDS) and hybridized overnight at 60°C in hybridization buffer containing 0.1 nM probe. After stringency washes (once for 30 min at 50°C in 2× SSC/0.1% SDS and once for 30 min at 50°C in 0.5× SSC and 0.1% SDS) blots were rinsed in PBST (PBS with 0.1% Tween-20) and blocked for 30 min at room temperature in PBST with 5% milk powder. Subsequently, blots were incubated for 1 h at room temperature with anti-DIG-AP antibody (Roche) in blocking buffer, washed six times for 15 min in PBST and twice for 5 min with AP-buffer (0.1 M Tris–HCl, pH 9.5, 50 mM MgCl2, 0.1 M NaCl and 0.1% Tween). Signal was detected by using CDP-star chemiluminescent substrate (Roche) and exposing the blots to X-ray films. Films were scanned and pictures were processed using Adobe photoshop 7.0 software. To control for equal loading, blots were hybridized for 2 h at 37°C with a radio-labeled probe against U6 snRNA. After washing twice in 2× SSC/0.2% SDS, blots were exposed to phosphor-imager screens.
RESULTS
Cloning of new miRNAs from zebrafish
Recently, miRNAs were profiled in zebrafish by cloning small-RNAs from different developmental stages and zebrafish cell lines (39). In addition to the miRNAs already known from other organisms, several new miRNAs were found. The total number of zebrafish miRNA genes in the miRNA registry is currently 369, corresponding to 168 different miRNAs (5). Computational identification, verification and cloning of miRNAs by our group and others has shown that the number of miRNAs in humans might be much higher and extend towards a thousand (E. Berezikov, R. H. A. Plasterk and E. Cuppen, unpublished data) (6,7).
In order to find more miRNAs in the zebrafish we performed sequencing of two newly generated small-RNA libraries derived from 5-day-old zebrafish larvae (zf-larvae) and adult zebrafish brain (zf-brain). These two sample types were chosen based on our previous in situ expression analysis of conserved vertebrate miRNAs in the zebrafish embryo, which revealed that strongest expression for most miRNAs is observed in later stages of development and that one-third of this set of miRNAs was found to be expressed in the zebrafish embryonic brain (36).
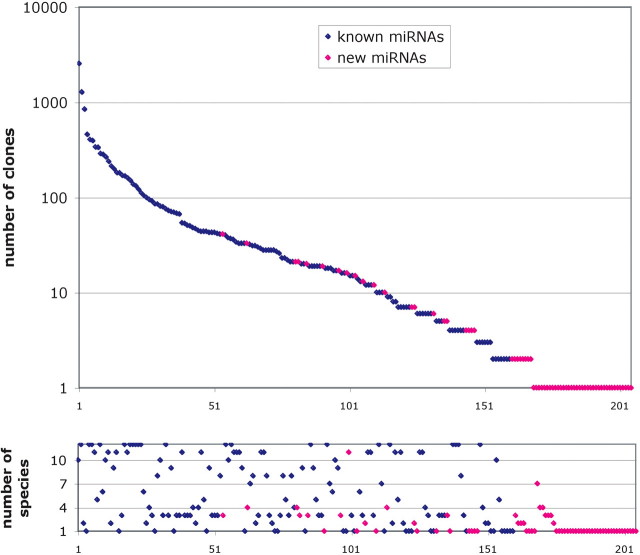
For each library 12 288 individual clones were sequenced. Of these 12 288 sequence reads 2182 from the zf-larvae library and 1231 reads from the zf-brain library were too short to be analyzed. The remaining sequence reads were selected based on the presence of a 3′ poly(A)-tail and a 5′ adapter sequence, both of which resulted from the cloning process. Clones containing inserts shorter than 18 bp were also removed from the dataset. The clones that fulfilled these criteria were analyzed using a computational pipeline that includes RNA secondary structure. Based on this analysis, 13 094 sequences could be annotated as miRNAs, representing 205 distinct miRNAs (Supplementary Tables S2 and S3). From these small-RNA sequences, 3,150 (zf-larvae) and 9,607 (zf-brain) could be annotated as known miRNAs from zebrafish. In total, we found 139 out of 168 known zebrafish miRNAs and 126 of these were found in both the zf-brain library and the zf-larvae library (Supplementary Figure S1 and Table S7). For 65 of the known miRNAs, we sequenced clones from both the 3p- and 5p-arm of the miRNA hairpin, i.e. both the miRNAand the miRNA star sequence were found (Supplementary Table S5). All of the known miRNAs that we found were represented by multiple clones in the library (Figure 1 and Supplementary Table S2). In addition, 2.6% of the clones represent 66 novel miRNAs (zf-larvae, 121 clones and zf-brain, 216 clones). These clones could be assigned to 116 different hairpins in the zebrafish genome, thus representing 116 potential new miRNA genes. For 11 of the new miRNAs, clones were sequenced from both the 5p- and the 3p-arm (Supplementary Table S6). Together, this limits the total number of unique miRNA hairpins to 66 (Supplementary Table S4). Of the new miRNAs 37 were found only once in one of both libraries. The overlap between the zf-larvae and zf-brain libraries for new miRNAs was 19 (Supplementary Figure S1 and Table S3). Although the majority of the known miRNAs have a clear homolog in other vertebrate species, many of the newly identified miRNAs are less conserved (Figure 1, Supplementary Tables S2 and S3). The first nucleotide of the newly identified miRNAs is most often a U, although this bias was less strong than for known miRNAs (25) (Supplementary Figure S2).
Figure 1.
Cloning frequency and conservation for all miRNAs cloned from zebrafish small-RNA cDNA libraries. The upper panel depicts the cloning frequency for all small RNAs that were found in the two libraries and that passed our computational pipeline. All 139 known miRNAs (blue data points) were cloned more than once, while 37 out of the 66 new miRNAs (pink dots) were represented by a single sequenced clone. The lower panel shows a scatter plot of the conservation of known (blue dots) and new (pink data dots) miRNAs in 12 vertebrate species (zebrafish, fugu, tetraodon, mouse, rat, human, dog, macaca, opossum, chicken, chimpanzee, cow). Forty-four of the new miRNAs were only found in zebrafish, while most of the known miRNAs were found in several species according to our conservation criteria.
In total, we found 66 novel miRNAs from zebrafish and these represent a new set of less conserved miRNAs.
In situ hybridization analysis of the spatial expression of known and new miRNAs in the zebrafish
To determine the spatial and temporal expression of known and new miRNAs during zebrafish development, we performed in situ hybridization and northern blotting using LNA probes (Supplementary Table S1, Tables 1 and 2). In total we analyzed the expression of 67 miRNAs, derived from three sources: 34 miRNAs cloned from the zf-larvae and the zf-brain libraries, one miRNA cloned from human (E. Berezikov, R. H. A. Plasterk and E. Cuppen. unpublished results) and 32 miRNAs that were found previously (7,39). Of the latter set, six were already in our previous in situ screen (36) (130a, 187, 101b 135, 193b, 301b), but the sequences of these probes turned out to contain some mismatches compared with the miRNA sequences cloned by Chen et al. (39), so that we decided to test new and correct probes.
Table 1.
Overview of expression data for known miRNAs
| miRNA | Expression timea | Expression adult tissuea | Expression in situ | Temporal expression in situ | |||||
|---|---|---|---|---|---|---|---|---|---|
| 12 h | 16 h | 24 h | 48 h | 72 h | 5 days | ||||
| dre-miR-101b | Adult | Ubiquitous | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-130a | 24 h–adult | Ubiquitous | Head (gills, brain, jaw) | 0 | 0 | 1 | 3 | 3 | 3 |
| dre-miR-130b | 24 h–adult | Ubiquitous | Ubiquitous | 0 | 0 | 1 | 3 | 3 | 3 |
| dre-miR-130c | 24 h–adult | Ubiquitous | Head (gills, brain, jaw) | 0 | 0 | 1 | 3 | 3 | 3 |
| dre-miR-135 | 48 h–adult | Brain | Structures in brain | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-187 | Not expressed | ND | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-193b | Adult | Stronger in muscle, fins, skin | Brain, jaw | 0 | 0 | 0 | 0 | 3 | 3 |
| dre-miR-202 | 24 h–adult | Not in any of the analyzed tissues | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-27c | 48 h–adult | Ubiquitous but not in liver | Weak ubiquitous, pharyngeal arches, jaw | 0 | 0 | 0 | 1 | 3 | 3 |
| dre-miR-301b | 48 h–adult | Ubiquitous but strongest in brain and eye | Ubiquitous | 0 | 0 | 0 | 2 | 2 | 2 |
| dre-miR-363 | 24 h–adult | Strongest in muscle, liver, skin | No expression | 0 | 0 | 0 | 0 | 1 | 1 |
| dre-miR-365 | 48 h–adult | Brain, eye, muscle, fins, skin | Weak brain | 0 | 0 | 0 | 0 | 3 | 3 |
| dre-miR-429 | 24 h–adult | Gills, fins, skin | Nose, neuromasts (hair and supporting cells), taste buds, proctodeum | 0 | 0 | 3 | 3 | 3 | 3 |
| dre-miR-430a | 24 h–5 days | ND | Ubiquitous in early stages | 2 | 2 | 2 | 2 | 1 | 1 |
| dre-miR-430b | 24 h–5 days | ND | Ubiquitous in early stages | 2 | 2 | 2 | 2 | 1 | 0 |
| dre-miR-430c | 24–48 h | ND | Ubiquitous in early stages | 2 | 2 | 2 | 2 | 1 | 0 |
| dre-miR-430i | 24–72 h | ND | Ubiquitous in early stages | 2 | 2 | 2 | 2 | 1 | 1 |
| dre-miR-430j | 24–5 days | ND | Ubiquitous in early stages | 2 | 2 | 2 | 2 | 1 | 1 |
| dre-miR-451 | 48 h–adult | Ubiquitous | Blood | 0 | 0 | 3 | 3 | 3 | 3 |
| dre-miR-454a | 48 h–adult | Ubiquitous, but stronger in brain and eye | Brain and pharyngeal arches | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-454b | 48 h–adult | Ubiquitous, but stronger in brain and eye | Brain and pharyngeal arches | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-455 | 72 h–5 days | Eye, muscle, fins, skin | Cartilage in head | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-456 | 24 h–adult | Weak brain | Weak brain | 0 | 0 | 1 | 1 | 3 | 3 |
| dre-miR-457a | 24 h–adult | Ubiquitous | Brain | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-457b | 24 h–adult | Ubiquitous | Brain | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-458 | 48 h–adult | Stronger in brain and eye | No expression | 0 | 0 | 0 | 1 | 1 | 1 |
| dre-miR-459 | 72 h–adult | Gut | Gut | 0 | 0 | 0 | 0 | 3 | 3 |
| dre-miR-460 | 72 h–adult | Fins | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-461 | Not expressed | ND | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-462 | Adult | Gills, fins, skin | Liver and head | 0 | 0 | 0 | 0 | 3 | 3 |
| dre-miR-92b | 24 h–adult | Ubiquitous | Outline of tectum and telencephalon | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-740 | Weak 5 days–adult | ND | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
ND = not determined; 0 = no expression; 1 = weak ubiquitous or background expression; 2 = strong ubiquitous expression; 3 = specific expression.
aDetermined by northern blot analysis.
Table 2.
Overview expression data new miRNAs
| New ID | Expression timea | Expression adult tissuea | Expression in situ | Temporal expression in situ | |||||
|---|---|---|---|---|---|---|---|---|---|
| 12 h | 16 h | 24 h | 48 h | 72 h | 5 days | ||||
| dre-miR-489b | No expression | Brain, eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_11 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-724-3pc | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-724-5p | No expression | Weak braind | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-725 | 48 h–adult | Gills, fins | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-726-3p | 72 h–adult | Eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-726-5pc | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_139 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_149 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_157 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-727-3p | No expression | Braind | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-727-5pc | No expression | Brain, eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-728 | 48 h–adult | Brain, eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-729 | 48 h–adult | Eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_21 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-190b | 48 h–adult | Eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_236 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-34c-3pc | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-34c-5p | 48 h–adult | Brain | Nose | 0 | 0 | 0 | 0 | 3 | 3 |
| dre-miR-722 | 24 h–adult | Brain, eye, gills, skin, liver | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-730-3pc | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-730-5p | 24 h–adult | Brain, eye, fins, skin | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_263 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_264 | no expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-731 | Adult | Gills, fins, skin, gut, heart | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_286 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-732 | No expression | Weak braind | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-733 | 24 h–adult | Eye, brain, muscle gills, fins, skin, gut | Ubiquitous, but more in yolk and gut | 2 | 2 | 3 | 3 | 3 | 3 |
| dre-miR-15c | 24 h–adult | Ubiquitous, not in liver | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_33 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-734 | No expression | Braind | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-735-3p | 24 h–adult | Ubiquitous but not in liver and gut | Ubiquitous (brain, neural tube, outline neuromasts) | 2 | 2 | 2 | 3 | 3 | 3 |
| ZF_nl_384 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-736 | 24 h–adult | Eye, gut | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-737-3p | No expression | Brain, eye | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-738 | 48 h–adult | Gills | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-739 | No expression | Brain | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| ZF_nl_51 | No expression | No expression | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-499 | 24 h–adult | Heart and muscles and fins | Heart and muscles in head | 0 | 0 | 0 | 3 | 3 | 3 |
| dre-miR-723-3p | 5 days–adult | Brain | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
| dre-miR-723-5pc | No expression | Weak braind | No expression | 0 | 0 | 0 | 0 | 0 | 0 |
aDetermined by northern blot analysis.
bPredicted based on verified mammalian sequence.
cRegarded as miRNA star sequence.
dOnly checked for expression in brain.
0 = no expression; 1 = weak ubiquitous or background expression; 2 = strong ubiquitous expression; 3 = specific expression.
First, we analyzed the expression of all 67 miRNAs by in situ hybridization on different zebrafish embryonic stages. Only a subset of 28 miRNAs could be detected in situ (Figure 2, Tables 1 and 2). For many of the miRNAs, the expression was restricted to specific tissues or cell types, although overall we observed less tissue-restricted expression as in our previous study for the conserved vertebrate miRNA set (Figure 2, Tables 1 and 2). Several miRNAs were expressed in (parts of) the brain (e.g. miR-92b, miR-500a/b and miR-135) and these have unique patterns as revealed by sectioning of the embryos (Figure 2b). Other examples of tissue-specific expression are miR-451, which is expressed in the blood cells, miR-455, which is expressed in the cartilage of the pharyngeal arches and head skeleton and miR-459, which is only expressed in the anterior part of the gut.
Figure 2.
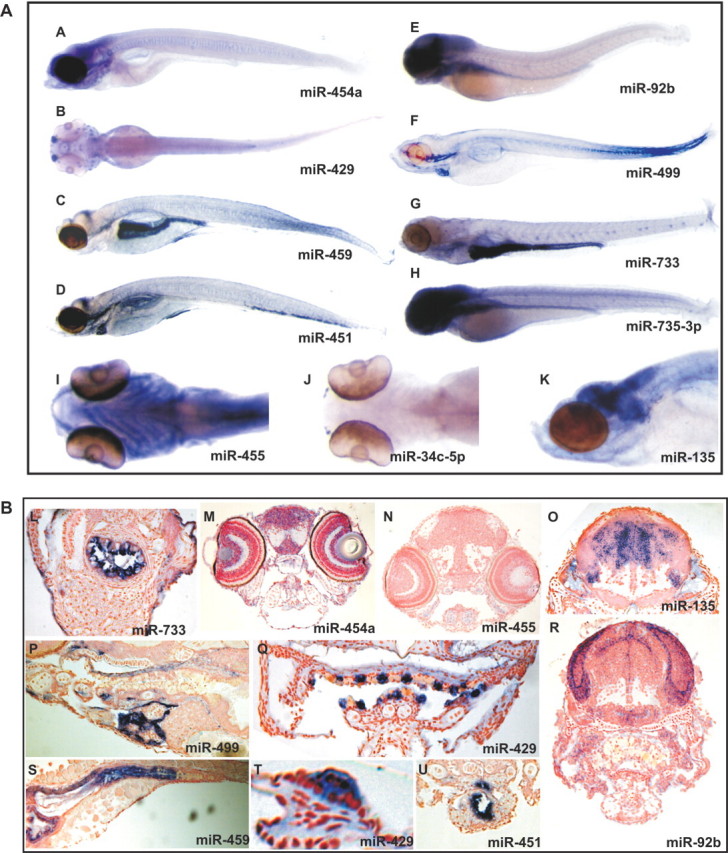
Examples of expression patterns of miRNAs in the zebrafish embryo as revealed by whole mount in situ hybridization. (A) Whole mount pictures of 72-h-old and 5-day-old larvae and (B) pictures of sections from 5-day-old larvae. (A and M) miR-454a is expressed in the brain, the pharyngeal arches and the jaw and the eye; (B, Q and T) miR-429 is expressed in the hair and supporting cells of the lateral line organ (T), the taste buds (Q), the nose and the epithelium of the lips; (C and S) miR-459 is expressed in the anterior gut; (D and U) miR-451 is expressed in the blood cells; (E and R) miR-92b is expressed in the proliferating zones of the preoptic region, optic tectum, tegmentum, telencephalon and octaval area; (F and P) miR-499 is expressed in the ventricle and atrium, the muscles of the head and the somitic muscles; (G and L) miR-733 is expressed ubiquitously but primarily in the intestine; (H) miR-735-3p is expressed ubiquitously, but primarily in the central nervous system; (I and N) miR-455 is expressed in the cartilage of the pharyngeal arches and head skeleton; (J) miR-34c-5p is expressed in the nose; (K and O) miR-135 is expressed in the pallium, optic ganglion, optic tectum, ventral telencephalon and at the beginning of the medulla oblongata.
The expression of several miRNAs was ubiquitous in the later stages of development (miR-130a/b/c and miR-301b), whereas all members from the miR-430 family of miRNAs were expressed ubiquitously only in the earlier stages up to 48 h of development, as described previously (29).
Of the 35 new miRNAs analyzed in this study, we could only detect four by in situ hybridization, which probably reflects the low abundance of these new miRNA as already indicated by the low cloning frequency. miR-34c-5p was expressed in the nose; miR-499 was expressed in the heart, the somitic muscles and the muscles of the head; miR-735-3p was expressed ubiquitously but primarily in the central nervous system; miR-733 was also expressed ubiquitously but more strongly in the gut and the cells surrounding the yolk.
Northern blot analysis of known miRNAs
As outlined above, we could observe a clear in situ expression pattern in the embryo for only 28 of the 67 miRNAs analyzed. We next went on analyzing the expression of all miRNAs by northern blot analysis, since this is a more sensitive method for detecting miRNAs, and it also determines the length of the RNA species, providing further evidence for the existence of a bonafide miRNA. Furthermore, the in situ hybridization analysis is only restricted to embryonic stages of development, whereas northern blot analysis enabled us to detect miRNAs in RNA derived from adult zebrafish tissues.
We first analyzed the temporal expression in five developmental stages ranging from 24 h to adult fish for the set of 32 miRNAs that was already cloned or predicted previously (Figure 3a and Table 1). While some miRNAs could mainly be detected in RNA from adult zebrafish (miR-202, miR-460), others were expressed at all time points analyzed (miR-130a/b/c) and some were also expressed mainly in embryonic stages, but at much lower levels in the adult fish (miR-363 and miR-301b). Again, all members of the miR-430 miRNA family were expressed abundantly in the early embryonic stages up to ∼72 h, but were absent in RNA from 5-day-old larvae and adult fish.
Figure 3.
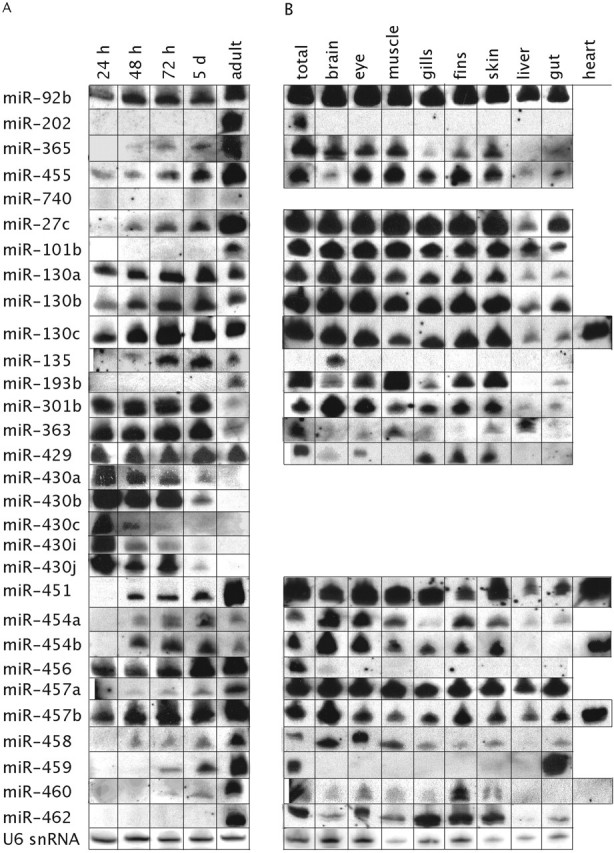
Northern blot analysis of the expression of known miRNAs from zebrafish. (A) Expression of miRNAs in five developmental stages: 24 h, 48 h, 72 h, 5 days and adult zebrafish. (B) Expression of miRNAs in RNA derived from 10 adult zebrafish tissues: total, brain, eye, muscle, gills, fins, skin, liver, gut and heart. For some miRNAs we did not analyze the expression in the heart, because we could not obtain enough heart tissue for analyzing all the miRNAs by northern blotting. U6 snRNA serves as a loading control.
Next, we compared the in situ patterns of miRNAs in the embryo with the spatial expression in the adult zebrafish (Figure 3b and Table 1). For the majority of miRNAs, the expression in RNA derived from adult tissues is similar to the expression in the embryo. For example, miR-135 is specific for the embryonic and the adult brain. Similarly, miR-459 is expressed in the embryonic anterior gut and in the adult fish it is expressed exclusively in the gut. However, for some miRNAs the expression in the embryo was different compared with the expression in the adult, e.g. miR-455 is expressed in the cartilage of the embryo (Figure 2a and b), but the expression in the adult is in many tissues. Overall, on northern blots we could detect 30 out of the 32 known miRNAs analyzed.
Northern blot analysis of newly cloned miRNAs
Although we were able to obtain in situ expression patterns for the majority of known miRNAs, we could only detect 4 out of the 35 novel miRNAs that were cloned in this study (Figure 2 and Table 2). We next went on analyzing the expression of these 35 newly identified miRNAs on northern blots using the same LNA probes as for in situ hybridization (Figure 4). First, we scanned the whole set of 35 miRNAs for presence in large quantities of total RNA (∼15 µg) from 24-h-old embryos and adult zebrafish (data not shown). For all miRNAs that gave a positive signal, we performed new northern blots with total RNA from different developmental stages and with total RNA from 10 dissected tissues from adult zebrafish. In total, we could detect 16 miRNAs on time series northern blots (Figure 4a). As for the known miRNAs analyzed in Figure 3a, some miRNAs were expressed throughout development (miR-15c and miR-736) and others were expressed only in the adult (miR-731). Another set of the new miRNAs was most abundant in 5-day-old larvae, while there was a strong drop in expression in the adult zebrafish (miR-726-3p, miR-729, miR-728 and miR-190b). Except for miR-726-3p, these have all been cloned primarily from the 5-day-old larvae library. An additional three miRNAs could be detected on northern blots containing RNA from dissected tissues (Figure 4b). For most miRNAs we could only detect an RNA species corresponding to the length of miRNAs, but for two we saw some additional bands corresponding to larger RNA molecules (miR-735-3p and miR-733), which is probably background.
Figure 4.
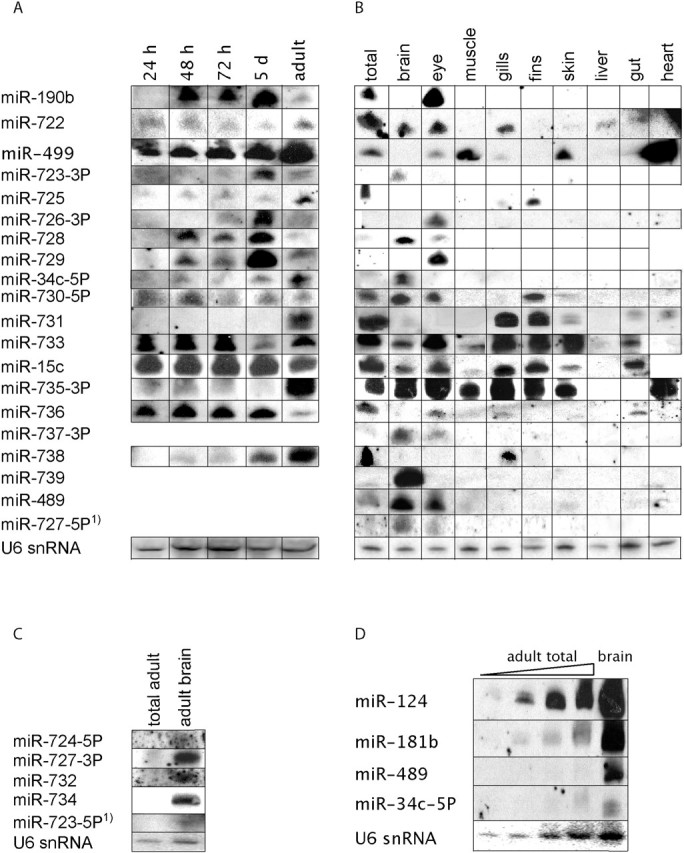
Northern blot analysis of newly cloned miRNAs from zebrafish. (A) Expression of miRNAs detected in five developmental stages: 24 h, 48 h, 72 h, 5 days and adult zebrafish. (B) Expression of miRNAs in 10 different tissues from adult zebrafish: total, brain, eye, muscle, gills, fins, skin, liver, gut and heart. (C) miRNAs detected by specifically probing RNA from adult fish brain. (D) Dilution series of adult fish RNA and one adult brain RNA sample. Blots were probed for two abundant and brain-specific miRNAs (miR-124 and miR-181b) and two new and brain specific miRNAs cloned in this study (miR-489 and miR-34c-5p). U6 snRNA serves as a loading control. 1, regarded as the miRNA star sequence.
There was a wide variety in expression in RNA from dissected tissues. For example, miR-499 is specific for the heart and the muscles. However, many newly cloned miRNAs were expressed in the adult brain (miR-739, miR-34c-5p, miR-728, miR-723-3p, miR-737-3p, miR-727-5p). Encouraged by this finding, we analyzed the expression of the miRNAs, which we did not initially detect in total RNA samples from 24-h-old embryos or adult fish, in RNA from adult fish brain. By doing this, we could detect four more miRNAs that were expressed in the adult brain sample, whereas hardly any signal was detected in the total RNA sample (Figure 4c). Thus, by increasing the amount of RNA on the northern blot and by looking in a specific tissue, we enriched enough to detect some additional new miRNAs. In total, we could detect 23 out of 35 newly cloned miRNAs by northern blotting.
To quantify the differential expression level of known and new miRNAs, we performed parallel northern blots using a dilution series of total adult fish RNA and an adult brain RNA sample. We probed these for two miRNAs that are easily detectable, also in situ, and that are expressed in the brain (miR-124 and miR-181b), with two new miRNAs that we found to be expressed at least in the adult fish brain (miR-489 and miR-34c-5p) (Figure 4d). Although miR-124 and miR-181b were easily detected also in the more diluted total RNA samples, miR-489 and miR-34c-5p could only be detected clearly in the adult brain samples. The expression level of miR-34c-5p in the undiluted total RNA sample is comparable to the expression level of miR-124 in the most diluted (27×) total RNA sample, showing that there is a ∼30 fold difference in abundance between these two miRNAs. These examples indicate that indeed many of the miRNAs that we cloned in addition to the already known miRNAs are much less abundant and therefore more difficult to detect.
Cloning and expression of miRNA star sequences
For 65 known and 11 new miRNAs we also cloned the other arm of the hairpin. The 65 star sequences of known miRNAs were represented by 621 clones (Supplementary Table S5) and the 11 star sequences of new miRNAs were represented by 45 clones (Supplementary Table S6). In most cases, the miRNA was cloned more often than the star sequence.
In our expression analysis, we included six star sequences of new miRNAs. We were unable to detect any of these by in situ hybridization. However, we detected miR-723-5p and miR-727-5p by northern blot analysis (Figure 4). As expected, the expression of these two miRNA star sequences overlaps in both cases with the expression of the miRNA.
DISCUSSION
Regulation of gene expression by miRNAs is an important process for development of multicellular organims, and organisms without miRNAs cannot live (29,30,44). This is strengthened further by recent insight into the abundance and conservation of miRNAs and the high number of miRNA targets found in animals (5,25,26).
Here we describe the cloning and expression of new miRNAs from the zebrafish. By deep sequencing two small-RNA cDNA libraries we found 66 new miRNAs and 11 star sequences corresponding to 116 potential miRNA hairpins in the zebrafish genome. The majority (97.4%) of small-RNAs that were found in the libraries corresponded to known miRNAs and 56% of the new miRNAs were represented by a single sequenced clone. However, only 13% of the new miRNAs that we detected on northern blots were cloned only once, indicating that these single cloned miRNAs are more difficult to detect and also that miRNA cloning frequency reflects miRNA abundance. In addition, 67% of the newly cloned miRNAs are, out of 12 different species, conserved only in zebrafish according to our conservation criteria (>90% identity for the mature miRNA and >70% identity for the precursor). Thus, our cloning and expression data show that although many miRNAs are abundantly expressed, there are also miRNAs that have much lower expression levels or that are expressed in only a few cells. For example, miR-34c-5p has a very restricted expression pattern in the nose of the embryo. Of the new miRNAs 37 were picked up only once, indicating that our sequencing did not reach saturation yet. Furthermore, we screened only two libraries derived from a limited amount of tissues. If some miRNAs are only expressed in specific adult tissues, and some of them are (Figures 3 and 4), these will be missed by our libraries, which do not contain all adult tissues. In order to determine the complete set of miRNAs that governs gene expression in an organism, one should perform saturated sequencing of small-RNA cDNA libraries from several tissues.
Current estimates, which are based on cloning and computational predictions of miRNAs from human, suggest that there might be up to a thousand miRNAs (6,7). Our data show that there are more miRNAs to be found, but predict that these will be lowly expressed and less conserved. The set of abundantly expressed miRNAs in zebrafish is, based on this study and previous work (36,39), limited to ∼150 different miRNAs and mostly contains conserved miRNAs.
Analysis of the spatial and temporal expression of miRNAs may shed light on their role in tissue specific processes. Many of the new miRNAs analyzed in this study have a cell type or tissue specific expression, providing a basis for understanding specific aspects of development that are under miRNA control. The differences in temporal expression of many miRNAs suggests that some play a role in early development during gastrulation and segmentation, whereas others may be required for organ morphogenesis in later stages of embryonic development and a few miRNAs are exclusively expressed in the adult fish. For some miRNAs there is a difference in expression in the embryo compared to the adult, suggesting differences in function. For example, miR-92b is expressed in the embryonic brain, but it is detected in many tissues in the adult fish.
In many cases, miRNA expression patterns correlate with miRNA function in the Drosophila embryo, where, for example, knockdown of miRNAs expressed in the peripheral or central nervous system induced nervous system defects (37,45). Careful examination of the phenotypes caused by depletion of specific miRNAs in the vertebrate embryo together with computational prediction of miRNA targets may further help in understanding their role in development.
Supplementary Material
Acknowledgments
We thank Brandon Ason and René Ketting for critically reading of the manuscript. This work was supported by the Netherlands Genomics Initiative and the European Union. Funding to pay the Open Access publication charges for this article was provided by the Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research.
Conflict of interest statement. None declared.
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors
REFERENCES
- 1.Ambros, V. 2004. The functions of animal microRNAs Nature 431350–355 [DOI] [PubMed] [Google Scholar]
- 2.Bartel, D.P. and Chen, C.Z. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs Nature Rev. Genet. 5396–400 [DOI] [PubMed] [Google Scholar]
- 3.Lee, R.C., Feinbaum, R.L., Ambros, V. 1993. The C.elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14 Cell 75843–854 [DOI] [PubMed] [Google Scholar]
- 4.Wightman, B., Ha, I., Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C.elegans Cell 75855–862 [DOI] [PubMed] [Google Scholar]
- 5.Griffiths-Jones, S. 2004. The microRNA Registry Nucleic Acids Res. 32D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentwich, I., Avniel, A., Karov, Y., Aharonov, R., Gilad, S., Barad, O., Barzilai, A., Einat, P., Einav, U., Meiri, E., et al. 2005. Identification of hundreds of conserved and nonconserved human microRNAs Nature Genet. 37766–770 [DOI] [PubMed] [Google Scholar]
- 7.Berezikov, E., Guryev, V., van de, B.J., Wienholds, E., Plasterk, R.H., Cuppen, E. 2005. Phylogenetic shadowing and computational identification of human microRNA genes Cell 12021–24 [DOI] [PubMed] [Google Scholar]
- 8.Cai, X., Hagedorn, C.H., Cullen, B.R. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs RNA. 101957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, Y., Kim, M., Han, J., Yeom, K.H., Lee, S., Baek, S.H., Kim, V.N. 2004. MicroRNA genes are transcribed by RNA polymerase II EMBO J. 234051–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, Y., Ahn, C., Han, J., Choi, H., Kim, J., Yim, J., Lee, J., Provost, P., Radmark, O., Kim, S., et al. 2003. The nuclear RNase III Drosha initiates microRNA processing Nature 425415–419 [DOI] [PubMed] [Google Scholar]
- 11.Lund, E., Guttinger, S., Calado, A., Dahlberg, J.E., Kutay, U. 2004. Nuclear export of microRNA precursors Science 30395–98 [DOI] [PubMed] [Google Scholar]
- 12.Yi, R., Qin, Y., Macara, I.G., Cullen, B.R. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs Genes Dev. 173011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C.elegans developmental timing Cell 10623–34 [DOI] [PubMed] [Google Scholar]
- 14.Bernstein, E., Caudy, A.A., Hammond, S.M., Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference Nature 409363–366 [DOI] [PubMed] [Google Scholar]
- 15.Hutvagner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex Science 2972056–2060 [DOI] [PubMed] [Google Scholar]
- 16.Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C.elegans Genes Dev. 152654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., Filipowicz, W. 2005. Inhibition of translational initiation by Let-7 microRNA in human cells Science 3091573–1576 [DOI] [PubMed] [Google Scholar]
- 18.Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., Parker, R. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies Nature Cell Biol. 7719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, J., Rivas, F.V., Wohlschlegel, J., Yates, J.R., III, Parker, R., Hannon, G.J. 2005. A role for the P-body component GW182 in microRNA function Nature Cell Biol. 71161–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehwinkel, J., Behm-Ansmant, I., Gatfield, D., Izaurralde, E. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing RNA 111640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwab, R., Palatnik, J.F., Riester, M., Schommer, C., Schmid, M., Weigel, D. 2005. Specific effects of microRNAs on the plant transcriptome Dev. Cell 8517–527 [DOI] [PubMed] [Google Scholar]
- 22.Brennecke, J., Stark, A., Russell, R.B., Cohen, S.M. 2005. Principles of microRNA-target recognition PLoS. Biol. 3e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doench, J.G. and Sharp, P.A. 2004. Specificity of microRNA target selection in translational repression Genes Dev. 18504–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kloosterman, W.P., Wienholds, E., Ketting, R.F., Plasterk, R.H. 2004. Substrate requirements for let-7 function in the developing zebrafish embryo Nucleic Acids Res. 326284–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, B.P., Burge, C.B., Bartel, D.P. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets Cell 12015–20 [DOI] [PubMed] [Google Scholar]
- 26.Lim, L.P., Lau, N.C., Garrett-Engele, P., Grimson, A., Schelter, J.M., Castle, J., Bartel, D.P., Linsley, P.S., Johnson, J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs Nature 433769–773 [DOI] [PubMed] [Google Scholar]
- 27.Farh, K.K., Grimson, A., Jan, C., Lewis, B.P., Johnston, W.K., Lim, L.P., Burge, C.B., Bartel, D.P. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution Science 3101817–1821 [DOI] [PubMed] [Google Scholar]
- 28.Stark, A., Brennecke, J., Bushati, N., Russell, R.B., Cohen, S.M. 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution Cell 1231133–1146 [DOI] [PubMed] [Google Scholar]
- 29.Giraldez, A.J., Cinalli, R.M., Glasner, M.E., Enright, A.J., Thomson, J.M., Baskerville, S., Hammond, S.M., Bartel, D.P., Schier, A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish Science 308833–838 [DOI] [PubMed] [Google Scholar]
- 30.Wienholds, E., Koudijs, M.J., van Eeden, F.J., Cuppen, E., Plasterk, R.H. 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development Nature Genet. 35217–218 [DOI] [PubMed] [Google Scholar]
- 31.Bernstein, E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn, H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., Hannon, G.J. 2003. Dicer is essential for mouse development Nature Genet. 35215–217 [DOI] [PubMed] [Google Scholar]
- 32.Sokol, N.S. and Ambros, V. 2005. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth Genes Dev. 192343–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao, Y., Samal, E., Srivastava, D. 2005. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis Nature 436214–220 [DOI] [PubMed] [Google Scholar]
- 34.Lai, E.C., Tam, B., Rubin, G.M. 2005. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs Genes Dev. 191067–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hornstein, E., Mansfield, J.H., Yekta, S., Hu, J.K., Harfe, B.D., McManus, M.T., Baskerville, S., Bartel, D.P., Tabin, C.J. 2005. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development Nature 438671–674 [DOI] [PubMed] [Google Scholar]
- 36.Wienholds, E., Kloosterman, W.P., Miska, E., Alvarez-Saavedra, E., Berezikov, E., de Bruijn, E., Horvitz, H.R., Kauppinen, S., Plasterk, R.H. 2005. MicroRNA expression in zebrafish embryonic development Science 309310–311 [DOI] [PubMed] [Google Scholar]
- 37.Aboobaker, A.A., Tomancak, P., Patel, N., Rubin, G.M., Lai, E.C. 2005. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development Proc. Natl Acad. Sci. USA 10218017–18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kloosterman, W.P., Wienholds, E., de Bruijn, E., Kauppinen, S., Plasterk, R.H. 2006In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes Nature Methods 327–29 [DOI] [PubMed] [Google Scholar]
- 39.Chen, P.Y., Manninga, H., Slanchev, K., Chien, M., Russo, J.J., Ju, J., Sheridan, R., John, B., Marks, D.S., Gaidatzis, D., et al. 2005. The developmental miRNA profiles of zebrafish as determined by small RNA cloning Genes Dev. 191288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ewing, B., Hillier, L., Wendl, M.C., Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment Genome Res. 8175–185 [DOI] [PubMed] [Google Scholar]
- 41.Aravin, A. and Tuschl, T. 2005. Identification and characterization of small RNAs involved in RNA silencing FEBS Lett. 5795830–5840 [DOI] [PubMed] [Google Scholar]
- 42.Hofacker, I.L. 2003. Vienna RNA secondary structure server Nucleic Acids Res. 313429–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J.D., Higgins, D.G., Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice Nucleic Acids Res. 224673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernstein, E., Kim, S.Y., Carmell, M.A., Murchison, E.P., Alcorn, H., Li, M.Z., Mills, A.A., Elledge, S.J., Anderson, K.V., Hannon, G.J. 2003. Dicer is essential for mouse development Nature Genet. 35215–217 [DOI] [PubMed] [Google Scholar]
- 45.Leaman, D., Chen, P.Y., Fak, J., Yalcin, A., Pearce, M., Unnerstall, U., Marks, D.S., Sander, C., Tuschl, T., Gaul, U. 2005. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development Cell 1211097–1108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.