Abstract
Within the past decade, imaging mass spectrometry has been increasingly recognized as an indispensable technique for studying biological systems. Its rapid evolution has resulted in an impressive array of instrument variations and sample applications, yet the tools and data are largely confined to specialists. It is therefore important that at this junction the IMS community begin to establish IMS as a permanent fixture in life sciences research thereby making the technology and/or the data approachable by non-mass spectrometrists, leading to further integration into biological and clinical research. In this perspective article, we provide insight into the evolution and current state of imaging mass spectrometry and propose some of the directions that IMS could develop in order to stay on course to become one of the most promising new tools in life science research.
1. Introduction
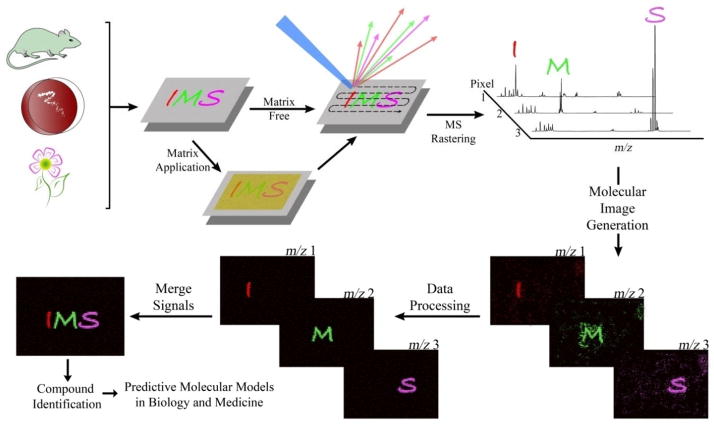
The ability to simultaneously visualize multiple molecular distributions across the surface of a sample without the need for chemical labels or antibodies is the main strength of imaging mass spectrometry (IMS) based research. The overall methodology behind IMS is fairly simple (Figure 1). A mass spectrometer need only be equipped with a computer controlled sample stage capable of motion about the x-y coordinate plane working in conjunction with a probe capable of producing ions from the sample surface, a process referred to as ionization. Through the utilization of specialized software, the motion of the sample stage can be programmed to allow discrete or continuous rastering across the sample area whereby individual mass spectra can be collected and assigned to a defined area of the sample therefore creating a pixel. Software is then used to assemble these pixels into an image where the relative intensity of an ion is displayed as a scaled false color allowing for visualization of the molecular distribution across the surface of the sample for any given observed mass. Granting researchers the ability to study a system with molecular eyes, IMS has many promising life science applications such as enabling the connection of molecular information with phenotypes and revealing entirely new molecular phenotypes that cannot be observed through classical histology.
Figure 1.
General overview of an IMS workflow. Samples (i.e. tissue sections, bacterial cultures, single cells and plant components) are mounted on a target surface and, if required, immediately followed by matrix application. The mass spectrometer is then programmed to raster across the surface of the sample collecting spectra at specified sampling locations which are compiled into a single dataset where the occurrence of any single mass can be visualized as a scaled false color overlay depicting the relative intensity of the ion across all sampled locations. This data can be further processed to improve its quality followed by merging with other data including optical images, additional observed IMS ions or biochemical data such as MRI or fluorescence images.
Despite the first examples of molecular images by mass spectrometers being obtained some 40 years ago1, only recently has IMS seen widespread use among mass spectrometrists. The advent of MALDI-MS based tissue imaging in the late 1990’s opened up a wide range of biological applications leading to rapid incorporation of the technology into new biochemical and medical research strategies2. Molecular histology by mass spectrometry based imaging became a promising new technology for candidate biomarker discovery3,4, drug metabolite profiling5,6, lipid analysis7–10 and proteomics11–14. Today, IMS is a constantly evolving technology that has been applied to not only the molecular histology of tissue and whole body sections15,16, but also bacterial thin films17–19, plant leafs20,21 and stems22, ancient glass23, quartz24, circuit boards25 and even fingerprints26–28 (Figure 3C). Advances in instrumentation and bioinformatics continue to drive the technology forward by widening the application range to which IMS can be applied and is resulting in a steadily decreasing gap between biology and chemistry as non-mass spectrometrists are beginning to get an appetite for the information that IMS provides.
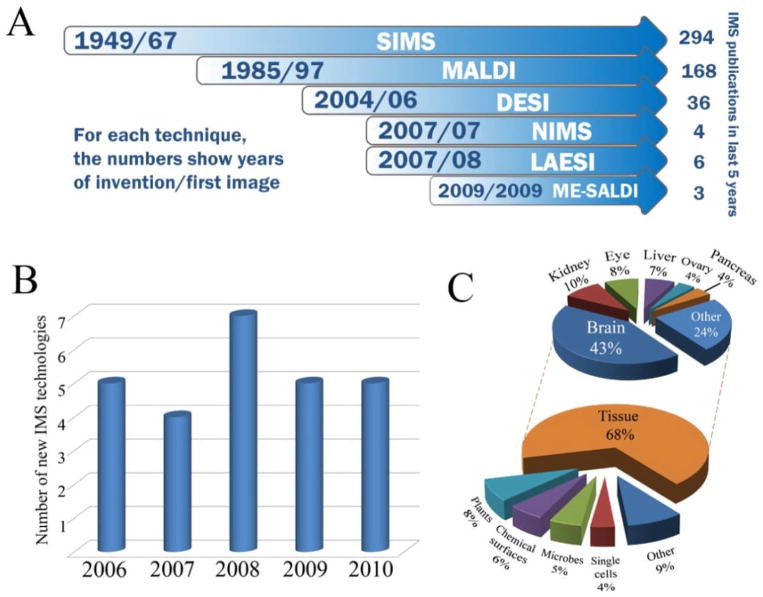
Figure 3.
Progression of IMS. A) Timeline of major and emerging advances in IMS instrumentation along with total number of publications within the last 5 years for each technique. B) Yearly totals of new IMS techniques developed since 2006. C) Distribution of IMS applications since 2006. Totals for A, B and C were determined by searching PubMed for research articles published within the past 5 years containing the words ‘imaging’, ‘mass’ and ‘spectrometry’ in the title or abstract. Search hits were archived manually. Totals for B represent the first reported use of a unique MS technology in collecting a molecular image.
Despite these promising attributes, the adoption rate of IMS in the scientific community is much slower than one would expect. In part this is due to the lack of specific workflows that provide answers to the questions that biologists have. Even within the mass spectrometry community there is a general skepticism of IMS since so few of the many observed signals can be accurately identified and/or interpreted. In principle, molecules observed by IMS are among the most abundant in the sample yet it is challenging -even when current proteomic and metabolomic tools are applied- to identify these signals and thus represents a general gap in knowledge of not just IMS but the entire mass spectrometry field as highlighted in recent articles29–36. While our ability to annotate signals is one area that must improve for IMS, a great strength of IMS is its ability to specifically pinpoint masses of interest that can be further investigated. Since many of the challenges facing IMS and its applications have been articulated very acutely in many recent reviews6, 37–44, this highlight will not cover such topics in great detail and we would encourage interested readers to consult these reviews for more information. Instead, this highlight will focus on describing the current state of the art of IMS and the direction where the technology can go in order to stay on course in becoming one of the most promising new tools in life science research.
2. The Current State of IMS
The Three Front Runners of IMS
IMS methodology has been adapted to many different types of mass spectrometers and ionization approaches; however, there are three methods (probes) for ionization that currently see the most amount of use across the widest range of applications (Figure 2). The oldest of these three, secondary ion mass spectrometry or SIMS, collected some of the first ion images of monolayers in the late 1960’s 1. SIMS uses a focused ion beam of individual or clusters of high energy particles, such as Bi+ 45,46, Au3+ 47,48 and C60 49, where upon impact of a sample surface causes emission of secondary ions that are typically analyzed via a time-of-flight (TOF) mass analyzer. The use of a primary ion beam allows SIMS instruments to collect nanometer size pixels, but usually limits its mass range in biological samples to below 1000 Da due to only a small percentage of molecules sputtered from the sample surface being ionized50 and to source induced fragmentation of surface molecules51. The absence of matrix in typical TOF-SIMS analysis also yields high spectral clarity in the low mass range allowing for imaging of small ions such as Na+, K+ and even H+. Matrix enhanced SIMS (ME-SIMS)52 and incorporation of cluster ion sources40 has dramatically increased secondary ion formation efficiency in biological samples to allow for sampling of peptides and oligonucleotides; however, the signal to noise ratio for molecules greater than 5kDa drops off dramatically in ME-SIMS compared to matrix assisted laser desorption ionization mass spectrometry (MALDI) 53.
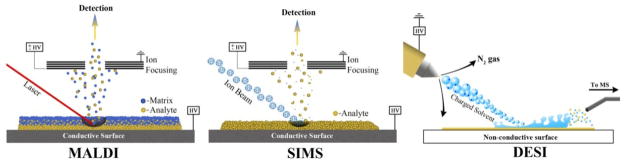
Figure 2.
Ionization mechanisms for MALDI, SIMS and DESI ionization sources. For MALDI experiments, a sample is mounted on a conductive target plate immediately followed by matrix application. The sample is then shot with a UV or IR wavelength laser whereby the energy is absorbed by the matrix and, via a black box mechanism, transferred to the analyte to induce ionization. Both positive and negative ions are generated by most matrices however certain matrices will favor positive or negative ion formation. In SIMS experiments, samples are mounted on a conductive surface and subjected to etching by a beam of high energy particles such as Ar+, Au3+, or C60. Upon collision with the sample surface, the energy of these primary ions is transferred to the analyte dislodging secondary ions. Samples for DESI are mounted on a non-conductive surface where a pneumatically assisted stream of charged solvent droplets is directed at the sample surface. During initial wetting of the surface, analyte is taken up by the solvent and subsequent collisions of charged droplets with the wetted surface produces secondary droplets which are taken up by the mass spectrometer inlet. The charge of the ions is determined largely by the solvent used.
The second and most widely used technique is MALDI, which first came onto the IMS scene in the late 1990’s where it was used to image peptides and proteins in biological tissue sections2. Instead of a focused ion beam, MALDI instruments utilize a high powered UV or IR laser to initiate compound ionization. Matrix, typically a low molecular weight light absorbing organic acid, is used to coat the surface of the sample for the purpose of improving ionization by utilizing absorbed energy from the laser to protonate or deprotonate the analyte via what is currently, despite intense studies54–59, a largely black box mechanism.
The necessity of covering samples with a chemical matrix somewhat limits the number of amendable sample types, spatial resolution and data clarity below 500 Da. However, the ability of the matrix to drastically increase ionization efficiency of compounds allows for less laser power resulting in an ionization mechanism gentle enough to monitor compounds up to 200k Da such as intact proteins12,60. Using new lasers with non-Gaussian beam profile and current on-market hardware, it is possible for pixel sizes in MALDI imaging be 10 μm while keeping high acquisition speed (1 kHz laser) with no significant loss of signal intensity up to 15 kDa for 20 μm pixel size61. 61.
The newest of the three is desorption electrospray ionization mass spectrometry or DESI62,63. This adaptation of electrospray ionization directs a pneumatically assisted stream of charged solvent droplets at a sample surface where, upon contact with the surface, analyte is taken up by the solvent64. Subsequent collisions of charged droplets with the wetted sample surface desorb secondary droplets which are taken up by a mass spectrometer and analyzed65,66. While the sensitivity and spatial resolution for most applications of DESI are less than what is attainable by SIMS or MALDI, DESI offers a few unique characteristics that allow it to analyze samples that are otherwise impossible to analyze. For instance, in addition to tissue63, bacterial17 and plant imaging67, analysis of wet or insulated samples is possible using DESI as well as reactive imaging where a chemical known to react with a compound of interest can be added to the spray solvent allowing you to monitor the product of the reaction68,69.
While MALDI, SIMS and DESI continue to be heavily utilized in the field of IMS, new instrumentation is constantly being developed with many being either combinations of existing ionization sources or improvements on existing technology (Figure 3B). Combinations of surface ablation/desorption and direct infusion techniques have led to technologies such as laser ablation electrospray ionization (LAESI)70, matrix assisted laser desorption electrospray ionization (MALDESI)71 and laser spray ionization72, all of which use laser ablation to desorb molecules from the sample surface and electrospray ionization to direct the released molecules into the mass analyzer. Trace metal analysis in biological samples with up to nanometer lateral resolution is possible by laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS)73. Novel sample surface technologies such as desorption/ionization on silicon (DIOS)74 and nanostructure initiator mass spectrometry (NIMS)75 allow for sensitive analysis without matrix using UV lasers (DIOS and NIMS) and primary ion beams (NIMS). New sample preparation approaches and ion sources combined with advancements in mass analyzers, such as ion mobility separation76, have created a dizzying number of instrument configurations that can be customized to fit a labs need with the easiest and most informative tools likely to become adopted by life scientists.
Despite impressive advancements in the number of available instrument combinations, 85% of all imaging MS publications in the last five years were performed using either MALDI-TOF or TOF-SIMS based approaches (Figure 3A) with most new probes seeing very little, if any, implementation outside their lab of invention. In order for emerging technologies to gain a foothold in biological and medical research, the needs of the end user (microbiologist, pathologist, botanist, etc.) have to be kept into consideration during the development cycle on the instrument77. This means that the instruments need to demonstrate a clear and unique area of application that was previously impossible or difficult to study before its invention, have a user friendly software package, have a simple and reproducible sample preparation protocol and eventually become commercially available. Many emerging techniques rarely achieve this level of distinction and even sometimes fail to demonstrate a clear advantage to their use. It is thus not uncommon to read about a new technique only to never see that technique used again in subsequent articles. New tools are only of use to other disciplines in biology and medicine if they provide information that is useful for that discipline and therefore it is important that medical, biological, analytical, computational and biochemical labs work together to create influential advancements in instrumentation and simple methodology workflows that are reproducible and provide meaningful results. Such advances would put IMS in prime position to tackle many of the current challenges in biology such as determining the function and distribution of signaling molecules, distributions of epigenetic markers within tissues, how the brains process information and how our microbiome is involved in controlling health and disease. Next we want to highlight the possibilities with informatics in this regard.
Informatics
At the present time, the development of computational methods for IMS is lagging behind its technological progress, especially informatic tools that are accessible to other non-IMS scientists. While some analytical labs tout their own private in-house data processing and analysis software, the majority of IMS users must rely on software provided by their instrument vendor. This is particularly true with non-mass spectrometry labs. While some vendor and open source software packages do come with a several data analysis options, these software packages typically provide only a basic interface with little more usability than allowing the user to visualize molecular distributions across the imaging area. Data processing tools beyond basic interpolations and normalizing, such as peak alignment78–80, intelligent peak picking78,81,82 and denoising83, are surprisingly sparse. Even basic statistical analyses to determine the quality of the acquired data are currently rarely, if at all, included in most software.
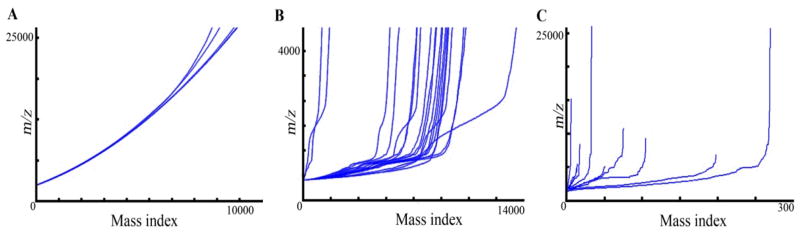
Although recently several reports of advanced data processing of clinical IMS data have been published84–87, there is still a need to establish standard procedures for data processing or at the least uniform incorporation of most of the critical data processing tools into vendor software packages to allow for better interpretation of collected data. For instance, using a simple statistical test, one can quickly judge the quality of a dataset based on the pixel to pixel variability of the data. We obtained three MALDI-IMS datasets from three different laboratories and indexed all observed masses for 10 to 20 randomly selected spectra where the smallest observed mass was given a mass index number of 1, the next largest a mass index number of 2 and so on (Figure 4). When the m/z values for each spectrum are plotted against their assigned index number, the baseline consistency of the data set can be easily observed with high quality data sets (Figure 4A) exhibiting similar total peak counts resulting in masses receiving very similar mass index values between spectra and lower quality data sets (Figure 4B and C) displaying highly variable index values for a given mass and large m/z gaps between adjacent mass index values. Simple tests like this one can be used as data is collected to determine the quality of the data set and, although not yet demonstrated, we speculate that such real-time data evaluation may even be used to allow the instrument to automatically adjust parameters during an imaging run to create “smart” data acquisition modes.
Figure 4.
Importance of evaluation of quality of IMS data. MALDI-IMS data sets (A, B and C) collected from three different laboratories were assessed for spectral consistency using a simple statistical test. For each data set, m/z values for 10 to 20 randomly selected spectra were automatically indexed (e.g. the first observed mass receives a mass index value of 1, the second with a value of 2 and so on). High quality data sets (A) demonstrate low pixel to pixel variability leading to consistent baselines and masses receiving the same or similar mass index value between spectra whereas lower quality data sets with high pixel to pixel variability will contain irregular baselines resulting in different mass index values for a given mass between spectra due to changes in total ion current (B and C) or large gaps in signal (C).
Even the most basic handling of raw data from IMS data sets needs to be revisited, especially in MALDI-IMS. In a modern IMS study, the dataset is typically viewed only as a collection of spectra with little to no consideration given to the spatial relationships between spectra. Instead, the more natural viewpoint would be to represent an IMS dataset as a multi-channel (or spectral) image with thousands of channels each corresponding to an individual ion mass. Such strategies are currently rarely transferred to IMS despite their obvious benefits as seen in other areas of science where multi-channel images are used in astronomy for hyper-spectral imaging88 and imaging spectroscopy89, Earth remote sensing90, life sciences and biomedicine (i.e. confocal Raman microscopy, near-infrared imaging)91.
Pre-processing of data is usually done hastily despite its heavy influence on subsequent data analysis. Consider the scarcely studied issue of normalization of spectra. In MALDI-imaging, there is still discussion on whether normalization should be applied and even which type of normalization should be applied (besides the popular total ion current method)92,93. It is our viewpoint that IMS is largely a discovery tool and different data processing methods that grant the biologist various ways to “visualize” a hypothesis are important uses of the data and as with any standard biological protocol these hypotheses formulated from IMS data need to be confirmed via alternate approaches. Advanced approaches to data processing such as selective suppression of spectra in certain areas of a sample, which proved to be useful in visualizing drug metabolites in whole body sections of rats16, can prove to be extremely useful in forming hypothesis and furthering biological research. We therefore would benefit if the IMS community began borrowing data processing concepts that are well established in many other non-mass spectrometry technologies, such as confocal microscopy, and even sharing ideas within the IMS community. For instance, normalization is well studied in SIMS (a.k.a. scaling of spectra) where, along with spectrum-wise normalization, more advanced ways of accounting for noise (i.e. Poisson scaling) are established94,95. There is currently little transfer of expertise from the SIMS to MALDI community and therefore there are boundless opportunities for sharing of knowledge between the two schools of thought.
A major and unavoidable challenge in IMS is pixel-to-pixel variability, which involves variation in peak height between spectra measured at different spatial points that are not due to differences in the concentration of the analyte of interest. This variability can be caused by differences in the ionization efficiency due to presence of ion suppressors such as proteins39, uneven matrix crystallization96, detector noise and charge accumulation97,98. Although this variability can be partially corrected at the stage of preprocessing with image processing methods, there are no foreseeable algorithmic solutions to this issue probably due to the mathematical complexity of image processing. However, IMS provides a unique possibility to study the noise properties since a typical dataset comprises thousands of spectra measured under the same instrumental conditions. Certainly, understanding and statistical modeling of IMS noise would lead to improved processing algorithms95.
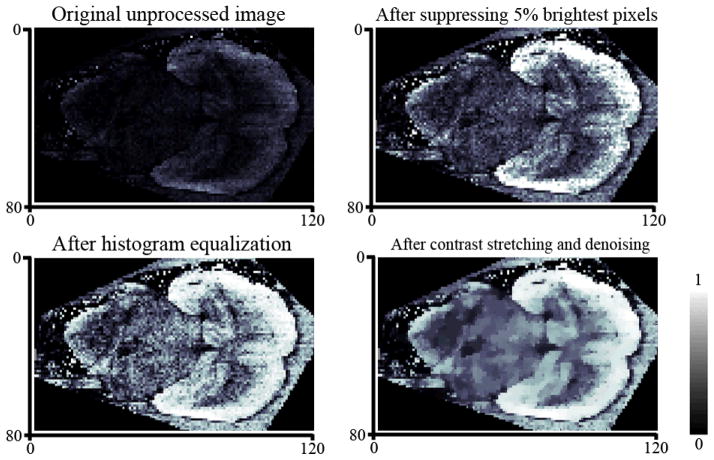
Visualization of IMS data sets can also appear to be deceptively straightforward whereas there are several little-known issues with the process that can have a significant negative impact on the results. For instance, individual spectra containing unusually high levels of analyte signal or noise (a.k.a. hot spots) commonly arise in MALDI-IMS due to uneven matrix distribution and crystallization. Note that a “hot spot” like effect can also occur in DESI or SIMS where a tiny discrete region of increased or decreased signal within the sample may arise due to changes in sample properties such as surface topography, texture or conductivity. Since most IMS software will scale an ion image based on the most intense pixels present, the resulting image will display only these few intense pixels and hide the rest of the image due to incorrect scaling (Figure 5A). Although this issue can be easily overcome, it highlights the need for statistical checks of spectra quality in IMS during data acquisition especially when using automated peak picking or sample segmentation. Along with IMS specific solutions, such as exclusion or suppression of bad spectra during data analysis, image contrast-enhancement methods99 can be used to significantly improve the visual quality and therefore the interpretability of the data (Figure 5). While pixel-to-pixel variability can be compensated for using image denoising, simple image denoising methods degrade the spatial structure of the image by smoothing out small details. Advanced image denoising edge-preserving methods, like those used by Alexandrov et al100, can be applied to maintain image resolution while simultaneously correcting for variability in image quality.
Figure 5.
Various strategies can be applied to visualization of IMS data. Background correction, normalization, denoising and contrast-enhancement are just a few popular methods for cleaning up and correcting analyte signal throughout a sample. A) Original image, MALDI-imaging dataset for a transverse section of mouse brain, fixed mass. B) After contrast-enhancement by correcting 5% of the brightest pixels. C) After contrast-enhancement by histogram equalization. D) After histogram equalization and edge-preserving denoising.
3. Opportunities For Development
Imaging mass spectrometry is a young and rapidly expanding technology with many current opportunities for optimization of current applications and development of new ones. By steadily working towards establishing new robust sample preparation protocols, automated data analysis software and improved instrumentation, IMS will see further adoption into the various niches of the scientific community helping to establish it as a dominant technology for studying biological systems. This will in turn fuel further development due to the growing user base, which will continue to improve the capabilities of IMS as well as the quality of research resulting from its use.
Improved sample preparation and instrumentation
The most obvious area of improvement is regarding the instruments themselves with most attention paid to increasing mass resolution, sensitivity and spatial resolution. Ambient ionization sources have the benefit of linking up with high mass resolution instruments such as FT-ICR’s; however, even after pairing with sensitive trap based instruments their sensitivity may suffer compared to experiments performed under vacuum101. TOF mass analyzers benefit from a very wide mass range and decent sensitivity with most capable of around 10k mass resolution and state of the art instruments achieving around 40k mass resolution. Improving spatial resolution and sensitivity is complicated by the fact that the two are inversely linked to one another in IMS studies42. Changes in pixel size will directly change the area that can be sampled by the mass spectrometer for each measurement. Therefore by decreasing the pixel size to achieve higher spatial resolution you decrease the amount of total analyte that will be sampled for each pixel necessitating more sensitive instruments. Despite these hurdles, improvements in instruments and sample preparation techniques have allowed for extremely small pixel sizes with subcellular imaging now achievable including imaging of ion transport within single cells102, microbial symbionts within their host cells103 and even imaging of individual human chromosomes104.
The increasing popularity of multi-modal data collection approaches, which utilize different mass spectrometry and other spectroscopy techniques to analyze a single sample, has also illustrated that the development of hybrid instruments may allow for more consolidated research protocols and more efficient data collection 6,24,105,107–112. Provided such instruments demonstrate similar technical capability of the individual stand alone systems, hybrid instruments may enable more streamlined workflows through incorporation of multiple MS imaging and histological modalities into a single commercially available instrument. Carado et al106 has constructed a hybrid SIMS-MALDI-TOF instrument capable of switching between SIMS and MALDI ion sources and Tsai et al has developed a dual polarity MALDI-TOF instrument capable of collecting positive and negative mode data simultaneously from a single sample107. These advances in conjunction with instruments now incorporating microscope type equipment complete with fluorescence filters on the mass spectrometer could allow for a one-instrument lab capable of collecting multiple layers of data across different modalities using a single instrument. Despite even incredible advances in instrumentation, sample preparation will always be a critical step to obtaining great data. At the present time data quality and reproducibility in IMS, compared to other imaging approaches, is very dependent on the quality of the sample preparation and for IMS be used in the clinic as a diagnostic tool, the analysis of the sample should always deliver results with little room for error.
In a typical IMS experiment thousands of individual measurements are taken from a sample demonstrating the need for homogeneity within a sample as well as protocols for systematic sample preparation. Poorly prepared samples can produce deviant signals and signal patterns. Signals such as false gradients in signal due to uneven sample or matrix thickness, distinct visual patterns in ion intensity caused not by analyte in the sample but uneven matrix crystallization or differences in conductivity, and drastic changes in sensitivity between consecutive sample preparations can occur due to inconsistent sample preparation. Automated sample preparation tools for matrix distribution3,113 and using a cryostat or microtome for preparation of tissue sections has greatly aided in sample reproducibility, but these instruments are applicable mainly only for imaging of tissue sections. With preparation of other types of samples such as plant tissue and bacterial thin films being done mostly by hand, sample reproducibility begins to rely less on the quality of the original sample and more in the expertise of the researcher preparing it.
Such uniform sample preparation techniques can help reduce spectral baseline variability throughout the image therefore improving the consistency of analyte signal; however, relative quantitation with IMS data is tricky as the visual nature of the data tends to cause researchers to treat intensities between different ions as differences in their relative concentration. While comparing the relative abundance of a single ion within a single sample is the main premise behind IMS, comparing signal for a single ion between multiple samples or signal between two different ions within a single sample becomes more of a grey area. As each unique compound has different ionization efficiencies and ways its signal can be suppressed, intensity differences between ions may not always reflect their relative concentrations39. Likewise, differences in instrument conditions and sample preparation from one sample to another make it unwise to compare the relative abundance of a single ion across multiple samples beyond a certain extent (i.e. one can generally assume a 100-fold change in signal is due to a change in concentration of the compound with all else being equal). Taking into account instrument parameters that effect signal such as total vacuum, plate voltage and detector cleanliness at each pixel may allow for in silico adjustment of spectra and therefore more consistent signal between sample preparations.
While a great number of hypotheses within biology can be formulated and/or confirmed using relative quantitation information from an IMS study, certain areas of biology, including pharmokinetic studies, would also benefit from robust quantitative imaging protocols. Non-linear detection has always made quantitation via mass spectrometry very difficult and is currently only well established with non-imaging analysis of peptides using special chemical labels such as SILAC114, iTRAC115 and AQUA116 labeling. While reports of quantitative IMS have shown promise (especially in SIMS and LA-ICP imaging)73, 117–120, the myriad of factors that affect reliable and reproducible quantitation in MS has resulted in slow progress towards establishing widely accepted protocols for quantitative imaging120–122. In fact, to the best of our knowledge there has yet to be a report of absolute quantitative MALDI imaging of biological samples of all the major molecules seen; a problem that is unsolved. One challenge of MALDI based IMS quantitation of signals region specific ion suppression within a single sample and reliability on matrix coverage. One would hope that possible solutions to this issue may lie in simply applying statistical analyses to adjust and correct for changes in analyte signal; however, such a long standing problem will likely take some time and truly creative thinking to resolve.
Improved Data Mining
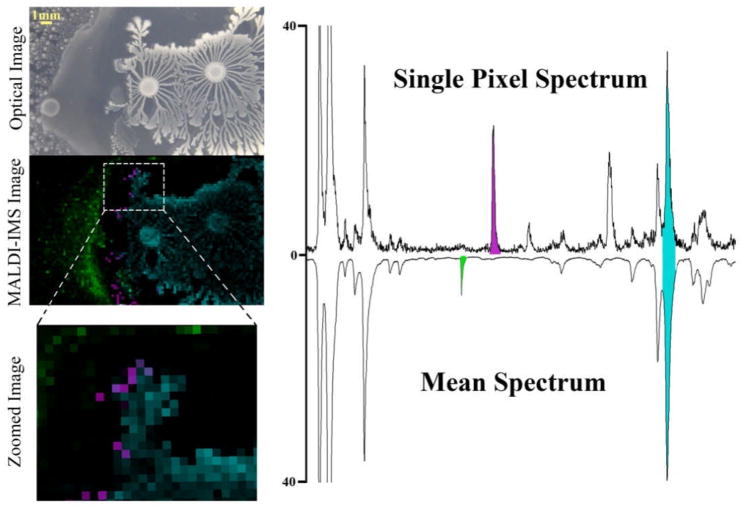
As discussed earlier, data mining of IMS data sets is currently a very time consuming endeavor as it is mostly done manually. An IMS data set collected from 300 to 10,000 Da using a 0.5 Da mass bin for each image will produce roughly 20,000 individual molecular images for a single sample. Currently, complete mining of such data requires the user to click through each image and look for distributions that may correlate to the morphology of the sample analyzed. All signal patterns that correlate and/or are localized to the sample area are catalogued before undergoing subsequent attempts at identification. While peak picking from a mean spectrum allows for easy cataloguing of abundant signals, highly localized signals present in only a small percentage of the sample area will not display in the mean spectrum and need to be mined manually if their mass is unknown (Figure 6). In many cases this could be simplified if there was an efficient approach to compare annotated data between laboratories. Though manual data mining is widely practiced, unsupervised data processing methods are quickly becoming commonplace in imaging laboratories.
Figure 6.
MALDI image of two bacteria interacting on nutrient agar illustrating how mining of IMS data from the mean spectrum will primarily only produce signals that are highly abundant or delocalized (green and blue signals) while less abundant or highly localized signals will not display in the mean spectrum (purple signal). These data were obtained by Jeramie Watrous together with Beth Shank.
Unsupervised processing methods, which do not rely on labeling of dataset elements, allow for automated extraction of data from a data set. Principal component analysis (PCA) is a typical unsupervised method where data is statistically represented in fewer dimensions123, whereas classification of spectra into several groups (i.e. “abnormal” versus “healthy” tissue) is considered supervised since it requires the user to label spectra in the training dataset as “abnormal” or “healthy”. Unsupervised methods are used to determine how data is organized. In IMS, unsupervised methods are of extreme importance since a dataset comprises a huge amount of spectra and has 3 dimensions: m/z value and spatial positions x and y with spatial another dimension in 3D MALDI IMS110. Other unsupervised methods exploited in IMS include variants of principal component analysis (PCA) accounting for variance124 and noise95 multivariate curve resolution125, and latent semantic analysis126. All these approaches transform the dataset into a small set of images showing main spatial features. Another approach is to do peak picking and show images corresponding to selected masses that, albeit unsophisticated, usually delivers more images. Peak picking can be straightforwardly done on the mean or quantile spectrum where for each mass the quantile, but not mean value, among all spectra is calculated, or in a spectrum-wise manner taking some consensus peaks afterwards100. The results of multivariate methods can be visualized as is or can be used in combination with other techniques (e.g. classification of PCA coordinates).
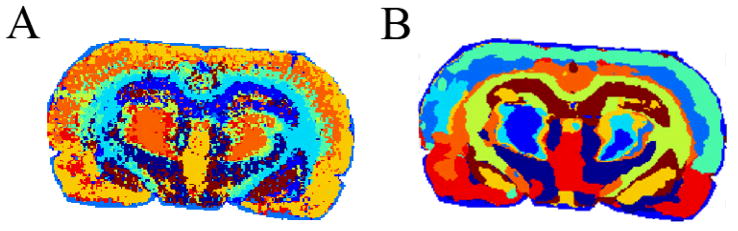
A recent unsupervised method, developed for MALDI IMS, is spatial segmentation of datasets by clustering of spectra127,128. The results of clustering spectra into several groups can be displayed as a spatial segmentation map (one image), coloring identically points grouped into one cluster. As shown in100, taking into account the spatial relations between spectra improves the segmentation maps considerably by suppressing the noise and pixel-to-pixel variability (Figure 7). Although the potential of the segmentation in IMS for improving the quality of data has been shown129, some development is still needed before it can be fully appreciated (i.e. reducing runtimes and memory consumption while keeping the segmentation maps detailed and robust to the pixel-to-pixel variability).
Figure 7.
Unsupervised segmentation of IMS data allows for rapid identification of spectrally unique regions within a sample. Shown are segmentation maps for a rat brain coronal section without (A) and with (B) taking into account spatial relations between spectra. Points are colored identically if they are grouped into the same cluster of spectra. Both images are for 10 clusters.
Supervised methods are currently widely used in IMS, especially in tissue imaging84–86. Typically a region within a single sample is manually designated as having one histological state (i.e. tumor tissue) while the remainder of the sample is classified as another histological state (i.e. healthy tissue). Software algorithms based on this information determine which ions are localized to each region and classify the corresponding spectra into categories. These classification methods are being developed mainly for MALDI for biomarker detection130 since once the classification is performed and evaluated to be successful, one can find discriminative masses. While supervised methods need to be used in classifying data, unsupervised methods (PCA, segmentation) can be exploited to select and label the regions for classification especially when the regional histology or expected distributions of the analyte are unknown.
Perhaps the biggest issue with most developed methods for spectral classification is that they operate based on analysis of individual spectra and do not consider the spatial relations between spectra. We hypothesize that by taking into account the spatial relation of analytes throughout the sample during processing can lead to better classifications including better identification of analytes that exhibit gradual gradient type distributions across histological boundaries which are hard to classify using current processing methods. Evaluation of the classification results is further complicated in IMS based data sets since even user labeled regions are usually heterogeneous making it misleading to target at 100% classification accuracy. It would therefore be more relevant to consider sensitivity/specificity in terms of obtained discriminative masses, but this is hardly possible since the discriminative masses are unknown. The heterogeneity of the spatial regions is important not only for evaluation, but also for classification as methods should consider noise in labels when they are being trained, which is a modern topic in machine learning.
One original supervised problem not yet studied in IMS is spatial regression, which would allow one to establish gradients of spatial concentrations of an analyte and finding masses showing these spatial intensity patterns. Although IMS is generally reputed to be not suitable for quantitative studies131,132, these signal gradients are visually observed in data and represent an ideal model for improving relative quantitation in IMS data. For instance, constant secretion of secondary metabolites from a bacterial colony on nutrient agar produces a uniform gradient of analyte decreasing in concentration with increasing distance from the colony or a disease specific signature for solid tumors often extend into healthy tissues via a gradient. Such a system can be used to optimize instrumental, matrix, and processing parameters possibly allowing for better linear, and therefore, quantitative detection of particular ions.
Improved annotation
Possibly the main pitfall of imaging mass spectrometry, as well as non-imaging mass spectrometry approaches, is currently the inability to annotate observed signals. Severe lack of database depth and nonexistent automated data annotation software for IMS data sets has resulted in very little return on time investment. Within a single IMS data set one might observe anywhere from hundreds to thousands of individual masses corresponding to hundreds of different compounds spatially localized to the sample area; however, identification of even the top 50 peaks is an impossible task at the present time with most published works typically annotating only a small fraction of the total signals observed31,122. In the end, metabolomic and proteomic workflows and the development of smarter annotation approaches will need to be merged with IMS to solve this problem. Perhaps community annotation is the most tractable annotation approach as discussed in the next section. There is also no single approach that can be used to compare data sets across laboratories to help expedite the process of annotation leading one to pursue more traditional methods of identification. Once a parent mass is identified from the imaging data set, fragmentation data is generally collected and subsequently identified via manual annotation or by comparison against known standards or published daughter ion spectra. Typically, complete annotation of daughter ion spectra is extremely difficult necessitating the need for biochemical purification followed by structural studies such as 2D NMR or detailed tandem mass spectrometry experiments. Lack of databases means that after this tedious process of purification and structure elucidation you may determine that your compound has already been discovered and archived elsewhere. Even with proper search algorithms in place for small metabolites, lipids, peptides and proteins, the lack of suitable databases to search against would result in little additional identifications. The IMS community as a whole contains a massive wealth of parent and daughter ion data that could be used to build impressive image databases, especially with similar samples (i.e. brain tissue sections).
Improved comparative analysis
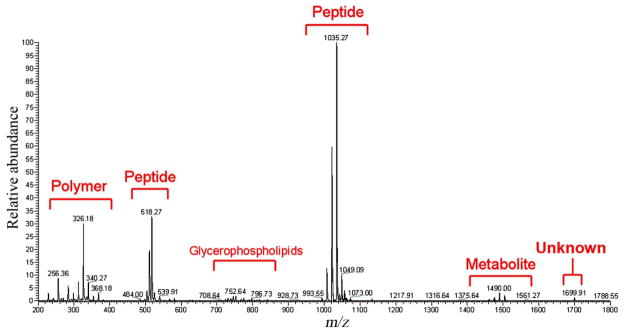
As with any area of science, the ability to compare results both within your own laboratory and between different laboratories is extremely beneficial. Newly collected data sets can be compared against a working archive of collected data in order to draw important conclusions such as cross patient/species compound conservation, environmental effects on metabolic output, or changes in data quality depending on the experimental protocol used. With most IMS data sets consisting of a combination of signals of everything from lipids to peptides to metabolites (Figure 8), cross lab comparative analysis also allows for communal annotation of data owing to the expertise of each laboratory. Such re-mining of data will not only provide insight into new research but will breathe new life into decades of old data. One labs metabolomic data set could be another labs lipidomic gold mine. Such transparency of data within the IMS community is an essential next step and it starts with the data formats in which the raw data is collected.
Figure 8.
IMS data sets typically contain information beyond the field of interest of the lab studying it. The above single spectra from a DESI image of bacterial thin films illustrates how a single data set can contain an abundance of information that, while not of use to the lab that collected the data, may prove extremely useful to another researchers field of study.
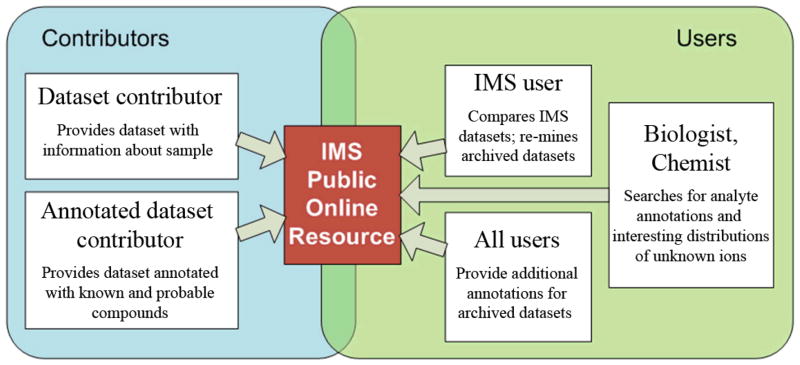
The development of an online data repository for published IMS data sets within and between laboratories is the next logical step for the IMS community (Figure 9). The Protein Data Bank (www.PDB.org) has greatly advanced the field of structure based analysis of proteins by not only making protein structural data freely available to any whom so wishes to view it, but by also increasing the integrity of data submitted for publication. In doing so, the data was no longer confined to specialists and therefore became widely implemented in the life sciences. IMS must undergo a similar evolution to get integrated into the wider community. By making data publicly available it can be re-mined to identify previously unknown compounds or cross correlated to discover new connections between systems133–135. However, viewing of IMS data will present unique challenges due to its multi-dimensional nature as well as the coming incorporation of data dependent based imaging approaches. This issue is further complicated with data regarding histological tissue sections as proper interpretation of such data is very dependent on the patient information and also on serial and subsequent tissue sections from the same experiment. Therefore, certain areas of IMS based research will likely find more utility in the generation of such a database than others and as a whole we feel that the IMS community, as well as its corresponding scientists, will benefit from such a resource being available. Now the question remains, is there the political willpower to do this? We have constructed a basic prototype online public server for viewing, uploading and downloading IMS datasets for use in our laboratory, which we will make available upon publication. Such a resource has been very useful for our lab in not only allowing us to share, access, and search stored data from anywhere in the world, but also for simultaneously analyzing hundreds of archived data sets (as new hypotheses surface) that would otherwise have to be mined manually one by one. Hopefully a server similar to this can act as a foundation for such a repository that will allow IMS and non-IMS labs around the world to view, share, annotate and analyze data sets to further the field as a whole. For example, there are very few comparisons that have been made between laboratories of images of brain sections, even though this is the most investigated tissue at this time (Figure 3C).
Figure 9.
A proposed workflow for an online public database for published IMS data sets. Database contributors can submit annotated and non-annotated datasets, which can then be searched by biologists for candidate compounds and by other IMS users for dataset comparison and cross species/phenotype correlations. A central online repository will also allow for assessment of published data quality and communal annotation of data sets.
Lastly, proprietary data formats between instrument manufacturers have limited open source software development and, in part, limited the sharing of data between labs. These proprietary formats have also haunted proteomics based search engines with certain search algorithms only applicable to certain formats. This inconvenient truth of mass spectrometry is pushing the community to demand a universal spectral format shared between all manufacturers. One such a format is the “.imzML” format which is currently used in some commercial software packages (www.maldi-msi.org) and will hopefully be incorporated into the next generation of software from other manufacturers as well. A universal data format would allow for faster and more reliable software development and aid in the development of data search engines and repositories.
4. Possible Future Applications of IMS
Imaging mass spectrometry has proven to be a very versatile technology whose application is limited only by the imagination of the scientist yielding it. So what does the future hold for such a bright young technology? Instead of discussing the obvious importance of improving sensitivity, spatial and mass resolution and speed, we would like to instead highlight some novel applications that we believe to be possible using IMS within the near future. The first of these is live cell imaging mass spectrometry.
Live cell IMS
Currently, all IMS based approaches on biological samples require that the sample be killed either before or during the imaging process. As ionization sources become gentler and more sensitive under atmospheric temperature and pressure, the ability to image an intact biological system without killing it becomes more and more possible. Even if sample preparation reaches a point of perfect reproducibility, controlling variability within biology itself remains very difficult and monitoring a system over time currently requires a new sample to be sacrificed and analyzed at each time point. The ability to follow a single sample through its life cycle by collecting multiple MS images at various time points would eliminate many speculations regarding validity of time course data taken from separate samples. Such protocols would be ideal in studying such systems as microbiome derived bacterial thin films and growing plants leaves.
From images to movies
One can even imagine further and envision mass spectrometers fast enough to collect an entire image in a matter of minutes on a living biological sample. Such instruments, if equipped with soft ionization sources as described above, could theoretically collect consecutive images of a single sample over the course of time (i.e. an image every 15 minutes for 4 hours). Such data sets could be used to generate molecular movies of the circadian rhythm of healthy and cancer cells, capture the destructive effects of a fungal toxin on neighboring cells or monitor bacterial swarming and matrix formation. An extension of real-time IMS would be tissue biopsies.
IMS tissue biopsies
One exciting application of IMS is the possibility that it can someday negate the need for certain tissue biopsies, such as a needle aspiration biopsy. The instrumental setup for such an application could be a remote probe equipped with a terminal needle which contains small sampling points along its shaft. The needle could then be inserted into the suspected tumor from the exterior of the body therefore passing through healthy and suspected unhealthy tissue. Once inserted, mass spectra can be taken at the sampling points along its shaft creating a 1-dimensional molecular image/movie where presence of cancer specific disease signatures could be visualized both in the healthy and tumor tissues. An alternative to this methodology could be a needle like probe with a single sampling point that continuously collects spectra as it is inserted into the body. Such a technology could provide immediate information on the identity of a lump or tissue mass in question or provide information to assess the progression of treatment through near surface tissue.
IMS in studying signaling interactions and community dynamics
Currently there is an absence of tools to study the molecules that cell-populations, such as bacteria, use to interact with their neighboring bacteria and host cells. These molecules, many of which are natural products, control biology by mediating such events as cellular cross talk, cellular metabolism, quorum sensing and growth and they are spatially defined within these communities. These molecules have been active in shaping modern healthcare with therapeutics such as penicillin, vancomycin and rapamycin all having origins in such molecules involved in metabolic exchange. Understanding the distribution of these molecules and changes over time is crucial for understanding the dynamics cellular communities within nature. Disease proliferation such as diabetes, cancer, infections and biofilm formation, biofouling on ships, plant maturation and growth all have been linked to the molecules that are produced for interspecies interactions. IMS is one of the first tools that can begin to capture the complex nature of such interspecies interactions and will become one of the frontline tools in such investigations.
5. Conclusion
The evolving field of imaging mass spectrometry has become a household name in biological and medical research and continues to do so thanks to regular advancements in instruments, applications, and data analysis tools. Its ability to simultaneously monitor hundreds of molecular distributions across a sample without the need for chemical tags or antibodies has allowed for a more complete analysis of biological systems that have resulted in the discovery of new disease specific signature candidates, drug candidates, and potentially towards personalized medicine and diagnostics. As with any growing technology, there remain certain areas of the science that progress slower than others; however, community awareness of these issues has encouraged their development and welcomed recent progress into emerging research. As the technology continues to evolve, its unique application for studying biological systems will be impossible to ignore by non-MS life science laboratories leading to more collaborative efforts within the scientific community allowing more fruitful and meaningful research.
Acknowledgments
Thanks to Yu-liang Yang and Michael Meehan for helping edit this report. This work was supported by Beckman Foundation, V-foundation, Hearst foundation, and National Institute of General Medical Sciences Grant NIH GM086283, GM094802 (P.C.D.), NIH Molecular Biophysics Training Grant NIH GM08326 (J.D.W.).
References
- 1.Liebl H. Ion microprobe mass analyzer. J Appl Phys. 1967;38:5277. [Google Scholar]
- 2.Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 3.Schuerenberg M, Luebbert C, Deininger S-O, Ketterlinus R, Suckau D. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat Methods. 2007;4 [Google Scholar]
- 4.Wong SCC, Chan CML, Ma BBY, Lam MYY, Choi GCG, Au TCC, Chan ASK, Chan ATC. Advanced proteomic technologies for cancer biomarker discovery. Exp Rev Prot. 2009;6:123. doi: 10.1586/epr.09.1. [DOI] [PubMed] [Google Scholar]
- 5.Sugiura Y, Setou M. Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: Toward in situ pharmacometabolomes. J Neuro Pharm. 2010;5:31. doi: 10.1007/s11481-009-9162-6. [DOI] [PubMed] [Google Scholar]
- 6.Solon E, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development. AAPS J. 2010;12:11. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson SN, Wang H-YJ, Woods AS. Direct profiling of lipid distribution in brain tissue using MALDI-TOF MS. Anal Chem. 2005;77:4523. doi: 10.1021/ac050276v. [DOI] [PubMed] [Google Scholar]
- 8.Amaya K, Monroe E, Sweedler J, Clayton D. Lipid imaging in the zebra finch brain with secondary ion mass spectrometry. Int J Mass Spectrom. 2007;260:121. [Google Scholar]
- 9.Tanaka H, Zaima N, Yamamoto N, Sagara D, Suzuki M, Nishiyama M, Mano Y, Sano M, Hayasaka T, Goto-Inoue N, Sasaki T, Konno H, Unno N, Setou M. Imaging mass spectrometry reveals unique lipid distribution in primary varicose veins. Eur J Vasc Endovasc. 2010 doi: 10.1016/j.ejvs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Brunelle A, Laprevote O. Lipid imaging with cluster time-of-flight secondary ion mass spectrometry. Anal Bioanal Chem. 2009;393:31. doi: 10.1007/s00216-008-2367-3. [DOI] [PubMed] [Google Scholar]
- 11.McDonnell LA, Corthals GL, Willems SM, van Remoortere A, van Zeijl RJM, Deelder AM. Peptide and protein imaging mass spectrometry in cancer research. J Proteomics. 2010;73:1921. doi: 10.1016/j.jprot.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 12.van Remoortere A, van Zeijl RJM, van den Oever N, Franck J, Longuespee R, Wisztorski M, Salzet M, Deelder AM, Fournier I, McDonnell LA. MALDI imaging and profiling MS of higher mass proteins from tissue. JASMS. 2010;21:1922. doi: 10.1016/j.jasms.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Chaurand P, Schwartz SA, Caprioli RM. Assessing protein patterns in disease using imaging mass spectrometry. J Prot Res. 2004;3:245. doi: 10.1021/pr0341282. [DOI] [PubMed] [Google Scholar]
- 14.Franck J, Arafah K, Elayed M, Bonnel D, Vergara D, Jacquet A, Vinatier D, Wisztorski M, Day R, Fournier I, Salzet M. MALDI imaging mass spectrometry: state of the art technology in clinical proteomics. Mol Cell Prot. 2009;8:2023. doi: 10.1074/mcp.R800016-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal Chem. 2006;78:6448. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 16.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int J Mass Spectrom. 2007;260:195. [Google Scholar]
- 17.Watrous J, Hendricks N, Meehan M, Dorrestein PC. Capturing bacterial metabolic exchange using thin film desorption electrospray ionization-imaging mass spectrometry. Anal Chem. 2010;82:1598. doi: 10.1021/ac9027388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y-L, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Bio. 2009;5:885. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu W-T, Yang Y-L, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, Pevzner PA, Pogliano J, Nizet V, Pogliano K, Dorrestein PC. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of. Bacillus subtilis PNAS. 2010;107:16286. doi: 10.1073/pnas.1008368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cha, Song Z, Nikolau BJ, Yeung ES. Direct profiling and imaging of epicuticular waxes on Arabidopsis thaliana by laser desorption/ionization mass spectrometry using silver colloid as a matrix. Anal Chem. 2009;81:2991. doi: 10.1021/ac802615r. [DOI] [PubMed] [Google Scholar]
- 21.Hamm G, Carre V, Poutaraud A, Maunit B, Frache G, Merdinoglu D, Muller JFF. Determination and imaging of metabolites from vitis vinifera leaves by laser desorption/ionisation time-of-flight mass spectrometry. Rapid Commun Mass Spectrom. 2010;24:335. doi: 10.1002/rcm.4395. [DOI] [PubMed] [Google Scholar]
- 22.Anderson DM, Carolan VA, Crosland S, Sharples KR, Clench MR. Examination of the distribution of nicosulfuron in sunflower plants by matrix-assisted laser desorption/ionisation mass spectrometry imaging. Rapid Commun Mass Sp. 2009;23:1321. doi: 10.1002/rcm.3973. [DOI] [PubMed] [Google Scholar]
- 23.Rutten FJM, Briggs D, Henderson J, Roe MJ. The application of time-of-flight secondary ion mass spectrometry (TOF-SIMS) to the characterization of opaque ancient glasses. Archaeometry. 2009;51:966. [Google Scholar]
- 24.Lehmann K, Berger A, Gotte T, Ramseyer K, Wiedenbeck M. Growth related zonations in authigenic and hydrothermal quartz characterized by SIMS-, EPMA-, SEM-CL- and SEM-CC-imaging. Mineral Mag. 2009;73:633. [Google Scholar]
- 25.Zoriy MV, Mayer D, Becker JS. Metal imaging on surface of micro- and nanoelectronic devices by laser ablation inductively coupled plasma mass spectrometry and possibility to measure at nanometer range. JASMS. 2009;20:883. doi: 10.1016/j.jasms.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Bailey MJ, Jones BN, Hinder S, Watts J, Bleay S, Webb RP. Depth profiling of fingerprint and ink signals by SIMS and MeV SIMS. Nucl Instrum Methods B. 2010;268:1929. [Google Scholar]
- 27.Ifa DR, Manicke NE, Dill AL, Cooks RG. Latent fingerprint chemical imaging by mass spectrometry. Science. 2008;321:805. doi: 10.1126/science.1157199. [DOI] [PubMed] [Google Scholar]
- 28.Tang H-W, Lu W, Che C-M, Ng K-M. Gold nanoparticles and imaging mass spectrometry: Double imaging of latent fingerprints. Anal Chem. 2010;82:1589. doi: 10.1021/ac9026077. [DOI] [PubMed] [Google Scholar]
- 29.Duncan MW, Aebersold R, Caprioli RM. The pros and cons of peptide-centric proteomics. Nat Biotech. 2010;28:659. doi: 10.1038/nbt0710-659. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell P. Proteomics retrenches. Nat Biotech. 2010;28:665. doi: 10.1038/nbt0710-665. [DOI] [PubMed] [Google Scholar]
- 31.Bowen BP, Northen TR. Dealing with the unknown: metabolomics and metabolite atlases. JASMS. 2010;21:1471. doi: 10.1016/j.jasms.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Pevzner PA, Kim S, Ng J. Comment on “protein sequences from mastodon and tyrannosaurus rex revealed by mass spectrometry”. Science. 2008;321:1040. doi: 10.1126/science.1155006. [DOI] [PubMed] [Google Scholar]
- 33.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJM. A HUPO test sample study reveals common problems in mass spectrometry based proteomics. Nat Method. 2009;6:423. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson SD. Data analysis: the Achilles heel of proteomics. Nat Biotech. 2003;21:221. doi: 10.1038/nbt0303-221. [DOI] [PubMed] [Google Scholar]
- 35.Aberg K, Alm E, Torgrip R. The correspondence problem for metabonomics datasets. Anal Bioanal Chem. 2009;394:151. doi: 10.1007/s00216-009-2628-9. [DOI] [PubMed] [Google Scholar]
- 36.Arita M. What can metabolomics learn from genomics and proteomics? Curr Opin Biotech. 2009;20:610. doi: 10.1016/j.copbio.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 37.Amstalden van Hove ER, Smith DF, Heeren RMA. A concise review of mass spectrometry imaging. J Chromatogr A. 2010;1217:3946. doi: 10.1016/j.chroma.2010.01.033. [DOI] [PubMed] [Google Scholar]
- 38.Francese S, Dani FR, Traldi P, Mastrobuoni G, Pieraccini G, Moneti G. MALDI mass spectrometry imaging, from its origins up to today: the state of the art. Comb Chem & HTS. 2009;12:156. doi: 10.2174/138620709787315454. [DOI] [PubMed] [Google Scholar]
- 39.Heeren RMA, Smith DF, Stauber J, Kukrer-Kaletas B, MacAleese L. Imaging mass spectrometry: Hype or hope? JASMS. 2009;20:1006. doi: 10.1016/j.jasms.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 40.Winograd N, Garrison BJ. Biological cluster mass spectrometry. Annu Rev Phys Chem. 2010;61:305. doi: 10.1146/annurev.physchem.040808.090249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry : looking beyond classical histology. Nat Rev Cancer. 2010;10:639. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 42.McDonnell LA, Heeren RM. Imaging mass spectrometry. Mass Spectrom Rev. 2007;26:606. doi: 10.1002/mas.20124. [DOI] [PubMed] [Google Scholar]
- 43.Svatos A. Mass spectrometric imaging of small molecules. Trends Biotechnol. 2010;28:425. doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Boxer SG, Kraft ML, Weber PK. Advances in imaging secondary ion mass spectrometry for biological samples. Annu Rev Biophys. 2009;38:53. doi: 10.1146/annurev.biophys.050708.133634. [DOI] [PubMed] [Google Scholar]
- 45.Kollmer F. Cluster primary ion bombardment of organic materials. Appl Surf Sci. 2004:231–232. 153. [Google Scholar]
- 46.Touboul D, Kollmer F, Niehuis E, Brunelle A, Laprevote O. Improvement of biological time-of-flight-secondary ion mass spectrometry imaging with a bismuth cluster ion source. JASMS. 2005;16:1608. doi: 10.1016/j.jasms.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 47.Benguerba M, Brunelle A, Dellanegra S, Depauw J, Joret H, Lebeyec Y, Blain M, Schweikert E, Assayag G, Sudraud P. Impact of slow gold clusters on various solids: nonlinear effects in secondary ion emission. Nucl Instrum Methods B. 1991;62:8. [Google Scholar]
- 48.Novikov A, Caroff M, Della-Negra S, Depauw J, Fallavier M, Le Beyec Y, Pautrat M, Schultz JA, Tempez A, Woods AS. The Au(n) cluster probe in secondary ion mass spectrometry: influence of the projectile size and energy on the desorption/ionization rate from biomolecular solids. Rapid Commun Mass Sp. 2005;19:1851. doi: 10.1002/rcm.1995. [DOI] [PubMed] [Google Scholar]
- 49.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. A C60 primary ion beam system for time of flight secondary ion mass spectrometry: its development and secondary ion yield characteristics. Anal Chem. 2003;75:1754. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 50.Benninghoven A, Hagenhoff B, Niehuis E. Surface MS: probing real-world samples. Anal Chem. 1993;65:630. [Google Scholar]
- 51.Diehnelt C, Van Stipdonka MJ, Schweikert EA. Effectiveness of atomic and polyatomic primary ions for organic secondary ion mass spectrometry. Int J Mass Spectrom. 2001;207:111. [Google Scholar]
- 52.Wu KJ, Odom RW. Matrix-enhanced secondary ion mass spectrometry: a method for molecular analysis of solid surfaces. Anal Chem. 1996;68:873. doi: 10.1021/ac950717i. [DOI] [PubMed] [Google Scholar]
- 53.Heeren R, McDonnell L, Amstalden E, Luxembourg S, Altelaar A, Piersma S. Why don’t biologists use SIMS? A critical evaluation of imaging MS. Appl Surf Sci. 2006;252:6827. [Google Scholar]
- 54.Chang W, Huang L, Wang Y, Peng W, Chang H, Hsu N, Yang W, Chen C. Matrix-assisted laser desorption/ionization (MALDI) mechanism revisited. Anal Chim Acta. 2007;582:1. doi: 10.1016/j.aca.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 55.Karas M, Kruger R. Ion formation in MALDI: the cluster ionization mechanism. Chem Rev. 2003;103:427. doi: 10.1021/cr010376a. [DOI] [PubMed] [Google Scholar]
- 56.Knochenmuss R, Vertes A. Time-delayed 2-pulse studies of MALDI matrix ionization mechanisms. J Phys Chem B. 2000;104:5406. [Google Scholar]
- 57.Knochenmuss R, Zenobi R. MALDI ionization: the role of in-plume processes. Chem Rev. 2003;103:441. doi: 10.1021/cr0103773. [DOI] [PubMed] [Google Scholar]
- 58.Knochenmuss R. A bipolar rate equation model of MALDI primary and secondary ionization processes, with application to positive/negative analyte ion ratios and suppression effects. Int J Mass Spectrom. 2009;285:105. [Google Scholar]
- 59.Yoon SH, Moon JH, Kim MS. A comparative study of in- and post-source decays of peptide and preformed ions in matrix-assisted laser desorption ionization time-of-flight mass spectrometry: Effective temperature and matrix effect. JASMS. 2010 doi: 10.1016/j.jasms.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka K, Waki H, Ido Y, Akita S, Yoshida Y, Yoshida T, Matsuo T. Protein and polymer analyses up to m/z 100 000 by laser ionization time-of-flight mass spectrometry. Rapid Commun Mass Sp. 1988;2:151. [Google Scholar]
- 61.Becker M, Deininger SO, Holle A, Hoehndorf J, Suckau D, Schrurenberg M, Pineau C. High definition MALDI imaging: New capabilities for protein biomarker discovery from tissue. Biomol Tech. 2010;21:S-33. [Google Scholar]
- 62.Takats Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:5695. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 63.Wiseman JM, Ifa DR, Song Q, Cooks RG. Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry. Angew Chem Int Edit. 2006;45:7188. doi: 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- 64.Takats Z, Wiseman JM, Cooks RG. Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology. J Mass Spectrom. 2005;40:1261. doi: 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- 65.Venter A, Sojka PE, Cooks RG. Droplet dynamics and ionization mechanisms in desorption electrospray ionization mass spectrometry. Anal Chem. 2006;78:8549. doi: 10.1021/ac0615807. [DOI] [PubMed] [Google Scholar]
- 66.Costa AB, Graham Cooks R. Simulated splashes: Elucidating the mechanism of desorption electrospray ionization mass spectrometry. Chem Phys Lett. 2008;464:1. [Google Scholar]
- 67.Talaty N, Takats Z, Cooks RG. Rapid in situ detection of alkaloids in plant tissue under ambient conditions using desorption electrospray ionization. Analyst. 2005;130:1624. doi: 10.1039/b511161g. [DOI] [PubMed] [Google Scholar]
- 68.Lane AL, Nyadong L, Galhena AS, Shearer TL, Stout EP, Parry RM, Kwasnik M, Wang MD, Hay ME, Fernandez FM, Kubanek J. Desorption electrospray ionization mass spectrometry reveals surface-mediated antifungal chemical defense of a tropical seaweed. PNAS. 2009;106:7314. doi: 10.1073/pnas.0812020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu C, Ifa DR, Manicke NE, Cooks RG. Rapid, direct analysis of cholesterol by charge labeling in reactive desorption electrospray ionization. Anal Chem. 2009;81:7618. doi: 10.1021/ac901003u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79:8098. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 71.Sampson JS, Hawkridge AM, Muddiman DC. Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) fourier transform ion cyclotron resonance mass spectrometry. JASMS. 2006;17:1712. doi: 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 72.Hiraoka K. Laser spray: electric field-assisted matrix-assisted laser desorption/ionization. J Mass Spectrom. 2004;39:341. doi: 10.1002/jms.621. [DOI] [PubMed] [Google Scholar]
- 73.Becker JS. Bioimaging of metals in brain tissue from micrometre to nanometre scale by laser ablation inductively coupled plasma mass spectrometry: State of the art and perspectives. Int J Mass Spec. 2010;289:65. [Google Scholar]
- 74.Lewis W. Desorption/ionization on silicon (DIOS) mass spectrometry: background and applications. Int J Mass Spectrom. 2003;226:107. [Google Scholar]
- 75.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordstrom A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 76.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom. 2007;42:1099. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 77.Chaurand P, Schwartz S, Billheimer D, Xu BJ, Crecelius A, Caprioli R. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 78.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 79.Broersen A, van Liere R, Altelaar AFM, Heeren RMA, McDonnell LA. Automated, feature-based image alignment for high-resolution imaging mass spectrometry of large biological samples. JASMS. 2008;19:823. doi: 10.1016/j.jasms.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 80.Yu W, He Z, Liu J, Zhao H. Improving mass spectrometry peak detection using multiple peak alignment results. J Prot Res. 2008;7:123. doi: 10.1021/pr070370n. [DOI] [PubMed] [Google Scholar]
- 81.Morris JS, Coombes KR, Koomen J, Baggerly KA, Kobayashi R. Feature extraction and quantification for mass spectrometry in biomedical applications using the mean spectrum. Bioinformatics. 2005;21:2005. doi: 10.1093/bioinformatics/bti254. [DOI] [PubMed] [Google Scholar]
- 82.Breen EJ, Hopwood FG, Williams KL, Wilkins MR. Automatic Poisson peak harvesting for high throughput protein identification. Electrophoresis. 2000;21:2243. doi: 10.1002/1522-2683(20000601)21:11<2243::AID-ELPS2243>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 83.Kwon D, Vannucci M, Song JJ, Jeong J, Pfeiffer RM. A novel wavelet-based thresholding method for the preprocessing of mass spectrometry data that accounts for heterogeneous noise. Proteomics. 2008;8:3019. doi: 10.1002/pmic.200701010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Groseclose MR, Massion PP, Chaurand P, Caprioli R. High-throuput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics. 2008;18:3715. doi: 10.1002/pmic.200800495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rauser S, Marquardt C, Balluff B, Deininger SO, Albers C, Belau E, Hartmer R, Suckau D, Specht K, Ebert MP, Schmitt M, Aubele M, Hofler H, Walch A. Classification of HER2 receptor status in breast cancer tissues by MALDI imaging mass spectrometry. J Proteome Res. 2010;9:1854. doi: 10.1021/pr901008d. [DOI] [PubMed] [Google Scholar]
- 86.Cazares LH, Troyer D, Mendrinos S, Lance RA, Nyalwidhe JO, Beydoun HA, Clements MA, Drake RR, Semmes OJ. Imaging mass spectrometry of a specific fragment mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin Cancer Res. 2009;15:5541. doi: 10.1158/1078-0432.CCR-08-2892. [DOI] [PubMed] [Google Scholar]
- 87.Willems SM, van Remoortere A, van Zeijl R, Deedler AM, McDonnell LA, Hogendoorn PC. Imaging mass spectrometry of myxoid sarcomas identifies proteins and lipids specific to tumor type and grade and reveals biochemical intratumor heterogeneity. J Pathology. 2010;222:400. doi: 10.1002/path.2771. [DOI] [PubMed] [Google Scholar]
- 88.Zakrzewski J, Didona K. Advances in hyperspectral imaging technologies for multichannel fiber sensing. SPIE. 2009;7316:73. [Google Scholar]
- 89.Molina R, Mateos J. Multichannel image restoration in astronomy. Vist AST S. 1997;41:373. [Google Scholar]
- 90.Goetz AFH, Vane G, Solomon JE, Rock BN. Imaging spectrometry for Earth remote sensing. Science. 1985;228:1147. doi: 10.1126/science.228.4704.1147. [DOI] [PubMed] [Google Scholar]
- 91.Sijtsema NM, Wouters SD, Grauw CJ, Otto C, Greve J. Confocal direct imaging Raman microscope: Design and applications in biology. Appl Spectrosc. 1998;52:348. [Google Scholar]
- 92.Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, Caprioli RM. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int J Mass Spectrom. 2007;260:212. doi: 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Deininger SO, Becker M, Wolski E, Kaminski H, Paape R, Cornett DS. Practical Considerations on Normalization in MALDI Imaging. Proceedings of the 58th ASMS Conference on Mass Spectrometry and Allied Topics; 2010, Mai 22–27.. [Google Scholar]
- 94.Keenan M. Optimal scaling of TOF-SIMS spectrum-images prior to multivariate statistical analysis. Appl Surf Sci. 2004:231–232. 240. [Google Scholar]
- 95.Keenan MR, Kotula PG. Accounting for Poisson noise in the multivariate analysis of TOF-SIMS spectrum images. Surf Interface Anal. 2004;36:203. [Google Scholar]
- 96.Schwartz SA, Reyzer ML, Caprioli RM. Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J Mass Spectrom. 2003;38:699. doi: 10.1002/jms.505. [DOI] [PubMed] [Google Scholar]
- 97.Ibanez AJ, Muck A, Svatos A. Dissipation of charge on MALDI-TOF polymeric chips using an electron-acceptor: analysis of proteins. J Mass Spectrom. 2007;42:634. doi: 10.1002/jms.1192. [DOI] [PubMed] [Google Scholar]
- 98.Wisztorski M, Franck J, Salzet M, Fournier I. MALDI direct analysis and imaging of frozen versus FFPE tissues: what strategy for which sample? Method Mol Bio. 2010;656:303. doi: 10.1007/978-1-60761-746-4_18. [DOI] [PubMed] [Google Scholar]
- 99.Russ JC, Woods RP. J Comp Assist Tom. 2. Vol. 19. 1995. The image processing handbook; p. 979. [Google Scholar]
- 100.Alexandrov T, Becker M, Deininger S-O, Ernst G, Wehder L, Grasmair M, von Eggeling F, Thiele H, Maass P. Spatial segmentation of imaging mass spectrometry data with edge-preserving image denoising and clustering. J Proteome Res. 2010 doi: 10.1021/pr100734z. in press. [DOI] [PubMed] [Google Scholar]
- 101.Mayrhofer C, Krieger S, Raptakis E, Allmaier G. Comparison of vacuum matrix-assisted laser desorption/ionization (MALDI) and atmospheric pressure MALDI (AP-MALDI) tandem mass spectrometry of 2-dimensional separated and trypsin-digested glomerular proteins for database search derived identification. J Prot Res. 2006;5:1967. doi: 10.1021/pr060165s. [DOI] [PubMed] [Google Scholar]
- 102.Chandra S. Imaging ion and molecular transport at subcellular resolution by secondary ion mass spectrometry. Int J Mass Spectrom. 1995;143:161. [Google Scholar]
- 103.Wagner M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
- 104.Levisetti R, Lebeau M. Cytogenetic applications of high resolution secondary ion imaging microanalysis: detection and mapping of tracer isotopes in human chromosomes. Biol Cell. 1992;74:51. doi: 10.1016/0248-4900(92)90008-o. [DOI] [PubMed] [Google Scholar]
- 105.Petit VW, Refregiers M, Guettier C, Jamme F, Sebanayakam K, Brunelle A, Laprevote O, Dumas P, Le Naour F. Multimodal spectroscopy combining time-of-flight-secondary ion mass spectrometry, synchrotron-FT-IR, and synchrotron-UV microspectroscopies on the same tissue section. Anal Chem. 2010;82:3963. doi: 10.1021/ac100581y. [DOI] [PubMed] [Google Scholar]
- 106.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal Chem. 2008;80:7921. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tsai S-TT, Chen CWW, Lora LCC, Huang M-CC, Chen C-HH, Wang Y-SS. Simultaneous mass analysis of positive and negative ions using a dual-polarity time-of-flight mass spectrometer. Anal Chem. 2006;78:7729. doi: 10.1021/ac061213v. [DOI] [PubMed] [Google Scholar]
- 108.Nyadong L, Harris GA, Balayssac S, Galhena AS, Malet-Martino M, Martino R, Parry RM, Wang MD, Fernandez FM, Gilard V. Combining two-dimensional diffusion-ordered nuclear magnetic resonance spectroscopy, imaging desorption electrospray ionization mass spectrometry, and direct analysis in real-time mass spectrometry for the integral investigation of counterfeit pharmaceuticals. Anal Chem. 2009;81:4803. doi: 10.1021/ac900384j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bradshaw JA, Ovchinnikova OS, Meyer KA, Goeringer DE. Combined chemical and topographic imaging at atmospheric pressure via microprobe laser desorption/ionization mass spectrometry-atomic force microscopy. Rapid Commun Mass Sp. 2009;23:3781. doi: 10.1002/rcm.4313. [DOI] [PubMed] [Google Scholar]
- 110.Sinha TK, Khatib-Shahidi S, Yankeelov TE, Mapara K, Ehtesham M, Cornett DS, Dawant BM, Caprioli RM, Gore JC. Integrating spatially resolved three-dimensional MALDI IMS with in vivo magnetic resonance imaging. Nat Methods. 2007;5:57. doi: 10.1038/nmeth1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eijkel GB, Kukrer Kaletas B, van der Wiel IM, Kros JM, Luider TM, Heeren RMA. Correlating MALDI and SIMS imaging mass spectrometric datasets of biological tissue surfaces. Surf Interface Anal. 2009;41:675. [Google Scholar]
- 112.Gholap DS, Izmer A, De Samber B, van Elteren JT, Selih VS, Evens R, De Schamphelaere K, Janssen C, Balcaen L, Lindemann I, Vincze L, Vanhaecke F. Comparison of laser ablation-inductively coupled plasma-mass spectrometry and micro-X-ray fluorescence spectrometry for elemental imaging in. Daphnia magna Anal Chim Acta. 2010;664:19. doi: 10.1016/j.aca.2010.01.052. [DOI] [PubMed] [Google Scholar]
- 113.Baluya DL, Garrett TJ, Yost RA. Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal Chem. 2007;79:6862. doi: 10.1021/ac070958d. [DOI] [PubMed] [Google Scholar]
- 114.Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Prot. 2002;1:376. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 115.Unwin RD, Pierce A, Watson RB, Sternberg DW, Whetton AD. Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol Cell Prot. 2005;4:924. doi: 10.1074/mcp.M400193-MCP200. [DOI] [PubMed] [Google Scholar]
- 116.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Prot. 2007;6:2212. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McMahon G, Glassner B, Lechene C. Quantitative imaging of cells with multi-isotope imaging mass spectrometry (MIMS): nanoautography with stable isotope tracers. Appl Surf Sci. 2006;252:6895. [Google Scholar]
- 118.Lechene C, Hillion F, McMahon G, Benson D, Kleinfeld AM, Kampf JP, Distel D, Luyten Y, Bonventre J, Hentschel D, Park KMM, Ito S, Schwartz M, Benichou G, Slodzian G. High-resolution quantitative imaging of mammalian and bacterial cells using stable isotope mass spectrometry. J Bio. 2006;5:20. doi: 10.1186/jbiol42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Becker JS, Matusch A, Depboylu C, Dobrowolska J, Zoriy MV. Quantitative imaging of selenium, copper, and zinc in thin sections of biological tissues (slugs-genus arion) measured by laser ablation inductively coupled plasma mass spectrometry. Anal Chem. 2007;79:6074. doi: 10.1021/ac0700528. [DOI] [PubMed] [Google Scholar]
- 120.Wagner MS, Shen M, Horbett TA, Castner DG. Quantitative analysis of binary adsorbed protein films by time of flight secondary ion mass spectrometry. J Biomed Mater Res. 2003;64:1. doi: 10.1002/jbm.a.10263. [DOI] [PubMed] [Google Scholar]
- 121.Elsila JE, de Leon NP, Zare RN. Factors affecting quantitative analysis in laser desorption/laser ionization mass spectrometry. Anal Chem. 2004;76:2430. doi: 10.1021/ac0354140. [DOI] [PubMed] [Google Scholar]
- 122.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 123.Van de Plas R, De Moor B, Waelkens E. Imaging mass spectrometry based exploration of biochemical tissue composition using peak intensity weighted PCA. IEEE Ind Applic Soc. 2007:209. [Google Scholar]
- 124.Klerk L, Broersen A, Fletcher I, Vanliere R, Heeren R. Extended data analysis strategies for high resolution imaging MS: New methods to deal with extremely large image hyperspectral datasets. Int J Mass Spectrom. 2007;260:222. [Google Scholar]
- 125.Keenan MR. Exploiting spatial-domain simplicity in spectral image analysis. Surf Interface Anal. 2009;41:79. [Google Scholar]
- 126.Hanselmann M, Kirchner M, Renard BY, Amstalden ER, Glunde K, Heeren RMA, Hamprecht FA. Concise representation of mass spectrometry images by probabilistic latent semantic analysis. Anal Chem. 2008;80:9649. doi: 10.1021/ac801303x. [DOI] [PubMed] [Google Scholar]
- 127.Deininger S-O, Ebert MP, Futterer A, Gerhard M, Rocken C. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J Prot Res. 2008;7:5230. doi: 10.1021/pr8005777. [DOI] [PubMed] [Google Scholar]
- 128.McCombie G, Staab D, Stoeckli M, Knochenmuss R. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal Chem. 2005;77:6118. doi: 10.1021/ac051081q. [DOI] [PubMed] [Google Scholar]
- 129.Walch A, Rauser S, Deininger S-O, Hoer H. MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem Cell Biol. 2008;130:421. doi: 10.1007/s00418-008-0469-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Alexandrov T, Decker J, Mertens B, Deelder AM, Tollenaar RAEM, Maass P, Thiele H. Biomarker discovery in MALDI-TOF serum protein profiles using discrete wavelet transformation. Bioinformatics. 2009;25:643. doi: 10.1093/bioinformatics/btn662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Valaskovic GA, Morrison GH. Quantitative imaging ion microscopy: a short review. Scanning Microscopy. 1992;6:305. [PubMed] [Google Scholar]
- 132.Pang R, Johnson P, Chan C, Kong E, Chan A, Sung J, Poon T. Technical evaluation of MALDI-TOF mass spectrometry for quantitative proteomic profiling; matrix formulation and application. Clin Prot. 2004;1:259. [Google Scholar]
- 133.Joosten RP, Salzemann J, Bloch V, Stockinger H, Berglund AC, Blanchet C, Rudlolf EB, Combet C, Da Costa AL, Deleage G, Diarena M, Fabbretti R, Fettahi G, Flegel V, Gisel A, Kasam V, Kervinen T, Korpelainen E, Mattila K, Pagni M, Reichstadt M, Breton V, Tickle IJ, Vriend G. PDB REDO: automated re-refinement of X-ray structure models in the PDB. J Appl Crystallogr. 2009;42:376. doi: 10.1107/S0021889809008784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lutteke T, Von Der Lieth C-W. The protein data bank (PDB) as a versatile resource for glycobiology and glycomics. Biocatal Biotransfor. 2006;24:147. [Google Scholar]
- 135.Oldfield TJ. Creating structure features by data mining the PDB to use as molecular-replacement models. Acta Crystallogr D. 2001;57:1421. doi: 10.1107/s0907444901012483. [DOI] [PubMed] [Google Scholar]