Abstract
Coenzyme Q10 is an essential cofactor in the respiratory chain and serves as a potent antioxidant in biological membranes. Recent studies in vitro and in vivo provide evidence that Coenzyme Q10 is involved in inflammatory processes and lipid metabolism via gene expression. To study these effects at the epigenomic level, C57BL6J mice were supplemented for one week with reduced Coenzyme Q10 (ubiquinol). Afterwards, gene expression signatures and DNA promoter methylation patterns of selected genes were analysed. Genome-wide transcript profiling in the liver identified 1112 up-regulated and 571 down-regulated transcripts as differentially regulated between ubiquinol-treated and control animals. Text mining and GeneOntology analysis revealed that the ”top 20” ubiquinol-regulated genes play a role in lipid metabolism and are functionally connected by the PPARα signalling pathway. With regard to the ubiquinol-induced changes in gene expression of about +3.14-fold (p≤0.05), +2.18-fold (p≤0.01), and −2.13-fold (p≤0.05) for ABCA1, ACYP1, and ACSL1 genes, respectively, hepatic DNA methylation analysis of 282 (sense orientation) and 271 (antisense) CpG units in the respective promoter islands revealed no significant effect of ubiquinol. In conclusion, ubiquinol affects the expression of genes involved in PPARα signalling and lipid metabolism without changing the promoter DNA methylation status in the liver of mice.
Keywords: ubiquinol, inflammation, lipid metabolism, methylation, gene expression
Introduction
Coenzyme Q10 (CoQ10) is an important cofactor in the respiratory chain. The reduced form of Coenzyme Q10 (ubiquinol) serves as a potent antioxidant in mitochondria and lipid.(1) More recently, studies in vitro,(2,3) in SAMP1 mice(4) and in humans(5) provide evidence that ubiquinol is involved in inflammatory processes and lipid metabolism via gene expression. Moreover, reducing effects of coenzyme Q10 (ubiquinone) have been also described on genes involved in inflammation and hepatic stress-associated processes in the liver of diet-induced obese mice.(6) However, when related to the oxidized form of coenzyme Q10 (ubiquinone), stronger effects have been observed for the reduced form (ubiquinol) both on the transcriptional(4,7) or protein level(8,9) in vitro and in vivo. To study the observed transcriptional effects of reduced coenzyme Q10 (ubiquinol) in more mechanistic detail, C57BL6J mice were supplemented for one week with 250 mg/kg BW/d ubiquinol. Afterwards, the resulting gene expression signatures were analyzed in the liver of mice, and DNA methylation patterns within the promoters of regulated genes were evaluated. Epigenetic mechanisms such as DNA methylation have been shown to entail heritable changes in gene expression.(10,11) Of note, impacts on DNA methylation processes have been already described for various dietary supplements and food nutrients so far.(12,13) DNA methylation consists of the addition of a methyl group to the fifth carbon-position of the cytosine pyrimidine ring in the context of a CpG dinucleotide. Although most genomic DNA in mammals is deficient in CpG sites, clusters of CpG dinucleotides (CpG islands) were described to be primarily located in promoter regions of genes.(14) Thus, the present study postulates that ubiquinol affects gene expression by modulating DNA methylation patterns in the respective CpG island promoter regions of the selected genes.
Materials and Methods
Animals and diet
C57BL6J mice were reared in the Biochemical and Medical Research Laboratories, KANEKA Corp. (Takasago, Japan), at 22 ± 2°C and a 12 h-light-dark cycle. Water and food intake were available ad libitum. At the beginning of the short-term supplementation study, eight 10-week old male C57BL6J mice were purchased from Charles River Lab. Inc. (Yokohama, Japan). To avoid fighting, mice were housed separately throughout 1-week acclimation period and ubiquinol-supplementation study. In the initial phase of the experiment, C57BL6J mice were randomly assigned to either the ubiquinol (n = 4) or control group (n = 4). Ubiquinol (KANEKA, QHTM) was added to a standard laboratory mouse diet (powdered CE-2, CLEA, Osaka, Japan) at a concentration of 0.2% with corn oil (1%, v/w). The only corn oil (1%, v/w) was used as a vehicle in the control diet. The mixture was incorporated in pellet-type chow by adding 30% ethanol solution (v/v), pressure shaping and drying. In general, body weights and food consumption of the mice were calculated every second day, while mice were inspected daily. After 7-day supplementation period with ubiquinol or the respective control diet, animals were sacrificed by anesthesia using isoflurane gas. Livers were removed and the outer left lobes were stored in RNA-later Storage Solution (Qiagen, Tokyo, Japan) until use for gene expression and methylation analyses.
Microarray analysis
Microarray analysis was conducted on four samples for each group, respectively, by using GeneChip® Mouse Genome 430 2.0 Array (Affymetrix, High Wycombe, United Kingdom) containing 45,100 probe sets. Initially, total RNA was extracted from liver tissues with the miRNeasy kit (Qiagen, Hilden, Germany). The following procedure was performed according to manufacturer’s instructions using Poly-A RNA Control Kit (Affymetrix) and One-Cycle cDNA Synthesis Kit (Affymetrix) for cDNA synthesis, Sample Cleanup Module (Affymetrix) for purification, and IVT Labeling Kit (Affymetrix) for synthesis of biotin-labeled cRNA. Fifteen µg of fragmented cRNA was hybridized to a Mouse Genome 430 2.0 Array for 16 h at 45°C at 60 rpm. Subsequently thereafter, arrays were washed and stained using Fluidics Station 450. Hybridization, washing and staining solutions were obtained from analogous GeneChip® kits. After hybridization and washing procedures, microarrays were scanned with the GeneChip® scanner 3000, using GCOS software. If not stated otherwise, all kits and equipment were purchased from Affymetrix. Fluorescence data were obtained in CEL file format. Quality control and normalization procedure of the files was performed with R software 2.7.1 and BioConductor 2.0.1 provided by the MADMAX database (https://madmax.bioinformatics.nl). Data were normalized with the GC-RMA algorithm. Only probe sets showing present calls for all arrays at one experimental group (intervention or control) were considered for further analysis. The complete datasets will be submitted to NCBI Gene Expression Omnibus (GEO).
qRT-PCR
Primer sequences for real-time quantitative RT-PCR (qRT-PCR) experiments were designed with Primer Express® Software 3.0 (Applied Biosystems, Darmstadt, Germany). Primer pairs (5'-3') for NELF (forward: GAACCCCGAGCCGAATG, reverse: CCGTTAGGGTTCCCCAGAAT) and GPX3 (forward: ACAGGAGCCAGGCGAGAA, reverse: CCACCTGGTCGAACATACTTGA) were obtained from MWG Biotech AG (Ebersberg, Germany). cDNA synthesis was initially carried out with the reverse transcriptase core kit (Eurogentec, Köln, Germany) on a thermocycler (Biometra, Göttingen, Germany). qRT-PCR amplification was performed with the Power SYBR® Green PCR Master Mix (Applied Biosystems, Darmstadt, Germany) on an Applied Biosystem 7300 qRT-PCR system. Ct-values of target genes were related to those of the corresponding housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Text mining analysis
For analysis of common pathways between regulated genes, Genomatix software 2010 (www. genomatix.de) was used. Probe set IDs of the selected genes were uploaded to BibliospherePathwayEdition (BSPE) software. This text mining tool identifies putative functional connections of genes based on co-citations with transcription factors and other genes in the network from NCBI PubMed. The most stringent co-citation filter restricted to sentences with expert curated information (level B4) was applied.
Promoter DNA-methylation analysis
The presence of CpG islands within the ABCA1, ACSL1 and ACYP1 gene promoters was predicted by EMBOSS CpGplot program. Quantitative methylation analysis of the three genes was performed with the MassARRAY® system (Sequenom, Hamburg, Germany). The MassCLEAVETM biochemistry was applied after bisulfite treatment of DNA samples and MALDI-TOF mass spectrometry for analyte detection according to the standard protocols recommended by the supplier. Genomic DNA was extracted from mice liver with the DNeasy Kit (Qiagen). One microgram DNA was treated with sodium bisulfate (DNA Bisulfite Treatment Kit, Sequenom, Hamburg, Germany) and target regions of the modified nucleic acid were amplified by PCR using methylation independent primers, designed by the MassARRAY platform specific EpiDesigner software (Table 2). The PCR products were then subjected to in vitro transcription with RNase A cleavage being used for the T-reverse reaction (Sequenom). The generated fragments were displayed based on their molecular weight in the mass spectrum, which was acquired after sample conditioning with a MassARRAY® Analyzer Compact. The resulting methylation calls were analyzed with EpiTyper Software (Sequenom) to generate quantitative results for each CpG site.
Statistics
If not stated otherwise, data are expressed as means ± SEM of four mice per each group, respectively. Differences between ubiquinol-treated and non-treated animals were analyzed by an unpaired two-tailed Students t test. p values less than or equal to 0.05 were considered as significant.
Results and Discussion
Ubiquinol supplementation reveals no impact on body weight changes
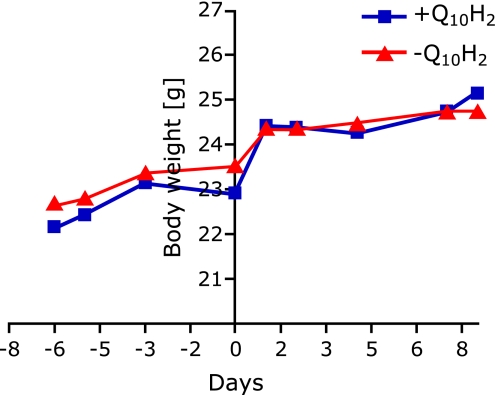
Eight male C57BL6J mice were received at the age of 10 weeks, and initially maintained for 1-week acclimation period. Immediately thereafter, mice were randomly grouped (n = 4 for each group) and either supplemented with ubiquinol (250 mg/kg/d) or a respective control diet for one week. As indicated in Fig. 1, body weights slightly increased both in the subsequently treated ubiquinol group and in control animals during acclimation period from day 1 (−6) to day 7 (0) from 22.15 ± 0.26 g and 22.68 ± 0.44 g to 22.9 ± 0.29 g (p = 0.0032) and 23.53 ± 0.60 g, respectively. Increases in body weights were obtained during grouping period (from day 0 to day 1) in both groups with total values of 24.43 ± 0.25 g in the ubiquinol group and 24.38 ± 0.11 g in control animals. No significant changes of body weights have been found during supplementation period from day 1 until sacrifice of mice (day 8) both in ubiquinol-treated and control animals. The final body weights were 25.15 ± 0.15 g and 24.73 ± 0.34 for treatment and control animals, respectively, and are thus comparable to age-matched C57BL6J mice described in the literature.(6) Finally, at any indicated time point, no significant differences in body weights were found between treatment and control group. Hence, subsequently described effects of ubiquinol supplementation on gene expression and promoter methylation patterns cannot be simply ascribed to changes in weight.
Fig. 1.
Body weight development of 10-week old C57BL6J mice during acclimation period and one-week supplementation period with ubiquinol (250 mg/kg/d) or a respective control diet. Body weights of Q10H2-treated and control animals increased slightly during acclimation period (from day 1 [−6] to day 7 [0]). Strong increases in body weights were obtained during grouping period (from day 0 to day 1) in both groups. No significant changes of body weights have been found during supplementation period from day 1 until sacrifice of mice (day 8) both in the ubquinol-treated (+Q10H2) and control group (−Q10H2).
Whole genome expression data reveal effects of ubiquinol supplementation on liver lipid metabolism
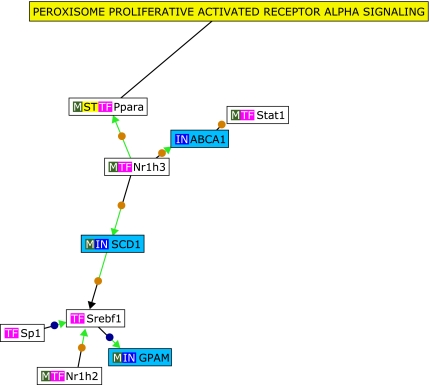
Microarray-based whole genome expression profiles were analyzed from liver samples of mice either supplemented with ubiquinol (250 mg/kg body weight/d) or a respective control diet for one week. Samples were taken from four mice per each group respectively, resulting in a total of eight microarrays. Differentially expressed genes between ubiquinol-treated and control animals were selected as follows: Initially transcripts with at least four present calls (100%) in one group (treatment or control) were chosen for further analysis. Secondly, transcripts showing at least a 1.5-fold increase or decrease in the ubiquinol-treated animals when related to controls (p⩽0.05, Students t test) were selected. Based on these criteria, 1112 and 571 transcripts were up- and down-regulated by ubiquinol, respectively. To study the observed effects of ubiquinol supplementation on gene expression in more detail, transcripts with the highest fold change values (”Top 20”, p⩽0.05) were selected. However, before unravelling putative functional connections among these genes, the accuracy of microarray data was verified by qRT-PCR for two selected ”top 20” genes, namely glutathione peroxidase 3 (GPX3) and nasal embryonic LHRH factor (NELF) (Fig. 2). In brief, GPX3 has been described to mediate PPARγ-associated antioxidant effects in human muscle cells and plasma.(15) Additionally, an influence on the regulation of local and systemic oxidative stress processes has been described.(16) In this context, effects of ubiquinol supplementation on hepatic oxidative stress-associated gene regulation has been already described previously.(6) The NELF gene is primarily expressed in olfactory sensory cells and LHRH cells during embryonic development,(17) and is therefore ascribed as an important factor in the developmental migration process. In adult tissue, NELF has been also shown to be present in the liver.(17) In summary we could show that the fold change values of the two selected ”top 20” genes glutathione peroxidase 3 (GPX3) and nasal embryonic LHRH factor (NELF) (Fig. 2) were in accordance to the microarray data (Table 1). To determine functional connections between the ”top 20” up- and down-regulated genes, text mining analysis was subsequently performed by the use of Genomatix BibliospherePathwayEdition Software including GFG level B4. Based on these stringent criteria, three ubiquinol-sensitive genes [ATP-binding cassette, sub-family A (ABC1), member (ABCA1), stearoyl-Coenzyme A desaturase 1 (SCD1) and glycerol-3-phosphate acyltransferase (GPAM)] were functionally connected with each other in the PPARα signalling pathway (Fig. 3). Moreover, when related to GeneOntology terms (Table 1), 6 (15%) of the ”top 20” up- and down-regulated genes were related to cholesterol or lipid metabolism. These results suggest a functional role of ubiquinol in cholesterol and/or lipid metabolism and are, inter alia in agreement to a previous performed supplementation study with ubiquinol in senescence accelerated mice prone (SAMP1). In this study, various genes with a functional role in cholesterol synthesis, fat assimilation and lipoprotein metabolism have been shown to be affected.(4) Moreover, these transcriptional effects were also translated into physiological readouts of cholesterol metabolism in vivo.(18) In summary, our present and previous data show both short and long-term effects of ubiquinol supplementation on genes involved in liver lipid and/or cholesterol metabolism.
Fig. 2.
Verification of selected ”top 20” genes from microarray experiments by qRT-PCR. For verification of microarray data, two ”top 20” up-regulated genes (GPX3, NELF) were further selected for qRT-PCR experiments. Finally, GPX3 (A) and NELF (B) genes were up-regulated about 2.0-fold (p = 0.0196) and 1.3-fold (p = 0.026) in the ubiquinol-treated (+Q10H2) animals, respectively, when related to control mice (−Q10H2).
Table 1.
Display of the ”top 20” up- and down-regulated genes in liver samples of mice supplemented with ubiquinol for 7 days
| Affymetrix Probe Set ID | FC | p value | Gene symbol | Gene name | GeneOntology Process/Function |
|---|---|---|---|---|---|
| up-regulated genes | |||||
| 1451488_at | 6.67 | 0.04 | FITM1 | fat storage-inducing transmembrane protein 1 | Lipid particle organization, positive regulation of sequestering of triglyceride |
| 1439398_x_at | 5.82 | 0.0001 | Nelf | Nasal embryonic LHRH factor | |
| 1452637_a_at | 5.09 | 0.02 | Bola1 | Bola-like 1 (E. coli) | biological process, molecular function |
| 1428554_a_at | 4.59 | 0.04 | 1810035L17Rik | RIKEN cDNA 1810035L17 gene | Biological process, regulation of transcription, RNA-binding, molecular function, nucleic acid binding, nucleotide binding |
| 1416439_at | 4.57 | 0.01 | Dctpp1 | dCTP pyrophosphatase 1 | Nucleoside triphosphate catabolic process, protein homotetramerization, dCTP diphosphatase activity, hydrolase activity, metal ion binding |
| 1428464_at | 4.48 | 0.05 | Ndufa3 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3 | Biological process, electron transport chain, transport, molecular function |
| 1420952_at | 4.26 | 0.04 | Son | Son cell proliferation protein | DNA binding, RNA binding, nucleic acid binding, protein binding |
| 1438403_s_at | 4.24 | 0.04 | — | — | |
| 1422608_at | 4.14 | 0.02 | Arpp19 | cAMP-regulated phosphoprotein 19 | positive regulation of Ras protein signal transduction, molecular function |
| 1449106_at | 4.1 | 0.05 | Gpx3 | glutathione peroxidase 3 | Glutathione metabolic process, hydrogen peroxide metabolic process, oxidation reduction, response to oxidative stress, glutathione binding, peroxidase activity, oxidoreductase activity |
| 1421374_a_at | 4.01 | 0.03 | Fxyd1 | FXYD domain-containing ion transport regulator 1 | Cellular calcium ion homeostasis, ion transport, muscle contraction, transport, chloride channel activity, ion channel activity |
| 1416217_a_at | 4 | 0.05 | Rpl37a | ribosomal protein L37a | biological process, molecular function |
| 1434823_x_at | 3.91 | 0.02 | Myeov2 | myeloma overexpressed 2 | biological process, molecular function |
| 1436757_a_at | 3.91 | 0.03 | Cox6b1 | cytochrome c oxidase, subunit VIb polypeptide 1 | cytochrome-c oxidase activity |
| 1420642_a_at | 3.91 | 0.05 | Romo1 | reactive oxygen species modulator 1 | biological process, molecular function |
| 1448685_at | 3.88 | 0.04 | 2900010M23Rik | RIKEN cDNA 2900010M23 gene | biological process, molecular function |
| 1438655_a_at | 3.86 | 0.04 | Rpl34 | ribosomal protein L34 | biological process, molecular function |
| 1431199_at | 3.84 | 0.04 | Ggnbp1 | gametogenetin binding protein 1 | Cell differentiation, mitochondrial fission, multicellular organismal development, spermatogenesis, protein binding |
| 1416285_at | 3.82 | 0.04 | Ndufc1 | NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1 | biological process, electron transport chain, transport, molecular function |
| 1429453_a_at | 3.8 | 0.02 | Mrpl55 | mitochondrial ribosomal protein L55 | biological process, molecular function |
| down-regulated genes | |||||
| 1452391_at | –3.37 | 0.002 | Cxadr | Coxsackie virus and adenovirus receptor | cardiac muscle fiber development, cell adhesion, cell-cell junction organization, heart development, mitochondrial organization, negative regulation of cardiac muscle cell proliferation, protein binding, receptor activity |
| 1450392_at | –3.15 | 0.02 | Abca1 | ATP-binding cassette, sub-family A (ABC1), member 1 | cholesterol efflux, cholesterol transport, lipoprotein biosynthetic process, phospholipids efflux, positive regulation of cholesterol efflux, protein amino acid lipidation, ATP binding, ATPase activity, cholesterol transporter activity |
| 1427408_a_at | –2.91 | 0.02 | Thrap3 | thyroid hormone receptor associated protein 3 | positive regulation of transcription from RNA polymerase II promoter, transcription coactivator activity |
| 1417015_at | –2.84 | 0.04 | Rassf3 | Ras association (RalGDS/AF-6) domain family 3 | biological process, signal transduction, molecular function, protein binding |
| AFFX-PyruCarbMur/L09192_5_at | –2.7 | 0.04 | Pcx | pyruvate carboxylase | gluconeogenesis, lipid biosynthetic process, metabolic process, oxaloacetate metabolic process, pyruvate metabolic process, ATP binding, biotin binding, metal ion binding, pyruvate carboxylase activity, nucleotide binding |
| 1425461_at | –2.67 | 0.02 | Fbxw11 | F-box and WD-40 domain protein 11 | Wnt receptor signalling pathway, biological process, cell cycle, rhythmic process, molecular function |
| 1420948_s_at | –2.61 | 0.002 | Atrx | alpha thalassemia/mental retardation syndrome X-linked homolog (human) | DNA repair, forebrain development, response to DNA damage stimulus, ATP binding, DNA binding, chromatin binding, metal ion binding |
| 1433515_s_at | –2.6 | 0.01 | Etnk1 | ethanolamine kinase 1 | biological process, phospholipid biosynthetic process, ATP binding, ethanolamine kinase activity, kinase activity, molecular function, nucleotide binding, transferase activity |
| 1425834_a_at | –2.6 | 0.05 | Gpam | glycerol-3-phosphate acyltransferase, mitochondrial | Acyl-CoA metabolic process, cellular response to insulin stimulus, defense response, fatty acid homeostasis, glycerophospholipid metabolic process, triglyceride metabolic process, phospholipid homeostasis, regulation of cytokine secretion, acyltransferase activity |
| 1430991_at | –2.56 | 0.03 | 1810014B01Rik | RIKEN cDNA 1810014B01 gene | biological process, molecular function |
| 1424484_at | –2.55 | 0.04 | Mobkl1b | MOB1, Mps One Binder kinase activator-like 1B (yeast) | metal ion binding, protein binding |
| 1448158_at | –2.49 | 0.04 | Sdc1 | syndecan 1 | canonical Wnt receptor signaling pathway, myoblast development, cytoskeletal protein binding, glycoprotein binding |
| 1422862_at | –2.45 | 0.02 | LOC669660 | similar to PDZ and LIM domain protein 5 (Enigma homolog) (Enigma-like PDZ and LIM domains protein) | |
| 1420864_at | –2.43 | 0.0003 | Zfp161 | zinc finger protein 161 | Regulation of transcription, DNA binding, metal ion binding, nucleic acid binding, protein binding, zinc ion binding |
| 1415965_at | –2.4 | 0.004 | Scd1 | stearoyl-Coenzyme A desaturase 1 | Brown fat cell differentiation, cholesterol esterification, fatty acid biosynthetic process, lipid metabolic process, oxidation reduction, white fat cell differentiation, oxidoreductase activity, stearoyl-CoA9-desaturase activity, iron ion binding |
| 1431096_at | –2.4 | 0.04 | Ints8 | integrator complex subunit 8 | biological process, molecular function |
| 1419816_s_at | –2.39 | 0.004 | Errfi1 | ERBB receptor feedback inhibitor 1 | lung alveolus development, negative regulation of epidermal growth factor receptor activity, regulation of keratinocyte differentiation, stress-activated protein kinase signalling cascade, kinase binding, protein binding, receptor activity |
| 1420927_at | –2.38 | 0.03 | St6gal1 | beta galactoside alpha 2,6 sialyltransferase 1 | metabolic process, protein amino acid glycosylation, sialyltransferase activity |
| 1448607_at | –2.37 | 0.004 | Nampt | nicotinamide phosphoribosyltransferase | NAD biosynthetic process, positive regulation of muscle cell proliferation, pyridine nucleotide biosynthetic process, cytokine activity, nicotinamide phosphoribosyltransferase activity, protein homodimerization activity |
| 1418279_a_at | –2.36 | 0.01 | Akap1 | A kinase (PRKA) anchor protein 1 | Negative regulation of NFAT protein import into nucleus, negative regulation of protein amino acid dephosphorylation, RNA binding, kinase activity, nucleic acid binding |
# presented by ≥2 probe set Ids in Top 20 gene list.
Fig. 3.
Bibliosphere network of ubiquinol-sensitive genes regulated in liver tissues of C57BL6J mice. Based on co-citations with transcription factors and other genes in the network (GFG level B4), 3 ubiquinol-inducible genes were connected with each other by BibliospherePathwayEdition Software. According to this, the uploaded genes seem to play a key role in PPAR-α signalling pathways. IN, input gene; TF, transcription factor; M, gene product is part of a metabolic pathway; ST, gene product is part of a Genomatix signal transduction pathway.
Ubiquinol-induced effects on gene expression are not mediated by changes in promoter methylation
With regard to the functional analysis of the microarray data from liver tissues of ubiquinol-supplemented mice, distinct effects on genes related to lipid and/or cholesterol metabolism were found (Table 1, Fig. 3). Because transcriptional variation has been correlated with CpG island variation(19) and previous literature reports have documented distinct roles of various micronutrients in the epigenetic regulation of gene expression,(20–23) CpG island regions in promoter regions of three regulated genes strongly related to lipid metabolism were analyzed for putative changes by using base-specific cleavage and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. In general, as an essential precondition, all three genes show CpG islands in their promoter regions and could be therefore selected for DNA methylation analysis. Based on these criteria and the results from microarray experiments, the following ubiquinol-regulated genes (p⩽0.05) were finally selected for subsequent methylation analysis: ABCA1 (ATP-binding cassette, sub-family A, member 1), ACSL1 (Acyl-CoA Synthetase long-chain family member 1) and ACYP1 (Acylphosphatase 1). These genes have been shown to be regulated about +3.14-fold (p⩽0.05), −2.13-fold (p⩽0.05) and +2.18-fold (p⩽0.01), respectively, when related to liver samples of control mice. In brief, the ABCA1 gene is a LXR target gene and plays an important role in reverse cholesterol transport.(24) ACSL1 plays an important role in fatty acid metabolism and triacylglycerol synthesis.(25) Disturbances of these pathways may result in e.g. dyslipidemia, one hallmark of the metabolic syndrome.(26) ACYP1 primarily mediates hydrolytic activity on e.g. acyl phosphates and aryl phosphate monoesters.(27) Overall, our DNA methylation analysis at 282 (sense orientation) and 271 (antisense orientation) CpG units, spanning nucleotides relatively to the gene start position from −748 to 1,391 for ABCA1 gene (Fig. 4A), from −552 to 819 for ACSL1 gene (Fig. 4B), and from −6,863 to –7,663 as well as −298 to 336 for ACYP1 gene (Fig. 4C) revealed no differences between Q10H2-treated and control animals. From these results, no significant differences in the promoter methylation patterns of ABCA1, ACSL1 and ACYP1 genes could be detected with regard to a short term supplementation period with ubiquinol in C57BL6J mice.
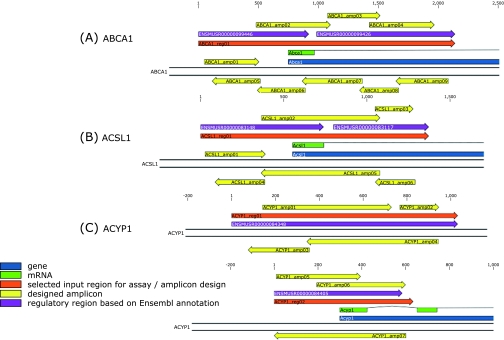
Fig. 4.
Schematic overview of amplicons processed for ABCA1 (A), ACSL1 (B) and ACYP1 (C) analyses in the genomic context. Positions are relative to the selected input region (indicated as reg) and correspond to Table 2 for each respective gene. The regions are located for ABCA1 (A) and ACSL1 (B) gene at position −748 to 1391 and −552 to 819, respectively, when related to the gene start position. For ACYP1 gene (C), region 01 (upper portion) and region 02 (lower portion) are located at position −6863 to −7663 and −298 to 336, respectively, when related to the gene start position. All genes are shown in their transcribed orientation, which is located for ABCA1 and ACYP1 on the antisense strand of chromosome 4 and 12, respectively. For ACSL1 gene, the transcribed orientation is shown on the sense strand of chromosome 8.
Table 2.
PCR primers for analysis of the methylation status of (A) ABCA1, (B) ACSL1 and (C) ACYP1 gene promoters. All positions are relative to the entire input sequences
| (A) ABCA1 | ||||||
|---|---|---|---|---|---|---|
| amplicon |
primer |
|||||
| start | end | length | CpGs | left | right | |
| forward direction | ||||||
| ABCA1_amp01 | 50 | 506 | 457 | 8 | GGTTTTTGGAAGGTAGAGATTTTTT | CCCTAACCCTAACCTACTAACCTTC |
| ABCA1_amp02 | 482 | 1101 | 620 | 40 | GAAGGTTAGTAGGTTAGGGTTAGGG | CCCTATACTATTTACATCCCCAAAAA |
| ABCA1_amp03 | 1076 | 1515 | 440 | 21 | TTTTTGGGGATGTAAATAGTATAGGG | CCCAAAAAACTCAAACAACAATAAC |
| ABCA1_amp04 | 1424 | 1967 | 544 | 22 | GATTGGGATTGTATGTTTTTGTTTT | CACCAATTTTAACCAAATTCACAAT |
| reverse direction | ||||||
| ABCA1_amp05 | 115 | 516 | 402 | 8 | TTTTTTGTAGTTTTGGTTTTGATTTG | AAACTAATATTCACTATCCATTCCACA |
| ABCA1_amp06 | 491 | 891 | 401 | 36 | GGGGGAAATAGGGAGTAGAGTAGTT | CAAATCAAAACCAAAACTACAAAAA |
| ABCA1_amp07 | 865 | 1367 | 503 | 23 | AGGATTTAGATGTGATATTTGTGGG | AAAACTACTCTACTCCCTATTTCCCC |
| ABCA1_amp08 | 1343 | 1669 | 327 | 7 | AGGGTGGATTGGGTATTTTAGTTT | CCCACAAATATCACATCTAAATCCT |
| ABCA1_amp09 | 1646 | 2084 | 439 | 17 | GGTTTGGGAGTTAGGAAATAAATAAA | AAACTAAAATACCCAATCCACCCT |
| (B) ACSL1 | ||||||
|---|---|---|---|---|---|---|
| amplicon |
primer |
|||||
| start | end | length | CpGs | left | right | |
| forward direction | ||||||
| ACSL1_amp01 | 27 | 390 | 364 | 13 | AGGATGGAAGAGTTAAAGGGTATTT | TATCTAAACTAATCCAAACCCCTCA |
| ACSL1_amp02 | 366 | 1078 | 713 | 78 | TGAGGGGTTTGGATTAGTTTAGATA | CAAAATCCCCAACAAACAAAA |
| ACSL1_amp03 | 1049 | 1278 | 230 | 22 | GAGAGGTGGTTTTGTTTGTTGG | TCTAAAAACTCCTCTTAAAAACACC |
| reverse direction | ||||||
| ACSL1_amp04 | 87 | 387 | 301 | 13 | TTGGGTTGATTTAAGTTTTTTAGATT | AAAACTTCCAAATTTACCCTTTACC |
| ACSL1_amp05 | 364 | 1077 | 714 | 78 | AGGGTTTTTAGTAAGTAGGATTATTTTT | TCTAAAAAACTTAAATCAACCCAAA |
| ACSL1_amp06 | 1050 | 1290 | 241 | 22 | GTGTGTAGTGGTTTTGGGAGTTT | AAAAATAATCCTACTTACTAAAAACCCT |
| (C) ACYP1 | ||||||
|---|---|---|---|---|---|---|
| amplicon |
primer |
|||||
| start | end | length | CpGs | left | right | |
| Region 01 | ||||||
| forward direction | ||||||
| ACYP1_amp01 | 142 | 729 | 588 | 29 | GAGAGGGGGTGATAGATATTTGAG | AATATAACCCCAAAAAAACCTCTCC |
| ACYP1_amp02 | 767 | 945 | 179 | 7 | AGATTTTAGGGTGTGTTTTTGTTTG | CCTAAAACTCTTACATAACCCTACCT |
| reverse direction | ||||||
| ACYP1_amp03 | 76 | 355 | 280 | 12 | TGGGGATTTTTTTTGAGTTGGAG | TCAAACCCTCATACAAAAATACTCTAT |
| ACYP1_amp04 | 344 | 943 | 600 | 26 | TGGAGTTTTTATATAGTTTTATTTGGGG | AAAAAATCCCCAAAACCTAATTTTC |
| Region 02 | ||||||
| forward direction | ||||||
| ACYP1_amp05 | 11 | 394 | 384 | 16 | TTTTAGGTATTTAGTAGGGTGAGTGGA | ATTTCTAAAACTTCCCCAAAAACC |
| ACYP1_amp06 | 195 | 599 | 405 | 26 | ATTTGTTTTGAAGTTTTATTTTTGGG | CTCCAATACCATAAAAACCTCTCC |
| reverse direction | ||||||
| ACYP1_amp07 | 1 | 601 | 601 | 29 | TTTTTTAATATTATGGGGGTTTTTT | ATACTAACAACCCTAAACACCCAA |
Although we could not found any significant effects of ubiquinol supplementation on CpG island promoter region methylation patterns of ABCA1, ACSL1 and ACYP1 genes, previous results in the literature show that coenzyme Q10 containing coenzyme supplementation is able to induce the disappearance of tamoxifen-induced RASSF1A DNA methylation patterns in breast cancer patients.(28) Moreover, this supplementary therapy has also been shown to counteract the tamoxifen-induced increased lipoprotein and lipid levels in these patients.(28) By the way, ubiquinol-induced reductions of LDL cholesterol levels have been also observed previously in healthy male volunteers.(5) From these results it seems that ubiquinol supplementation may modulate lipid metabolism through an impact on DNA methylation patterns. Although we could not find any ubiquinol-induced supplementary effects in the present study, it can not be ruled out that changes in the methylation pattern other than in the promoter region of ABCA1, ACSL1 and ACYP1 genes have led to the observed differences on gene expression. In fact, effects on global methylation patterns have been already described for several other dietary supplements and micronutrients in the literature.(20,21,29–32) Thus, a global methylation analysis would be required to finally answer the question of a ubiquinol-dependent epigenetic regulation of ABCA1, ACSL1 and ACYP1 genes. Moreover, the observed changes in gene expression might be also mediated through modifications of histones by acetylation, methylation, phosphorylation, ubiquitination, sumoylation or isomerisation.(22,23) The ability of dietary compounds to influence epigenetic processes via histone modification has been already described in vitro and in vivo.(32,33) However, the translation of these findings into clinical health applications is still a remaining challenge for further studies in the future.
In summary, we could show that one-week supplementation with ubiquinol (250 mg/kg BW/d) induces a gene expression pattern in liver tissues of C57BL6J mice with a functional role in PPARα signalling, lipid and/or cholesterol metabolism. Because transcriptional variation has been correlated with CpG island variation, CpG island regions in promoter regions of three regulated genes (ABCA1, ACSL1 and ACYP1) with a relevant impact in lipid and/or cholesterol metabolism were analyzed. However, promoter DNA methylation analysis of ABCA1, ACSL1 and ACYP1 promoters in the liver of C57BL6J mice revealed no differences between ubiquinol-treated and control mice. Thus, ubiquinol affects the expression of genes involved in PPARα signalling and lipid metabolism without changes in promoter DNA methylation in the liver of mice.
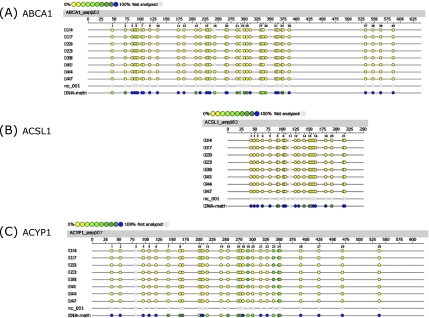
Fig. 5.
Epigram of quantitative methylation analysis of promoter regions in ABCA1, ACSL1 and ACYP1 genes. Genomic DNA isolated from liver samples of Q10H2 supplemented (14, 17, 20, 23) and non-supplemented control mice (38, 41, 44, 47) was analyzed for methylation status of 553 (sense and antisense) CpG sites of the (A) ABCA1, (B) ACSL1 and (C) ACYP1 gene promoter. Exemplary data for one amplicon of each gene is shown. The coloured dots indicate the software determined methylation ratio at each analyzed CpG-unit for each sample. nc_001 represents the reaction negative control (water) and DNA-meth the artificially completely methylated control DNA. The reference sequence above the epigram corresponds to the genomic sequence of the analyzed strand. Displayed is the sense orientation in 5' → 3' direction. Base numbering in the epigram refers to the analyzed amplicon.
Acknowledgments
This work was supported by KANEKA Corporation, Japan.
References
- 1.Littarru GP, Tiano L. Bioenergetic and antioxidant properties of coenzyme Q10: recent developments. Mol Biotechnol. 2007;37:31–37. doi: 10.1007/s12033-007-0052-y. [DOI] [PubMed] [Google Scholar]
- 2.Schmelzer C, Döring F. Identification of LPS-inducible genes down-regulated by ubiquinone in human THP-1 monocytes. Biofactors. 2010;36:222–228. doi: 10.1002/biof.93. [DOI] [PubMed] [Google Scholar]
- 3.Schmelzer C, Kohl C, Rimbach G, Döring F. The reduced form of coenzyme Q10 decreases the expression of lipopolysaccharide-sensitive genes in human THP-1 cells. J Med Food. 2011;14:391–397. doi: 10.1089/jmf.2010.0080. [DOI] [PubMed] [Google Scholar]
- 4.Schmelzer C, Kubo H, Mori M, et al. Supplementation with the reduced form of coenzyme Q10 decelerates phenotypic characteristics of senescence and induces a peroxisome proliferator-activated receptor-alpha gene expression signature in SAMP1 mice. Mol Nutr Food Res. 2010;54:805–815. doi: 10.1002/mnfr.200900155. [DOI] [PubMed] [Google Scholar]
- 5.Schmelzer C, Niklowitz P, Okun JG, Haas D, Menke T, Döring F. Ubiquinol-induced gene expression signatures are translated into altered parameters of erythropoiesis and reduced low density lipoprotein cholesterol levels in humans. IUBMB Life. 2011;63:42–48. doi: 10.1002/iub.413. [DOI] [PubMed] [Google Scholar]
- 6.Sohet FM, Neyrinck AM, Pachikian BD, et al. Coenzyme Q10 supplementation lowers hepatic oxidative stress and inflammation associated with diet-induced obesity in mice. Biochem Pharmacol. 2009;78:1391–1400. doi: 10.1016/j.bcp.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Schmelzer C, Okun JG, Haas D, et al. The reduced form of coenzyme Q10 mediates distinct effects on cholesterol metabolism at the transcriptional and metabolite level in SAMP1 mice. IUBMB Life. 2010;62:812–818. doi: 10.1002/iub.388. [DOI] [PubMed] [Google Scholar]
- 8.Schmelzer C, Lorenz G, Rimbach G, Döring F. Influence of coenzyme Q10 on release of pro-inflammatory chemokines in the human monocytic cell line THP-1. Biofactors. 2007;31:211–217. doi: 10.1002/biof.5520310308. [DOI] [PubMed] [Google Scholar]
- 9.Schmelzer C, Lorenz G, Rimbach G, Döring F. In vitro effects of the reduced form of coenzyme Q(10) on secretion levels of TNF-alpha and chemokines in response to LPS in the human monocytic cell line THP-1. J Clin Biochem Nutr. 2009;44:62–66. doi: 10.3164/jcbn.08-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silahtaroglu A, Stenvang J. MicroRNAs, epigenetics and disease. Essays Biochem. 2010;48:165–185. doi: 10.1042/bse0480165. [DOI] [PubMed] [Google Scholar]
- 11.Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151:4756–4764. doi: 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stidley CA, Picchi MA, Leng S, et al. Multivitamins, folate, and green vegetables protect against gene promoter methylation in the aerodigestive tract of smokers. Cancer Res. 2010;70:568–574. doi: 10.1158/0008-5472.CAN-09-3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Engeland M, Weijenberg MP, Roemen GM, et al. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–3137. [PubMed] [Google Scholar]
- 14.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung SS, Kim M, Youn BS, et al. Glutathione peroxidase 3 mediates the antioxidant effect of peroxisome proliferator-activated receptor gamma in human skeletal muscle cells. Mol Cell Biol. 2009;29:20–30. doi: 10.1128/MCB.00544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YS, Kim AY, Choi JW, et al. Dysregulation of adipose glutathione peroxidase 3 in obesity contributes to local and systemic oxidative stress. Mol Endocrinol. 2008;22:2176–2189. doi: 10.1210/me.2008-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer PR, Wray S. Nasal embryonic LHRH factor (NELF) expression within the CNS and PNS of the rodent. Brain Res Gene Expr Patterns. 2001;1:23–26. doi: 10.1016/s1567-133x(01)00004-7. [DOI] [PubMed] [Google Scholar]
- 18.Schmelzer C, Okun JG, Haas D, et al. The reduced form of coenzyme Q10 mediates distinct effects on cholesterol metabolism at the transcriptional and metabolite level in SAMP1 mice. IUBMB Life. 2010;62:812–818. doi: 10.1002/iub.388. [DOI] [PubMed] [Google Scholar]
- 19.Shen L, Kondo Y, Guo Y, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3:2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uekawa A, Katsushima K, Ogata A, et al. Change of epigenetic control of cystathionine beta-synthase gene expression through dietary vitamin B12 is not recovered by methionine supplementation. J Nutrigenet Nutrigenomics. 2009;2:29–36. doi: 10.1159/000165374. [DOI] [PubMed] [Google Scholar]
- 21.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 22.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 23.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 24.Cavelier C, Lorenzi I, Rohrer L, von Eckardstein A. Lipid efflux by the ATP-binding cassette transporters ABCA1 and ABCG1. Biochim Biophys Acta. 2006;1761:655–666. doi: 10.1016/j.bbalip.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Li LO, Ellis JM, Paich HA, et al. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and beta-oxidation and alters phospholipid fatty acid composition. J Biol Chem. 2009;284:27816–27826. doi: 10.1074/jbc.M109.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miura Y, Fukumoto Y, Shiba N, et al. Prevalence and clinical implication of metabolic syndrome in chronic heart failure. Circ J. 2010;74:2612–2621. doi: 10.1253/circj.cj-10-0677. [DOI] [PubMed] [Google Scholar]
- 27.Paoli P, Pazzagli L, Giannoni E, et al. A nucleophilic catalysis step is involved in the hydrolysis of aryl phosphate monoesters by human CT acylphosphatase. J Biol Chem. 2003;278:194–199. doi: 10.1074/jbc.M206918200. [DOI] [PubMed] [Google Scholar]
- 28.Yuvaraj S, Premkumar VG, Vijayasarathi K, Gangadaran SG, Sachdanandam P. Ameliorating effect of coenzyme Q10, riboflavin and niacin in tamoxifen-treated postmenopausal breast cancer patients with special reference to lipids and lipoproteins. Clin Biochem. 2007;40:623–628. doi: 10.1016/j.clinbiochem.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Fischer A, Gaedicke S, Frank J, Döring F, Rimbach G. Dietary vitamin E deficiency does not affect global and specific DNA methylation patterns in rat liver. Br J Nutr. 2010;104:935–940. doi: 10.1017/S0007114510001649. [DOI] [PubMed] [Google Scholar]
- 30.Waterland RA. Assessing the effects of high methionine intake on DNA methylation. J Nutr. 2006;136 (Suppl 6):1706S–1710S. doi: 10.1093/jn/136.6.1706S. [DOI] [PubMed] [Google Scholar]
- 31.Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J. 2007;21:3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
- 32.Davis CD, Uthus EO, Finley JW. Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr. 2000;130:2903–2909. doi: 10.1093/jn/130.12.2903. [DOI] [PubMed] [Google Scholar]
- 33.Caudill MA, Wang JC, Melnyk S, et al. Intracellular S-adenosylhomocysteine concentrations predict global DNA hypomethylation in tissues of methyl-deficient cystathionine beta-synthase heterozygous mice. J Nutr. 2001;131:2811–2818. doi: 10.1093/jn/131.11.2811. [DOI] [PubMed] [Google Scholar]