Abstract
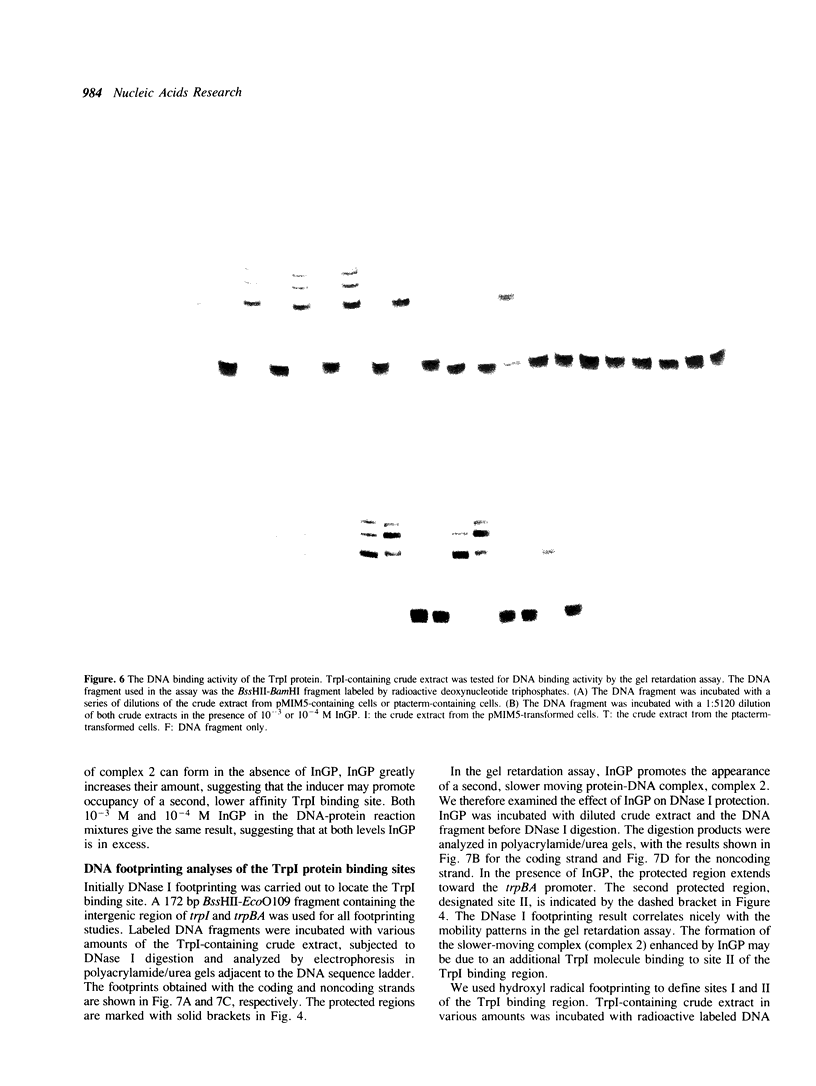
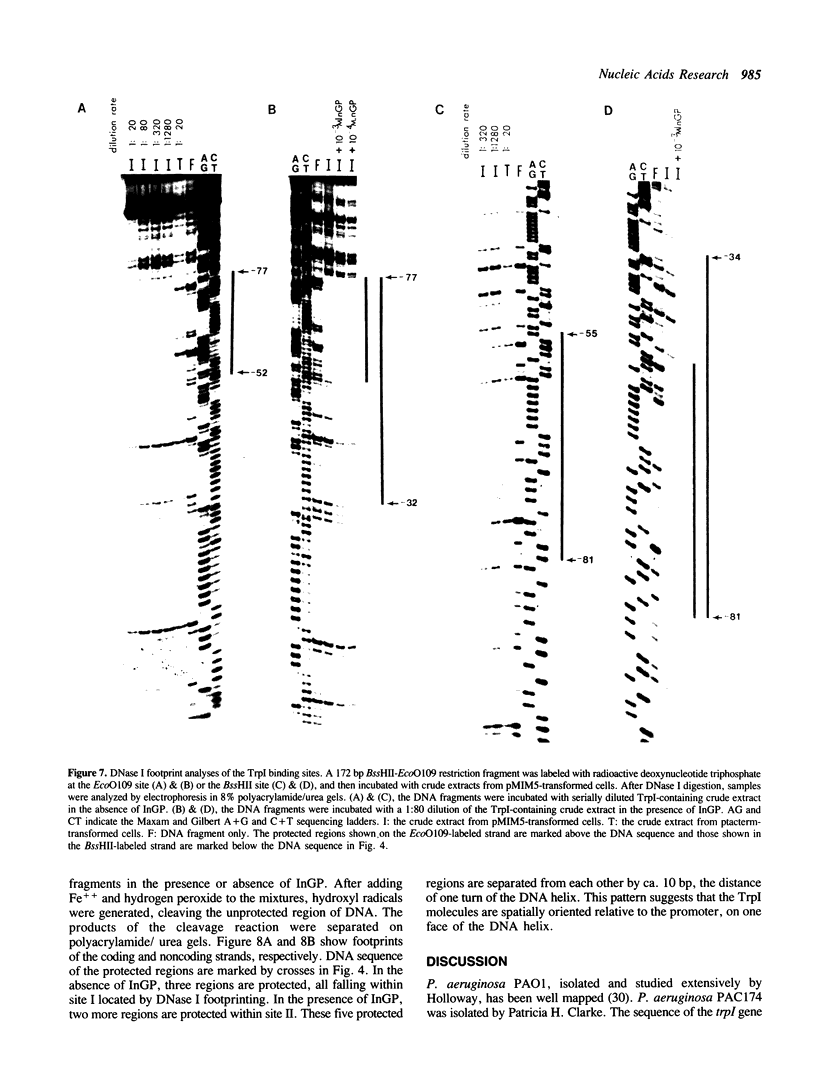
The TrpI protein belongs to the LysR-family of procaryotic regulatory proteins. Members of this family share a characteristic similarity of their N-terminal amino acid sequences, and many of them are activators of divergently transcribed genes or operons. In Pseudomonas aeruginosa, the genes for tryptophan synthase, trpBA, are regulated by indoleglycerol phosphate (InGP) and TrpI. We demonstrate here that in the absence of InGP, the binding site of TrpI is located in the -52 to -77 region of the trpBA promoter; in the presence of InGP, the binding region is extended to the -32 region. In addition, two major, slow moving protein-DNA complexes are seen in gel retardation assays: the faster moving complex is formed in the absence of InGP and the amount of the slower moving complex is greatly enhanced in the presence of InGP. These results suggest that the binding of a second TrpI protein molecule, promoted by InGP, plays a crucial role in activating the expression of the trpBA gene pair.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker R. F., Yanofsky C. The periodicity of RNA polymerase initiations: a new regulatory feature of transcription. Proc Natl Acad Sci U S A. 1968 May;60(1):313–320. doi: 10.1073/pnas.60.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun D. H., Pierson D. L., Jensen R. A. The regulation of tryptophan biosynthesis in Pseudomonas aeruginosa. Mol Gen Genet. 1973 Mar 1;121(2):117–132. doi: 10.1007/BF00277526. [DOI] [PubMed] [Google Scholar]
- Chang M., Hadero A., Crawford I. P. Sequence of the Pseudomonas aeruginosa trpI activator gene and relatedness of trpI to other procaryotic regulatory genes. J Bacteriol. 1989 Jan;171(1):172–183. doi: 10.1128/jb.171.1.172-183.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford I. P., Eberly L. DNA sequence of the tryptophan synthase genes of Pseudomonas putida. Biochimie. 1989 Apr;71(4):521–531. doi: 10.1016/0300-9084(89)90183-1. [DOI] [PubMed] [Google Scholar]
- Crawford I. P., Gunsalus I. C. Inducibility of tryptophan synthetase in Pseudomonas putida. Proc Natl Acad Sci U S A. 1966 Aug;56(2):717–724. doi: 10.1073/pnas.56.2.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graña D., Gardella T., Susskind M. M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988 Oct;120(2):319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadero A., Crawford I. P. Nucleotide sequence of the genes for tryptophan synthase in Pseudomonas aeruginosa. Mol Biol Evol. 1986 May;3(3):191–204. doi: 10.1093/oxfordjournals.molbev.a040388. [DOI] [PubMed] [Google Scholar]
- Hedges R. W., Jacob A. E., Crawford I. P. Wide ranging plasmid bearing the Pseudomonas aeruginosa tryptophan synthase genes. Nature. 1977 May 19;267(5608):283–284. doi: 10.1038/267283a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B. W., Morgan A. F. Genome organization in Pseudomonas. Annu Rev Microbiol. 1986;40:79–105. doi: 10.1146/annurev.mi.40.100186.000455. [DOI] [PubMed] [Google Scholar]
- Lagrimini L. M., Brentano S. T., Donelson J. E. A DNA sequence analysis package for the IBM personal computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):605–614. doi: 10.1093/nar/12.1part2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manch J. N., Crawford I. P. Genetic evidence for a positive-acting regulatory factor mediating induction in the tryptophan pathway of Pseudomonas aeruginosa. J Mol Biol. 1982 Mar 25;156(1):67–77. doi: 10.1016/0022-2836(82)90459-4. [DOI] [PubMed] [Google Scholar]
- Manch J. N., Crawford I. P. Ordering tryptophan synthase genes of Pseudomonas aeruginosa by cloning in Escherichia coli. J Bacteriol. 1981 Apr;146(1):102–107. doi: 10.1128/jb.146.1.102-107.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Paluh J. L., Yanofsky C. High level production and rapid purification of the E. coli trp repressor. Nucleic Acids Res. 1986 Oct 24;14(20):7851–7860. doi: 10.1093/nar/14.20.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M. D., Stauffer G. V. Characterization of the Escherichia coli gene for serine hydroxymethyltransferase. Gene. 1983 Apr;22(1):9–18. doi: 10.1016/0378-1119(83)90059-8. [DOI] [PubMed] [Google Scholar]
- Renault P., Gaillardin C., Heslot H. Product of the Lactococcus lactis gene required for malolactic fermentation is homologous to a family of positive regulators. J Bacteriol. 1989 Jun;171(6):3108–3114. doi: 10.1128/jb.171.6.3108-3114.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Schell M. A., Sukordhaman M. Evidence that the transcription activator encoded by the Pseudomonas putida nahR gene is evolutionarily related to the transcription activators encoded by the Rhizobium nodD genes. J Bacteriol. 1989 Apr;171(4):1952–1959. doi: 10.1128/jb.171.4.1952-1959.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]