Imaging small molecules in real time in living cells is usually accomplished with genetically encoded sensors, which are typically fluorescent proteins flanking a ligand-binding domain. Ligand binding induces conformational changes that reorient the fluorescent proteins, which is detected by changes in Förster resonance energy transfer (FRET). However, sensor development is difficult since proteins that undergo conformational changes upon binding a desired target molecule are usually not available. Here we describe fluorescence imaging of small molecules using RNA. These RNA-based sensors comprise a ligand-binding RNA aptamer and Spinach, an aptamer that binds and switches on the fluorescence of a small molecule fluorophore. We describe sensors that detect a variety of small molecules in vitro, and allow imaging of the dynamic changes and cell-to-cell variation in the intracellular levels of adenosine 5-diphosphate (ADP) and S-adenosylmethionine (SAM) in E. coli.
Recently we described RNAs that bind and switch on the fluorescence of molecules that resemble the fluorophore of green fluorescent protein (GFP) (1). These fluorophores do not exhibit nonspecific fluorescence in cells or cytotoxicity (1). The brightest RNA-fluorophore complex is an RNA termed Spinach and the fluorophore 3,5-difluoro-4-hydroxybenzylidene imidazolinone (DFHBI). Trafficking of Spinach-tagged RNAs can be imaged in cells (1). Because RNA aptamers that selectively bind essentially any small molecule can be rapidly generated using in vitro selection (2), we sought to develop RNA sensors that comprise small molecule-binding aptamers linked to Spinach so that ligand binding induces binding of Spinach to DFHBI, leading to fluorescence.
Spinach contains three stem-loops encircling a central loop (fig. S1A–B) (1). Mutagenesis revealed that one stem-loop has an essential structural role in Spinach fluorescence (fig S1C–D). We therefore used this stem-loop as an entry point for the insertion of small molecule-binding aptamers. Many aptamers are unstructured until they bind their targets (3). If a small molecule-binding aptamer and Spinach share the critical stem required for Spinach fluorescence, small molecule binding can fold the aptamer, stabilize the stem, resulting in fluorescence (Fig 1A).
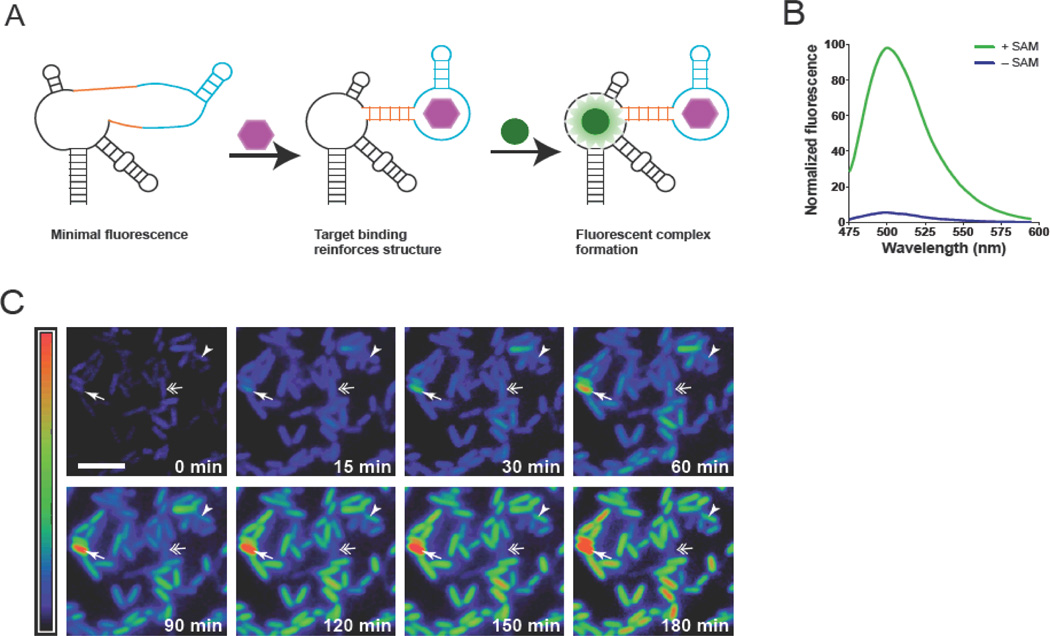
Fig. 1. Imaging S-adenosylmethionine in living cells using RNA.
(A) The sensor comprises Spinach (black), a transducer (orange), and target-binding aptamer (blue). Target binding to the aptamer promotes stabilization of the transducer stem, enabling Spinach to fold and activate DFHBI fluorescence. (B) Emission spectra of the SAM sensor in the presence or absence of SAM. (C) Distinct patterns of SAM accumulation after adding methionine to E. coli expressing the SAM sensor RNA. Some cells exhibited higher than average (arrow) or slow increases (arrowhead) in SAM (arrowhead). A cell that first increases and then decreases its SAM levels is indicated by a double arrow. Images are pseudocolored to show the fold increase in fluorescence at each time point relative to 0 min (0-11.2-fold). Scale bar, 5 µm.
We fused aptamers that bind adenosine, ADP, SAM, guanine, or guanosine 5-triphosphate (table S1) to Spinach via a stem sequence that functioned as a “transducer” (fig. S1E–F). We designed transducers so that stem hybridization is thermodynamically unfavorable because the stem is (1) short, (2) composed of weak basepairs, such as A-U or G-U, or (3) contains mismatched basepairs. We tested sensors containing different transducers, and assayed for ligand-induced fluorescence (fig. S2, table S1). The optimal adenosine, ADP, SAM, guanine, and GTP sensors exhibited 20-, 20-, 25-, 32-, and 15-fold increases in fluorescence, respectively, upon binding their cognate ligand (Fig. 1B, fig. S3A–D). The fluorescence increases were linear in physiological concentration ranges (fig. S3E–I). Most sensors detected the intended target, but not related metabolites, and exhibited rapid fluorescence activation and deactivation kinetics (fig. S3J–R). Notably, there are no obvious approaches for designing FRET-based sensors for these metabolites.
We next used these RNAs to monitor metabolite dynamics in live cells. DFHBI-treated E. coli expressing the SAM sensor exhibited minimal fluorescence when deprived of meth-ionine, the SAM precursor (Fig. 1C). Provid-ing methionine increased fluorescence ~6-fold over 3h (fig. S4C–D), matching increases measured biochemically. SAM levels exhibited cell-to-cell variability following methionine treatment, with most cells exhibiting continuous increases, but others briefly increasing and then decreasing, or rapidly increasing their SAM levels (fig. S4–S5, movie S1). SAM is regenerated by recycling the SAM byproduct S-adenosyl-homocysteine (SAH), through the SAH hydrolase or nucleosidase pathways (4). Selective inhibition of SAH nucleosidase markedly reduced the inter-cell variability after adding methion-ine to cells, indicating a role for this pathway in SAM metabolic variability in E. coli (fig. S5–S6).
Similarly, dynamic changes in ADP levels in E. coli could be detected using the ADP sensor (fig. S7), demonstrating the versatility of these RNA-based sensors. These sensors produce ~20-fold increases in fluorescence upon metabolite binding, unlike FRET sensors which typically exhibit 30–100% increases (5). Because RNA aptamers can be readily generated against any biomolecule (2), the strategies described here should enable the design of sensors to image essentially any molecule.
Supplementary Material
ACKNOWLEDGEMENTS
We thank V. Schramm for inhibitors and M. Cohen, A. Deglincerti, W. Ping and S. Blanchard for suggestions. Supported by the McKnight Foundation, NIH-EB010249 and T32CA062948.
Footnotes
SUPPORTING ONLINE MATERIAL
Supplementary Discussion, Materials and Methods Figs. S1–S8, Movie S1, Table S1
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
REFERENCES
- 1.Paige JS, Wu KY, Jaffrey SR. Science. 2011;333:642. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho EJ, Lee J-W, Ellington AD. Annu. Rev. Anal. Chem. 2009;2:241. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 3.Hermann T, Patel DJ. Science. 2000;287:820. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 4.Lu SC. Int. J. Bioch. Cell Biol. 2000;32:391. doi: 10.1016/s1357-2725(99)00139-9. [DOI] [PubMed] [Google Scholar]
- 5.Lemke EA, Schultz C. Nat. Chem. Biol. 2011;7:480. doi: 10.1038/nchembio.620. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.