Abstract
Understanding how the human gut microbiota and host are impacted by probiotic bacterial strains requires carefully controlled studies in humans and in mouse models of the gut ecosystem where potentially confounding variables that are difficult to control in humans can be constrained. Therefore, we characterized the fecal microbiomes and metatranscriptomes of adult female monozygotic twin pairs through repeated sampling 4 weeks prior to, 7 weeks during, and 4 weeks following consumption of a commercially available fermented milk product (FMP) containing a consortium of Bifidobacterium animalis subsp. lactis, two strains of Lactobacillus delbrueckii subsp. bulgaricus, Lactococcus lactis subsp. cremoris, and Streptococcus thermophilus. In addition, gnotobiotic mice harboring a 15-species model human gut microbiota whose genomes contain 58,399 known or predicted protein-coding genes were studied prior to and after gavage with all five sequenced FMP strains. No significant changes in bacterial species composition or in the proportional representation of genes encoding known enzymes were observed in the feces of humans consuming the FMP. Only minimal changes in microbiota configuration were noted in mice following single or repeated gavage with the FMP consortium. However, RNA-Seq analysis of fecal samples and follow-up mass spectrometry of urinary metabolites disclosed that introducing the FMP strains into mice results in significant changes in expression of microbiome-encoded enzymes involved in numerous metabolic pathways, most prominently those related to carbohydrate metabolism. B. animalis subsp. lactis, the dominant persistent member of the FMP consortium in gnotobiotic mice, upregulates a locus in vivo that is involved in the catabolism of xylooligosaccharides, a class of glycans widely distributed in fruits, vegetables and other foods, underscoring the importance of these sugars to this bacterial species. The human fecal metatranscriptome exhibited significant changes, confined to the period of FMP consumption, that mirror changes in gnotobiotic mice, including those related to plant polysaccharide metabolism. These experiments illustrate a translational research pipeline for characterizing the effects of fermented milk products on the human gut microbiome.
INTRODUCTION
Our physiology and physiological differences are not only manifestations of our human genes and epigenomes, but also a reflection of the genes and genetic variations that exist in our resident microbial communities (microbiomes). Our microbiomes contain at least 100 times more genes than our human genomes (1). Dramatic increases in DNA sequencing capacity have led to an explosive increase in the number and types of culture-independent metagenomic studies of intra- and interpersonal variations in human microbial ecology---as a function of our human lifecycle, cultural traditions, and health status (2–7). Long-term goals of this quest to understand the genomic and metabolic underpinnings of our mutually beneficial relationships with microbes include using our symbionts as a new class of biosensors and biomarkers of wellness, and devising safe and effective ways to deliberately manipulate the structure and functions of our microbiome in order to optimize our health, as well as to treat various diseases.
A necessary starting point for assessing how the structure and functions of the human microbiome are related to our biology is to characterize the normal variations that occur in these communities, their gene pools, and their gene expression profiles both within and between individuals. This requires carefully designed studies where potentially confounding variables such as host genotype, diet and various environmental exposures can be controlled and systematically manipulated. Monozygotic (MZ) twins represent one way to constrain some of these variables, given that they have more similar genotypes and have experienced more similar dietary and other early environmental exposures than any other combination of individuals. A complementary approach is to use germ-free mice colonized at various points in their life with defined collections of microbes, with sequenced genomes, that represent major phylogenetic lineages encountered in the body habitats of human populations of interest. Gnotobiotic mice harboring ‘synthetic’ model human microbiomes, where all component organisms and microbial genes are known, can be reared under conditions where a number of the variables that confound human studies are extremely well controlled. Insights gleaned from these gnotobiotic animals can be applied back to humans (8).
Common intended or unintentional disturbances to our microbiomes include changes in our diets, consumption of antibiotics, and ingestion of live microbial strains posited to improve health. The latter include commercially available probiotics that are incorporated into fermented milk products (FMPs). With increasing regulatory pressure to validate the composition and health claims of probiotics and ‘functional’ foods, it is particularly important to develop informative translational medicine pipelines so that proof-of-concept clinical trials can be performed using validated biomarkers for quantitative phenotyping of subjects and of their responses. The present study demonstrates one type of approach. It uses adult MZ twin pairs and metagenomic methods to first define temporal fluctuations in the organismal and gene content and gene expression profiles of their fecal microbial communities as a function of administration of a widely used commercial FMP. It then takes the five sequenced strains present in the FMP and introduces them as a consortium, at a dose analogous to that experienced by humans, into gnotobiotic mice containing a model human microbiome composed of 15 sequenced human gut symbionts. Quantitative analyses of temporal changes in the proportional representation of microbial species and genes, and of microbiome gene expression and metabolism before and after an ecological ‘invasion’ with the 5-member FMP microbial consortium, has provided insights into the ways that FMP strains and the indigenous model gut community respond to one another. The transcriptional responses were used as biomarkers to interrogate metatranscriptome datasets obtained from the MZ twins’ fecal specimens.
RESULTS
Human studies
Study design and assessment of intrapersonal and interpersonal variations in the fecal microbiota of monozygotic twin pairs over a four month period
Details concerning the seven adult female MZ twin pairs recruited for this study are provided in Table S1. All had been vaginally delivered; none consumed antibiotics in the 6 months prior to and during their participation in the present study; none had a history of gastrointestinal diseases (including irritable bowel syndrome) or any other acute or chronic medical conditions; none were consuming dietary supplements or probiotics at the time of enrollment; and none had a history of gluten sensitivity or other food allergies, nor were any vegans or lacto-vegetarians.
Fresh lots of a FMP were shipped every two weeks to the participants’ homes from the same pilot production facility; strain-specific qPCR-based assays indicated that at the time of shipment each gram of the FMP contained on average 3.2×107 genome equivalents (GE) of Bifidobacterium animalis subsp. lactis (strain CNCM I-2494) and 6.3×107 GE of Lactobacillus delbrueckii subsp. bulgaricus (CNCM I-1632, CNCM I-1519). These results were consistent with previous measurements of the number of colony-forming units (cfu) in a typical cup of the FMP [4.9×107 cfu/g (B. animalis subsp. lactis), 8.4×107 cfu/g (L. delbrueckii subsp. bulgaricus)].
Three fecal samples were obtained over the course of a 4-week period prior to initiation of FMP consumption (‘pre-treatment phase’; see Fig. 1A). Each co-twin then consumed two servings of the FMP per day for 7 weeks (breakfast and dinner). Four fecal samples were collected at defined intervals during this treatment period, while two additional samples were collected during the 4 weeks following cessation of FMP consumption (‘post-treatment phase’; Fig. 1A). Participants kept a daily log of their FMP consumption and stool parameters including frequency. Statistical analyses of this log indicated that in this population, FMP consumption was associated with significantly softer stools but no significant changes in stool frequency (see Supplementary Material). However, based on existing regulatory criteria, our study of this small cohort was insufficiently powered to draw clinical conclusions about these stool parameters. Moreover, the MZ twin population recruited was comprised entirely of healthy individuals, so these data cannot be used to make statements about the impact of FMP consumption on stool softness in unhealthy patient populations.
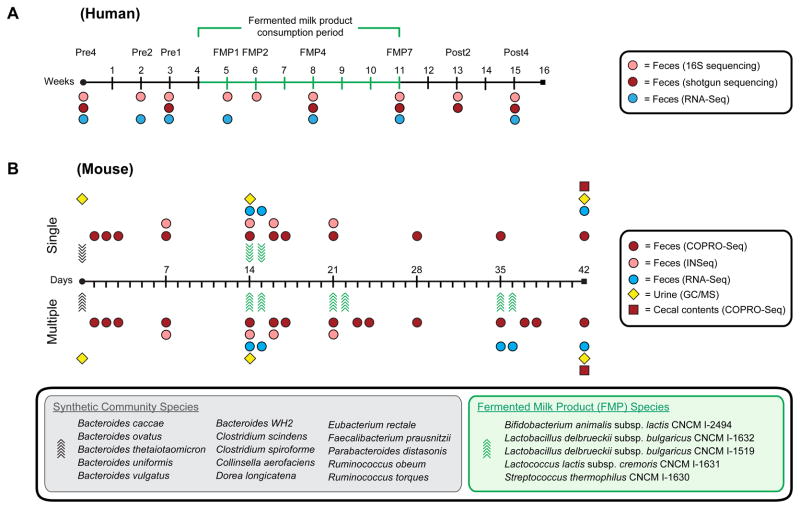
Figure 1. Experimental design for human and mouse studies.
(A) Human study. Seven healthy lean MZ female twin pairs were sampled before, during, and after FMP consumption. (B) Gnotobiotic mouse study. Two groups of five germ-free mice were colonized by oral gavage at 6–8 weeks of age with a 15-member microbial consortium constituting a model human gut microbiota (day of gavage denoted by black arrows). Two weeks later, the five species FMP strain consortium was administered by oral gavage to each group of mice twice over two days (denoted by green arrows). Mice in the single treatment group underwent no further manipulations, whereas animals in the multiple treatment group received additional two-day gavages one and three weeks following the first gavage. Samples were collected at the indicated time points for profiling bacterial community membership (shotgun and 16S rRNA gene sequencing for human fecal samples, COPRO-Seq for mouse fecal and cecal samples), gene expression profiling (microbial RNA-Seq) and metabolite analysis (urines, GC/MS). The species comprising the model 15-member human community and the 5-member FMP consortium are listed in the gray and green boxes, respectively.
All fecal samples collected during the three phases of this study were frozen at −20°C within 30 min of their passage, and maintained at this temperature during overnight shipment to a biospecimen repository where they were subsequently stored at −80°C prior to metagenomic analyses. To assess intra- and interpersonal variations in microbial community structure, we performed multiplex 454 FLX pyrosequencing of amplicons generated from variable region 2 (V2) of bacterial 16S rRNA genes present in fecal DNA. A total of 431,700 sequencing reads were obtained from 126 fecal samples (3,426 ± 2,665 sequences per sample, Table S2A). Noise due to PCR and pyrosequencing artifacts was removed from this dataset using software incorporated into the QIIME suite of 16S rRNA analysis tools (9). De-noised reads were binned into species-level phylogenetic types (phylotypes), with a species defined as isolates that share ≥97% identity in their 16S rRNA gene sequence. To ensure even coverage across samples, each of the 126 datasets was subsampled to 1,640 reads per fecal microbiota. A phylogenetic tree was built from one representative sequence from each phylotype using FastTree’s approximately maximum-likelihood implementation (10) and communities were compared using unweighted UniFrac (11): the UniFrac metric measures community similarity based on the degree to which their members share branch length on a reference phylogenetic tree of Bacteria.
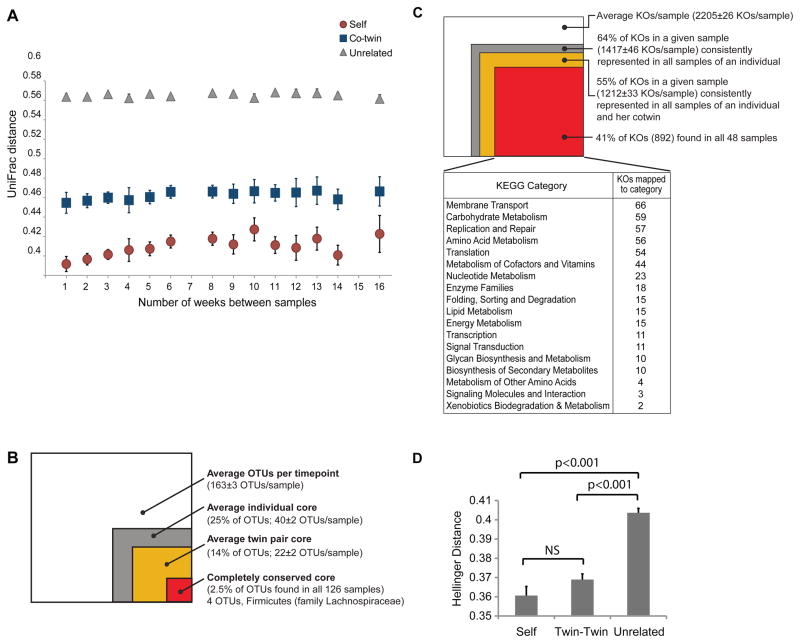
To quantify temporal variation in community composition within and between MZ twins, we generated a matrix of unweighted UniFrac distances for all pairwise comparisons of all 126 fecal samples obtained from the twins in our study. This matrix allowed us to compare any two fecal communities separated by all possible time intervals between sampling for each individual in each of the 7 twin pairs (Fig. 2A). The results indicated that no matter how far apart in time sample collection occurred (1 week to 4 months), the phylogenetic distance between communities from the same individual was less than the distance between communities between co-twins or unrelated individuals. UniFrac distances between samples harvested from a given individual increased modestly as a function of time during the 4-month period, although the changes were not statistically significant (Fig. 2A).
Figure 2. Metagenomic studies of human fecal microbiomes sampled over time.
(A) 16S rRNA-based time course study of intra- and interpersonal variations in fecal bacterial community structure during the course of the 4-month study. Unweighted UniFrac measurements of community distances, from pairwise comparisons of all samples obtained from a given individual, from co-twins, and from unrelated individuals are plotted as mean values ±SEM. (B) Colored boxes represent the proportion of bacterial phylotypes that were consistently present within an individual over time (gray), between co-twins over time (orange), and in all 126 fecal samples (red). The white box represents the average number of species-level phylotypes found in a given sample. All measures of spread provided in parentheses represent ±SEM. (C) KEGG Orthology groups (KOs) consistently present within the fecal microbiome of an individual over time (gray), between co-twins over time (orange), and in all 48 microbiomes analyzed from the 4 sets of MZ twins during the 4 month study (red). The white box indicates the average number of unique KOs (±SEM) identified in a particular sample. All measures of spread provided in parentheses represent ±SEM. (D) Hellinger distance measurements of fecal microbiomes based on their KO content. Tests of statistical significance are based on 1000 permutations of a Hellinger distance matrix. Mean values (±SEM) are shown for the three types of comparisons (self-self; co-twin-co-twin; unrelated-unrelated individual).
Each sample contained 163±3 (mean±SEM) observed species-level phylotypes. Four of the total 1,673 phylotypes identified in our dataset were found in all 126 samples; all belonged to the family Lachnospiraceae (order Clostridiales; phylum Firmicutes) and represented 2.5±0.04% of the 16S rRNA sequences in each sample. 24.6±0.4% of species-level phylotypes observed in a given sample were consistently represented in all 9 samples from that individual (Fig. 2B): the family-level taxa to which these species belong consist principally of Lachnospiraceae, Ruminococcaceae, and Veillonellaceae (phylum Firmicutes), the Bacteroidaceae and Rikenellaceae (phylum Bacteroidetes), and Coriobacteriaceae (phylum Actinobacteria). 13.7±0.2% of the observed phylotypes were represented in all samples from both co-twins (Fig. 2B).
Impact of FMP consumption on fecal bacterial community composition
A qPCR assay disclosed that 1 week after initiation of FMP consumption, the level of representation of B. animalis subsp. lactis (CNCM I-2494) was 107 cell equivalents (CE)/g feces; this level was sustained in all 14 individuals throughout the ensuing 7 weeks of FMP consumption (i.e., there were no statistically significant differences between the 1, 2, 4 and 7 week time points as determined by Friedman test with post-hoc correction). The Spearman correlation test revealed no significant effect of human family membership on the levels of B. animalis subsp. lactis during FMP consumption. Levels fell to below the limits of detection of the assay in all but 4 participants within 2 weeks of cessation of FMP consumption (Fig. S1); two of these individuals represented a twin pair, while the other two individuals were unrelated to each other.
Co-occurrence analysis (see Supplementary Material) indicated that with the FMP dosing schedule used no species-level phylotypes present in the pre-treatment microbiota exhibited a statistically significant change in their proportional representation in feces in any individual, during or following the period of FMP consumption. In addition, no species-level taxa that were undetected in the pre-treatment period appeared and persisted during and/or after treatment in any individual (paired t-test, ANOVA). Of course, it is possible that with even deeper sampling differences might be revealed in feces, or may exist in more proximal regions of the gut. Further details of this co-occurrence analysis, including the results of tests at the genus and family level, plus deeper sequencing of a subset of twin pairs are provided in Supplementary Material.
Effects of FMP consumption on the functional gene repertoire of the fecal microbiome
To determine the effects of FMP consumption on the representation of gene functions in the microbiome, we performed shotgun sequencing on 48 of the fecal DNA preparations generated from 4 of the MZ twin pairs (n=6 samples/individual; 2 fecal samples obtained before, 2 during, and 2 after cessation of FMP consumption; see Fig. 1A). Two of these twin pairs lived together, while two pairs lived 3 and 932 miles apart. A 634 Mb dataset was generated (60,863±28,775 sequences per sample, average length 238 nt; Table S2B). A BLASTX search against version 54 of the Kyoto Encyclopedia of Genes and Genomes (KEGG) GENES database (12–14) yielded a total of 2,205±26 (mean±SEM) KEGG Orthology identifiers (KOs) per microbiome sample: 64% of the KOs in a given sample (1,417±46) were consistently represented in all 6 samples from that individual; 55% were consistently represented in all samples from both co-twins; 892 KOs (41% of the total KOs in a given sample) were identified in all 48 samples forming a core set of shared fecal microbiome-associated functions. Fig. 2C provides a visual representation of this conserved set of 892 KOs: 38% of the 892 belong to six KEGG categories - ‘membrane transport’, ‘carbohydrate metabolism’, ‘DNA replication and repair’, ‘amino acid metabolism’, ‘translation’, and ‘metabolism of co-factors and vitamins’.
The proportional representation of KEGG pathways and their component KOs was subsequently calculated for each of the 48 microbiomes. The microbiomes were then subjected to all possible pairwise comparisons based on these two classification schemes. The results, quantified using the Hellinger distance metric, disclosed that over time, unlike the UniFrac-based 16S rRNA comparisons of community bacterial species composition, there was no significant difference in the degree of similarity of microbiome functional profiles for a given co-twin compared to the degree of similarity that existed between co-twins (i.e., intrapersonal variation was not significantly different from interpersonal variation between co-twins). However, as with the UniFrac results, individual and twin pair microbiomes were significantly more similar to one another than those from unrelated individuals (Fig. 2D). No KEGG pathways or KOs exhibited a statistically significant change in their relative abundance in response to FMP consumption in any of the subjects at any of the time points (Student’s paired t-test and 2-way ANOVA with Bonferroni post-hoc testing).
At this point in our analysis, the human studies indicated that exposure of a healthy individual’s resident gut microbiota to the FMP strains did not produce a detectable perturbation in fecal bacterial species composition, nor did it have a broad effect on the functional profile of fecal microbiome genes. To help guide further analysis of the human datasets, we turned to a simplified in vivo model of the human gut microbiota. We based our selection of model community members on several criteria. All members of this model community, or their close relatives, would be represented in the fecal microbiota of the MZ twins and other sampled human populations. They would encompass the three major bacterial phyla present in this host habitat (Firmicutes, Bacteroidetes, Actinobacteria), and would have deep draft or finished genome sequences available. Gnotobiotic mice harboring such a model human microbiome would be used to characterize the impact of FMP strain introduction on the community’s species and microbial gene abundances, as well as the microbiome’s transcriptional profile, and to ascertain the impact of the model community on the abundance and gene expression profiles of the FMP strains whose genome sequences were also known. The knowledge gleaned would then be used to help guide further analysis of the human fecal microbiome datasets, including microbial RNA-Seq datasets generated from a subset of the human fecal samples.
Studies in gnotobiotic mice
A community of 15 sequenced human gut-derived microbes containing a total of 58,399 known or predicted protein-coding genes was constructed (Fig. 1B, Table S3). Fig. S2 uses assigned KOs to provide evidence of the functional similarity of this model human microbiome to a collection of 127 genomes generated from cultured members of the human gut microbiota, a deeply sampled set of fecal microbiomes obtained from 124 unrelated Europeans (1), and deeply sampled microbiomes from a pair of obese MZ co-twins (15).
Fig. 1B presents the study protocol. Two groups of adult 6–8 week old germ-free C57Bl/6J male mice were colonized with a single gavage of the 15-member community (6×106 cfu/member, total of 9×107 cfu). Each group (n=5 animals) was maintained on a low fat, plant polysaccharide-rich diet. Fourteen and fifteen days after gavage with the 15-member community, both groups of mice were inoculated with a mixture of the five FMP strains. One group received a second pair of gavages of the five strains 7d and 8d later, and a third pair 21d and 22d after the first inoculation of the FMP consortium (multiple treatment group). Each gavage consisted of a community composed of 2×107 cfu: 25% (5×106 cfu) S. thermophilus; 25% B. animalis subsp. lactis, and 25% L. lactis subsp. cremoris, with the remaining 25% split between the two L. delbrueckii subsp. bulgaricus strains (12.5% each; 2.5×106 cfu/strain). Dosing was based on the following considerations: (i) a daily dose of two cups of the FMP contains ~1010 cfu of B. animalis subsp. lactis; (ii) assuming ~1014 bacteria in the human gut, the ratio of the number of input B. animalis subsp. lactis cfu to the human gut symbiont population is approximately 10−4; (iii) to maintain this ratio of 10−4 in mice, and assuming 1011–1012 organisms in the mouse gut, we administered a total of 107 B. animalis subsp. lactis cfu per gavage period (one period equals two gavages within 24h); (iv) the difference in cfu between the least and most abundant microbial species in the FMP product remains ≤2-fold during manufacture and storage; therefore, each species in the gavage was represented at equivalent levels.
FMPs contain defined collections of microbes, a dairy matrix, and products of microbial metabolism of this matrix generated during manufacturing and storage. In principle, the dairy matrix and microbial metabolic products could be responsible for some of the effects observed when humans consume a FMP. Nonetheless, we chose to administer the strain consortium to mice directly by gavage, rather than by giving them the corresponding commercial FMP. This allowed us to more precisely control dosing. It also allayed concerns about possible unintended colonization of the gnotobiotic mice with microbial species (other than strains deliberately introduced during manufacturing) that could be introduced during handling of a FMP prior to gavage.
The repertoire of carbohydrate active enzymes (CAZymes) in members of the FMP consortium and model human gut microbial community
The genomes of the five FMP strains in this study were sequenced, either completely (B. animalis subsp. lactis) or at a deep draft level (other four strains) for subsequent analyses of their representation in the model community after gavage of gnotobiotic mice and so we could define their in vivo patterns of gene expression (Table S3).
Analysis of the 48 CAZyme families (16) identified in the five FMP strains, and the 126 CAZyme families identified in the 15-member model human microbiota disclosed that 23 of the 24 CAZyme glycoside hydrolase (GH) families, 11 of the 12 glycosyltransferase (GT) families, 4 of the 4 carbohydrate esterase (CE) families, and 4 of the 8 carbohydrate binding modules (CBMs) represented in the former were also represented in the latter. The FMP consortium contains only six CAZyme families that were not represented in the model human gut community. Three of these are associated with L. lactis subsp. cremoris: of these, two are predicted to play roles in the binding and metabolism of chitin (Fig. S3; Table S4). The other three are from B. animalis subsp. lactis: BALAC2494_01193 encodes a GT39 family mannose transferase involved in O-glycosylation of proteins; BALAC2494_01288 specifies a predicted beta-mannanase carrying a C-terminal CBM10 carbohydrate-binding module predicted to bind cellulose; BALAC2494_01971 gives rise to a protein with a CBM23 module predicted to bind mannan.
Introducing the FMP strain consortium produces minimal changes in the species representation of the 15-member model human gut microbiota
We used COmmunity PROfiling by Sequencing (COPRO-Seq), a generally applicable method based on highly parallel DNA sequencing (17), to quantify the proportional representation of each component of the 15 member microbiota and of the FMP consortium in our gnotobiotic mice. Sequencing reads generated from fecal DNA samples collected before, during and after introduction of the FMP strains were analyzed as described in Fig. S4A. Briefly, “informative” tags (i.e., reads that can be mapped uniquely to a single genome) were first identified. Informative tags were then summed by species to generate digital “counts” of abundance. In cases where a read could not be assigned with certainty during COPRO-Seq analysis, it was ignored. To account for this fact, species-specific counts were normalized using their “informative genome size” (defined as the percent of all possible k-mers a genome can produce that are unique multiplied by the total genome length). Multiplex sequencing using the Illumina GA-IIx instrument yielded sufficient numbers of reads per sample so that an organism comprising ≥0.003% of the community could be detected: for a mouse colonized at 1011–12 cfu/ml cecal contents or feces, this represents 106 cfu/ml.
COPRO-Seq produced several notable findings. First, community assembly prior to introduction of the FMP strains occurred in a highly reproducible manner, both within and between the two groups of animals (Fig. S5A, Table S5A). This reproducibility ensured that animals in both treatment groups harbored communities with structures comparable to one another at the time of administration of the five-member FMP strain consortium. Second, within 1 week of introducing the FMP strains, either as a single treatment or in multiple treatments, B. animalis subsp. lactis and L. lactis were detectable in the fecal microbiota (Fig. S5B; Table S5A). These two species persisted in the gut throughout the study. Importantly, B. animalis subsp. lactis was the most prominently represented member of the FMP consortium in the model human gut microbiota, exhibiting a progressive increase in its representation during the 28 days following initial introduction, and reaching comparable levels in both the single and multiple treatment groups (up to 1.1%; see Fig. S5B). In contrast, S. thermophilus and the two strains of L. delbrueckii subsp. bulgaricus were undetectable or intermittently just over the limit of detection in both the single- or multi-treatment groups. Third, as with the MZ twin pairs, introduction of the consortium led to minimal rearrangements in overall community structure, whether or not the consortium was administered twice in a two-day period or on two subsequent occasions (see Table S5B for the results of Mann-Whitney tests of significance for each species at each time point surveyed relative to the time point just before initial introduction). Collinsella aerofaciens, the lone Actinobacteria in the 15-member community, showed a significant reduction in its abundance in both treatment groups immediately following FMP strain introduction (Fig. S5C) that persisted through later time points, raising the possibility of a competitive relationship between this organism and B. animalis subsp. lactis, the only Actinobacteria in the FMP strain consortium.
The B. thetaiotaomicron component of the 15-member human gut microbiota was composed of a library of 34,544 randomly inserted transposon mutant strains covering 3,435 of the organism’s 4,779 genes (72%). As noted in Supplementary Material and Table S6, by comparing the representation of mutants in fecal samples before and after introducing the FMP strains, we were able to determine that their presence did not affect the profile of B. thetaiotaomicron’s genetic determinants of fitness in the distal gut.
Microbial RNA-Seq analysis of the response of B. animalis subsp. lactis to the gut environment and members of the 15-member community to the FMP strain consortium
Moving beyond COPRO-Seq based structural analysis, we performed microbial RNA-Seq analysis to determine the functional impact of exposing the established model human community to the FMP strains, and to ascertain which FMP consortium genes are most highly expressed in the intestines of these animals. B. animalis subsp. lactis attained sufficient abundance in gnotobiotic mice to allow profiling of its transcriptome at late time points (days 35, 36, and 42). When its in vivo patterns of gene expression were compared with those documented during mid-log and stationary-phase in MRS medium and in the commercial FMP (see Supplementary Material and Table S7), we noted that the BALAC2494_00604-BALAC2494_00614 locus, encoding enzymes involved in the catabolism of xylooligosaccharides (18), was strongly upregulated in vivo (average across the locus; 27-fold at the day 42 time point compared to mid-log phase in MRS monoculture; 128-fold compared to the FMP, Table S7). Xylose is the main building block of dietary hemicelluloses. Addition of this pentose sugar is also one of the first steps in O-glycosylation of host mucins. These results support previous observations suggesting xylooligosaccharides may serve as potent ‘bifidogenic factors’, whose consumption increases the densities of Bifidobacteria in the gut (19, 20).
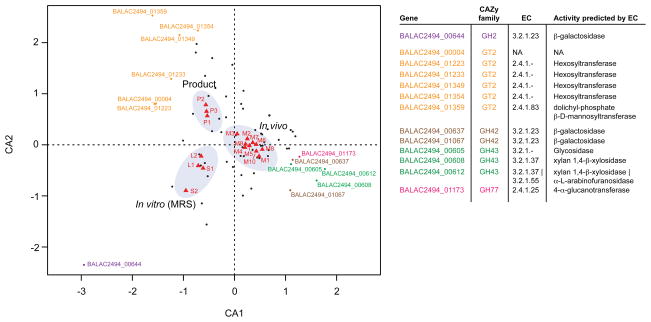
Ordination of samples and B. animalis subsp. lactis CAZyme gene expression patterns by correspondence analysis identified additional CAZymes that correlate strongly with the in vivo state (Fig. 3), including members of families expected to play roles in the degradation of dietary plant polysaccharides [GH43 (xylan beta-xylosidases), GH77 (4-alpha-glucanotransferases)]. The analysis revealed sets of B. animalis subsp. lactis CAZymes that corresponded well with each growth condition (i.e., MRS medium, commercial dairy matrix, and mice). Within each growth condition, the expressed groups of CAZymes often had related functions (Fig. 3).
Figure 3. Correspondence analysis of B. animalis subsp. lactis CAZyme gene expression.
RNA-Seq data for all B. animalis subsp. lactis genes encoding known or predicted CAZymes were subjected to unconstrained correspondence analysis using the ‘vegan’ package in R. Correspondence analysis (CA) allows for the generation of biplots in which samples and genes can be plotted in the same ordinate space to reveal associations/anti-associations between the two. Circles represent individual CAZymes (genes). The genes ordinating furthest from the origin in the direction of one of the sample clusters (treatment groups) are labeled according to their locus number and are colored based on CAZyme family assignment (see Table to the right of the Figure for details; the abbreviation NA refers to no designation). Red triangles represent samples and are labeled according to the following nomenclature: LX, logarithmic phase cells in MRS with X being the technical replicate number (e.g. L1 refers to the first technical replicate harvested in log phase); SX, stationary phase cells in MRS with accompanying replicate number; MX, feces from designated gnotobiotic animals obtained 4 weeks after the initial invasion with the FMP strain consortium; PX, samples obtained after fermentation in the FMP dairy matrix. Each cluster of samples from a particular treatment is associated with a functionally related set of expressed CAZymes.
We next examined the impact of the FMP strain consortium on expression of genes in the 15-member community. In a ‘top-down’ analysis, genes were binned by function and the community’s metatranscriptome evaluated in aggregate, ignoring the species from which each transcript arose. A complementary ‘bottom-up’ analysis allowed us to determine how each species in the community responded to the introduction of the FMP consortium.
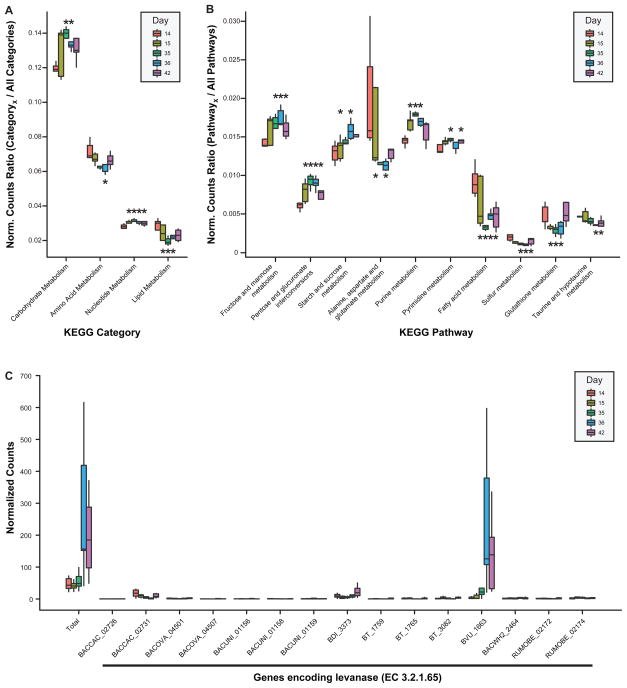
Top-down analysis of the impact of the FMP strains on the community metatranscriptome revealed significant increases in expression of genes falling within the KEGG categories ‘carbohydrate metabolism’, and ‘nucleotide metabolism’, while decreases were observed in ‘amino acid metabolism’ and ‘lipid metabolism’ (Fig. 4A, Table S8). Peak responses in both treatment groups occurred 3 weeks following the first gavage of the FMP strains, corresponding to the time of highest representation of B. animalis subsp. lactis in the community.
Figure 4. ‘Top-down’ analysis of the effects of the FMP strain consortium on the model 15-member community’s metatranscriptome.
RNA-Seq reads were mapped to the sequenced genomes of the 15 community members. Transcript counts were normalized [reads per kb of gene length per million reads (RPKM), see Supplementary Material] and binned using the hierarchical levels of functional annotation employed by KEGG. For each KEGG category (A) or pathway (B) shown, boxplots depict the proportion of normalized read counts assignable to that annotation out of all reads which could be assigned annotations for that hierarchical level. Data shown correspond to the ‘multiple’ treatment group of mice (the group for which the most time points were collected), however, data for all mice are provided in Table S8. (C) Illustration of how a model community’s functional response (e.g., the increased expression of levanase-encoding genes) can be dissected to identify the subset of genes/species driving the response. Boxes denote top quartile, median, and bottom quartile. Whisker length represents 1.5x inter-quartile range (IQR), except where there are no outliers; in these situations, whiskers span the range from minimum to maximum values. Box color denotes the day fecal samples were obtained (day 14 is the pre-treatment timepoint immediately preceding gavage of the FMP strain consortium). When an asterisk is centered over a box, it indicates that there was a statistically significant change following administration of the FMP consortium relative to the pre-treatment timepoint (p<0.05 by paired, two-tailed Student’s t-test). The positioning of asterisks above versus below a box emphasizes the direction of change (above, upregulation; below, downregulation).
The genes that exhibited the highest fold-change in expression were heavily skewed towards the KEGG categories ‘carbohydrate metabolism’ and ‘membrane transport’. The latter includes a number of ABC- and PTS-type carbohydrate transporters (Table S9). When these KEGG category-level responses were subsequently broken down into KEGG pathways (Fig. 4B), it was apparent that the most significant responses in the ‘carbohydrate metabolism’ category involved increases in ‘starch and sucrose metabolism’, ‘fructose and mannose metabolism’, and ‘pentose and glucuronate interconversions’.
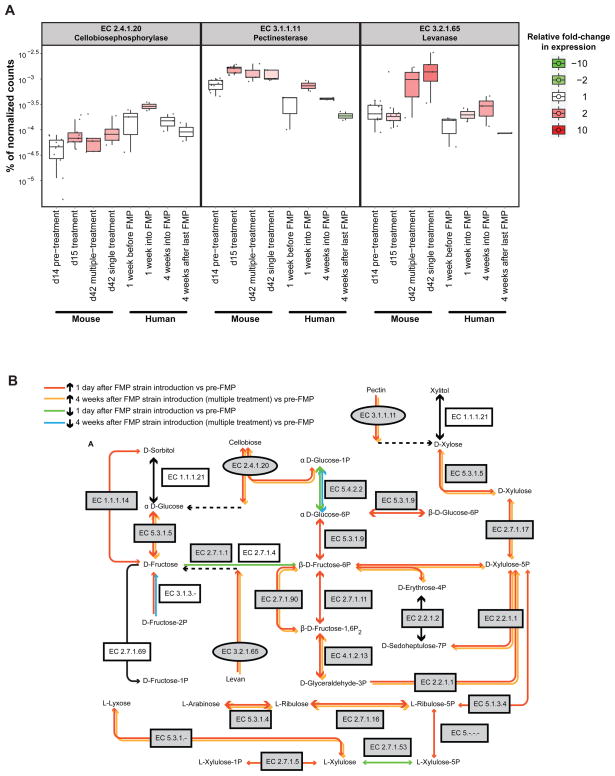
Transcript data were subsequently binned by enzyme commission (EC) number. The levels of mRNAs encoding these ECs at each time point were compared using ShotgunFunctionalizerR, an R-based statistical and visualization tool originally designed to identify genes significantly enriched or depleted in environmental microbiomes (21, 22). Using this approach, we were able to determine that the ‘starch and sucrose metabolism’ pathway response to the FMP strains was driven by significant upregulation of genes encoding three enzymes involved in metabolism of dietary plant polysaccharides: (i) EC 3.2.1.65 (levanase; Fig. 4C), which cleaves 2,6-beta-D-fructofuranosidic linkages in 2,6-beta-D-fructans (levans); (ii) EC 3.1.1.11 (pectinesterase), which de-esterifies pectin to pectate and methanol; and (iii) EC 2.4.1.20 (cellobiose phosphorylase), which uses cellobiose formed from partial hydrolysis of cellulose as its substrate to generate alpha-D-glucose-1-phosphate and D-glucose. The genes encoding these ECs, which catalyze early steps in three entry points of the ‘starch and sucrose metabolism’ KEGG pathway, underwent significant increases in their expression within 24 hours after introduction of the FMP consortium (Fig. 5A). The levels of expression of these genes either increased further (levanase) or were sustained (the other two ECs) in both the single and multiple treatment groups through the remaining 4 weeks of the experiment (Fig. 5A). The levanase response showed remarkable species specificity: this gene is represented in 8 members of the 15-member community, yet the community’s transcriptional response is driven almost exclusively by the levanase in Bacteroides vulgatus (BVU_1663; Fig. 4C). In contrast, the pectinesterase response was distributed across 6 members of the community (B. caccae, B. ovatus, B. thetaiotaomicron, B. vulgatus, B. WH2, C. aerofaciens), with changes in transcription largely due to pectinesterase genes found in B. ovatus (BACOVA_03576, BACOVA_03581, BACOVA_04902), B. thetaiotaomicron (BT_4109, BT_4110), B. vulgatus (BVU_1116), and B. WH2 (BACWH2_3569, BACWH2_3615). Increases in the proportional abundance of cellobiose phosphorylase transcripts reflected the contributions of three community members: B. uniformis, E. rectale, and R. obeum (Table S8).
Figure 5. Mouse and human communities share transcriptional responses to the FMP strain consortium involving ECs related to carbohydrate metabolism.
(A) Box plots of the proportion of all RPKM-normalized reads in mouse and human fecal metatranscriptomes represented by three ECs involved in plant biomass degradation. Individual samples are shown as black dots (n=2–10). Boxes are also colored by fold-change, as determined by comparing mean values at a given time point to the value at the pre-treatment time point [for gnotobiotic mice pre-treatment refers to day 14; in the case of humans, pre-treatment refers to the fecal sample collected 1 week prior to initiation of FMP consumption (sample ‘Pre1’ in Fig. 1A)]. Statistical significance was determined using the ShotgunFunctionalizeR package in R and an adjusted p-value cutoff of <0.01. Pre-treatment time points, and subsequent time points where expression levels were not significantly different from the pre-treatment mean are colored white. (B) Components of KEGG ‘starch and sucrose metabolism’, ‘pentose and glucuronate interconversions’ and ‘pentose phosphate’ pathways whose expression in the 15-member model community changed compared to pre-treatment values when the 5-member FMP strain consortium was introduced. Gray indicates that the fold-change was statistically significant (adjusted p-value <0.01). Ovals highlight the three enzymes shown in panel A. Dashed arrows indicate that multiple enzymatic reactions lead from these ECs and their indicated substrates to the products shown. These intermediate reactions have been omitted for clarity or because the omitted ECs did not manifest significant changes in their expression.
The KEGG ‘starch and sucrose metabolism’, ‘pentose and glucuronate interconversions’ and ‘pentose phosphate’ pathways process products generated by these three enzymes. Fig. 5B shows that many of the other components of these pathways are upregulated in the 15-member community when the FMP strain consortium is introduced. ShotgunFunctionalizeR also identified significant increases in the expression of genes encoding five ECs that participate in the generation of propionate and succinate: the induction occurred within 24 hours after the FMP strains were introduced and involved acetate kinase (EC 2.7.2.1; catalyzes a bidirectional reaction between propanoyl phosphate and propionate), phosphate acetyltransferase (EC 2.3.1.8), methylmalonylCoA decarboxylase (EC 4.1.1.41), propionylCoA carboxylase (EC 6.4.1.3) and methylmalonylCoA mutase (EC 5.4.99.2, yields succinylCoA as its product) (Fig. S6). Only a single treatment with the FMP consortium was required to produce a sustained response involving the enzymes that can yield propionate (Fig. S6).
A breakdown of RNA-Seq reads by the community member genome to which they mapped revealed that the abundance of a species in the 15-member community did not necessarily correlate with its contribution to the community transcript pool. At the time point sampled immediately prior to invasion (d14), two of the most extreme outliers were B. WH2 (comprised 39.6±1.6%; mean±SD) of the community but only contributed 15.4±2% of the raw reads to the total RNA-Seq read pool) and R. obeum (2.1±0.4 % of the community; 18.2±4.4% of the transcript pool) (Fig. S7). These observations indicate that community-level transcriptional responses can be driven by species representing small fractions of the microbiota.
Our ‘bottom-up’ analysis is summarized in Fig. S8 and Table S10, and disclosed early- and later-responding species. Specifically, there were more significantly highly-regulated R. obeum transcripts within the community metatranscriptome 1d after gavage than would be expected based on its community representation, and more highly regulated R. obeum genes in the comparison between day 14 (just before gavage) versus day 15 metatranscriptomes than between day 14 versus day 42 metatranscriptomes. In contrast, B. WH2, Clostridium scindens, and B. uniformis were defined as late responders to the FMP consortium.
Identifying predictive features from the model community metatranscriptome data using a Random Forests classifier
Machine learning techniques employing the random forests classifier can be applied to metagenomic data (23) to learn a function that maps a set of input values or predictors (in this case relative abundance of KEGG categories, KEGG pathways or ECs in a community) to a discrete output value (here, the presence/absence of the FMP strains). KEGG categories, KEGG pathways and ECs were all able to predict pre-/post- treatment status with low estimated generalization error (KEGG categories: 6.7%, ECs: 13.3%, KEGG pathways: 10.0%). In all cases, these generalization error rates were less than half of the baseline error rate of 33% (i.e. that achieved by always predicting the largest category). There were 11 predictive and 5 highly predictive KEGG categories, 35 moderately predictive ECs, and 27 predictive and 4 highly predictive KEGG pathways (Table S11). The predictive ECs identified using our supervised classification approach include a number of carbohydrate metabolism-related functions that were also identified using ShotgunFunctionalizeR in our top-down analysis.
Metabolomic analyses
To evaluate the impact of invasion with the 5-member FMP consortium on microbial-host co-metabolism, we performed untargeted gas chromatography-mass spectrometry (GC/MS) on urine samples collected at multiple time points (days 0, 14 and 42) from members of the single- and multi-treatment groups (Fig. 1B). A metabolite profile was constructed for each urine sample (n=19) using the spectral abundances of all identifiable metabolites. A total of 198 metabolites met our reverse match score cutoff of 65% and were present in at least 50% of samples at one or more time points (for an explanation of the reverse match score, see (24) and Table S12). Comparing day 0 and 14 samples revealed 39 metabolites whose levels were significantly higher or lower following colonization with the defined 15- member community (see Table S12A). The changes included decreases in the levels of oligosaccharides we would expect to be consumed by members of the microbiota [melibiose (87% decline); raffinose/maltotriose (98%); note that oligosaccharides are by their nature difficult to identify with certainty with the present, non-targeted GC/MS technique, and our annotations of these metabolites as melibiose, and raffinose/maltotriose are provisional]. The observed 3.4-fold increase in pyrogallol, a polyphenol, is consistent with the known ability of many gut microbes to cleave these molecules from polyphenols present in dietary plant material. A 4.4-fold increase in taurine following the initial colonization of mice was also noted, probably a result of microbial deconjugation of taurine from bile compounds.
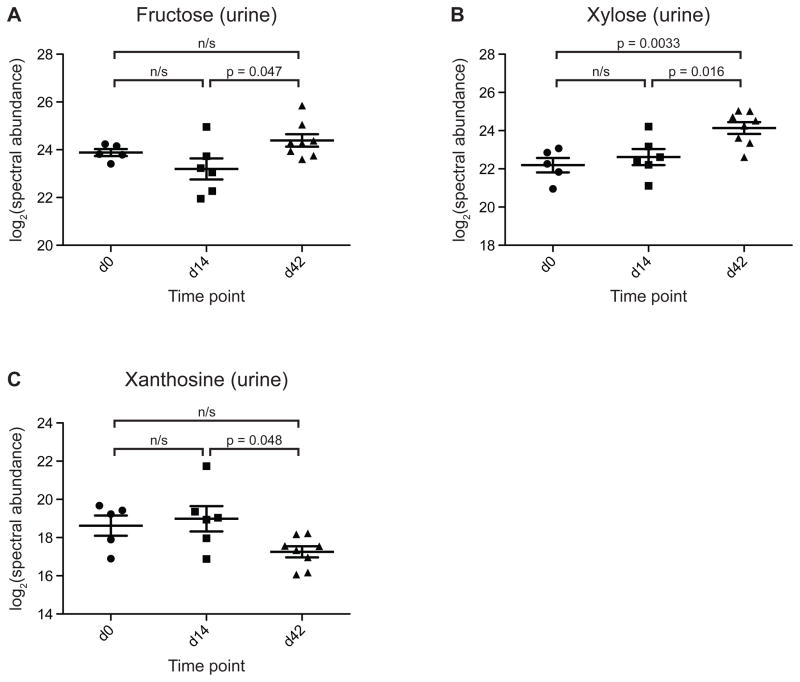
Table S12B lists urinary metabolites that change significantly after introducing the five FMP strains (compare day 14 versus 42 in Table S12B). Fructose and xylose were not significantly affected by introduction of the defined 15-member community but increased significantly following introduction of the FMP strain consortium (2.3- and 2.9-fold, respectively; Fig. 6A,B). Increases in fructose may reflect an enhanced capacity of the community to liberate this monosaccharide from levan and other polyfructans via levanase-catalyzed reactions. Increases in xylose might be explained by the additional xylanase activity introduced by B. animalis subsp. lactis (Fig. 3), or alternatively by the induction of microbiome genes encoding xylan-degrading enzymes (e.g. BACOVA_04387 and BACOVA_04390, which were upregulated 5.2- and 11.0-fold, respectively, following introduction of the FMP strains, Table S10). Changes in other metabolites such as xanthosine (Fig. 6C), a purine metabolite, suggest that the metabolic consequences of FMP strain introduction extend beyond the processing of carbohydrates.
Figure 6. Select urinary metabolites whose levels are altered after the introduction of the FMP strain consortium into mice harboring a defined model human gut microbiota.
The statistical significance in pairwise comparisons shown in panels A-C was evaluated using a two-tailed Student’s t-test on the log-transformed spectral abundance of the metabolite in each sample. Values for the statistical significance of differences between time points as evaluated by one-way ANOVA, followed by FDR-correction and a post-hoc Tukey HSD test are also provided in Table S12. Horizontal bars represent group means, vertical bars represent ± SEM. n/s; not significant.
Collectively, our transcriptional and metabolite analyses indicated that introducing FMP strains that constitute a small fraction of a defined model human gut microbiota signals the microbiota to change its metabolic activities, including activities related to carbohydrate metabolism. With this information in hand, we returned to the human fecal samples to determine the extent to which observations made in our gnotobiotic mouse model were applicable to humans.
Microbiome transcriptional responses to FMP strains that are shared by gnotobiotic mice and humans
Microbial RNA-Seq analysis was performed on human fecal samples obtained 1 week prior to FMP consumption, 1 and 4 weeks into the consumption period, and 4 weeks following cessation (both co-twins from family 1; one co-twin from family 3; see Table S1). Using an analysis pipeline comparable to the one employed for the mouse data, we first aligned all RNA-Seq reads against a reference set of 131 microbial genomes plus the five FMP strain genomes, binned the aligned transcripts based on their EC annotations, and used ShotgunFunctionalizeR to identify ECs whose abundances were significantly changed as a function of FMP exposure (Benjamini-Hochberg adjusted p-value <0.01).
Categorical analysis of the responses of the human fecal community to FMP consumption revealed that significantly upregulated ECs were principally distributed among the KEGG categories ‘carbohydrate metabolism’, ‘amino acid metabolism’, and ‘metabolism of cofactors and vitamins’ (see Table S13 for a complete list of ECs identified from the various pairwise comparisons of time points).
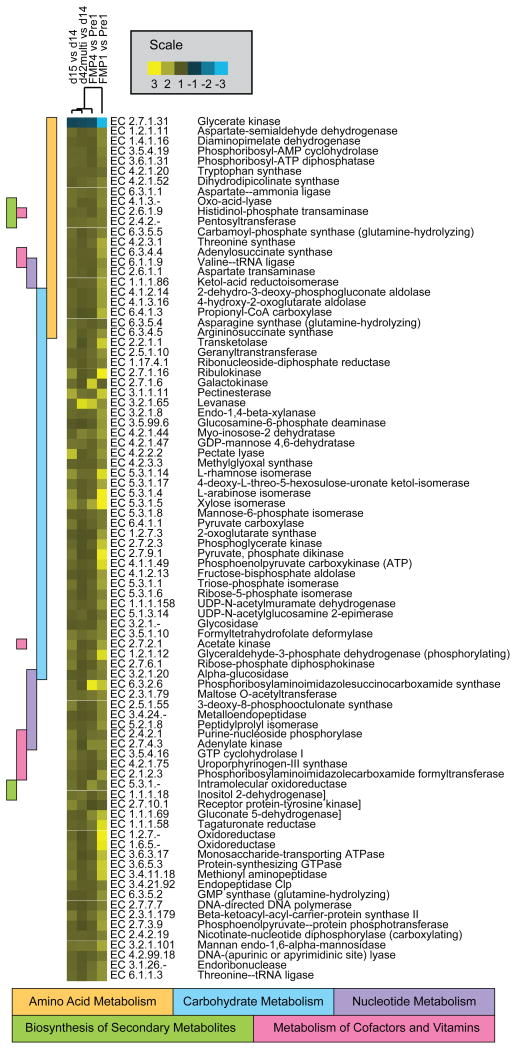
Fig. 7 highlights the 86 ECs that were significantly changed (p<0.01) in the same direction in all humans and in all sampled mice as a function of exposure to the FMP strain consortium. Similar to our findings in mice, the most prominently represented KEGG category among up-regulated gene functions in all comparisons of human metatranscriptomes was ‘carbohydrate metabolism’ (Fig. 7). The three ECs involved in entry points in the KEGG ‘starch and sucrose metabolism’ pathway [levanase (EC 3.2.1.65); pectinesterase (EC 3.1.1.11), and cellobiose phosphorylase (EC 2.4.1.20)] were significantly upregulated within 1 week after FMP consumption was initiated in the humans surveyed. This transcriptional response was sustained in the case of levanase and pectinesterase and ceased (fell to below the limits of detection) within 4 weeks after FMP administration was stopped (Fig. 5A).
Figure 7. Shared transcriptional responses to FMP strain exposure in mice and humans.
The heatmap shows ECs that exhibit a statistically significant change in their expression (ShotgunFunctionalizeR, adjusted p < 0.01) and manifest a consistent direction of change in their expression in all four comparisons shown. Comparisons include those where the pre-treatment timepoint was compared with a timepoint shortly after FMP strains were introduced (mouse: ‘d15 vs d14’, human: ‘FMP1 vs Pre1’) and those where the pre-treatment period was compared to a timepoint several weeks after strain introduction (mouse: ‘d42multi vs d14’, human: ‘FMP4 vs Pre1’). ‘d42multi’ indicates the multiple-treatment group at day 42 of the mouse experiment. The colored boxes correspond to the KEGG categories that contain the ECs shown to the right of the heatmap. The scale refers to fold-difference in the mean of relative abundance of each EC between treatment and pre-treatment groups based on the mean number of normalized reads (RPKM) of transcripts assigned to a given EC. The 18 ECs shown at the bottom of the Figure are not associated with the five prominent KEGG categories listed. Their assigned categories are provided in Table S13.
ECs involved in succinate and propionate metabolism (EC 2.7.2.1 and EC 6.4.1.3) were also upregulated in the human fecal metatranscriptome within 1 week of the initiation of FMP consumption (FMP1 versus Pre1, Fig. 7). As with levanase, pectinesterase and cellobiose phosphorylase, this response was sustained during, and subsided after the period of FMP consumption (see ‘FMP4 versus Pre1’ and ‘FMP1 versus Pre1’ in Fig. 7 and Table S13).
Human fecal transcripts were detected that mapped to the B. animalis subsp. lactis genome (see Supplementary Material). The presence of these transcripts was limited to the period of FMP consumption, supporting the notion that they emanated from the FMP strain rather than from a related species present within the microbiota (Fig. S9). This clear linkage to FMP consumption was not evident in the case of other members of the consortium, so we could not confidently analyze their patterns of gene expression in vivo. The highest number of mapped reads to the B. animalis subsp. lactis genome was obtained 1 week after FMP administration began: among the 4,000 reads, we were able to detect transcripts from all but 1 of the 10 genes in the BALAC2494_00604-BALAC2494_00614 locus that encodes enzymes involved in the catabolism of xylo-oligosaccharides, leading us to conclude that this locus is highly expressed in the distal human gut, just as it is in our mouse model.
DISCUSSION
Repeated sampling of seven healthy MZ adult twin pairs over a 4-month period emphasized that intrapersonal variation in bacterial community structure was less than interpersonal variation, with co-twins having significantly more similar phylogenetic and taxonomic structure in their fecal microbiota compared to those from unrelated individuals (9, 25, 26). The results also showed that (i) consumption of a fermented milk product containing 5 bacterial strains was not associated with a statistically significant change in the proportional representation of resident community members within and between individuals; (ii) the appearance and disappearance of strains comprising the FMP consortium did not exhibit familial patterns in the fecal microbiota; and (iii) B. animalis subsp. lactis CNCM I-2494 was the most prominent assayed member of the consortium represented in the microbiota during the 7 week period of FMP consumption. Analyses of the fecal gene repertoire over the course of the 16 weeks of the experiment indicated that (i) variations in the functional features of the (fecal) microbiome were less than the variations in bacterial species composition; (ii) there was no significant difference in the degree of similarity in representation of KEGG orthology group functions for a given co-twin at each time point compared to the degree of similarity that existed between co-twins, while individual and twin pair microbiomes were significantly more similar to one another than those from unrelated individuals; and (iii) there were no statistically significant changes in the representation of these functions when the FMP strain consortium was being consumed. With these findings in mind, and with each individual as well as each genetically identical co-twin serving as a control, we concluded that at least at the depth and frequency of sampling employed for this small healthy cohort, the bacterial species and gene content of their fecal microbiota/microbiome was not an informative biomarker for understanding whether or how this commercial fermented milk product impacted microbial community properties.
Gnotobiotic mice harboring a model 15-member gut microbial community that represented the three principal bacterial phyla present in the human gut microbiota, and whose 58,399 known or predicted protein-coding genes encompassed many of the prominent functions present in the normal adult human fecal microbiome, provided a means for characterizing the impact of the 5-member FMP strain consortium on expressed gut microbial community functions, and then applying the results to the human fecal specimens collected for this study. As with the MZ twins, introduction of the 5-member strain consortium did not significantly affect the representation of the 15 species comprising the model human microbiota. As with the MZ twins, B. animalis subsp. lactis exhibited the greatest fitness of the five FMP strains in the gut, as judged by its prominence and persistence. Unlike the human arm of the study, where all subjects consumed the FMP twice daily, the design of the mouse study, with its single versus multiple treatment regimens, allowed us to directly compare the persistence of FMP consortium members. Only B. animalis subsp. lactis and L. lactis subsp. cremoris were able to maintain a foothold in the gut ecosystem at detectable levels for the entire 4 week monitoring period after a single dose. In addition, colonization levels were not affected by the number of times the FMP strains were administered to mice.
An advantage of constructing the model human gut microbiome was that its entire predicted gene repertoire was known. This allowed us to define the impact of introducing the FMP strain consortium on the functions expressed by the overall community as well as by its individual components. A major theme emanating from our analysis was the effect of introducing the FMP consortium on carbohydrate metabolism by the community, as well as the effect of the community on a feature of carbohydrate metabolism by B. animalis subsp. lactis. The model 15-member community responded to the FMP consortium by inducing genes encoding enzymes involved in catalyzing reactions that represent the three entry points into the KEGG ‘starch and sucrose metabolic pathway’, as well as enzymes that catalyze fermentation of carbohydrates to propionate. The mechanism by which the FMP strains elicit this response is unclear at present, but the effect is rapid (occurring within the first 24h after invasion) and was persistent whether the consortium was introduced in a single set of gavages during a 1-day period, or with subsequent repeated gavage over a several week period. The persistence of both the carbohydrate pathway response, and of B. animalis subsp. lactis, suggests but does not prove that the latter may be instrumental in instigating and maintaining the former.
Intriguingly, the carbohydrate response showed features of ‘differentiation’. As noted in Results, the levanase response was driven almost entirely by changes in transcription in just a single species (B. vulgatus), the pectinesterase response by 6 community members (B. caccae, B. ovatus, B. thetaiotaomicron, B. vulgatus, B. WH2, C. aerofaciens) and the cellobiose phosphorylase response by three components of the defined model human gut microbiota (B. uniformis, E. rectale, and R. obeum). Of the 50 genes with predicted xylan-degrading capacity in the model microbiome (i.e. those encoding enzymes in ECs 3.2.1.37 and 3.2.1.8), only BACOVA_04387 and BACOVA_04390 (both from B. ovatus) were significantly upregulated after FMP strain introduction (this is ignoring xylanase genes encoded by FMP strains like B. animalis subsp. lactis). This upregulation in a limited subset of the model community coincides with an increase in urinary xylose.
The ability to attribute EC-level changes to individual genes in specific bacterial species was not possible with our RNA-Seq analysis of the human fecal samples. The differentiation of carbohydrate responses among bacterial species documented in gnotobiotic mice emphasizes a challenge and opportunity that can be addressed in these models: namely, to further delineate the niches, interactions and adaptive resource switching behaviors of community members by intentional addition, removal or substitution of taxa, and/or by their modification through genetic manipulation. Although requiring significantly more animals and loss of the ability to use an animal as its own control, future studies could be expanded to include sampling of community gene expression in different segments of the small intestine.
The increased expression of genes encoding enzymes involved in the interconversion of propionate and succinate is intriguing given the fact that this short chain fatty acid has been linked in some reports to effects on gastrointestinal transit time. However, work in this area has yielded varying results and conclusions, perhaps because of the diversity of models and methodological approaches used (27–30). Propionate may also link the gut microbiota and human physiology through its effects on hepatic and adipose tissue metabolism (31). Notably, another group has reported that in the T-bet−/−Rag2−/− mouse model of colitis, consumption of a fermented milk product containing a dairy matrix plus the same strains used in this study led to increased cecal propionate levels and a reduction in intestinal inflammation (32).
The extent of translatability of data from gnotobiotic mouse models harboring collections of sequenced representatives of the human gut microbiota to humans themselves needs to be tested further, not only at the transcriptional level but also at the level of community-host co-metabolism. Although current models can and should be evolved to embrace more of the diversity present in our gut communities, even with current limitations they can serve as part of a pre-clinical discovery pipeline designed to identify candidate biomarkers and mediators of the effects of existing or new probiotic strains on the properties of microbial communities and their hosts. They also represent an analytical tool for characterizing the effects of specified dietary components on the indigenous gut community and on probiotic species that are deliberately consumed. The results could yield new candidate prebiotics that may impact the representation and metabolic properties of probiotic species or entrenched members of our gut microbiota and provide the proof-of-mechanism and proof-of-principle observations needed to justify, direct and interpret human studies.
MATERIALS AND METHODS
Human studies
Subject recruitment
Seven MZ female twin pairs aged 21–32 years with BMIs ranging from 20–25 kg/m2 were recruited for this study. These twins were long-standing participants in the Missouri Adolescent Female Twin Study (MOAFTS; (26, 33)). Procedures for obtaining consent, for providing fecal samples, and for maintaining diaries of FMP consumption, and stool frequency and consistency were approved by the Washington University Human Studies Committee.
Other procedures
Methods used for the production and distribution of the FMP to study participants, analysis of the effects of FMP consumption on stool parameters, qPCR analysis of fecal levels of FMP strains, multiplex pyrosequencing of 16S rRNA genes in fecal samples and the FMP, co-occurrence analysis, and shotgun sequencing of human fecal microbiomes are described in the Methods section of Supplementary Material.
Studies in gnotobiotic mice
Colonization of germ-free mice
The justification for using mice and the protocols employed for treating them were approved by the Washington University Animal Studies Committee. Animals belonging to the C57Bl/6J inbred strain were maintained in plastic flexible film gnotobiotic isolators, and fed a standard autoclaved chow diet (B&K rat and mouse autoclavable chow #7378000, Zeigler Bros, Inc) ad libitum. Two groups of 6–8 week-old germ-free male animals (n=5/group) were colonized with a single gavage of 500 μl of supplemented TYG medium (TYGs; (34)) containing 15 sequenced human gut-derived bacterial symbionts (6×106 cfu/strain; total of 9×107 cfu for the community). The B. thetaiotaomicron component of this community was composed of a library of 34,544 transposon mutants prepared as described (34). Fourteen and fifteen days later, both groups of mice were gavaged with a mixture of the five FMP strains (each species at 5 × 106 cfu) in 300 ul of TYGs. One group of mice received a second pair of gavages 7d and 8d later, and a third pair of gavages 21d and 22d after the initial FMP strain introduction.
Other procedures
Methods used for sampling animals, COPRO-Seq, INSeq, microbial RNA-Seq and non-targeted metabolomics via gas chromatography/mass spectrometry (GC/MS) are described in the Methods section of Supplementary Material, as are methods for sequencing and annotating FMP strain genomes.
Supplementary Material
Figure S1. Levels of B. animalis subsp. lactis (CNCM I-2494) in human fecal samples collected prior to, during, and after consumption of a FMP.
Figure S2. KEGG pathway coverage ratios suggest that the model human gut microbiome encodes many of the functions present in more complex human fecal communities.
Figure S3. CAZyme profiles of the 20 bacterial strains introduced into gnotobiotic mice.
Figure S4. Summary of analysis pipelines utilized in this study. (A) COPRO-Seq. (B) RNA-Seq.
Figure S5. COPRO-Seq-based time series analysis of the abundance of members of the model human microbiota and of the FMP strain consortium in the feces of gnotobiotic mice.
Figure S6. Top-down analysis of the model community’s transcriptional response to the FMP strain consortium reveals upregulation of genes involved in interconversion of propionate and succinate.
Figure S7. A species’ contribution to the metatranscriptome is not necessarily proportional to its abundance in the 15-member community.
Figure S8. Bottom-up analysis of genes whose expression changes significantly following introduction of the FMP strain consortium.
Figure S9. The number of RNA-Seq reads obtained from human fecal samples that map to genomes in the FMP strain consortium peaks shortly after FMP consumption begins.
Table S1. Characteristics of adult female monozygotic (MZ) twins enrolled in study.
Table S2. Summary of human fecal metagenomic datasets.
Table S3. Features of the microbial genomes in the 5-member FMP strain consortium and the 15-member model human gut microbiota.
Table S4. Carbohydrate active enzyme (CAZy) annotation data.
Table S5. COPRO-Seq analysis of bacterial species abundance in mouse fecal samples.
Table S6. INSeq analysis.
Table S7. Differentially expressed B. animalis subsp. lactis (CNCM I-2494) genes.
Table S8. Top-down function-level analysis of the impact of the FMP strain consortium on the model human gut microbiota’s metatranscriptome.
Table S9. Model human gut microbiota membrane transport genes demonstrating ≥4-fold increases or decreases in their expression following introduction of the FMP strain consortium.
Table S10. Bottom-up (gene-level) analysis of the impact of the FMP strain consortium on the model community’s metatranscriptome.
Table S11. Results of Random Forests supervised classification analysis.
Table S12. Urine metabolites whose levels change significantly in transitions between colonization states.
Table S13. ShotgunFunctionalizeR analysis of EC-level changes in the metatranscriptome as a function of FMP strain introduction into mice and humans.
Table S14. Primers and amplification conditions used for quantitative PCR assays of FMP consortium strains in fecal DNA.
Table S15. List of 136 microbial genomes used to analyze human fecal RNA-Seq data.
Overline: Human Genomics.
Single Sentence Summary
Metagenomic, metatranscriptomic, and metabolomic studies of gnotobiotic mice containing a model human gut microbiome provide insights about the effects of consuming a five member microbial consortium, found in a popular commercial fermented dairy product, on the gut communities of healthy human monozygotic twins.
300-word accessible Summary
With increasing regulatory pressure to validate the composition and health claims of probiotics and ‘functional’ foods, it is particularly important to develop informative, relevant animal models so that proof-of-concept and proof-of-mechanism studies can be used to direct and interpret studies of their effects on humans. The present study demonstrates one type of approach.
We studied seven healthy adult female identical twin pairs before they consumed, while they were consuming, and after they stopped consuming a widely used commercial fermented milk product (FMP) that contains a microbial consortium of five bacterial strains. The bacterial and gene composition, as well as the gene expression patterns of their gut microbial communities were defined. The results were compared to those obtained in mice reared under conditions where the only microbes they harbored were 15 prominent, sequenced human gut bacterial symbionts. The five bacterial strains present in the FMP were then given orally to these ‘humanized’ mice at a dose analogous to that given to the twins. We found that:
Repeated sampling of the twins over a 4-month period showed that the species and gene content of their gut bacterial communities remained stable and was not appreciably perturbed by consuming the FMP.
Exposure of the humanized mice to the five FMP strains did not produce marked changes in the proportional representation of their human symbiotic bacterial species or genes, mirroring the results seen in the twins.
A subset of FMP strains successfully colonized the guts of mice harboring the model human microbiome, allowing us to characterize the lifestyle of the most prominently represented of these strains, Bifidobacterium animalis subsp. lactis.
Analysis of gut bacterial gene expression profiles and of urinary metabolites in the mice disclosed that introducing the FMP strains results in significant changes in many metabolic pathways, most prominently those related to carbohydrate processing.
Many of the changes in metabolism first detected in mice were also found to take place in humans.
Mice containing a sequenced model human gut microbiome can serve as part of a pre-clinical discovery pipeline designed to identify the effects of existing or new probiotic species on the properties of the gut microbiome and human host.
Acknowledgments
We thank Jill Manchester, Jessica Hoisington-López for assistance with DNA sequencing, Maria Karlsson, David O’Donnell and Sabrina Wagoner for help with gnotobiotic mouse husbandry, Su Deng for assistance in preparing Illumina DNA libraries, Stacy Marion and Deborah Hooper for their contributions to the human study, Deanna Carlsen for coordination of FMP production and logistics, Stephan Baumann and Steven Fischer (Agilent Corp) for kindly providing the Fiehn GC/MS Metabolomics RTL library used for metabolomics analyses, members of the Gordon lab for valuable suggestions during the course of this work, and Gerard Denariaz for his continued support. We are also grateful to Integrated Genomics for generating the draft genome sequences of B. animalis subsp. lactis (CNCM I-2494) and S. thermophilus (CNCM I-1630).
Funding: Supported by grants from the NIH (DK30292, DK70977) and Danone Research. Maintenance of the MOAFTS twin cohort is supported by NIH grants AA09022, AA11998, AA17915 and HD49024. Author contributions: N.P.M., T.Y., A.C.H., and J.I.G. designed experiments; N.P.M., A.H., J.J.F., A.L.G., J.R.B., and M.J.M. performed experiments involving mice while T.Y., J.J.F., R.O., S.C-P., and G.G. conducted analyses of human biospecimens; N.P.M., T.Y., A.H., B.D.M., A.L.G., B.H., C.C., D.K., C.A.L., R.K., A.E.D., and C.B.N. analyzed the data; N.P.M., A.H., T.Y., and J.I.G. wrote the paper.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Accession numbers: The complete genome sequence of Bifidobacterium animalis subsp. lactis (CNCM I-2494) has been deposited at GenBank under accession number CP002915. The Whole Genome Shotgun projects for Lactobacillus delbrueckii subsp. bulgaricus (CNCM I-1632, CNCM I-1519), Streptococcus thermophilis CNCM I-1630, and Lactococcus lactis subsp. cremoris CNCM I-1631 have been deposited at DDBJ/EMBL/GenBank under accession numbers AGFO00000000, AGHW00000000, AGFN00000000, and AGHX00000000, respectively. The version of each of these genomes described in this paper is the first version (CP002915.1, AGFO01000000, AGHW01000000, AGFN01000000, and AGHX01000000, respectively). COPRO-Seq and RNA-Seq data are deposited in GEO (accession numbers GSE31943 and GSE31670, respectively), while 16S rRNA pyrosequencing reads and shotgun pyrosequencing reads of human fecal community DNA are deposited in MG-RAST (accession numbers qiime:803 and 4473933-4473980, respectively).
References
- 1.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Jian M, Zhou Y, Li Y, Zhang X, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Bork P, Ehrlich SD. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol. 2002;68:219. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M’Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faith JJ, Rey FE, O’Donnell D, Karlsson M, McNulty NP, Kallstrom G, Goodman AL, Gordon JI. Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 2010;4:1094. doi: 10.1038/ismej.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone C, Hamady M, Knight R. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107:7503. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilad O, Jacobsen S, Stuer-Lauridsen B, Pedersen MB, Garrigues C, Svensson B. Combined transcriptome and proteome analysis of Bifidobacterium animalis subsp. lactis BB-12 grown on xylo-oligosaccharides and a model of their utilization. Appl Environ Microbiol. 2010;76:7285. doi: 10.1128/AEM.00738-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu CK, Liao JW, Chung YC, Hsieh CP, Chan YC. Xylooligosaccharides and fructooligosaccharides affect the intestinal microbiota and precancerous colonic lesion development in rats. J Nutr. 2004;134:1523. doi: 10.1093/jn/134.6.1523. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki M, Fujikawa S, Matsumoto N. Effect of xylooligosaccharide on the growth of bifidobacteria. Bifidobacteria and Microflora. 1990;9:77. [Google Scholar]
- 21.Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- 22.R. D. C. Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. [Google Scholar]
- 23.Knights D, Costello EK, Knight R. Supervised classification of human microbiota. FEMS Microbiol Rev. 2011;35:343. doi: 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 24.Stein SE. An Integrated Method for Spectrum Extraction and Compound Identification from Gas Chromatography/Mass Spectrometry Data. J Am Soc Mass Spectrom. 1999;10:770. [Google Scholar]
- 25.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature. 2009;457:480. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherbut C, Ferrier L, Roze C, Anini Y, Blottiere H, Lecannu G, Galmiche JP. Short-chain fatty acids modify colonic motility through nerves and polypeptide YY release in the rat. Am J Physiol. 1998;275:G1415. doi: 10.1152/ajpgi.1998.275.6.G1415. [DOI] [PubMed] [Google Scholar]
- 28.Kamath PS, Hoepfner MT, Phillips SF. Short-chain fatty acids stimulate motility of the canine ileum. Am J Physiol. 1987;253:G427. doi: 10.1152/ajpgi.1987.253.4.G427. [DOI] [PubMed] [Google Scholar]
- 29.Marteau P, Cuillerier E, Meance S, Gerhardt MF, Myara A, Bouvier M, Bouley C, Tondu F, Bommelaer G, Grimaud JC. Bifidobacterium animalis strain DN-173 010 shortens the colonic transit time in healthy women: a double-blind, randomized, controlled study. Aliment Pharmacol Ther. 2002;16:587. doi: 10.1046/j.1365-2036.2002.01188.x. [DOI] [PubMed] [Google Scholar]
- 30.Squires PE, Rumsey RD, Edwards CA, Read NW. Effect of short-chain fatty acids on contractile activity and fluid flow in rat colon in vitro. Am J Physiol. 1992;262:G813. doi: 10.1152/ajpgi.1992.262.5.G813. [DOI] [PubMed] [Google Scholar]
- 31.Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. 2010;1801:1175. doi: 10.1016/j.bbalip.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Veiga P, Gallini CA, Beal C, Michaud M, Delaney ML, DuBois A, Khlebnikov A, van Hylckama Vlieg JE, Punit S, Glickman JN, Onderdonk A, Glimcher LH, Garrett WS. Bifidobacterium animalis subsp. lactis fermented milk product reduces inflammation by altering a niche for colitogenic microbes. Proc Natl Acad Sci U S A. 2010;107:18132. doi: 10.1073/pnas.1011737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath AC, Howells W, Bucholz KK, Glowinski AL, Nelson EC, Madden PA. Ascertainment of a mid-western US female adolescent twin cohort for alcohol studies: assessment of sample representativeness using birth record data. Twin Res. 2002;5:107. doi: 10.1375/1369052022974. [DOI] [PubMed] [Google Scholar]
- 34.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Levels of B. animalis subsp. lactis (CNCM I-2494) in human fecal samples collected prior to, during, and after consumption of a FMP.
Figure S2. KEGG pathway coverage ratios suggest that the model human gut microbiome encodes many of the functions present in more complex human fecal communities.
Figure S3. CAZyme profiles of the 20 bacterial strains introduced into gnotobiotic mice.
Figure S4. Summary of analysis pipelines utilized in this study. (A) COPRO-Seq. (B) RNA-Seq.
Figure S5. COPRO-Seq-based time series analysis of the abundance of members of the model human microbiota and of the FMP strain consortium in the feces of gnotobiotic mice.
Figure S6. Top-down analysis of the model community’s transcriptional response to the FMP strain consortium reveals upregulation of genes involved in interconversion of propionate and succinate.
Figure S7. A species’ contribution to the metatranscriptome is not necessarily proportional to its abundance in the 15-member community.
Figure S8. Bottom-up analysis of genes whose expression changes significantly following introduction of the FMP strain consortium.
Figure S9. The number of RNA-Seq reads obtained from human fecal samples that map to genomes in the FMP strain consortium peaks shortly after FMP consumption begins.
Table S1. Characteristics of adult female monozygotic (MZ) twins enrolled in study.
Table S2. Summary of human fecal metagenomic datasets.
Table S3. Features of the microbial genomes in the 5-member FMP strain consortium and the 15-member model human gut microbiota.
Table S4. Carbohydrate active enzyme (CAZy) annotation data.
Table S5. COPRO-Seq analysis of bacterial species abundance in mouse fecal samples.
Table S6. INSeq analysis.
Table S7. Differentially expressed B. animalis subsp. lactis (CNCM I-2494) genes.
Table S8. Top-down function-level analysis of the impact of the FMP strain consortium on the model human gut microbiota’s metatranscriptome.
Table S9. Model human gut microbiota membrane transport genes demonstrating ≥4-fold increases or decreases in their expression following introduction of the FMP strain consortium.
Table S10. Bottom-up (gene-level) analysis of the impact of the FMP strain consortium on the model community’s metatranscriptome.
Table S11. Results of Random Forests supervised classification analysis.
Table S12. Urine metabolites whose levels change significantly in transitions between colonization states.
Table S13. ShotgunFunctionalizeR analysis of EC-level changes in the metatranscriptome as a function of FMP strain introduction into mice and humans.
Table S14. Primers and amplification conditions used for quantitative PCR assays of FMP consortium strains in fecal DNA.
Table S15. List of 136 microbial genomes used to analyze human fecal RNA-Seq data.