Abstract
Maintenance and manipulation of large DNA and RNA virus genomes had presented an obstacle for virological research. BAC vectors provided a solution to both problems as they can harbor large DNA sequences and can efficiently be modified using well-established mutagenesis techniques in Escherichia coli. Numerous DNA virus genomes of herpesvirus and pox virus were cloned into mini-F vectors. In addition, several reverse genetic systems for RNA viruses such as members of Coronaviridae and Flaviviridae could be established based on BAC constructs. Transfection into susceptible eukaryotic cells of virus DNA cloned as a BAC allows reconstitution of recombinant viruses. In this paper, we provide an overview on the strategies that can be used for the generation of virus BAC vectors and also on systems that are currently available for various virus species. Furthermore, we address common mutagenesis techniques that allow modification of BACs from single-nucleotide substitutions to deletion of viral genes or insertion of foreign sequences. Finally, we review the reconstitution of viruses from BAC vectors and the removal of the bacterial sequences from the virus genome during this process.
1. Introduction
For many years, genetic manipulation of the genomes of large DNA viruses was extremely tedious and dependent on rare recombination events in susceptible eukaryotic cells. Infected cells were transfected with plasmids or linear DNA fragments containing a selection marker flanked by sequences homologous to the target locus. This process allowed the deletion or mutation of the gene of interest in the virus genome. However, purification of resulting recombinant viruses was laborious and often required several passages. Deletion of essential viral genes was usually not feasible due to the strong selection for progeny viruses that efficiently replicate in vitro. Constant selective pressure during serial virus passage often resulted in compensatory mutations in the viral genome. To overcome the obstacle of essentiality, transcomplementing cell lines were generated that would compensate for the absence of the gene in the virus context. This solution would work in most cases but was not always possible due to the toxicity of some viral proteins. A solution to the problems was the maintenance and modification of virus genomes in bacteria where the accuracy of the bacterial polymerase allows clonal maintenance of viral sequences in E. coli. As viral promoters are not functional in bacteria, there is no selective pressure on the virus genome in E. coli. In rare cases, high-copy plasmids containing virus sequences can be toxic for bacteria; however, this problem could also be overcome by the use of single or low-copy vectors, such as bacterial artificial chromosomes (BACs). Another advantage of BAC replicons is the high capacity of up to 300 kbp that is required for the cloning of large DNA and RNA virus genomes. Maintenance and faithful replication of the BAC construct in E. coli are facilitated by components encoded in the minimal fertility factor replicon (mini-F), the backbone of the BAC vector. Replication of the BAC is initiated at the origin of replication S (oriS) and stringently regulated by the repE and repF gene products encoded in the mini-F. Resulting copies of the replicon are subsequently allocated to the two daughter cells. This dynamic process is facilitated by the mini-F-encoded proteins SopA and SopB and the centromere region sopC [1]. Bacterial antibiotic resistance cassettes present in the BAC vector backbone allow the selection in E. coli. Besides the essential sequences required for replication and maintenance, many BAC vectors harbor a variety of expression cassettes that allow visualization of BAC-containing sequences in transfected cells selection in mammalian cells and, hence, ease the generation of recombinant viruses that contain the mini-F.
The major advantage of the maintenance of virus genomes in BAC vectors is the availability of well-established mutagenesis techniques in E. coli. Over the last decade, a number of methods have been developed that allow the generation of virtually any mutation in a virus genome. Within a few days, genes or sequence elements can be removed. Foreign sequences can readily be inserted into the genome, allowing detection of viral proteins via epitope tags, or expression of foreign genes for development of vector vaccines [2–7]. Furthermore, reporter genes such as the green fluorescent protein (GFP) can be incorporated into the viral genome to visualize infected cells in vitro and in vivo or fused to a viral protein to determine its localization in living cells [3, 6, 8]. In addition, luciferase reporter genes can be inserted which allows tracking of the virus in live animals [9, 10]. Even single nucleotides can be edited in the virus genome, a modification that is almost impossible with conventional virus mutagenesis in mammalian cells.
Until now, BACs have been generated for a large number of DNA but also some RNA viruses. In this paper we provide an overview on strategies for the generation of virus BAC clones for both DNA and RNA viruses. Furthermore, we review the viral BAC systems that are currently available to the research community. As modification of virus genomes is an important tool in virological research, we outline the available mutagenesis techniques for viral BAC vectors. Last but not least, we describe various techniques that can be used for the removal of the mini-F from viral genomes upon reconstitution in eukaryotic cells.
2. Generation of Bacterial Artificial Chromosomes (BACs)
2.1. Homologous Recombination in Mammalian Cells
One of the most common methods for the insertion of the mini-F vector into the genomes of DNA viruses utilizes the recombination machinery of mammalian cells. For this approach, a transfer vector is generated that harbors the mini-F cassette flanked by sequences identical to the insertion site in the virus genome. The choice of the mini-F insertion site is critical as essential genes may not be affected in the process [11]. Circular or linearized transfer plasmid is either transfected into virus-infected cells or is co-transfected with virus genomic DNA. In a small portion of transfected cells, a recombination event facilitated by cellular recombinases allows the insertion of the mini-F sequences into the virus genome. Upon virus reconstitution, cells producing recombinant viruses can be visualized by fluorescent markers such as green fluorescent protein (GFP), which is often introduced in standard mini-F plasmids. Alternatively, cells infected with recombinant viruses can be enriched using selection markers such as gpt, hygromycin, or neomycin that are encoded by resistance genes present in the mini-F vector. Upon purification of recombinant viruses harboring mini-F vector sequences, susceptible cells are infected and circular virus DNA is isolated. In many cases, viral DNA is prepared using a method previously published by Hirt (1967), which uses fractionated precipitation with SDS and NaCl to separate viral from cellular DNA [12]. Residual cellular DNA can be fragmented using restriction enzymes that do not cut in the virus genome. Resulting linearized DNA may be removed by incubation with λ-exonuclease, an enzyme that exclusively digests linear DNA, leaving circular DNA intact [13]. This approach allows an enrichment of circular virus DNA. Isolated viral DNA harboring the mini-F is then electroporated into E. coli K12 strains, and BAC-containing bacteria are selected using antibiotic resistance genes present in the mini-F backbone [14]. Resulting BAC clones are routinely analyzed by restriction fragment length polymorphism (RFLP) to ensure the integrity of the virus genome. In addition, the insertion site of the mini-F is sequenced to determine if mutations that are present may have occurred in the recombination process. Recently, sequencing of entire BAC clones has become an attractive alternative, as the cost of high-throughput sequencing has dropped dramatically.
2.2. Cosmid-Based Approach
An alternative strategy often used for BAC generation of cell-associated viruses utilizes cosmid vectors to initially maintain overlapping parts of the genome of DNA viruses. The mini-F is subsequently inserted into one of the cosmids by ligation or homologous recombination in E. coli. Transfection of the overlapping cosmids into eukaryotic cells results in recombination between homologous sequences and reconstitution of infectious virus. During the process, the cosmid containing the mini-F cassette is incorporated into the virus genome, all resulting viruses harbor the mini-F, and no laborious selection steps to obtain recombinant clones are required. As described above, circular virus DNA is then isolated and transformed into E. coli and clones are screened for the integrity of the virus genomes they contain [13].
2.3. In Vitro Ligation
Recently, it has been shown that the mini-F replicon can be inserted into herpesvirus genomes by direct ligation [29]. For this purpose, concatemeric virus DNA is isolated from herpesvirus infected-cells and cleaved with a restriction enzyme that cuts only in a single locus within the virus genome. The resulting full-length viral genome is then ligated with a linearized mini-F vector containing compatible DNA ends. To avoid ligation of the mini-F with cellular fragments, restriction enzymes that recognize an interrupted palindrome and allow the generation of desired directional sticky ends such as SfiI or BstXI can be used. This strategy has been successfully applied to the generation of a BAC system for human herpesvirus 6A (HHV-6A) [29]. There are, however, several disadvantages to this method. Firstly, the strategy requires a completely sequenced virus genome in order to determine potential restriction sites that can be used for the ligation procedure. Secondly, many virus genomes do not possess a unique restriction site that is suitable for the strategy. Thirdly, the mini-F insertion site is limited to the location of the unique restriction site. Insertion into open reading frames (ORFs) or promoters of the virus genome can impair or abrogate infectivity of BAC-derived viruses. Last but not least, ligation and transformation procedures for large BAC vectors are very inefficient, therefore hampering cloning attempts.
2.4. Strategy for Poxviruses
As described in Section 2.1, insertion of the mini-F sequences into the poxvirus genome can be facilitated by the cellular recombination machinery in mammalian cells [40–43]. However, unlike herpesviruses, poxviruses do not produce a circular form of the virus genome during replication. This poses a major hurdle for the transfer of the recombinant poxvirus constructs into E. coli. To overcome the problem, infected cells are treated with isatin-β-thiosemicarbazone that promotes accumulation of nonresolved, genomic concatemers [40–44]. For the generation of some poxvirus BAC clones, it was sufficient to transform E. coli with concatemeric DNA, a procedure that likely resulted in a recombination event allowing circularization of the replicon [40, 43]. Alternatively, isolated poxvirus DNA was circularized prior to transformation of E. coli using the Cre/loxP- or Flp/FRT-recombination system that will be further described in Section 4.3 [41, 42].
2.5. Generation of BACs for RNA Viruses
BAC vectors can also be used to maintain and modify the genome of RNA viruses. This has been successfully shown for several members of the Coronaviridae and Flaviviridae [45–50]. To generate a full-length BAC clone for these nonsegmented RNA viruses, viral RNA is isolated from infected cells and cDNA synthesized using reverse transcriptase. Full-length genomic cDNA can be purified by gel electrophoresis and subsequently cloned into a mini-F vector by a standard ligation reaction. If synthesis of a full-length cDNA is not possible due to the size of the virus genome, fragments of the genome can be transcribed into cDNA and subsequently combined by ligation to obtain a complete virus genome [51]: similar to the situation for DNA viruses, some RNA viruses can be reconstituted by transfection of (cloned) viral nucleic acid into eukaryotic cells. Expression of viral genomic RNA can be facilitated by cellular RNA polymerase II under the control of the major immediate-early promoter of human cytomegalovirus (HCMV) [45, 46]. Alternatively, a T7 promoter can be employed to drive the expression of viral RNA, where T7 polymerase is either stably expressed in the cell line used for virus reconstitution or delivered in trans by transfection or using vaccinia virus as a delivery vehicle [52, 53]. Correct processing of the 3′-ends of the virus genomes is often achieved by the bovine growth hormone (BGH) terminator and sequences of the hepatitis delta virus (HDV) ribozyme [45, 46, 49]. Full-length viral RNA can also be transcribed in vitro using a T7 promoter/polymerase system with the virus BAC clone as a template. Transfection into susceptible cells of the viral RNA transcribed in vitro usually allows virus reconstitution [51].
Currently, several laboratories are working on the generation of BAC systems for segmented RNA viruses. For this approach, cDNA clones of viral RNA segments are combined into a BAC vector. Alternatively, segments of the virus genome can be generated by de novo synthesis [54]. Transcription of genomic RNA of segmented viruses utilizes similar promoter/terminator systems as described above for nonsegmented RNA viruses. For example, all eight influenza A virus genome segments were recently cloned into a single high-capacity vector. This system allows a more efficient reconstitution of the virus in eukaryotic cells and could be used for the production of recombinant influenza vaccines [55].
3. BACs Available for Members of Various Herpesvirus Families
3.1. Herpesvirales
Over the last two decades, BAC vectors have become an important tool for herpesvirus research. In 1997, the first virus BAC system was developed by Messerle and colleagues for the murine cytomegalovirus (MCMV), one of the largest herpesviruses with a genome size of 230 kbp [14]. Shortly after the generation of the MCMV BAC, the technology was applied to many other species in the order Herpesvirales. These include numerous members of the Herpesviridae with species from all three subfamilies, Alpha-, Beta- and Gammaherpesvirinae (Table 1). Until now, BAC clones have been generated for all human herpesviruses with the exception of human herpesvirus 7 (HHV-7). Besides the Herpesviridae, two members of the Alloherpesviridae, koi herpesvirus and channel catfish herpesvirus, have been cloned into mini-F vectors [38, 39]. All in all, BAC systems are available for at least 27 herpesvirus species that infect hosts as diverse as fish, birds, and humans (Table 1). For many of these herpesviruses, several strains have been cloned as BACs, allowing the maintenance and manipulation of laboratory strains and clinical isolates with varying virulence. The plethora of BAC-based genetic systems has eased the analysis of herpesvirus-encoded genes and has immensely contributed to our understanding of the viruses' life cycles and pathogenesis.
Table 1.
Overview of published BAC systems for species of the order Herpesvirales. Common species names, corresponding acronyms, taxon names, genome sizes, and references for the first BAC construct of every species are given. *Common names and acronyms are as they were used in the listed reference.
| Virus | Acronym* | Taxon name | Genome size | Reference |
|---|---|---|---|---|
| Common name* | ||||
| Herpesviridae | ||||
| Alphaherpesvirinae | ||||
| Bovine herpesvirus 1 | BoHV-1 | Bovine herpesvirus 1 | 135 kbp | [15] |
| Canine herpesvirus | CHV | Canid herpesvirus 1 | 160 kbp | [16] |
| Equine herpesvirus 1 | EHV-1 | Equid herpesvirus 1 | 150 kbp | [17] |
| Equine herpesvirus 4 | EHV-4 | Equid herpesvirus 4 | 146 kbp | [18] |
| Felid herpesvirus 1 | FeHV-1 | Felid herpesvirus 1 | 136 kbp | [19] |
| Herpes simplex virus 1 | HSV-1 | Human herpesvirus 1 | 152 kbp | [20] |
| Herpes simplex virus 2 | HSV-2 | Human herpesvirus 2 | 155 kbp | [21] |
| Herpesvirus of turkey | HVT | Meleagrid herpesvirus 1 | 160 kbp | [22] |
| Marek's disease virus | MDV | Gallid herpesvirus 2 | 178 kbp | [23] |
| Pseudorabies virus | PRV | Suid herpesvirus 1 | 142 kbp | [24] |
| Simian varicella virus | SVV | Cercopithecine herpesvirus 9 | 125 kbp | [25] |
| Varicella-zoster virus | VZV | Human herpesvirus 3 | 125 kbp | [26] |
| Betaherpesvirinae | ||||
| Guinea pig cytomegalovirus | GPCMV | Caviid herpesvirus 2 | 220 kbp | [27] |
| Human cytomegalovirus | HCMV | Human herpesvirus 5 | 229 kbp | [28] |
| Human herpes virus 6A | HHV-6A | Human herpesvirus 6 | 159 kbp | [29] |
| Mouse cytomegalovirus | MCMV | Murid herpesvirus 1 | 230 kbp | [14] |
| Rhesus cytomegalovirus | RhCMV | Macacine herpesvirus 3 | 221 kbp | [30] |
| Gammaherpesvirinae | ||||
| Bovine herpesvirus 4 | BoHV-4 | Bovine herpesvirus 4 | 171 kbp | [31] |
| Epstein-Barr virus | EBV | Human herpesvirus 4 | 172 kbp | [32] |
| Herpesvirus saimiri | HVS | Saimiriine herpesvirus 2 | 113 kbp | [33] |
| Kaposi's sarcoma-associated herpesvirus | KSHV | Human herpesvirus 8 | 137 kbp | [34] |
| Murine gammaherpesvirus 68 | MHV-68 | Murid herpesvirus 4 | 119 kbp | [8] |
| Rhesus lymphocryptovirus | rhLCV | Macacine herpesvirus 4 | 171 kbp | [35] |
| Rhesus rhadinovirus | RRV | Macacine herpesvirus 5 | 133 kbp | [36] |
| Unassigned | ||||
| Duck enteritis virus | DEV | Anatid herpesvirus 1 | 158 kbp | [37] |
| Alloherpesviridae | ||||
| Channel catfish herpesvirus | CCV | Ictalurid herpesvirus 1 | 134 kbp | [38] |
| Koi Herpesvirus | KHV | Cyprinid herpesvirus 3 | 295 kbp | [39] |
3.2. Poxviridae
So far, several full-length poxvirus genomes of the genus Orthopoxvirus have been cloned into BAC vectors. The first BAC construct was generated for the vaccinia virus strain Western Reserve [41, 56]. After this proof-of-principle for poxvirus BAC generation, two BAC vectors were developed for the highly attenuated modified vaccinia virus Ankara (MVA) and one for its more virulent parental strain, chorioallantois vaccinia virus Ankara (CVA). The MVA and CVA BAC constructs allowed an analysis of the effect of six major deletions present in MVA genome on poxvirus pathogenesis and the differences in cellular tropisms of MVA and CVA [40, 42]. Recently, a BAC clone was established for cowpox virus (CPXV), a zoonotic, rodent-borne poxvirus that has the largest and most complete genome of all orthopoxviruses [43]. The broad spectrum of mutagenesis techniques described in Section 4 facilitated many studies that shed light on the role of viral genes in the poxvirus lifecycle and allowed the establishment of recombinant next-generation vector vaccines [40, 43, 56, 57].
3.3. RNA Viruses
The establishment of stable genetic systems for RNA viruses was, and in some cases still is, one of the major challenges. Cloning of cDNA sequences into expression vectors allows maintenance and manipulation of the RNA virus genomes. However, in case of large nonsegmented RNA viruses, the capacity of regular plasmids is often not sufficient. Furthermore, high-copy vectors containing virus-derived cDNA fragments can be instable or exhibit toxic effects on the bacterial host. To circumvent the obstacles, cDNAs of several nonsegmented viruses were inserted into mini-F vectors. The first BAC of an RNA virus was developed by Almazán and colleagues in 2000 for transmissible gastroenteritis coronavirus (TGEV), a member of the Coronaviridae [45]. The large TGEV genome of almost 29 kbp was transcribed into cDNA and was successfully cloned and efficiently maintained as a BAC construct. In the following years, BAC systems were generated for other coronaviruses such as human coronavirus (HCoV) and SARS-related coronavirus (SARS-CoV) [46, 49]. Similarly, the cDNA sequence of Japanese encephalitis virus (JEV), a member of the Flaviviridae, was cloned into a mini-F vector in 2003 [50]. Since then, BAC-based genetic systems have been generated for the pestiviruses bovine viral diarrhea virus (BVDV) and classical swine fever virus (CSFV) (Table 2) [47, 48].
Table 2.
Overview of published BAC systems for members of the Poxviridae, Flaviviridae, and Coronaviridae family. Common species names, corresponding acronyms, taxon names, genome sizes, and references for the first BAC construct of every species are given.
| Virus | Acronym* | Taxon name | Genome size | Reference |
|---|---|---|---|---|
| Common name* | ||||
| Poxviridae | ||||
| Cowpox virus | CPXV | Cowpox virus | 224 kbp | [43] |
| Modified vaccinia virus Ankara | MVA | Vaccinia virus | 178 kbp | [40] |
| Vaccinia virus | VAC | Vaccinia virus | 195 kbp | [41] |
| Flaviviridae | ||||
| Japanese encephalitis virus | JEV | Japanese encephalitis virus | 11 kbp | [50] |
| Bovine viral diarrhea virus | BVDV | Bovine viral diarrhea virus 1 | 12 kbp | [47] |
| Classical swine fever virus | CSFV | Classical swine fever virus | 12 kbp | [48] |
| Coronaviridae | ||||
| Human coronavirus (OC43) | HCoV | Betacoronavirus 1 | 31 kbp | [49] |
| Severe acute respiratory syndrome coronavirus | SARS-CoV | Severe acute respiratory syndrome-related coronavirus | 30 kbp | [46] |
| Transmissible gastroenteritis coronavirus | TGEV | Alphacoronavirus 1 | 29 kbp | [45] |
*Common names and acronyms are as they were used in the listed reference.
4. BAC Mutagenesis
4.1. Transposon Mutagenesis
One major advantage of virus BACs systems is the availability of well-established genetic tools that allow random and specific modifications of the virus genome in E. coli. One method that allows modification of BACs constructs in a nontargeted fashion is transposon-mediated mutagenesis. Random integration of transposable elements (Tn) into virus BACs can result in the interruption of viral genes and sequence elements. The integration of Tn is mediated by the transposase (tnpA) and resolvase (tnpR) gene products of the transposon system [58, 59]. Transposon vectors were generated which contain all required components for the mutagenesis system. Insertion of various antibiotic resistance cassettes into the Tn sequence allows the selection of BAC clones that harbor a transposon insertion. A temperature-sensitive origin of replication can be utilized for a rapid removal of the transposon vector. To ensure that Tn transposition occurs in the BAC construct and not in the cellular genome, transposons such as Tn1721 have been developed with a strong preference for circular DNA. The optimized transposon system can be used to generate recombinant BAC libraries. The phenotype of mutagenized, BAC-derived viruses can be subsequently analyzed and can be used for genome-wide screens. Such screens are dependent on the available readout systems, but have been performed for viral genes involved in replication, immune evasion, and other processes important for completion of life cycles of viruses [60].
4.2. RecA-Based Mutagenesis
To allow a more detailed characterization of viral genes, targeted sequence modifications are necessary. Due to the large size of the BAC constructs, ligation and transformation procedures are usually very inefficient [61]. To overcome the obstacles, most modifications of BAC constructs are facilitated by homologous recombination techniques in E. coli. The two well-established RecA and Red/RecET recombination systems allow rapid and convenient modifications of BAC constructs [7, 62, 63]. The RecA system utilizes cellular RecA recombinase expressed in bacteria. In order to introduce the desired modification through RecA, long homologous sequences of 500 bp to 3 kbp are required for recombination events [28, 64]. However, as repetitive sequences are present in many viruses such as herpesviruses, RecA expression can lead to rapid destabilization of viral BAC clones. This often results in the loss of portions of the virus genome. For this reason, BACs are usually maintained in RecA-deficient E. coli strains, and the recombinase is only transiently expressed during the mutagenesis procedure [14, 28, 64].
A common RecA-based mutagenesis technique is termed shuttle mutagenesis. The system makes use of vectors that harbor the desired modifications flanked by sequences homologous to the target site in the BAC (Figure 1) [28, 64]. Recombination of homologous sites is facilitated by the RecA that usually is encoded by the shuttle vector [28]. The plasmid regularly harbors a positive and negative selection marker and often a temperature-sensitive origin of replication. The plasmid is then transformed into E. coli containing the desired BAC construct for mutagenesis to proceed, and bacteria harboring the shuttle vector are selected using the positive selection marker. Expression of RecA facilitates recombination of sequences that are identical in the shuttle plasmid and the target site in the BAC construct. Replication of the plasmid can be repressed in a temperature-dependent manner, and enrichment is achieved of bacteria in which the plasmid sequences were integrated into the BAC. In a second recombination event, the backbone of shuttle vector is excised from the BAC construct. This can occur with either homologous flank present in shuttle vector sequences. If the second recombination utilizes the homologous sequence that was not utilized in the first step, then the desired modification is maintained in the BAC vector (Figure 1) [28, 64]. Negative selection markers such as rspL [65], sacB [66], and tetR [67] in the shuttle vector backbone can be used to suppress nonresolved BAC clones.
Figure 1.

Schematic illustration of shuttle mutagenesis. In a first step, a shuttle plasmid is inserted into the target sequence via RecA-mediated recombination of homologous sequences. Replication of the shuttle plasmid containing a temperature sensitive origin (oriTS) is repressed by a temperature increase to 42°C. Positive co-integrates are selected with corresponding antibiotics. In a second step, vector sequences are excised from co-integrates by another recombination. Negative selection markers can be used to select BAC constructs that lost the shuttle plasmid [28, 64]. Dotted lines symbolize recombination events.
One of the major advantages of shuttle mutagenesis is that it can be used for the generation of a wide variety of mutations without leaving any unwanted selection markers or other bacterial sequences behind. Deletions or modification of the target sequence in the BAC can be introduced within a matter of days. Standard shuttle vectors for specific target sites can be generated and rapidly modified to be used for introduction of various mutations into a single locus. Furthermore, the system allows the insertion of short and long sequences that can be utilized for the development of vector vaccines. However, the RecA system has also a number of disadvantages. The major problem poses the instability of the BAC clones upon induction of the recombination system, often resulting in the loss of large portions of the BAC. The frequency of unwanted recombination events is increased when negative selection markers are used. Finally, construction of the shuttle plasmids can be very laborious if several loci in a BAC construct are targeted for mutagenesis.
4.3. Recombineering of BAC Constructs
An alternative system for the modification of BACs in E. coli is the well-established Red and RecE/T recombination systems that are derived from bacteriophage λ and the Rac prophage, respectively [7, 63]. Both systems utilize double-strand DNA (dsDNA) ends as substrate for the recombination reaction and consist of three components [7, 63, 68]. The first component is the Gam protein that protects dsDNA ends from degradation in bacteria [69]. A second component is the 5′–3′ exonuclease, alpha or RecE, that generates single-strand 3′ DNA overhangs despite the presence of Gam [70–72]. The last component is the single-strand binding protein Beta or RecT. The protein can bind and protect single-strand DNA from degradation [73–75]. In addition, Beta/RecT aids in annealing of single-strand DNA to complementary sequences and in invasion into replication forks with a preference for lagging DNA strands [76, 77]. The major advantage of the Red or RecE/T recombination system is that only short homologous sequences of 30 to 50 bp are required for the recombination to proceed. Furthermore, unwanted recombination events or rearrangements occur rarely, as only homologous double-strand ends can be used as a substrate. For the recombination in bacteria, components of the Red or RecE/T recombination system can be delivered in trans by plasmids such as pKD46 that allow inducible expression of Alpha, Beta and Gam. Once the mutagenesis procedure is completed, pKD46 can be cured from bacteria by its temperature-sensitive replication mechanism [7, 78]. A more convenient alternative is the use of bacteria containing a chromosomally encoded λ prophage, such as E. coli strain DY380 and its derivates. In this case Alpha, Beta, and Gam can be induced in a temperature-dependent manner [77, 79].
The Red or RecE/T mutagenesis system usually utilizes PCR products that contain a positive selection marker and sequences homologous to the target site in the BAC at either end as substrate for recombination. The short homologous sequences needed for targeted insertion into the BAC construct can readily be inserted by 5′ overhangs of the primers used for PCR amplification of the selection marker. The PCR products are then electroporated into E. coli that harbor the desired BAC construct. Clones that incorporate the cassette can be selected for the presence of the selection marker. Several mutagenesis techniques have been developed that often combine Red or RecE/T with other recombination systems and strategies that allow the removal of unwanted sequences.
One of these recombination systems that utilize specific recognition sites is the Cre/loxP system of bacteriophage P1. It consists of the Cre recombinase that facilitates recombination between two 34 bp loxP sites [80]. An orthologous system is based on Flp recombinase derived from Saccharomyces cerevisiae that utilizes FRT recognition sites for recombination [81]. The presence of two loxP or FRT sites within a vector results in the excision of sequences flanked by the recognition sites and can be used for the removal of unwanted bacterial sequences or selection markers that were introduced by Red or RecE/T recombination. The Cre/loxP and FLP/FRT system can also be used for insertion of sequences. A recombination event between one loxP or FRT in the BAC construct and the donor sequence allows insertion of the desired sequences [82, 83]. Expression plasmids can be used to transiently express the recombinases; alternatively, a number of E. coli strains, such as EL250 and EL350, that harbor an inducible form of the cre or flp gene in the chromosome are available [7, 79].
Besides recombination at specific recognition sites, other strategies have been developed that allow complete removal of the introduced marker sequences. One approach utilizes a combination of positive and negative selection markers [7]. In a first Red recombination step, the dual selection cassette is inserted into the target site. Resulting clones are selected for the presence of the positive selection marker. In a second step, a PCR product with the desired modification and flanking homologous sequences for the Red recombination results in the replacement of the positive and the negative selection marker. Desired clones can then be enriched by negative selection, but one disadvantage of the system is the low efficiency of counterselection. In addition, mutations in the negative selection marker can result in clones that are resistant to counterselection. Similarly, illegitimate recombination events removing the counterselection marker without the insertion of the sequence modification can result in many false positive clones.
An alternative approach uses two-way selectable markers, such as galactokinase (GalK) [84]. The galK gene can be used as a positive selection marker in E. coli strains such as SW102 that contain the galactose operon but lack cellular galK and, therefore, cannot utilize galactose as a carbon source. In the first recombination step, the galK cassette is introduced into the BAC construct and desired clones are selected on minimal media containing only galactose as an energy source. However, galK can also be used as a negative selection marker as it converts 2-deoxy-galactose (DOG) to a toxic metabolite, 2-deoxy-galactose-1-phosphate, that suppresses bacterial growth [84]. This property is used in the second recombination step as clones can be selected in which galK is replaced with the desired sequence modification. Besides galK, two other two-way selection markers, thyA and tolC, can be used in bacteria lacking the corresponding gene product [85, 86].
There is an additional Red-based recombination method, en passant mutagenesis, that allows removal of an initially introduced selection marker cassette [6, 77]. The method is based on the insertion of a positive selection marker with an adjacent 18 bp I-SceI restriction site and short-sequence duplications. Short homologous sequences at either end or the linear marker cassette allow the insertion into the target site by Red recombination as described above. Next, expression of the homing endonuclease I-SceI is induced that allows linearization of the BAC construct by the cleavage of the I-SceI restriction site E. coli. The resulting dsDNA ends of the BAC serve as a substrate for a second Red recombination of the duplicated sequences, resulting in the complete removal of all foreign sequences including the selection marker [6, 77]. Inducible expression of the I-SceI enzyme can be accomplished by the use of expression plasmids or E. coli strains that harbor the I-SceI cassette chromosomally (e.g., GS1783) [77].
4.4. Deletion of Sequences in BACs
As discussed in Section 4.3, Red or RecE/T recombination systems provide a stable and more efficient alternative for the generation of BAC mutants. For the deletion of sequences from a BAC construct, a positive selection marker is amplified by PCR using primers with 30 to 50 bp extensions that are homologous to the target site in the BAC (Figure 2(a), light blue and orange boxes) [7]. This PCR product is electroporated into recombination-competent bacteria that harbor the BAC clone. The short homologous sequences at the end of the PCR product are used for two individual Red recombination events that result in the replacement of the target sequence with the selection marker. The resistance gene cassette remains in the BAC and cannot be used for the mutagenesis of a second locus in the construct (Figure 2(a)).
Figure 2.
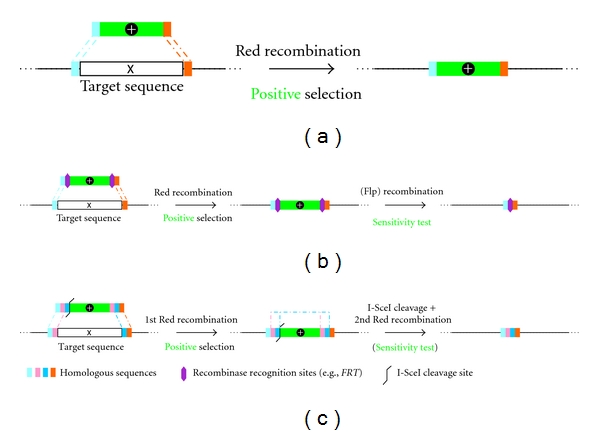
Overview of strategies for the Red-mediated deletion of sequences. Boxes of same color represent identical sequences.
Another strategy that allows the removal of the resistance marker utilizes the Cre/loxP and FLP/FRT recombination systems as described in Section 4.3. For this approach, a positive selection marker flanked by loxP or FRT sites is amplified and introduced into the BAC by homologous recombination. Induction of the Cre or FLP recombinase subsequently results in the removal of the marker cassette from the BAC construct, while only a single loxP or FRT site remains at the site of deletion (Figure 2(b)). Due to this residual recognition site, the system can only be applied once in one and the same BAC construct.
The complete removal of all foreign sequences can be accomplished by en passant mutagenesis [6, 77]. As described in Section 4.3, a cassette is generated that contains a positive selection marker with an adjacent I-SceI restriction site. The cassette is amplified with primers that not only contain homologous sequences for the insertion into the target site, but also a sequence duplication (Figure 2(c), pink and dark blue boxes) in the 5′ overhang. This duplication contains the site to be deleted and allows the removal of entire marker cassette in a second Red recombination step [6, 77]. As all foreign sequences are removed from the site of deletion, en passant mutagenesis can be utilized for consecutive modifications of any locus in a BAC construct.
4.5. Insertion of Sequences
Several strategies that allow the insertion of a sequence of interest (soi) including reporter genes, fluorescent tags, or foreign, viral antigens into BAC constructs have been developed (Figure 3). One approach utilizes a transfer construct that contains the soi and a positive selection marker flanked again by two loxP or FRT recognition sites. The construct is inserted into the target site by Red recombination. In a second step, the selection marker can be removed by the induction of the Flp or Cre recombinase, while one recognition site remains in the BAC construct downstream of the soi (Figure 3(a)) [7].
Figure 3.
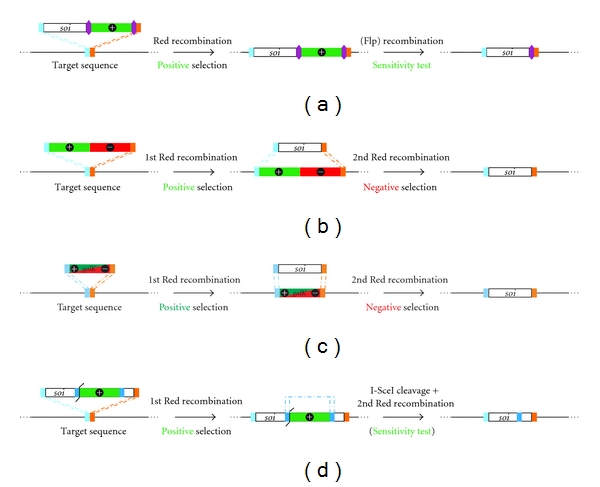
Overview of techniques that facilitate the insertion of a sequence of interest (soi) into a target site. Boxes of same color represent identical sequences.
Alternatively, a cassette that allows positive and negative selection or a two-way marker can be inserted at the target side in a first Red recombination event. Subsequently, the soi with sequences homologous to the target sequence at either end of the soi facilitates the replacement of the dual marker cassette with the soi. Clones that have incorporated the soi can be selected for the absence of the negative selection marker (Figures 3(b) and 3(c)) [7, 84].
Another method that allows the insertion of a soi into BAC constructs is en passant mutagenesis [6, 77]. Here, a transfer construct is generated by the insertion of a positive selection marker, an adjacent I-SceI site, and a sequence duplication into a unique restriction site within the soi. Upon insertion of the transfer cassette, expression of I-SceI is induced which results in the linearization of the BAC construct. The sequence duplications allow a second Red recombination resulting in the removal of all operational sequences [6, 77]. Existing transfer constructs can be used for the insertion of soi into any target sequence (Figure 3(d)). The advantage of the en passant techniques described in this section is that they allow the removal of selection marker and can be used to introduce multiple modifications in a BAC construct.
4.6. Sequence Editing
The compact organization of viral genomes is often a challenge for genetic manipulation of virus BACs. Overlapping coding sequences and regulatory elements require a strategy that allows the introduction of minimal sequence modifications that do not interfere with the expression and function of neighboring genes or sequences. In this section, we describe the available techniques that can be used for specific, minimal editing of sequences. The methods allow the manipulation of BACs on the nucleotide level; they include start codon mutation or insertion of a stop codon to abrogate gene expression as well as modification of functional domains of proteins or regulatory elements.
Sequence editing can be achieved by techniques described in Section 4.3. As described above, a dual or two-way selection marker is initially inserted into the target site in the BAC. In addition, a construct that contains the target sequence with the desired sequence modification is generated. In a second Red recombination step, the modified target sequence is used to replace the negative selection marker. Counterselection allows the enrichment of clones that contain the desired mutation (Figures 4(a) and 4(b)) [7, 84]. The advantage of the system is that the same intermediate clones obtained upon the insertion of the selection marker cassette can be used to introduce a variety of modifications into the same locus. However, false positive clones can present a problem for the selection of correct clones as discussed in Section 4.3 [7, 84].
Figure 4.

Overview of strategies that allow sequence editing of the target sequences. Boxes of same color represent identical sequences.
An alternative technique that allows sequence editing of the target site is en passant mutagenesis, in which the selection marker with an adjacent I-SceI is amplified by PCR using primers that contain homologous sequences for the insertion into the target sequence and duplications for the removal of the marker cassette. The desired modifications are included in the center of the duplicated sequences that are present within primers used for PCR amplification of the selectable marker (Figure 4(c), pink and dark blue boxes). In the first Red recombination, the marker cassette including the sequence duplications and modifications are incorporated into the target site. Upon induction of I-SceI expression, the sequence duplications allow the removal of the marker cassette while one duplicated sequence with the desired sequence modifications remains in the final construct (Figure 4(c)). The technique can be used to insert modifications of up to 50 bp in length. Larger modification can be facilitated by the en passant approach described in Section 4.5 and Figure 3(d) [6, 77].
5. Reconstitution of Virus and Mini-F Removal
5.1. Reconstitution of Recombinant Viruses
After mutagenesis of the viral genomes in E. coli, recombinant virus can be reconstituted by transfection of purified virus BAC DNA or in vitro transcribed RNA into susceptible eukaryotic cells. Upon uptake of the virus genome into transfected cells, virus proteins are expressed and virus replication is initiated. In some cases, cotransfection of BAC DNA and expression plasmids encoding transcriptional activators are required to stimulate virus replication [13]. For poxviruses, infection of susceptible cells with helperviruses prior to or after transfection is required to achieve reconstitution of recombinant virus [40–43]. As discussed in Section 2.5, expression of full-length RNA virus genomes from transfected BAC DNA can be dependent on the presence of T7 polymerase that is provided to the transfected cell in trans.
5.2. Site-Specific Excision of Mini-F Sequences
In case of DNA viruses, BAC sequences usually remain in the genome upon reconstitution of the virus. This can have negative effects on virus replication as, for example, the additional mini-F sequences might challenge the packaging capacity of the herpesvirus capsid [87]. In addition, residual bacterial sequences are often unfavorable for some applications including the development and licensing of life-attenuated vaccines. For this reason, several strategies have been developed that allow the excision of the mini-F sequences. One frequently used approach utilizes the Cre/loxP or FLP/FRT recombination system (see Section 4.3). For removal of the mini-F sequences, loxP or FRT sites are inserted at either end of the mini-F sequences. Cotransfection of the virus BAC with a Cre or FLP expression plasmid allows the transient production of the recombinase genes. Upon transfection, Cre or FLP facilitates removal of the mini-F sequences, only leaving a scar of a single loxP or FRT site of 34 bp (Figure 5(a)) [8, 14].
Figure 5.

Overview of strategies that allow mini-F removal upon virus reconstitution. Positive selection marker is portion of the BAC vector backbone. Boxes of same color represent identical sequences.
5.3. Delivery of Homologous Sequences In Trans
A second method that allows the removal of the mini-F uses the recombination machinery of eukaryotic cells as described in Section 2. For this approach, a repair vector (or linear PCR product) is generated that contains a 1–4 kb fragment representing the original locus that was used for the insertion of the mini-F. The construct is then cotransfected with virus BAC DNA into susceptible cells. In transfected cells, homologous sequences in the repair vector can recombine with sequences up- and downstream of the mini-F resulting in the removal of all vector sequences. Virus plaques that lost the mini-F sequences can be detected by the loss of visual markers encoded in the mini-F such as GFP [88]. Mini-F negative virus is then isolated, amplified, and used for further experiments. The major advantage of this method is that all bacterial sequences are removed, and no loxP or FRT sites remain in the virus genome. However, laborious purification steps are required to obtain clonal virus in which mini-F sequences were removed. This problem can be minimized by the insertion of the mini-F vector in an essential gene thereby generating a growth advantage for viruses that eliminate bacterial sequences (Figure 5(b)) [11, 88].
5.4. Removal of Mini-F via Duplications
Another method that allows complete removal of the mini-F replicon and does not require laborious purification steps utilizes sequence duplications that are inserted into the virus genome. Viral sequences flanking one end of the mini-F backbone are directly duplicated at the other end. An intramolecular recombination event during virus DNA replication allows the removal of all bacterial sequences, thus restoring the original insertion locus in the virus. However, one disadvantage of direct duplications is that they can serve as a substrate for bacterial recombinases, resulting in the instability of the BAC construct in E. coli (Figure 5(c)) [87]. To circumvent this problem, sequence duplications can be inserted in an inverted orientation. For this approach, a 1 to 3 kbp fragment corresponding to the original insertion site of the mini-F vector is inserted in inverse orientation between the mini-F replicon and the antibiotic resistance gene of the backbone. This antiparallel duplication allows stable maintenance and modification of the BAC in E. coli, while the mini-F sequences are completely removed from the virus genome by two intra- or intermolecular recombination events facilitated by recombinases in eukaryotic cells (Figure 5(d)) [13, 43, 57, 89].
6. Conclusions
Since the establishment of the first BAC system in 1997, BAC technology has contributed substantially to our understanding of the life cycle of large DNA and RNA viruses. Several techniques have been developed that facilitate the insertion of mini-F sequences into the virus genome. The methods allowed the generation of BAC systems for a plethora of virus species including members of the Herpesvirales, Poxviridae, Coronaviridae, and Flaviviridae. Well-established mutagenesis techniques described in this paper facilitate a site-specific manipulation of the virus genome in E. coli. Several strategies can be used to introduce any desired modification including deletions of viral or insertions of foreign sequences. Reconstitution of recombinant viruses can be accomplished by transfection of purified BAC DNA into susceptible mammalian cells, while in some cases helperviruses or additional expression vectors are required in this process. Finally, various techniques have been established that allow the excision of the mini-F sequences from the virus genomes without leaving unwanted sequence behind.
Acknowledgments
The authors thank Matthias Sieber and Nikolaus Osterrieder for editing the manuscript. This work was supported by the DFG Grant TI732/1-1 and unrestricted funding from the Freie Universität Berlin.
References
- 1.Adachi S, Hori K, Hiraga S. Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. Journal of Molecular Biology. 2006;356(4):850–863. doi: 10.1016/j.jmb.2005.11.088. [DOI] [PubMed] [Google Scholar]
- 2.Borst E, Messerle M. Development of a cytomegalovirus vector for somatic gene therapy. Bone Marrow Transplantation. 2000;25(supplement 2):S80–S82. doi: 10.1038/sj.bmt.1702361. [DOI] [PubMed] [Google Scholar]
- 3.Neubauer A, Rudolph J, Brandmuller C, Just FT, Osterrieder N. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology. 2002;300(2):189–204. doi: 10.1006/viro.2002.1488. [DOI] [PubMed] [Google Scholar]
- 4.Rosas CT, König P, Beer M, Dubovi EJ, Tischer BK, Osterrieder N. Evaluation of the vaccine potential of an equine herpesvirus type 1 vector expressing bovine viral diarrhea virus structural proteins. Journal of General Virology. 2007;88(3):748–757. doi: 10.1099/vir.0.82528-0. [DOI] [PubMed] [Google Scholar]
- 5.Rosas CT, Tischer BK, Perkins GA, Wagner B, Goodman LB, Osterrieder N. Live-attenuated recombinant equine herpesvirus type 1 (EHV-1) induces a neutralizing antibody response against West Nile virus (WNV) Virus Research. 2007;125(1):69–78. doi: 10.1016/j.virusres.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques. 2006;40(2):191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nature Genetics. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 8.Adler H, Messerle M, Wagner M, Koszinowski UH. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. Journal of Virology. 2000;74(15):6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kovacs F, Mettenleiter TC. Firefly luciferase as a marker for herpesvirus (pseudorabies virus) replication in vitro and in vivo. Journal of General Virology. 1991;72(12):2999–3008. doi: 10.1099/0022-1317-72-12-2999. [DOI] [PubMed] [Google Scholar]
- 10.Luker GD, Bardill JP, Prior JL, Pica CM, Piwnica-Worms D, Leib DA. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. Journal of Virology. 2002;76(23):12149–12161. doi: 10.1128/JVI.76.23.12149-12161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapp S, Osterrieder N, Keil GM, Beer M. Mutagenesis of a bovine herpesvirus type 1 genome cloned as an infectious bacterial artificial chromosome: analysis of glycoprotein E and G double deletion mutants. Journal of General Virology. 2003;84(2):301–306. doi: 10.1099/vir.0.18682-0. [DOI] [PubMed] [Google Scholar]
- 12.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. Journal of Molecular Biology. 1967;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 13.Tischer BK, Kaufer BB, Sommer M, Wussow F, Arvin AM, Osterrieder N. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. Journal of Virology. 2007;81(23):13200–13208. doi: 10.1128/JVI.01148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski UH. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(26):14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahony TJ, McCarthy FM, Gravel JL, West L, Young PL. Construction and manipulation of an infectious clone of the bovine herpesvirus 1 genome maintained as a bacterial artificial chromosome. Journal of Virology. 2002;76(13):6660–6668. doi: 10.1128/JVI.76.13.6660-6668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strive T, Hardy CM, French N, Wright JD, Nagaraja N, Reubel GH. Development of canine herpesvirus based antifertility vaccines for foxes using bacterial artificial chromosomes. Vaccine. 2006;24(7):980–988. doi: 10.1016/j.vaccine.2005.08.078. [DOI] [PubMed] [Google Scholar]
- 17.Rudolph J, O’Callaghan DJ, Osterrieder N. Cloning of the genomes of equine herpesvirus type 1 (EHV-1) strains KyA and racL11 as bacterial artificial chromosomes (BAC) Journal of Veterinary Medicine B. 2002;49(1):31–36. doi: 10.1046/j.1439-0450.2002.00534.x. [DOI] [PubMed] [Google Scholar]
- 18.Azab W, Kato K, Arii J, et al. Cloning of the genome of equine herpesvirus 4 strain TH20p as an infectious bacterial artificial chromosome. Archives of Virology. 2009;154(5):833–842. doi: 10.1007/s00705-009-0382-0. [DOI] [PubMed] [Google Scholar]
- 19.Costes B, Thirion M, Dewals B, et al. Felid herpesvirus 1 glycoprotein G is a structural protein that mediates the binding of chemokines on the viral envelope. Microbes and Infection. 2006;8(11):2657–2667. doi: 10.1016/j.micinf.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 20.Saeki Y, Ichikawa T, Saeki A, et al. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Human Gene Therapy. 1998;9(18):2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 21.Meseda CA, Schmeisser F, Pedersen R, Woerner A, Weir JP. DNA immunization with a herpes simplex virus 2 bacterial artificial chromosome. Virology. 2004;318(1):420–428. doi: 10.1016/j.virol.2003.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baigent SJ, Petherbridge LJ, Smith LP, Zhao Y, Chesters PM, Nair VK. Herpesvirus of turkey reconstituted from bacterial artificial chromosome clones induces protection against Marek’s disease. Journal of General Virology. 2006;87(4):769–776. doi: 10.1099/vir.0.81498-0. [DOI] [PubMed] [Google Scholar]
- 23.Schumacher D, Tischer BK, Fuchs W, Osterrieder N. Reconstitution of marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. Journal of Virology. 2000;74(23):11088–11098. doi: 10.1128/jvi.74.23.11088-11098.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith GA, Enquist LW. Construction and transposon mutagenesis in Escherichia coli of a full- length infectious clone of pseudorabies virus, an alphaherpesvirus. Journal of Virology. 1999;73(8):6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray WL, Zhou F, Noffke J, Tischer BK. Cloning the simian varicella virus genome in E. coli as an infectious bacterial artificial chromosome. Archives of Virology. 2011;156(5):739–746. doi: 10.1007/s00705-010-0889-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagaike K, Mori Y, Gomi Y, et al. Cloning of the varicella-zoster virus genome as an infectious bacterial artificial chromosome in Escherichia coli. Vaccine. 2004;22(29-30):4069–4074. doi: 10.1016/j.vaccine.2004.03.062. [DOI] [PubMed] [Google Scholar]
- 27.McGregor A, Schleiss MR. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Molecular Genetics and Metabolism. 2001;72(1):15–26. doi: 10.1006/mgme.2000.3102. [DOI] [PubMed] [Google Scholar]
- 28.Borst EM, Hahn G, Koszinowski UH, Messerle M. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli a new approach for construction of HCMV mutants. Journal of Virology. 1999;73(10):8320–8329. doi: 10.1128/jvi.73.10.8320-8329.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borenstein R, Frenkel N. Cloning human herpes virus 6A genome into bacterial artificial chromosomes and study of DNA replication intermediates. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):19138–19143. doi: 10.1073/pnas.0908504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang WL, Barry PA. Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. Journal of Virology. 2003;77(9):5073–5083. doi: 10.1128/JVI.77.9.5073-5083.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillet L, Daix V, Donofrio G, et al. Development of bovine herpesvirus 4 as an expression vector using bacterial artificial chromosome cloning. Journal of General Virology. 2005;86(4):907–917. doi: 10.1099/vir.0.80718-0. [DOI] [PubMed] [Google Scholar]
- 32.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(14):8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White RE, Calderwood MA, Whitehouse A. Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. Journal of General Virology. 2003;84(12):3393–3403. doi: 10.1099/vir.0.19387-0. [DOI] [PubMed] [Google Scholar]
- 34.Delecluse HJ, Kost M, Feederle R, Wilson L, Hammerschmidt W. Spontaneous activation of the lytic cycle in cells infected with a recombinant Kaposi’s sarcoma-associated virus. Journal of Virology. 2001;75(6):2921–2928. doi: 10.1128/JVI.75.6.2921-2928.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohashi M, Orlova N, Quink C, Wang F. Cloning of the epstein-barr virus-related rhesus lymphocryptovirus as a bacterial artificial chromosome: a loss-of-function mutation of the rhBARF1 immune evasion gene. Journal of Virology. 2011;85(3):1330–1339. doi: 10.1128/JVI.01411-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Estep RD, Powers MF, Yen BK, Li H, Wong SW. Construction of an infectious rhesus rhadinovirus bacterial artificial chromosome for the analysis of Kaposi’s sarcoma-associated herpesvirus-related disease development. Journal of Virology. 2007;81(6):2957–2969. doi: 10.1128/JVI.01997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J, Osterrieder N. Generation of an infectious clone of duck enteritis virus (DEV) and of a vectored DEV expressing hemagglutinin of H5N1 avian influenza virus. Virus Research. 2011;159(1):23–31. doi: 10.1016/j.virusres.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 38.Kunec D, Hanson LA, Van Haren S, Nieuwenhuizen IF, Burgess SC. An overlapping bacterial artificial chromosome system that generates vectorless progeny for channel catfish herpesvirus. Journal of Virology. 2008;82(8):3872–3881. doi: 10.1128/JVI.02152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Costes B, Fournier G, Michel B, et al. Cloning of the koi herpesvirus genome as an infectious bacterial artificial chromosome demonstrates that disruption of the thymidine kinase locus induces partial attenuation in Cyprinus carpio koi. Journal of Virology. 2008;82(10):4955–4964. doi: 10.1128/JVI.00211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cottingham MG, Andersen RF, Spencer AJ, et al. Recombination-mediated genetic engineering of a bacterial artificial chromosome clone of modified vaccinia virus Ankara (MVA) Plos One. 2008;3(2) doi: 10.1371/journal.pone.0001638. Article ID e1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domi A, Moss B. Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(19):12415–12420. doi: 10.1073/pnas.192420599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meisinger-Henschel C, Spath M, Lukassen S, et al. Introduction of the six major genomic deletions of modified vaccinia virus Ankara (MVA) into the parental vaccinia virus is not sufficient to reproduce an MVA-like phenotype in cell culture and in mice. Journal of Virology. 2010;84:9907–9919. doi: 10.1128/JVI.00756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roth SJ, Hoper D, Beer M, et al. Recovery of infectious virus from full-length cowpox virus (CPXV) DNA cloned as a bacterial artificial chromosome. Veterinary Research. 2011;42(1):p. 3. doi: 10.1186/1297-9716-42-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merchlinsky M, Moss B. Resolution of vaccinia virus DNA concatemer junctions requires late-gene expression. Journal of Virology. 1989;63(4):1595–1603. doi: 10.1128/jvi.63.4.1595-1603.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almazan F, Gonzalez JM, Penzes Z, et al. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(10):5516–5521. doi: 10.1073/pnas.97.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Almazan F, Dediego ML, Galan C, et al. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. Journal of Virology. 2006;80(21):10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fan ZC, Bird RC. An improved reverse genetics system for generation of bovine viral diarrhea virus as a BAC cDNA. Journal of Virological Methods. 2008;149(2):309–315. doi: 10.1016/j.jviromet.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rasmussen TB, Reimann I, Uttenthal Å, et al. Generation of recombinant pestiviruses using a full-genome amplification strategy. Veterinary Microbiology. 2010;142(1-2):13–17. doi: 10.1016/j.vetmic.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 49.St.-Jean JR, Desforges M, Almazan F, Jacomy H, Enjuanes L, Talbot PJ. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. Journal of Virology. 2006;80(7):3670–3674. doi: 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yun SI, Kim SY, Rice CM, Lee YM. Development and application of a reverse genetics system for Japanese encephalitis virus. Journal of Virology. 2003;77(11):6450–6465. doi: 10.1128/JVI.77.11.6450-6465.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yount B, Curtis KM, Baric RS. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. Journal of Virology. 2000;74(22):10600–10611. doi: 10.1128/jvi.74.22.10600-10611.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boehme KW, Ikizler M, Kobayashi T, Dermody TS. Reverse genetics for mammalian reovirus. Methods. 2011;55(2):109–113. doi: 10.1016/j.ymeth.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kobayashi T, Antar AA, Boehme KW, et al. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host and Microbe. 2007;1(2):147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Gennip RG, Veldman D, Van De Water SG, Van Rijn PA. Genetic modification of Bluetongue virus by uptake of “synthetic” genome segments. Virology Journal. 2010;7, article 261 doi: 10.1186/1743-422X-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neumann G, Fujii K, Kino Y, Kawaoka Y. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(46):16825–16829. doi: 10.1073/pnas.0505587102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Domi A, Moss B. Engineering of a vaccinia virus bacterial artificial chromosome in Escherichia coli by bacteriophage λ-based recombination. Nature Methods. 2005;2(2):95–97. doi: 10.1038/nmeth734. [DOI] [PubMed] [Google Scholar]
- 57.Cottingham MG, Gilbert SC. Rapid generation of markerless recombinant MVA vaccines by en passant recombineering of a self-excising bacterial artificial chromosome. Journal of Virological Methods. 2010;168(1-2):233–236. doi: 10.1016/j.jviromet.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 58.Haas R, Kahrs AF, Facius D, Allmeier H, Schmitt R, Meyer TF. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130(1):23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 59.Kahrs AF, Odenbreit S, Schmitt W, Heuermann D, Meyer TF, Haas R. An improved TnMax mini-transposon system suitable for sequencing, shuttle mutagenesis and gene fusions. Gene. 1995;167(1-2):53–57. doi: 10.1016/0378-1119(95)00671-0. [DOI] [PubMed] [Google Scholar]
- 60.Brune W, Menard C, Hobom U, Odenbreit S, Messerle M, Koszinowski UH. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nature Biotechnology. 1999;17(4):360–364. doi: 10.1038/7914. [DOI] [PubMed] [Google Scholar]
- 61.Brune W, Messerle M, Koszinowski UH. Forward with BACs: new tools for herpesvirus genomics. Trends in Genetics. 2000;16(6):254–259. doi: 10.1016/s0168-9525(00)02015-1. [DOI] [PubMed] [Google Scholar]
- 62.Smith GR. Homologous recombination in procaryotes. Microbiological Reviews. 1988;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zagursky RJ, Hays JB. Expression of the phage λ recombination genes exo and bet under lacPO control on a multi-copy plasmid. Gene. 1983;23(3):277–292. doi: 10.1016/0378-1119(83)90018-5. [DOI] [PubMed] [Google Scholar]
- 64.Posfai G, Koob MD, Kirkpatrick HA, Blattner FR. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157: H7 genome. Journal of Bacteriology. 1997;179(13):4426–4428. doi: 10.1128/jb.179.13.4426-4428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnston DM, Cannon JG. Construction of mutant strains of Neisseria gonorrhoeae lacking new antibiotic resistance markers using a two gene cassette with positive and negative selection. Gene. 1999;236(1):179–184. doi: 10.1016/s0378-1119(99)00238-3. [DOI] [PubMed] [Google Scholar]
- 66.Blomfield IC, Vaughn V, Rest RF, Eistenstein BI. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Molecular Microbiology. 1991;5(6):1447–1457. doi: 10.1111/j.1365-2958.1991.tb00791.x. [DOI] [PubMed] [Google Scholar]
- 67.Yang XW, Model P, Heintz N. Homologous recombination based modification in Esherichia coli and germline transmission in transgenic mice of a bacterial artificial chromsome. Nature Biotechnology. 1997;15(9):859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 68.Murphy KC. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. Journal of Bacteriology. 1998;180(8):2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakaki Y, Karu AE, Linn S, Echols H. Purification and properties of the γ protein specified by bacteriophage λ: an inhibitor of the host RecBC recombination enzyme. Proceedings of the National Academy of Sciences of the United States of America. 1973;70(8):2215–2219. doi: 10.1073/pnas.70.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kovall R, Matthews BW. Toroidal structure of λ-exonuclease. Science. 1997;277(5333):1824–1827. doi: 10.1126/science.277.5333.1824. [DOI] [PubMed] [Google Scholar]
- 71.Kushner SR, Nagaishi H, Clark AJ. Isolation of exonuclease VIII: the enzyme associated with the sbcA indirect suppressor. Proceedings of the National Academy of Sciences of the United States of America. 1974;71(9):3593–3597. doi: 10.1073/pnas.71.9.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weissbach A, Korn D. The effect of lysogenic induction on the deoxyribonucleases of Escherichia coli K12 lambda. The Journal of Biological Chemistry. 1962;237:C3312–C3314. [PubMed] [Google Scholar]
- 73.Hall SD, Kane MF, Kolodner RD. Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. Journal of Bacteriology. 1993;175(1):277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kmiec E, Holloman WK. β-protein of bacteriophage λ promotes renaturation of DNA. Journal of Biological Chemistry. 1981;256(24):12636–12639. [PubMed] [Google Scholar]
- 75.Wu Z, Xing X, Bohl CE, Wisler JW, Dalton JT, Bell CE. Domain structure and DNA binding regions of β protein from bacteriophage λ. Journal of Biological Chemistry. 2006;281(35):25205–25214. doi: 10.1074/jbc.M512450200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ellis HM, Yu D, DiTizio T, Court DL. High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(12):6742–6746. doi: 10.1073/pnas.121164898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: a two step markerless red recombination system. Methods in Molecular Biology. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 78.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee EC, Yu D, Martinez De Velasco J, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 80.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. Journal of Molecular Biology. 1981;150(4):467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 81.McLeod M, Craft S, Broach JR. Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2 microns circle. Molecular and Cellular Biology. 1986;6(10):3357–3367. doi: 10.1128/mcb.6.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fiering S, Kim CG, Epner EM, Groudine M. An “in-out” strategy using gene targeting and FLP recombinase for the functional dissection of complex DNA regulatory elements: analysis of the β- globin locus control region. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(18):8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods in Enzymology. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- 84.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Research. 2005;33(4, article e36) doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong QN, Ng VC, Lin MC, Kung HF, Chan D, Huang JD. Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli. Nucleic Acids Research. 2005;33(6, article e59) doi: 10.1093/nar/gni059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Devito JA. Recombineering with tolC as a selectable/counter-selectable marker: remodeling the rRNA operons of Escherichia coli. Nucleic Acids Research. 2008;36(1, article e4) doi: 10.1093/nar/gkm1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wagner M, Jonjic S, Koszinowski UH, Messerle M. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. Journal of Virology. 1999;73(8):7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rudolph J, Osterrieder N. Equine herpesvirus type 1 devoid of gM and gp2 is severely impaired in virus egress but not direct cell-to-cell spread. Virology. 2002;293(2):356–367. doi: 10.1006/viro.2001.1277. [DOI] [PubMed] [Google Scholar]
- 89.Wussow F, Fickenscher H, Tischer BK. Red-mediated transposition and final release of the mini-f vector of a cloned infectious herpesvirus genome. Plos One. 2009;4(12) doi: 10.1371/journal.pone.0008178. Article ID e8178. [DOI] [PMC free article] [PubMed] [Google Scholar]


