Abstract
Aims
Heterozygous mutations in KCNQ1 cause type 1 long QT syndrome (LQT1), a disease characterized by prolonged heart rate-corrected QT interval (QTc) and life-threatening arrhythmias. It is unknown why disease penetrance and expressivity is so variable between individuals hosting identical mutations. We aimed to study whether this can be explained by single nucleotide polymorphisms (SNPs) in KCNQ1's 3′ untranslated region (3′UTR).
Methods and results
This study was performed in 84 LQT1 patients from the Academic Medical Center in Amsterdam and validated in 84 LQT1 patients from the Mayo Clinic in Rochester. All patients were genotyped for SNPs in KCNQ1's 3′UTR, and six SNPs were found. Single nucleotide polymorphisms rs2519184, rs8234, and rs10798 were associated in an allele-specific manner with QTc and symptom occurrence. Patients with the derived SNP variants on their mutated KCNQ1 allele had shorter QTc and fewer symptoms, while the opposite was also true: patients with the derived SNP variants on their normal KCNQ1 allele had significantly longer QTc and more symptoms. Luciferase reporter assays showed that the expression of KCNQ1's 3′UTR with the derived SNP variants was lower than the expression of the 3′UTR with the ancestral SNP variants.
Conclusion
Our data indicate that 3′UTR SNPs potently modify disease severity in LQT1. The allele-specific effects of the SNPs on disease severity and gene expression strongly suggest that they are functional variants that directly alter the expression of the allele on which they reside, and thereby influence the balance between proteins stemming from either the normal or the mutant KCNQ1 allele.
Keywords: Long QT syndrome, Single nucleotide polymorphism, 3′ untranslated region, KCNQ1, QTc
Introduction
Long QT syndrome (LQTS) is the most common heritable cardiac channelopathy, and can cause premature sudden death due to life-threatening arrhythmias in relation to prolongation of the heart rate-corrected QT interval (QTc).1 The most prevalent form, LQT1, is caused by loss-of-function mutations in the KCNQ1-encoded Kv7.1 potassium channel (IKs).2,3
LQT1 is characterized by incomplete penetrance and variable expressivity whereby family members carrying identical mutations have profound differences in their QTc and clinical course.4,5 The cause for this heterogeneity remains largely elusive. Kv7.1 is a tetrameric channel resulting from the post-translational assembly of four KCNQ1-encoded subunits. Therefore, patients heterozygous for an LQT1-causative mutation combine the translated products from normal and LQT1-mutation-containing alleles to form tetrameric channels. If both alleles are similarly expressed, one would predict that 2/16 of the Kv7.1 channels stem solely from either the normal or the mutated allele. The remaining channels are hybrids of the healthy and the mutated alleles. This reasoning predicts that factors that influence the balance between normal and mutant allele expression affect the proportion of mutated protein in the Kv7.1 channels, and thereby modify disease severity.
We hypothesized that single nucleotide polymorphisms (SNPs) in KCNQ1's 3′ untranslated region (3′UTR) modify the relative expression of normal vs. LQT1-mutation-containing allele and thereby contribute to disease variability. The 3′UTR is known to play an important regulatory role in gene expression, in particular by controlling mRNA stability and translation.6,7 Heterozygosity for SNPs in the 3′UTR may alter the regulatory effects of the 3′UTR on gene expression in an allele-specific manner, and this is expected to be especially relevant when one allele contains a disease-causing mutation. In the case of LQT1, 3′UTR SNP variants residing on either the healthy or the mutated allele may alter the relative number of Kv7.1 channel subunits stemming from either allele. We therefore hypothesized that SNPs in KCNQ1's 3′UTR could affect 3′UTR function, and thereby modify the disease phenotype in an allele-specific fashion. We initially assessed this in LQT1 patients from the Academic Medical Center (AMC) in Amsterdam (the Netherlands) and validated our findings in patients from the Mayo Clinic (MC) in Rochester (MN, USA).
Methods
Patient inclusion
From both institutions (AMC and MC), we included families where a mutation in KCNQ1 was identified in at least three members, and at least one affected member displayed QTc prolongation. Genomic DNA (if available) was used for sequencing the 3′UTR of KCNQ1. This data were used to determine whether 3′UTR SNP variants occur in trans (opposite allele) or in cis (on the same allele) as the pathogenic KCNQ1 mutation (as defined by analysis of phase in the families). From each family, at least three members were genotyped (unless patients were homozygous for the SNP variants). Next, we included those patients in whom the KCNQ1 mutation was the only mutation present in a LQTS-linked gene, and in whom a 12-lead ECG and clinical data were available for analysis. All patients (and their family members) with double and compound mutations in major LQT-susceptibility genes (i.e. KCNH2, SCN5A, KCNE1, and KCNE2) were excluded. Individuals with acquired cardiac diseases, electrolyte abnormalities, or repolarization prolonging medication were also excluded. The following clinical parameters were obtained from medical records: age, gender, proband status, family history of sudden cardiac death at age <45 years, incidence of Torsades des Pointes, VF, and/or syncope, and type of KCNQ1 mutation. The researchers who obtained clinical parameters (and those who analysed ECGs) were blind to the 3′UTR SNP haplotypes. The study was approved by the institutional review committees and the subjects had given informed consent.
Genetic analysis
LQT1-associated KCNQ1 mutations had been identified previously using standard protocols.8 For this study, the final exon and the 3′UTR of KCNQ1 were additionally sequenced. The region to be analysed comprised of three amplicons, and was amplified with polymerase chain reaction (PCR) using oligonucleotide pairs: 5′-GGCACCTTCCCTTCTCTGG-3′ with 5′-ACCACCATGCCAGTGATGTC-3′, 5′-CACAGCCTGCACTTGGG-3′ with 5′-CAGGGCTCCTCTCCAGC-3′, and 5′-CAGTCTCACCATTTCCCCAG-3′ with 5′-GCCCAGAACAGGAGCGAC -3′.
ECG analysis
Twelve-lead electrocardiograms were enlarged to facilitate manual analysis. QT duration (lead II or V5) was corrected for heart rate using Bazett's formula (QTc = QT/√RR, where RR is the interval, measured in seconds, from the onset of 1 QRS complex to the onset of the following QRS complex). To measure QTc within each individual, the mean value of five consecutive beats was calculated.
The effect of SNPs on QTc in the general population
To study the effect of SNPs rs2519184, rs8234, and rs10798 in the general population, we contacted the investigators of the KORA Study to evaluate whether the SNPs rs2519184, rs8234, or rs10798 are correlated with the QTc duration in the general population. The KORA Study is a series of independent population-based epidemiological surveys of participants residing in the region of Augsburg, Southern Germany. All survey participants had German nationality, were identified through the registration office, and were examined in 1994–95 (KORA S3) and 1999–2001 (KORA S4). In 2004–05, 3006 subjects participated in a 10-year follow-up examination of S3 (KORA F3). Individuals for genotyping in KORA F3 and KORA S4 were randomly selected.
Luciferase reporter assay
Plasmid construction
Luciferase reporter plasmids were constructed by PCR amplification of the 3′UTR of KCNQ1 from a patient heterozygous for the SNPs rs2519184, rs8234, and rs10798 with the primers: 5′-ACTGACTAGTCATGGACCATGCTGTCTG-3′ and 5′-ACTGGAGCTCCAGCCTGTGATTCTCCACG-3′. This 878bp fragment was cloned into the pMIR-REPORTTM Luciferase vector (Ambion) downstream of the luciferase coding region, creating luciferase KCNQ1-3′UTR-G-A-A (ancestral haplotype) and luciferase KCNQ1-3′UTR-A-G-G (derived haplotype). The plasmids luciferase KCNQ1-3′UTR-A-A-A, luciferase KCNQ1-3′UTR-G-G-A, and luciferase KCNQ1-3′UTR-G-A-G were generated from the luciferase KCNQ1-3′UTR-G-A-A plasmid by PCR-based mutagenesis.
Cell isolation and preparation
Neonatal rat cardiac myocytes, immortalized with a temperature-sensitive SV40 T antigen (H10 cells),9 were cultured in Dulbeccos modified Eagles medium supplemented with 10% Fetal Calf Serum (Gibco-BRL) and glutamine at 33°C. One day prior to transfection, cells were seeded in a 24-well plate at a density of 1.4 × 106 cells per plate. Cardiomyocytes from 1 to 2-day-old Lewis neonatal rats were isolated and cultured as described previously.10
Transfection and luciferase assay
H10 cells were transiently transfected per well with 5 ng Renilla luciferase plasmid, phRL vector (Promega), and 5 ng or 10 ng of either luciferase KCNQ1-3′UTR-G-A-A, luciferase KCNQ1-3′UTR-A-A-A, luciferase KCNQ1-3′UTR-G-G-A, luciferase KCNQ1-3′UTR-G-A-G, or luciferase KCNQ1-3′UTR-A-G-G using GeneJammer (Agilent Technologies). Neonatal rat cardiomyocytes were transfected with 50 ng Renilla luciferase plasmid, phRL vector, and 100 ng of either luciferase KCNQ1-3′UTR-G-A-A, luciferase KCNQ1-3′UTR-A-A-A, luciferase KCNQ1-3′UTR-G-G-A, luciferase KCNQ1-3′UTR-G-A-G, or luciferase KCNQ1-3′UTR-A-G-G using lipofectamine 2000 reagent (Invitrogen). At 48 h after transfection, cells were lysed and assayed for luciferase and Renilla luciferase activity with a luminometer (Glomax multi detection system, Promega) by using the Renilla reporter assay system (Promega). Renilla luciferase activity was assayed to normalize luciferase results for cell densities and transfection efficiency. The luciferase assays were performed at least three times, and in every experiment plasmids were isolated from three different colonies per haplotype. Per plasmid, four wells were transfected and the average signal of these four wells was used for the statistical analysis.
Statistical methods
QTc was compared between various LQT1 genotype and 3′UTR SNP haplotype combinations. We used a linear mixed effect regression model from the kinship package in R.11 Differences in symptom prevalence were analysed with a logistic regression model using generalized estimation equations. In both models, a correction for the relatedness among individuals was applied. Furthermore, age at the time of ECG, gender, and proband status were included as covariates and genotype effects were modelled as additive effects. For the in vitro experiments, differences between groups were compared using analysis of variance (ANOVA) or Student's t-test, where appropriate. Values are expressed as mean ± standard error. We regarded P-values < 0.05 as statistically significant.
RESULTS
Patient inclusion and KCNQ1 3′UTR SNP detection
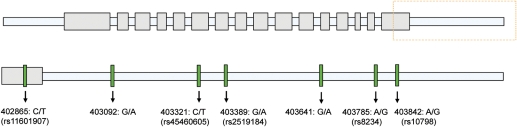
From AMC, 84 LQT1-positive subjects from 24 families were included. From MC, 84 LQT1-positive subjects from 17 families were included. Demographic data and details of the identified mutations are displayed in Table 1 and Supplementary material online, Table S1, respectively. In all subjects, the final exon and the complete 3′UTR of KCNQ1 were sequenced. In the AMC cohort, seven SNPs were found of which six SNPs were in the 3′UTR. Single nucleotide polymorphisms at nucleotide positions 403092 and 403641 were not reported previously (NCBI build 36, hg18). Except for the SNP at nucleotide position 403092, all SNPs (and no additional SNPs) were also found in the MC cohort (Figure 1; see Table 2 for SNP frequencies). Single nucleotide polymorphisms rs8234 and rs10798 were in complete linkage disequilibrium in all patients in this study.
Table 1.
Clinical characteristics, mutation types, and ECG parameters of the study populations from the AMC and the Mayo Clinic
| Variable | AMC | Mayo |
|---|---|---|
| Clinical characteristics | ||
| Patients, n (%) | 84 (100) | 84 (100) |
| Male/female, n | 34/50 | 38/46 |
| Age, years | 34 ± 3 | 27 ± 2 |
| Proband, n (%) | 22 (26) | 13 (15) |
| Family history for sudden death, n (%) | 40 (48) | 58 (69) |
| Syncope, n (%) | 18 (21) | 18 (21) |
| Documented TdP/VF, n (%) | 5 (6) | 1 (1) |
| Mutation type | ||
| Missense, n (%) | 63 (75) | 67 (80) |
| Frameshift, n (%) | 7 (8) | 1 (1) |
| Splice site, n (%) | 11 (13) | 4 (5) |
| Deletion, n (%) | 3 (4) | 12 (14) |
| ECG parameters | ||
| RR interval (ms) | 859 ± 19 | 917 ± 24 |
| Males | 879 ± 26 | 968 ± 41 |
| Females | 844 ± 28 | 875 ± 27 |
| QT duration (ms) | 411 ± 7 | 426 ± 6 |
| Males | 405 ± 8 | 429 ± 10 |
| Females | 416 ± 10 | 425 ± 8 |
| QTc (ms) | 446 ± 7 | 451 ± 4 |
| Males | 435 ± 6 | 445 ± 6 |
| Females | 454 ± 7a | 457 ± 4a |
aStatistical significance compared with males.
Values are expressed as number of patients (percentage of total), or as mean ± standard error of the mean (SEM).
Figure 1.
Genetic variation in the 3′ untranslated region of KCNQ1. Single nucleotide polymorphisms (SNPs) found in the study cohorts of the Academic Medical Center Amsterdam (AMC) and the Mayo Clinic (MC). Position of the nucleotide change is starting from the ATG start codon (NCBI build 36, hg18).
Table 2.
Single nucleotide polymorphisms found in the 3′untranslated region of KCNQ1
| Genetic location | Nucleotide change position | Allele (ancestral/derived) | SNP id | Genotype | AMC (n = 84) |
Mayo (n = 84) |
||
|---|---|---|---|---|---|---|---|---|
| N (%) | Minor allele frequency | N (%) | Minor allele frequency | |||||
| Exon 16 | 402865 | C/T | rs11601907 | CC | 38 (45) | 0.327 | 50 (60) | 0.214 |
| CT | 37 (44) | 32 (38) | ||||||
| TT | 9 (11) | 2 (2) | ||||||
| 3′UTR | 403092 | G/A | Not available | GG | 81 (96) | 0.018 | 84 (100) | 0.000 |
| GA | 3 (4) | 0 | ||||||
| AA | 0 | 0 | ||||||
| 3′UTR | 403321 | C/T | rs45460605 | CC | 81 (96) | 0.018 | 83 (99) | 0.006 |
| CT | 3 (4) | 1 (1) | ||||||
| TT | 0 | 0 | ||||||
| 3′UTR | 403389 | G/A | rs2519184 | GG | 78 (93) | 0.036 | 50 (60) | 0.202 |
| GA | 6 (7) | 34 (40) | ||||||
| AA | 0 | 0 | ||||||
| 3′UTR | 403641 | G/A | Not available | GG | 83 (99) | 0.006 | 83 (99) | 0.006 |
| GA | 1 (1) | 1 (1) | ||||||
| AA | 0 | 0 | ||||||
| 3′UTR | 403785 | A/G | rs8234 | AA | 52 (62) | 0.208 | 31 (37) | 0.351 |
| AG | 29 (34) | 47 (56) | ||||||
| GG | 3 (4) | 6 (7) | ||||||
| 3′UTR | 403842 | A/G | rs10798 | AA | 52 (62) | 0.208 | 31 (37) | 0.351 |
| AG | 29 (34) | 47 (56) | ||||||
| GG | 3 (4) | 6 (7) | ||||||
Position of the nucleotide change is starting from the ATG start codon (NCBI build 36, hg18). The SNP id denotes single nucleotide polymorphism identity from public databases. N (%) denotes number of patients (percentage of total).
Association between KCNQ1 3′UTR SNPs and QTc in LQT1 patients
Next, we studied whether the three most frequent SNPs, for which group sizes in both the AMC and MC cohorts were large enough for statistical analysis, were associated with QTc. We hypothesized that SNP variants would modify the QTc depending on whether they occur in trans (opposite allele) or in cis (on the same allele) as the pathogenic KCNQ1 mutation as defined by analysis of phase in the families.
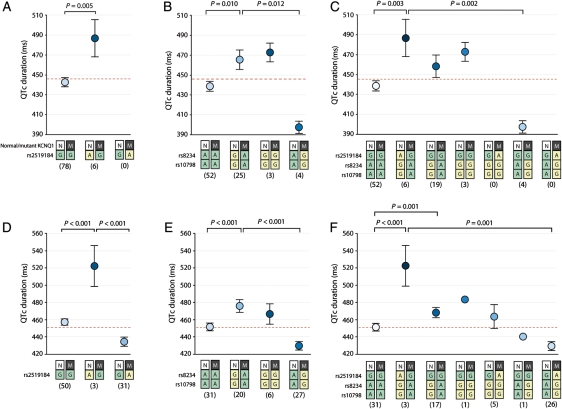
The presence of the derived variant of rs2519184 (A) in trans to the mutated allele was indeed associated with a marked increase in QTc by 46 ± 16 ms in the AMC cohort (P = 0.005; Figure 2A). This was corroborated by similar findings in the MC cohort where the derived variant in trans was associated with 60 ± 16 ms longer QTc (P < 0.001; Figure 2D). Notably, in the MC cohort, we also identified subjects where this derived variant resided on the LQT1-mutation containing allele. Here, location of this variant in cis was associated with 16 ± 8 ms shorter QTc (P < 0.001).
Figure 2.
Allele-specific effects of single nucleotide polymorphisms (SNPs) rs2519184, rs8234, and rs10798 on QTc. Allele-specific effects of SNPs rs2519184, rs8234, and rs10798 on QTc are displayed in panels (A and B) and (C) for the AMC population, and (D and E), and (F) for the MC population. Effects of SNP rs2519184 are displayed in (A) and (D). Effects of SNPs rs8234 and rs10798 are displayed in (B) and (E). Allele-specific haplotype analysis of the three SNPs is displayed in (C) and (F). The red line represents mean QTc of all individuals regardless of the specific LQT1-causative mutation and the 3′ untranslated region SNP status. Numbers below genotypes denote group sizes. Data are presented as mean; I bars represent standard errors. N, normal KCNQ1 allele; M, mutant KCNQ1 allele; green box, ancestral SNP variant; yellow box, derived SNP variant.
The same effects were seen for SNPs rs8234 and rs10798. When the derived variants of rs8234 and rs10798 (G-G) were in trans to the mutated allele, QTc was increased by 26 ± 10 ms (P = 0.010; Figure 2B). Again, this was confirmed in the MC cohort where the derived variants located in trans increased the QTc by 33 ± 7 ms (P < 0.001; Figure 2E). The opposite location of the derived variants (i.e. in cis to the mutation) was related with shorter QTc (−41 ± 25 ms; not significant), which was confirmed in the MC population (reduction by 9 ± 9 ms; not significant).
Allele-specific haplotype analysis revealed that location of the derived variants of the SNPs (A-G-G) in trans to the mutation was associated with 49 ± 16 ms longer QTc in the AMC cohort (P = 0.003) and 60 ± 14 ms longer QTc in the MC cohort (P < 0.001; Figure 2C and F). Subjects where the A-G-G haplotype was in cis to the mutation were only present in the MC cohort. Indeed, in these subjects, the anticipated opposite effect on QTc was seen, as the QTc was attenuated by 12 ± 9 ms (not significant). Various allele-specific haplotype combinations of KCNQ1 and the derived SNP variants, as shown in Figure 2C and F, displayed an intermediate effect on QTc. Finally, the effects of SNPs rs2519184, rs8234, and rs10798 on QTc in the combined study population (both AMC and MC) are shown in the Supplementary material online, Figure S1.
Since our study cohorts included LQT-1 patients with different types of mutation in KCNQ1, we analysed whether the effect of the 3′UTR SNPs on QTc is significantly different for different mutation types or whether this effect is different between carriers of a missense mutation vs. carriers of a frameshift, splice site, or deletion mutation. The effect of the SNPs on QTc was not significantly different for different mutation types (P = 0.118 for rs2519184, P = 0.493 for rs8234 and rs10798, and P = 0.148 for all three SNPs), and was also not different between carriers of a missense mutation vs. carriers of a frameshift, splice site, or deletion mutation (P = 0.118 for rs2519184, P = 0.493 for rs8234 and rs10798, and P = 0.148 for all three SNPs).
Association between KCNQ1 3′ untranslated region single nucleotide polymorphisms and QTc in single families
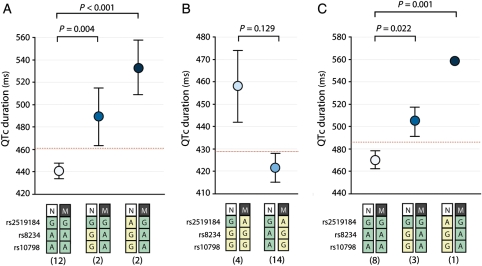
Our data suggest that assessing these 3′UTR SNPs can predict disease severity of a given KCNQ1 mutation in one family. To test this, we analysed the allele-specific haplotype of SNPs rs2519184, rs8234, and rs10798 with regard to QTc in three families with more than 10 members affected, including one family with the KCNQ1-R243C mutation (14 affected), one family with the KCNQ1-I235N mutation (20 members), and one family with the KCNQ1-339delF mutation (12 affected). Indeed, the KCNQ1-R243C carriers where the derived variants of the SNPs (A-G-G or G-G-G) were found in trans to the mutation exhibited markedly longer QTc than their KCNQ1-R243C-positive relatives who did not carry these variants (P < 0.001; Figure 3A, see Supplementary material online, Figure S2a for family pedigree). The opposite was seen in the family with the KCNQ1-I235N mutation, where all affected members carried the derived variants of the 3′UTR SNPs (A-G-G) on their mutation-containing allele. Family members with the derived SNP variants in cis to the mutation displayed mostly normal to near-normal QTc values, while members who were homozygous for the derived variants of SNPs rs8234 and rs10798 (thus carrying these variants also on their healthy allele) displayed longer QTc values (P = 0.129; Figure 3B, see Supplementary material online, Figure S2b for family pedigree). Finally, in the KCNQ1-339delF family, members with the derived SNP variants (A-G-G or G-G-G) in trans to the mutation exhibited longer QTc than their relatives who were homozygous for the ancestral SNP variants (P = 0.001; Figure 3C; see for family pedigree Supplementary material online, Figure S2c).
Figure 3.
Allele-specific effects of single nucleotide polymorphisms (SNPs) rs2519184, rs8234, and rs10798 on QTc in three single families. Allele-specific haplotype analysis of SNPs rs2519184, rs8234, and rs10798 with regard to QTc are displayed for the three largest families: one family carrying the KCNQ1-R243C mutation (A), one family with the KCNQ1-I235N mutation (B), and one family carrying the KCNQ1-339delF mutation (C). Numbers below genotypes denote group sizes. Data are presented as mean; I bars represent standard errors. N, normal KCNQ1 allele; M, mutant KCNQ1 allele; green box, ancestral SNP variant; yellow box, derived SNP variant.
Association between KCNQ1 3′ untranslated region single nucleotide polymorphisms and symptomatology in LQT1 patients
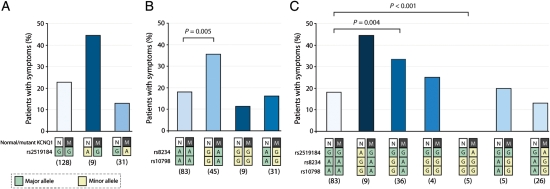
Next, we assessed in the total combined study population (AMC and MC) whether allele-specific location of the derived SNP variants in the KCNQ1's 3′UTR also modifies the known cardiac sequelae of LQT1 [unexplained syncope, documented torsades de pointes ventricular tachycardia/ventricular fibrillation, and/or (aborted) sudden death]. Location of the derived haplotype of SNPs rs8234 and rs10798 (G-G-G) in trans to the mutation was associated with a statistically significant increased occurrence of symptoms (P = 0.004). Inversely, the opposite location of the derived SNP haplotype (i.e. in cis to the mutation) tended to be related to fewer symptoms (not significant; Figure 4). Additionally, we corrected the effects of the 3′UTR SNPs on symptomatology also for QTc (thus not only for age, sex, and proband status). Interestingly, after correction for QTc, the 3′UTR haplotype was still associated with the occurrence of symptoms (Supplementary material online, Figure S3).
Figure 4.
Allele-specific effects of single nucleotide polymorphisms (SNPs) rs2519184, rs8234, and rs10798 on symptomatology. Allele-specific effects of SNPs rs2519184, rs8234, and rs10798 with regard to the occurrence of cardiac symptoms [unexplained syncope, documented torsades de pointes ventricular tachycardia/ventricular fibrillation, and/or (aborted) sudden death] are shown in the total combined study population (AMC and MC). Effects of SNP rs2519184 are displayed in (A). Effects of SNPs rs8234 and rs10798 are displayed in (B). Allele-specific haplotype analysis of the three SNPs with regard to the occurrence of symptoms is displayed in (C). Numbers below genotypes denote group sizes. Data are presented as mean; I bars represent standard errors. N, normal KCNQ1 allele; M, mutant KCNQ1 allele; green box, ancestral SNP variant; yellow box, derived SNP variant.
The effect of single nucleotide polymorphisms rs2519184, rs8234, and rs10798 on QTc in the general population
To study the effect of SNPs rs2519184, rs8234, and rs10798 in the general population, we evaluated whether the SNPs rs2519184, rs8234, or rs10798 are correlated with the QTc duration in two populations of the KORA study (KORA F3 and KORA S4). The data about rs2519184 have been published previously.12 This study indeed found a correlation between the SNP and QTc duration in their screening sample of 689 individuals, but this association was not found in a confirmation sample of 3277 individuals. Our novel analysis in the KORA study populations showed that rs8234 and rs10798 were not associated with QTc duration in KORA F3 and KORA S4 (Table 3).
Table 3.
The effect of single nucleotide polymorphisms rs8234 and rs10798 on QTc in the general population
| SNP | Population* | Number | Ancestral allele/haplotype | Derived allele/haplotype | MAF | Effect on QTc Beta (ms) | P-value |
|---|---|---|---|---|---|---|---|
| rs8234 | KORA F3 | 1459 | A | G | 0.3041 | −0.573105 | 0.52 |
| rs10798 | KORA F3 | 1459 | A | G | 0.304 | −0.572674 | 0.52 |
| rs8234 | KORA S4 | 975 | A | G | 0.3216 | 1.383768 | 0.13 |
| rs10798 | KORA S4 | 975 | A | G | 0.3209 | 1.42896 | 0.12 |
| Haplotype | KORA F3 | 1459 | AA | GG | 0.3047 | 0.569323 | 0.52 |
| Haplotype | KORA S4 | 975 | AA | GG | 0.3185 | 0.611701 | 0.42 |
Number, number of individuals within the screening sample; MAF, minor allele frequency. The effect on the QTc duration refers to the dosage (i.e. expected number of copies) of the major allele. The QTc duration is calculated by the Bazett's formula, and is corrected for age and gender.
In vitro effects of KCNQ1 3′ untranslated region single nucleotide polymorphism on expression
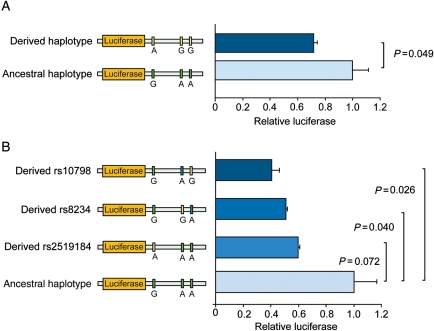
Since the above shows that the derived variants of SNPs rs2519184, rs8234, and rs10798 attenuate QTc prolongation when they are located in cis (i.e. on the same allele) to the LQT1 mutation, but aggravate QTc prolongation when located in trans (i.e. on the normal allele) to the mutation, we hypothesized that these SNP variants suppress the expression of the alleles on which they reside. Thus, the occurrence of these ‘suppressive’ 3′UTR variants in cis to the LQT1 mutation may attenuate QTc prolongation by decreasing the abundance of mutant protein available for tetrameric assembly, while ‘suppressive’ 3′UTR variants in trans to the mutation exerts the opposite effects on QTc by decreasing expression of the normal allele and thereby increasing the relative amount of mutant Kv7.1 subunits. To test this hypothesis, we cloned KCNQ1's 3′UTR with the ancestral 3′UTR haplotype (G-A-A) or the derived haplotype (A-G-G) of SNPs rs2519184, rs8234, and rs10798 downstream of the luciferase coding region in the pMIR-REPORTTM plasmid (Ambion), and transfected these plasmids into both primary neonatal rat cardiomyocytes (Figure 5A) and heart-derived H10 cells (Supplementary material online, Figure S4a). The plasmid containing the derived 3′UTR haplotype (A-G-G) had significantly lower luciferase activity in both cell types (P = 0.049 and P = 0.032, respectively). Moreover, introduction of either derived variant of SNP rs2519184, rs8234, or rs10798 into the ancestral haplotype was sufficient to decrease luciferase activity, indicating that those three nucleotides can separately suppress the expression of KCNQ1 (Figure 5A and Supplementary material online, Figure S4b).
Figure 5.
The functional effects of genetic variation in the 3′untranslated region (3′UTR) of KCNQ1 in vitro. (A) Luciferase assays in primary neonatal rat cardiomyocytes transfected with two independent reporter plasmids containing either the ancestral or the derived haplotype of single nucleotide polymorphisms (SNPs) in KCNQ1's 3′UTR. Observed differences in luciferase activity between the ancestral and derived haplotype suggest that expression is inhibited. (B) Luciferase assays in neonatal rat cardiomyocytes transfected with a reporter plasmid containing either the ancestral haplotype of the 3′UTR of KCNQ1 or the same reporter where the SNPs rs2519184, rs8234, and rs10798 were separately changed to the derived allele. Introducing either of the derived SNP variants was sufficient to decrease the luciferase activity. Data are presented as means; I bars denote standard errors. Green box, ancestral SNP variant; yellow box, derived SNP variant.
Discussion
This study shows that SNPs rs2519184, rs8234, and rs10798 in the 3′UTR of KCNQ1 modify the clinical effects of mutations in LQT1. The modifying effects of these SNPs on disease severity are robust and depend on the KCNQ1 allele on which they reside. When the SNPs are on the allele that contains the mutation, disease is less severe. Oppositely, when they reside on the healthy allele (free of the LQT1-causing mutation), the disease is clearly aggravated. Our in vitro experiments indicate that the derived variants of the SNPs suppress expression. This suggests that these SNPs modify disease severity in an allele-specific manner, by suppressing the allele on which they reside. The clear effects obtained by the isolated SNPs in vitro strongly suggest that they are functional and that the clinical effect is a direct consequence of these SNPs rather than being caused by distant genetic variation. Taken together, our data suggest that with the SNPs on the mutated allele, this mutated allele is suppressed and the expressed LQT1 phenotype is less severe. When the SNPs reside in trans to the mutation, i.e. on the allele opposite to the mutation, then this healthy allele is suppressed and the LQT1 phenotype is more severe (Figure 6). Furthermore, the 3′UTR SNPs were still associated with the occurrence of symptoms even after correction for QTc, suggesting that the SNPs may play a role in the occurrence of symptoms not only by modifying the baseline QTc but possibly also by modifying the response to arrhythmogenic triggers.
Figure 6.
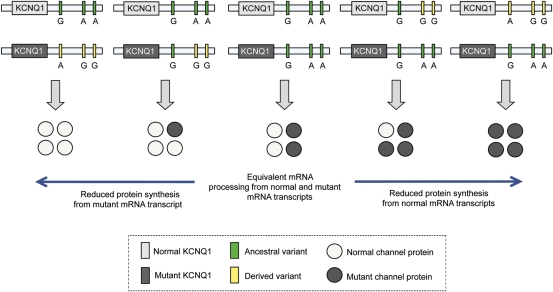
The postulated mechanism where single nucleotide polymorphisms (SNPs) in the 3′UTR of KCNQ1 modulate the assembly of the Kv7.1 potassium channel in type 1 long QT syndrome. Individuals with LQT1 are heterozygous for the disease-causing mutation in KCNQ1. Four KCNQ1-encoded subunits co-assemble to form one tetrameric channel. Single nucleotide polymorphisms in this region may influence repolarization by altering the balance and composition of the Kv7.1 tetramers derived from expression of the normal allele and the mutant allele. If SNPs exert no effect on expression, the balance between normal and mutant subunits within each channel is equal. However, if the derived variants of the SNPs cause reduced expression, then the balance between normal and mutant subunits within each channel depends on whether the ‘suppressive’ SNP variants reside on the normal allele or the mutant allele. If the ‘suppressive’ SNP variants reside on the normal allele, the number of normal subunits in the channels would decrease. Inversely, if the ‘suppressive’ SNP variants reside on the mutant allele, this would decrease the relative number of mutant subunits and shift the Kv7.1 tetramers to a greater percent of normal allele-derived monomeric subunits.
In line with our data, it has been previously shown in lymphoblastoid cell lines that the KCNQ1 allele containing the ancestral variant of SNP rs10798 is expressed three-fold higher than the allele with the derived variant of the SNP.13 Although in this study, differences in allelic expression of KCNQ1 were attributed to genetic imprinting (i.e. preferential expression of the maternal allele), genetic imprinting seems less likely to explain the cardiac effects as Lee et al.14 have shown that KCNQ1 is not imprinted in the heart. Moreover, if imprinting would explain the allele-specific disease-modifying effects of the 3′UTR SNPs, LQT1-positive subjects who have inherited the mutation from their father would be expected to have less severe phenotype. However, this was not the case for our LQT1 patients (as exemplified in Supplementary material online, Figures S2).
Our findings imply that LQT1 genetic testing should include analysis and cis/trans phase determination of suppressive 3′UTR SNPs in KCNQ1. More generally, our study shows that genetic variation in the 3′UTR may be an important source for clinical variability by altering the balance of expression between two alleles. This is the first clinical confirmation of previous predictions that genetic variation in the 3′UTR may be an essential source for ‘Darwinian fitness’, and hence individual variation in disease severity.7 Earlier reports have linked 3′UTR SNPs to various acquired diseases but could not establish allele-specific effects of the SNPs.15,16 Furthermore, other genetic factors have been shown to modify QTc in LQT1, e.g. SNPs in intronic regions of KCNQ1,12 and variants in genes encoding (putative) ion channel regulatory subunits, such as NOS1AP which encodes a nitric oxide synthase adaptor protein.17 However, it is noteworthy that the size of the effects on QTc and symptoms described here go well beyond what has been shown for genetics modifiers described previously.
Since the suppressive SNPs are expected to decrease overall Kv7.1 levels, we explored whether SNPs rs2519184, rs8234, and rs10798 were related to QTc in two populations from the KORA study.12 However, no relation with QTc was found in these populations, suggesting that the suppressive effects of these SNPs do not lower Kv7.1 sufficiently to induce QTc prolongation in healthy individuals.
In conclusion, we show that naturally occurring SNPs in KCNQ1's 3′UTR suppress gene expression in an allele-specific manner. In KCNQ1-mutation carriers, these functional SNPs may alter the balance between the two alleles. This may explain an important part of the incomplete penetrance and variable expressivity associated with LQT1. Our findings suggest that the clinical effects of a pathogenic mutation may be determined by naturally occurring genetic variation in its 3′UTR which results in the alteration of normal or mutant allele expression. This finding also suggests that in autosomal dominant diseases like LQTS, disease severity can be importantly modified by allelic imbalance induced by sequence variation in the 3′UTR stemming from the unaffected parent.
Supplementary material
Supplementary material is available at European Heart Journal online.
Funding
This work was supported by the Netherlands Heart Foundation (2007B167 to Y.M.P.), the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program (to M.J.A.), the Foundation Leducq (05 CVD) Alliance against Sudden Cardiac Death (to A.A.M.W.), the Academic Medical Center (to A.A.M.W. and A.S.A.), and CTMM COHFAR Center for Translational Molecular Medicine (to A.S.A.), and the National Institutes of Health (HL 106993 to J.R.G.).
Conflict of interest: M.J.A. and A.A.M.W. are consultants for Transgenomic that has released the FAMILIONTM genetic test for cardiac ion channel abnormalities, and Mayo Clinic Health Solutions receives royalties from PGxHealth. However, PGxHealth did not provide financial support for this study.
Supplementary Material
Acknowledgements
We are grateful for the help of Dr A.V. Postma from the ICIN Durrer Center with gathering DNA samples.
References
- 1.Roden DM. Clinical practice. Long-QT syndrome. N Engl J Med. 2008;358:169–176. doi: 10.1056/NEJMcp0706513. [DOI] [PubMed] [Google Scholar]
- 2.Splawski I, Shen J, Timothy KW, Lehmann MH, Priori S, Robinson JL, Moss AJ, Schwartz PJ, Towbin JA, Vincent GM, Keating MT. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, and KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 3.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 4.Vincent GM, Timothy KW, Leppert M, Keating M. The spectrum of symptoms and QT intervals in carriers of the gene for the long-QT syndrome. N Engl J Med. 1992;327:846–852. doi: 10.1056/NEJM199209173271204. [DOI] [PubMed] [Google Scholar]
- 5.Moss AJ, Shimizu W, Wilde AA, Towbin JA, Zareba W, Robinson JL, Qi M, Vincent GM, Ackerman MJ, Kaufman ES, Hofman N, Seth R, Kamakura S, Miyamoto Y, Goldenberg I, Andrews ML, McNitt S. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, Rajewsky N. Natural selection on human microRNA binding sites inferred from SNP data. Nat Genet. 2006;38:1452–1456. doi: 10.1038/ng1910. [DOI] [PubMed] [Google Scholar]
- 8.Bhuiyan ZA, Momenah TS, Amin AS, Al-Khadra AS, Alders M, Wilde AA, Mannens MM. An intronic mutation leading to incomplete skipping of exon-2 in KCNQ1 rescues hearing in Jervell and Lange-Nielsen syndrome. Prog Biophys Mol Biol. 2008;98:319–327. doi: 10.1016/j.pbiomolbio.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Jahn L, Sadoshima J, Greene A, Parker C, Morgan KG, Izumo S. Conditional differentiation of heart- and smooth muscle-derived cells transformed by a temperature-sensitive mutant of SV40 T antigen. J Cell Sci. 1996;109:397–407. doi: 10.1242/jcs.109.2.397. [DOI] [PubMed] [Google Scholar]
- 10.Leenders JJ, Wijnen WJ, Hiller M, van der Made I, Lentink V, van Leeuwen RE, Herias V, Pokharel S, Heymans S, de Windt LJ, Høydal MA, Pinto YM, Creemers EE. Regulation of cardiac gene expression by KLF15, a repressor of myocardin activity. J Biol Chem. 2010;285:27449–27456. doi: 10.1074/jbc.M110.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R Development Core Team. R: a language and environment for statistical computing. 2009 Available at: http://www.r-project.org . [Google Scholar]
- 12.Pfeufer A, Jalilzadeh S, Perz S, Mueller JC, Hinterseer M, Illig T, Akyol M, Huth C, Schöpfer-Wendels A, Kuch B, Steinbeck G, Holle R, Näbauer M, Wichmann HE, Meitinger T, Kääb S. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circ Res. 2005;96:693–701. doi: 10.1161/01.RES.0000161077.53751.e6. [DOI] [PubMed] [Google Scholar]
- 13.Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, Fan JB, Hudson TJ. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee MP, Hu RJ, Johnson LA, Feinberg AP. Human KVLQT1 gene shows tissue-specific imprinting and encompasses Beckwith-Wiedemann syndrome chromosomal rearrangements. Nat Genet. 1997;15:181–185. doi: 10.1038/ng0297-181. [DOI] [PubMed] [Google Scholar]
- 15.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saetrom P, Biesinger J, Li SM, Smith D, Thomas LF, Majzoub K, Rivas GE, Alluin J, Rossi JJ, Krontiris TG, Weitzel J, Daly MB, Benson AB, Kirkwood JM, O'Dwyer PJ, Sutphen R, Stewart JA, Johnson D, Larson GP. A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res. 2009;69:7459–7465. doi: 10.1158/0008-5472.CAN-09-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crotti L, Monti MC, Insolia R, Peljto A, Goosen A, Brink PA, Greenberg DA, Schwartz PJ, George AL., Jr NOS1AP is a genetic modifier of the long-QT syndrome. Circulation. 2009;120:1657–1663. doi: 10.1161/CIRCULATIONAHA.109.879643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.