Abstract
Fgfrl1 (fibroblast growth factor receptor-like 1) is a transmembrane receptor that is essential for the development of the metanephric kidney. It is expressed in all nascent nephrogenic structures and in the ureteric bud. Fgfrl1 null mice fail to develop the metanephric kidneys. Mutant kidney rudiments show a dramatic reduction of ureteric branching and a lack of mesenchymal-to-epithelial transition. Here, we compared the expression profiles of wildtype and Fgfrl1 mutant kidneys to identify genes that act downstream of Fgfrl1 signaling during the early steps of nephron formation. We detected 56 differentially expressed transcripts with 2-fold or greater reduction, among them many genes involved in Fgf, Wnt, Bmp, Notch, and Six/Eya/Dach signaling. We validated the microarray data by qPCR and whole-mount in situ hybridization and showed the expression pattern of candidate genes in normal kidneys. Some of these genes might play an important role during early nephron formation. Our study should help to define the minimal set of genes that is required to form a functional nephron.
Introduction
The mammalian kidney is a complex organ comprising thousands of nephrons that are connected by a branched collecting duct system [1]. The nephrons, the functional units of the kidney, filter the blood through a basement membrane and drain the filtrate via the collecting ducts to the bladder. In the mouse, nephron development is initiated at E10.5 when a caudal portion of the Wolffian duct near the hindlimbs bulges out and forms the ureteric bud. Signals from the metanephric mesenchyme induce the ureteric bud to branch in a stereotypical fashion to form the highly branched collecting duct system. The ureteric bud in turn releases signals that induce the metanephric mesenchyme to condense around the tips of the ureteric bud and to form the cap mesenchyme. Some cells of the cap mesenchyme undergo a mesenchymal-to-epithelial transition and develop into renal vesicles. These vesicles elongate, form s-shaped bodies and finally mature into nephrons.
Several signaling pathways are involved in the development of the nephron, including the Fgf/Fgfr, the Wnt/ß-catenin and the Notch/Presenilin pathway. To induce nephron formation, Wnt9b is secreted from the ureteric bud into the adjacent mesenchyme where it binds to Frizzled receptors and activates the canonical ß-catenin pathway [2], [3]. In response, the metanephric mesenchyme expresses the morphogens Fgf8 [4], [5] and Wnt4 [6]. Fgf8 is required for cell survival at different stages of nephrogenesis. Wnt4 induces cells from the cap mesenchyme to undergo the mesenchymal-to-epithelial transition, which finally leads to the formation of renal vesicles. Signaling by Notch and Presenilin is then needed to pattern the proximal tubule of the nephron [7], [8].
Recently, we have identified Fgfrl1 as a novel receptor that is essential for nephron development [9]. Fgfrl1 belongs to the Fgfr (fibroblast growth factor receptor) family of single transmembrane receptors (for review see [10]). Its extracellular domain resembles those of the classical Fgfrs in amino acid sequence and in that it contains three Ig-like loops. However, the intracellular domain differs from the classical receptors and does not possess any tyrosine kinase activity [11], [12]. The extracellular domain of Fgfrl1 interacts with heparin [13] and with Fgf ligands, primarily Fgf-2, -3, -4, -8, and -22 [14]. The intracellular domain binds to members of the Sprouty/Spred family that are known as negative regulators of the growth factor-mediated activation of the Ras/Raf/Erk signaling pathway [15]. During embryonic development, Fgfrl1 is expressed in tissues of the musculoskeletal system, including cartilage, bone and muscles [13], but also in the lung and the kidneys [9]. Information about the function of Fgfrl1 was gained from studies with mice, in which the Fgfrl1 gene was deleted by targeted inactivation [16], [17]. Fgfrl1 knock-out mice die shortly after birth due to malformation of the diaphragm. The mutant diaphragm muscle obviously is not strong enough to inflate the lungs after birth. However, the most striking phenotype of the Fgfrl1 deficient mice is the nearly complete absence of the metanephric kidneys. Utilizing organ cultures and different staining techniques, we demonstrated that Fgfrl1 deficiency leads to a dramatic reduction of ureteric branching and to a lack of mesenchymal-to-epithelial transition in the nephrogenic mesenchyme [9]. As a result, the mutant embryos lack any renal vesicles in their developing kidneys.
In the present study we used the DNA microarray profiling technique to identify genes that act downstream of Fgfrl1 signaling in the regulatory hierarchy of genes required for early nephron development. We confirmed reduced expression of Wnt4 and Fgf8 in the kidneys of the Fgfrl1 deficient mice. In addition, we identified more than 50 genes that are expressed at significantly reduced levels in our mutant mice. Many of these genes are involved in the Fgf/Fgfr, Wnt/ß-catenin, Bmp, Notch, and Six/Eya/Dach signaling pathway.
Results
Fgfrl1 is expressed throughout metanephric kidney development
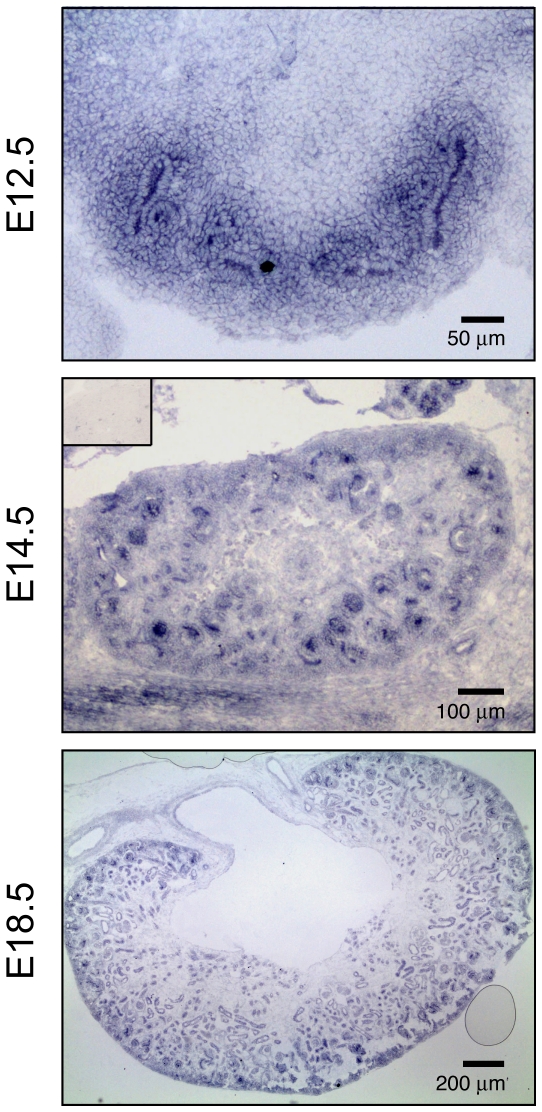
In a previous study we have used polyclonal antibodies on thin sections of E15.5 mouse kidneys to demonstrate that Fgfrl1 is expressed in nephrogenic structures of the cortical zone, with strong expression in renal vesicles, comma- and s-shaped bodies. Weaker staining was found in the undifferentiated mesenchyme and in the ureteric epithelium [9]. To verify these findings and to gain more information about Fgfrl1 expression during kidney development, we performed in situ hybridization on thin sections (SISH) at three developmental stages (Fig. 1). At E12.5, Fgfrl1 mRNA was highly expressed in the metanephric mesenchyme and in the ureteric bud. At E14.5, strong Fgfrl1 signal was detected in nascent nephrons and in the metanephric mesenchyme. At E18.5, Fgfrl1 signal was primarily found in tubules and nephrons. These results demonstrate that Fgfrl1 is expressed in developing nephrons and in the ureteric bud throughout kidney development, thus confirming our previous results obtained by immunohistochemistry.
Figure 1. Expression of Fgfrl1 in the developing mouse kidney.
Thin sections of embryonic mouse kidneys at E12.5, E14.5 and E18.5 were hybridized with a digoxigenin-labeled anti-sense RNA probe for Fgfrl1. After hybridization, the sections were incubated with alkaline phosphatase-conjugated antibodies against digoxigenin and the signal was developed with BM purple. Expression of Fgfrl1 was observed in the ureteric bud and in all nephrogenic structures. The inset of the panel at E14.5 shows a control section hybridized with the sense probe for Fgfrl1.
Transcriptional profiling of Fgfrl1 deficient kidneys
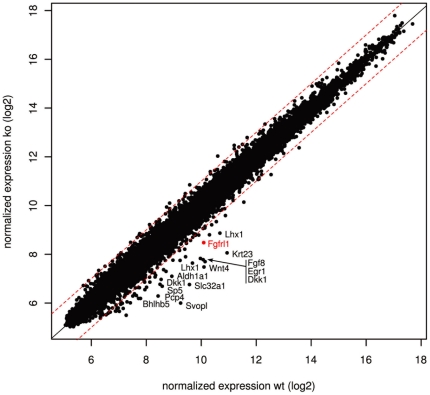
To determine how the absence of Fgfrl1 signaling would affect kidney development, we used Agilent DNA microarrays and compared mRNA levels between wildtype and Fgfrl1 deficient kidneys at E12.5. This time point was chosen because the first phenotypic differences between wildtype and Fgfrl1−/− kidneys are observed at E12.5 and because the first nephrogenic structures become visible at this developmental stage. Our efforts led to the identification of significant alterations in the transcriptome of the Fgfrl1 deficient kidneys. Of the ∼20,000 genes analyzed, 17 genes showed an up-regulation ≥2 fold and 56 genes showed a down-regulation ≥2 fold. None of the genes was up-regulated ≥3 fold and only 14 genes were down-regulated ≥3 fold (Fig. 2). Among the most strongly affected genes (indicated in the scatter plot of Fig. 2 by gene symbols) were Wnt4, Fgf8, Lhx1 and Fgfrl1, which had already been discovered as down-regulated markers in our previous study [9]. Expression of Eya1 and Six2, which define the uninduced mesenchyme, was barely affected by the absence of Fgfrl1 (fold change 1.3 and 1.0, respectively). Thus, the microarray data demonstrate the validity of our approach to identify mRNAs that may act downstream of Fgfrl1 in the regulatory hierarchy of genes required for nephron development.
Figure 2. Identification of transcripts that are differentially expressed in Fgfrl1 deficient kidneys.
The scatter plot shows average normalized signal intensities from three independent experiments using E12.5 kidneys from wildtype and Fgfrl1 knock-out mice. Each dot represents an individual gene. Dashed lines correspond to a fold change of 2. Transcripts that are down-regulated more than 3-fold are given by their gene symbol.
Arbitrarily, we have chosen a minimal fold change >1.9 and a p-value cutoff <0.05 to generate a table of the most critical genes that were down-regulated in Fgfrl1 deficient kidneys (Table 1, for a list of all genes see GEO Series accession number GSE32013). This table included all the genes that had previously been found to be affected in the Fgfrl1 knock-out mouse (indicated in bold in Tables 1 and 2). Besides Wnt4, Pax8, Fgf8 and Lhx1, our list included 60 other genes that might contribute to the phenotype of the Fgfrl1 knock-out mice. Among these hits are many genes that have previously been described to be essential for normal kidney development, such as Spry1 [18], Etv4 [19] and Itga8 [20], but there are also several genes whose function in kidney development has not been appreciated so far, including Fzd10, Frzb, Il17rd and Dach1 (Table 1).
Table 1. Genes with reduced expression in Fgfrl1 deficient kidneys.
| Nr | Gene Symbol | Gene Description | Gene ID | pValue | Fold Change | Verification Method | GUDMAP | |
| 1 | Svopl | SV2 related protein homolog-like | 320590 | 0.0022 | 9.5 | qPCR | 8950;7412 | |
| 2 | Krt23 | keratin 23 | 94179 | 0.0014 | 7.4 | qPCR, | WISH | |
| 3 | Slc32a1 | solute carrier family 32 member 1 | 22348 | 0.0015 | 7.0 | qPCR, | WISH | 13958 |
| 4 | Wnt4 | wingless-related MMTV integration site 4 | 22417 | 0.0013 | 6.2 | qPCR, | WISH | 8208;11295 |
| 5 | Dkk1 | dickkopf homolog 1 | 13380 | 0.0106 | 5.4 | qPCR, | WISH | 9041 |
| 6 | Egr1 | early growth response 1 | 13653 | 0.0077 | 4.9 | qPCR | 6106;11301 | |
| 7 | Pcp4 | immunoglobulin superfamily member 5 | 18546 | 0.0043 | 4.4 | qPCR, | WISH | |
| 8 | Fgf8 | fibroblast growth factor 8 | 14179 | 0.0015 | 4.4 | qPCR, | WISH | |
| 9 | Lhx1 | LIM homeobox protein 1 | 16869 | 0.0028 | 4.1 | qPCR, | WISH | 5384;7928 |
| 10 | Sp5 | trans-acting transcription factor 5 | 64406 | 0.0185 | 3.7 | WISH | ||
| 11 | Aldh1a1 | aldehyde dehydrogenase family 1, subfamily A1 | 11668 | 0.0069 | 3.5 | qPCR | ||
| 12 | Clec18a | C-type lectin domain family 18 member A | 353287 | 0.0055 | 3.4 | qPCR, | WISH | |
| 13 | Fgfrl1 | fibroblast growth factor receptor-like 1 | 116701 | 0.0035 | 3.1 | qPCR, | WISH | |
| 14 | Bhlhb5 | basic helix-loop-helix domain containing, class B5 | 59058 | 0.0002 | 3.0 | 5911 | ||
| 15 | Hes5 | hairy and enhancer of split 5 | 15208 | 0.0011 | 3.0 | 5928 | ||
| 16 | Fzd10 | frizzled homolog 10 | 93897 | 0.0180 | 2.9 | 8488 | ||
| 17 | Gpx6 | glutathione peroxidase 6 | 75512 | 0.0116 | 2.9 | |||
| 18 | Alx1 | ALX homeobox 1 | 216285 | 0.0104 | 2.8 | 5336 | ||
| 19 | Lmcd1 | LIM and cysteine-rich domains 1 | 30937 | 0.0001 | 2.7 | 6340 | ||
| 20 | Cck | cholecystokinin | 12424 | 0.0247 | 2.7 | |||
| 21 | Amph | amphiphysin | 218038 | 0.0158 | 2.7 | |||
| 22 | Aldh1a7 | aldehyde dehydrogenase family 1, subfamily A2 | 26358 | 0.0164 | 2.6 | |||
| 23 | Galntl2 | polypeptideN-acetylgalactosaminyltransferase-like 2 | 78754 | 0.0419 | 2.6 | |||
| 24 | Jag1 | jagged 1 | 16449 | 0.0269 | 2.5 | qPCR | 8532;11379 | |
| 25 | Hey1 | hairy/enhancer-of-split related with YRPW motif 1 | 15213 | 0.0101 | 2.4 | qPCR | 5912 | |
| 26 | Dll1 | delta-like 1 | 13388 | 0.0162 | 2.4 | qPCR | 11371 | |
| 27 | Plekhg6 | pleckstrin homology domain-containing family G6 | 213522 | 0.0065 | 2.4 | |||
| 28 | Akr1b7 | aldo-keto reductase family 1, member B7 | 11997 | 0.0225 | 2.4 | |||
| 29 | Greb1 | gene regulated by estrogen in breast cancer protein | 268527 | 0.0017 | 2.3 | 8529;8891 | ||
| 30 | Cxcr4 | chemokine (C-X-C motif) receptor 4 | 12767 | 0.0233 | 2.3 | qPCR | ||
| 31 | Uncx | UNC homeobox | 22255 | 0.0060 | 2.3 | qPCR | 5729 | |
| 32 | Notum | notum pectinacetylesterase homolog | 77583 | 0.0061 | 2.3 | |||
| 33 | Bmp2k | BMP2 inducible kinase | 140780 | 0.0434 | 2.3 | qPCR | ||
| 34 | C1qdc2 | family with sequence similarity 132, member A | 67389 | 0.0101 | 2.3 | 9273 | ||
| 35 | Ism1 | isthmin 1 homolog | 319909 | 0.0106 | 2.3 | |||
| 36 | Lef1 | lymphoid enhancer binding factor 1 | 16842 | 0.0071 | 2.3 | qPCR | 5539 | |
| 37 | Msx2 | homeobox, msh-like 2 | 17702 | 0.0358 | 2.2 | qPCR | 5365 | |
| 38 | Pax8 | paired box gene 8 | 18510 | 0.0202 | 2.2 | qPCR, | WISH | 10742;11179 |
| 39 | Etv4 | ets variant gene 4 (E1A enhancer binding protein) | 18612 | 0.0414 | 2.2 | 5486;12534 | ||
| 40 | Bmp2 | bone morphogenetic protein 2 | 12156 | 0.0020 | 2.2 | qPCR | 8949 | |
| 41 | B3galt5 | beta-1,3-galactosyltransferase 5 | 93961 | 0.0131 | 2.2 | |||
| 42 | Apom | apolipoprotein M | 55938 | 0.0127 | 2.1 | 10784 | ||
| 43 | Cxcl14 | chemokine (C-X-C motif) ligand 14 | 57266 | 0.0175 | 2.1 | 8425 | ||
| 44 | Ankrd56 | mus musculus ankyrin repeat domain 56 | 78088 | 0.0165 | 2.1 | |||
| 45 | Frzb | frizzled-related protein | 20378 | 0.0102 | 2.1 | qPCR, | WISH | |
| 46 | Naaa | N-acylethanolamine acid amidase | 67111 | 0.0077 | 2.1 | |||
| 47 | Osr2 | odd-skipped related 2 | 107587 | 0.0094 | 2.0 | qPCR | 6335;13623 | |
| 48 | Il17rd | interleukin 17 receptor D | 171463 | 0.0003 | 2.0 | qPCR, | WISH | |
| 49 | Cpa2 | carboxypeptidase A2, pancreatic | 232680 | 0.0072 | 2.0 | |||
| 50 | Lbx2 | ladybird homeobox homolog 2 | 16815 | 0.0400 | 2.0 | qPCR | 6590 | |
| 51 | Spry1 | sprouty homolog 1 | 24063 | 0.0183 | 2.0 | WISH | ||
| 52 | Col13a1 | collagen type XIII, alpha 1 | 12817 | 0.0034 | 2.0 | 8082 | ||
| 53 | Dusp2 | dual specificity phosphatase 2 | 13537 | 0.0003 | 2.0 | |||
| 54 | Rbm20 | RNA binding motif protein 20 | 73713 | 0.0107 | 2.0 | |||
| 55 | Cdh4 | cadherin 4 | 12561 | 0.0221 | 2.0 | 7763 | ||
| 56 | Chrdl2 | chordin-like 2 | 69121 | 0.0349 | 2.0 | |||
| 57 | Gldc | glycine decarboxylase | 104174 | 0.0207 | 1.9 | |||
| 58 | Itga8 | integrin alpha 8 | 241226 | 0.0062 | 1.9 | qPCR | ||
| 59 | Car4 | carbonic anhydrase 4 | 12351 | 0.0006 | 1.9 | |||
| 60 | Metap2 | methionine aminopeptidase 2 | 56307 | 0.0234 | 1.9 | |||
| 61 | Unc93a | unc-93 homolog A | 381058 | 0.0076 | 1.9 | |||
| 62 | Gdnf | glial cell line derived neurotrophic factor | 14573 | 0.0445 | 1.9 | qPCR | ||
| 63 | Dach1 | dachshund 1 | 13134 | 0.0062 | 1.9 | qPCR, | WISH | |
| 64 | Hs3st3b1 | heparan sulfate(glucosamine)3-O-sulfotransferase3B1 | 54710 | 0.0342 | 1.9 | 11741 | ||
Table 2. Verification of differential gene expression by qPCR.
| Gene | Fold change wt/ko qPCR | Fold change wt/ko | |||
| E11.5 | E12.5 | E14.5 | E16.5 | array E12.5 | |
| Slc32a1 | 0.8 | >20 | 13.4 | 11.7 | 7 |
| Krt23 | 7.5 | 17.4 | 1.5 | 3.6 | 7.4 |
| Pcp4 | 2.7 | 17 | 6 | 1 | 4.4 |
| Wnt4 | 2.3 | 13.3 | 3.8 | 2.3 | 6.2 |
| Pax8 | 1.5 | 9.6 | 3.9 | 4.7 | 2.2 |
| Svopl | 11.1 | 9.4 | 10.6 | >20 | 9.5 |
| Fgf8 | 3.2 | 9.2 | 16.4 | >20 | 4.4 |
| Il17rd | 2.3 | 8.5 | 5.9 | 4.9 | 2 |
| Lef1 | 2.4 | 8.2 | 0.8 | 2.1 | 2.3 |
| Lhx1 | 0.7 | 7.7 | 12.7 | >20 | 4.1 |
| Jag1 | 2 | 7.4 | 5.7 | 3.9 | 2.5 |
| Uncx | 1.6 | 7 | 10.1 | 17.2 | 2.3 |
| Itga8 | 3.1 | 6.9 | 2.6 | 2.1 | 1.9 |
| Fgfrl1 | >20 | 6.8 | 7.5 | >20 | 3 |
| Frzb | 2.5 | 6.1 | 2.9 | 3.4 | 2.1 |
| Dach1 | 1.7 | 6 | 4 | 5.1 | 1.9 |
| Bmp2 | 0.8 | 5.2 | 7.9 | 6.3 | 2.2 |
| Osr2 | 2.9 | 4.8 | 11.7 | >20 | 2 |
| Egr1 | 1 | 4.6 | 2.1 | 1.5 | 4.9 |
| Aldh1a1 | 9.8 | 4 | 6.3 | 3.7 | 3.5 |
| Gdnf | 3.5 | 3 | 3.1 | 4.1 | 1.9 |
| Cxcr4 | 1.7 | 3.4 | 3.7 | 3.1 | 2.3 |
| Dkk1 | 2.5 | 3 | 13.1 | >20 | 5.4 |
| Clec18a | 2.1 | 2.3 | >20 | >20 | 3.4 |
| Lbx2 | 0.8 | 2.1 | 2.1 | 1.5 | 2 |
| Bmp2k | 1 | 2 | 1 | 1 | 2.3 |
| Dll1 | 1.1 | 1.7 | 7.4 | 13.6 | 2.4 |
| Msx2 | 0.4 | 1.6 | 1.2 | 0.7 | 2.2 |
| Hey1 | 1.6 | 1.4 | 3.3 | 5 | 2.4 |
| Tcfcp2l1 | 3.1 | 1.4 | 1.2 | 3.6 | 0.5 |
| Cxcl12 | 2.2 | 1.3 | 0.7 | 1.2 | 0.5 |
| Rps9 | 1.9 | 1.2 | 1.8 | 2.4 | 1.1 |
| Gapdh | 1 | 1 | 1 | 1 | 1 |
| Col1a1 | 0.8 | 1 | 0.7 | 0.5 | 0.5 |
Validation by quantitative PCR
In order to confirm the results of the DNA microarray, we quantified the mRNA levels of selected genes by RT-PCR. For this purpose, we focused on hits that had yielded large differences between wildtype and mutant kidneys. In this way, 29 down-regulated genes, two control genes (Rps9 and Gapdh) and 3 up-regulated genes were analyzed by qPCR (Table 2). For the majority of the down-regulated hits, the qPCR results were found to be in good agreement with the microarray data, although the differences (fold changes) observed by qPCR were larger than those determined by microarray analysis. Only three genes (Dll1, Msx2, Hey1) showed a minimal fold change <1.9 by qPCR, which had been selected as cutoff above.
In sharp contrast to the down-regulated transcripts, none of the up-regulated transcripts (Tcfcp2l1, Cxcl12, Col1a1) could be confirmed by qPCR as the fold changes observed by qPCR were between 1.0 and 1.4. We may therefore conclude that the lack of Fgfrl1 expression in the mutant kidneys was barely compensated for by the up-regulation of other genes. In particular, the classical receptors Fgfr1-Fgfr4 did not show altered expression in the mutant kidneys.
For the microarray experiment, we had used E12.5 kidneys because overt differences in the phenotype between wildtype and Fgfrl1 deficient kidneys appear at this developmental stage. However, changes in gene expression may occur prior to the actual appearance of an altered phenotype and these changes may or may not persist throughout kidney development. We therefore analyzed the 29 down-regulated genes, the 3 up-regulated genes and the two control genes also at three additional time points during kidney development, one prior to and two after the stage used for the microarray experiment (E11.5, E14.5, E16.5). These data are also included in Table 2. Although some of the results might be difficult to interpret, the majority of the genes can clearly be grouped into two different categories. One category includes genes whose fold changes show a bell-shaped curve during embryonic development (e.g. Slc32a1, Pcp4, Wnt4, Lef1, Itga8, Egr1). These genes might preferentially be required during the initial stages of nephron development. The other category includes genes whose fold changes steadily increase throughout development (e.g. Fgf8, Lhx1, Uncx, Osr2, Dkk1, Clec18a, Dll1). It is likely that these genes are particularly needed at later stages of nephron development.
Validation by WISH
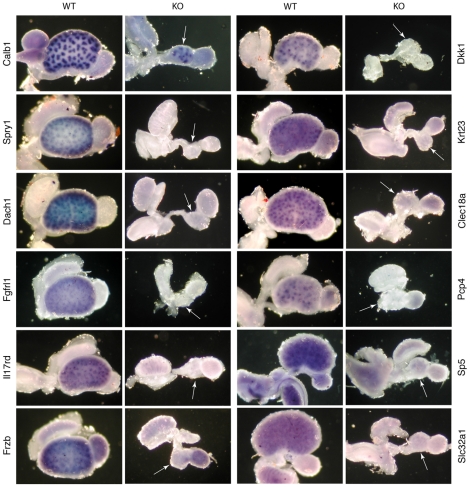
To further validate our data and to examine the spatial expression pattern of the genes in the kidney, we performed whole-mount in situ hybridization experiments (WISH). For this purpose, we used E14.5 kidneys because samples at this stage revealed a clearer expression pattern than samples at E12.5 due to the increased size and the generally stronger gene expression level [9]. We primarily focused on genes, which had shown a difference >3 and which had not yet been annotated in the GUDMAP database (www.gudmap.org). In total 12 genes were tested, including the positive control Calb1 (Fig. 3). Calb1 revealed the expected expression pattern in the ureteric bud and its derivatives, in both the wildtype and the Fgfrl1 knock-out kidneys. In contrast, expression of the other 11 genes was significantly reduced or even absent in the Fgfrl1 deficient kidneys (Fig. 3). Likewise, expression of the four nephrogenic marker genes Wnt4, Fgf8, Lhx1 and Pax8 that had been analyzed by WISH in our previous study was not detected in the Fgfrl1−/− kidneys [9]. Thus, our in situ hybridization data confirmed the microarray and the qPCR results.
Figure 3. Whole mount in situ hybridization (WISH) of selected marker genes in E14.5 kidneys.
Kidneys from wildtype and Fgfrl1 knock-out mice were hybridized with digoxigenin-labeled RNA probes and the probes were detected with alkaline phosphatase conjugated antibodies against digoxigenin. All probes produced clear expression patterns in the wildtype kidneys. In contrast, the same probes did not display any pattern in knock-out kidneys, with the exception of Calb1 that was included as a positive control. The white arrows indicate the kidney rudiments of the knock-out embryos.
It is of interest to note that most of the genes detected by WISH in the wildtype kidneys were expressed in the ureteric bud and/or in developing nephrogenic structures, such as renal vesicles and s-shaped bodies. These genes included Wnt4, Fgf8, Lhx1 and Pax8 from the previous study and Spry1, Dach1, Dkk1, Il17rd, Frzb, Krt23, Clec18a and Pcp4 from the present study (Fig. 3). In contrast, expression of the transcription factor Sp5 and the amino acid transporter Slc32a1 was rather diffuse, revealing no distinct pattern. We may therefore conclude that the majority of the differentially expressed genes identified by microarray analysis and confirmed by WISH showed expression in renal vesicles and s-shaped bodies, structures that are missing in our Fgfrl1 deficient mice.
Discussion
In the present study we demonstrated that Fgfrl1 is expressed in the ureteric bud and in all nephrogenic structures of wildtype mice, including renal vesicles and comma- and s-shaped bodies. Mice lacking the Fgfrl1 gene do not develop any nephrogenic structures. A careful analysis of the kidney transcriptome from wildtype and knock-out mice allowed us to identify more than 50 genes that act - directly or indirectly - downstream of Fgfrl1 in the regulatory cascade of genes required for early nephron development. Many of these genes appear to be involved in well-established signaling pathways. However, it cannot be deduced from our study whether all these genes are directly involved in the same signaling pathway as Fgfrl1. Since Fgfrl1 deficient mice do not develop any renal vesicles it is also possible that some of the identified mRNAs lack in the Fgfrl1 null mice simply because they are normal constituents of renal vesicles. To minimize such “secondary” effects, we analyzed kidneys at stage E12.5 where the first nephrogenic structures become visible and where the first phenotypic differences between wildtype and Fgfrl1−/− mice are observed.
FGF signaling pathway
By virtue of our microarray approach, we found five down-regulated genes (Fgf8, Spry1, Il17rd, Ism1, Etv4) that are involved, directly or indirectly, in Fgf signaling. Fgf8 is essential for nephron formation as mice lacking this gene do not progress beyond the renal vesicle stage [4], [5]. Interestingly, Fgf8 is one of the best ligands for Fgfrl1 as demonstrated by a ligand dot blot assay [14]. Spry1 is an antagonist of Fgf signaling, which is crucial for normal outgrowth of the ureteric bud as Spry1−/− embryos possess supernumerous ureteric buds [18]. Il17rd (also termed Sef), Ism1 and Etv4 belong to the Fgf synexpression group [20]–[22]. This group comprises several genes that show a similar spatiotemporal expression pattern and that may serve a similar function during development. Fgfrl1 [10], Il17rd [21], Spry2 and Spry4 [23] have been reported to act as negative regulators of Fgf signaling. The exact function of Fgfrl1 is not yet known, but we have speculated that it might act as a decoy receptor, which sequesters Fgf ligands away from the actively signaling receptors, or as a dominant negative binding partner, which interacts with the other receptors and inhibits transphosphorylation of the intracellular domains [13]. If this were true, one would expect up-regulation of genes that act downstream of FGF signaling, such as Fgfr1, Fgfr2 or FGF8, resulting in increased numbers of ureteric buds and nephrogenic structures. In sharp contrast, Fgfrl1 null mice have a phenotype with renal dysplasia [9] very similar to mice with a conditional disruption of Fgf8 [4], [5] or a compound disruption of the two receptors Fgfr1 and Fgfr2 [24]. This observation suggests that Fgfrl1 might act as a positive regulator of FGF signaling during kidney development and not as a decoy receptor.
WNT signaling pathway
The Wnt signaling pathway is often activated in concert with Fgf signaling during developmental processes [25]. We found by our microarray approach at least five genes that have been implicated in Wnt signaling, namely Wnt4, Fzd10, Frzb, Lef1 and Sp5. Among these hits, Wnt4 ranked at the top with a 6-fold expression difference between wildtype and knock-out mice when measured by microarray and 13-fold when verified by qPCR experiments. Wnt4 is usually expressed in the metanephric mesenchyme, where it induces the mesenchymal-to-epithelial transition. Therefore mice lacking Wnt4 activity show a greatly reduced number of nascent nephrogenic structures [6].
In a recent study, Valerius & McMahon [26] performed a transcriptional profiling screen using Wnt4 deficient kidneys. The authors identified 236 genes with differential expression levels between wildtype and Wnt4−/− kidneys. Interestingly, several genes exhibiting reduced expression in their study were also found to be down-regulated in our study, such as Dll1, Pcp4, Dkk1, Pax8, Fgf8, Lhx1, Hes5, Hey1 and Egr1. This result can be explained by the fact that Fgfrl1 acts upstream of Wnt4 in the cascade of regulatory genes as Fgfrl1 deficient mice lack Wnt4 expression in their kidney rudiments.
The other members of the Wnt signaling cascade that were significantly down-regulated in our study appear to have diverse functions. Fzd10 is one of the receptors for Wnt ligands. Frzb is a secreted, frizzled-related receptor that interferes with Wnt signaling. Lef1 is a transcription factor participating in Wnt signaling. Canonical Wnt signaling leads to the stabilization of ß-catenin, which - after translocation to the nucleus - interacts with transcription factors of the Lef/Tcf family to induce expression of target genes [27]. Sp5 is a member of the Sp1 transcription factor family. It ranks among the ten best hits of our microarray analysis. Fujimura et al. [28] presented evidence that Sp5 is involved in Wnt signaling since constitutive activation of the Wnt/ß-catenin pathway resulted in the up-regulation of Sp5 expression in the mouse telencephalon.
Bmp signaling
With our microarray, we found reduced expression of Bmp2 and Bmp2k. The Bmps play a key role in the development of the skeleton, but they are also involved in patterning of the metanephric kidney [29]. Bmp2 is expressed in the distal renal vesicle as shown by Georgas et al. [30]. The authors suggested that Bmp2 might be involved - together with other factors - in the fusion of the renal vesicle with the ureteric tip. Bmp2k is a serine/threonine protein kinase whose expression is induced after addition of Bmp2 to prechondroblastic cells in order to trigger their differentiation [31]. However, the role of Bmp2k during nephron formation has not yet been investigated.
Notch signaling pathway
Notch signaling is required to pattern the distal and proximal tubule of the nephron [32]. In our microarray, the Notch ligands Jag1 and Dll1 as well as the downstream effector Hey1 were down-regulated. However, Hey1 and Dll1 showed only mild reduction when validated by qPCR (1.4- and 1.7-fold, respectively), in contrast to Jag1, whose expression was reduced 7.4-fold. Jag1 is known to interact with Notch2 and other Notch receptors [33]. It regulates ureteric budding and branching by crosstalk with Gdnf/Ret signaling [34]. It is therefore likely that downregulation of Jag1 in the Fgfrl1 deficient kidneys reduces Notch signaling and hence interferes with normal nephron development.
Six-Eya-Dach signaling pathway
Six (sine oculis), Eya (eyes absent) and Dach are transcription factors that constitute the “retinal determination network”, whose loss inhibits eye development and whose forced expression leads to ectopic eye formation [35]. It is believed that the three factors form a complex that binds to the promoter region of target genes to control eye formation in mammals and insects. Besides their function in eye formation, Six1 and Eya1 are also involved in the development of the metanephric kidneys of mammals [36] as targeted deletion of either one of these genes leads to severe kidney malformation. Targeted disruption of Dach1, however, does not seem to have any obvious effect on kidney or eye formation, although mice without Dach1 exhibit postnatal lethality [37].
In our gene array, Six1 and Eya1 were not differentially expressed (fold change 1.1 and 1.3, respectively). This is in contrast to Dach1, which was significantly down-regulated (qPCR 6.0-fold, Table 2). It is therefore likely that Six1 and Eya1 act upstream, while Dach1 acts downstream of Fgfrl1 in the same regulatory network of genes that are required for kidney development. As a matter of fact, the phenotypes of Fgfrl1, Eya1 and Six1 deficient mice look intriguingly similar. All of them show kidney and bone malformations [9], [38], [39]. In addition, Six1 and Fgfrl1 knock-out mice exhibit defects in the diaphragm [16], [40]. Dach1 is expressed in developing nephrons, primarily in comma- and s-shaped bodies [41]. This is consistent with our WISH experiments where a prominent signal for Dach1 was found in the metanephric mesenchyme surrounding the ureteric tips (Fig. 3). Brunskill et al. [42] observed strong upregulation of Dach1 during nephron formation, especially when the stage of the renal vesicle was compared with that of the s-shaped body (46-fold). It is likely that Fgfrl1 is involved in this upregulation since our Fgfrl1 deficient kidneys show strongly reduced Dach1 expression.
Conclusions
We have identified a number of genes that act downstream of Fgfrl1 signaling in the regulatory hierarchy of genes required for early nephron development. Several of these genes are involved in Fgf/Fgfr, Wnt/ß-catenin, Bmp, Notch, and Six/Eya/Dach signaling. The downregulation of these genes might be responsible for the lack of nephrogenesis observed in Fgfrl1 knock-out mice. For some of the identified genes, a potential involvement in the development of the metanephric kidneys has not yet been appreciated (e.g. Fzd10, Frzb, Il17rd and Dach1). Our study should therefore help to define the minimal set of genes that is required for normal nephron formation.
Materials and Methods
Animals
All animal work was conducted according to the relevant national guidelines and was approved by the Amt für Landwirtschaft und Natur of Bern (approval number 69/09). The Fgfrl1 deficient mice have previously been described [9]. Littermates of wildtype and Fgfrl1 knock-out mice were used for all experiments. For an exactly timed pregnancy, the noon of the day, at which a vaginal plug was detected, was considered as E0.5.
DNA Microarray analysis
Kidney rudiments were dissected in parallel from wildtype and Fgfrl1 knock-out mice of stage E12.5. Total RNA was extracted from pooled kidney rudiments (n = 6–9) with the GeneElute miniprep kit from Sigma-Aldrich Co. The quality of the RNA was assessed with an Agilent 2100 Bioanalyser (Agilent Technologies, Palo Alto, CA, USA). Three individual RNA preparations from wildtype mice and three individual RNA preparations from Fgfrl1 knock-out mice were separately transcribed into double stranded cDNA in the presence of RNA poly-A controls (RNA Spike-In Kit, Agilent 5188-5279). The cDNAs were transcribed with T7 polymerase into cRNA utilizing cyanine 3-CTP (Cy3) for knock-out and cyanine 5-CTP (Cy5) for wildtype samples, respectively (Agilent 5190-0444). No amplification step was performed. The three pairs of fluorescently labeled cRNA were hybridized for 17 h at 65°C to an Agilent gene expression array (Whole Mouse Genome Microarray 4×44 K, G4122F) according to the instructions of the manufacturer utilizing reagents from the Gene Expression Hybridization Kit (Agilent 5188-5242). The slides were washed and scanned using an Agilent G2565BA microarray scanner. Signals were extracted from images using the Agilent Feature Extraction software version 10. Data analysis was performed on the R platform for statistical computing with packages from the Bioconductor project [43]. Gene annotation and identifier conversions were retrieved from the Mouse Genome Database (MGD, http://www.informatics.jax.org). All microarray data were deposited in the GEO database and comply with MIAME standards (accession number GSE32013).
Quantitative PCR analysis
Quantitative PCR was performed as previously described [9]. In brief, the RNA was transcribed into first strand cDNA with reverse transcriptase from Moloney Murine Leukemia Virus. The cDNAs were quantified by real time PCR on an ABI 7500 platform using the primer pairs listed in Table S1.
In situ hybridization
Whole-mount in situ hybridization (WISH) was performed as described before [9]. Different hybridization probes were generated by PCR utilizing cDNA prepared from E16.5 kidneys and the primer pairs listed in Table S2.
In situ hybridization on thin sections (SISH) was performed according to Koch et al. [44] with minor modifications. Sense and anti-sense riboprobes for Fgfrl1 were prepared by reverse transcription from the full-length cDNA cloned into the vector pcDNA3.1 (+/−) using the digoxigenin RNA labeling kit from Roche Diagnostics (Rotkreuz, Switzerland). Dissected kidneys were fixed with paraformaldehyde (4% PFA in PBS, overnight at 4°C) and incubated in a sucrose solution (30% sucrose in PBS, overnight at 4°C). Equilibrated samples were embedded in Tissue-Tek, frozen on dry ice and cut to 12 µm sections. Cryosections were fixed with PFA (4% in PBS, 20 min at RT) and digested with proteinase K (10 µg/ml, 10 min at RT). After refixation (4% PFA, 20 min), sections were acetylated (0.25% acetic anhydride, 0.1 M triethanolamine, pH 8.0, 10 min). Acetylated sections were prehybridized in hybridization buffer (50% formamide, 4× SSC, 2× Denhardt's solution, 5% dextran sulfate, 100 µg/ml yeast tRNA, 5 h at RT) and hybridized with the Fgfrl1 probe (overnight at 68°C with 1 µg/ml of the digoxigenin-labeled riboprobe). After a series of washing steps (wash 1: 0.2× SSC, 30 min at 60°C; wash 2: 50% formamide, 2× SSC, 30 min at RT; wash 3: 0.2× SSC, 10 min at RT; wash 4: 0.1 M maleic acid, 0.15 M NaCl, 0.1% Tween-20, pH 7.4, 15 min at RT) the sections were blocked with BSA (3% in Tris buffered saline, 2 h at RT) and incubated with anti-digoxigenin antibodies (alkaline phosphatase-conjugated Fab fragments from Roche, diluted 1∶2000, overnight at 4°C). The slides were rinsed and equilibrated with detection buffer (0.1 M Tris, 0.1 M NaCl, 50 mM MgCl2, pH 9.5). Finally, the hybridization signal was developed with BM Purple (Roche, 12–24 h at RT) and the slides were photographed under an Olympus BX-51 microscope.
Supporting Information
Primers used for RT-PCR.
(DOC)
Primers used for WISH. Underlined nucleotides indicate restriction sites used for subcloning.
(DOC)
Acknowledgments
We thank Sabina Hafen-Wirth and Andrea Patrignani (Functional Genomic Center Zurich) for their help with the Agilent gene expression arrays.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Swiss National Science Foundation (grant 31003A-127046) and the Swiss Foundation for Research on Muscular Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Costantini F, Kopan R. Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev Cell. 2010;18:698–712. doi: 10.1016/j.devcel.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP. Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell. 2005;9:283–292. doi: 10.1016/j.devcel.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Park JS, Valerius MT, McMahon AP. Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development. 2007;134:2533–2539. doi: 10.1242/dev.006155. [DOI] [PubMed] [Google Scholar]
- 4.Grieshammer U, Cebrian C, Ilagan R, Meyers E, Herzlinger D, et al. FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development. 2005;132:3847–3857. doi: 10.1242/dev.01944. [DOI] [PubMed] [Google Scholar]
- 5.Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni-Underwood S, et al. Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development. 2005;132:3859–3871. doi: 10.1242/dev.01945. [DOI] [PubMed] [Google Scholar]
- 6.Stark K, Vainio S, Vassileva G, McMahon AP. Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature. 1994;372:679–683. doi: 10.1038/372679a0. [DOI] [PubMed] [Google Scholar]
- 7.Wang P, Pereira FA, Beasley D, Zheng H. Presenilins are required for the formation of comma- and s-shaped bodies during nephrogenesis. Development. 2003;130:5019–5029. doi: 10.1242/dev.00682. [DOI] [PubMed] [Google Scholar]
- 8.Cheng HT, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, et al. Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development. 2007;134:801–811. doi: 10.1242/dev.02773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerber SD, Steinberg F, Beyeler M, Villiger PM, Trueb B. The murine Fgfrl1 receptor is essential for the development of the metanephric kidney. Dev Biol. 2009;335:106–119. doi: 10.1016/j.ydbio.2009.08.019. [DOI] [PubMed] [Google Scholar]
- 10.Trueb B. Biology of FGFRL1, the fifth fibroblast growth factor receptor. Cell Mol Life Sci. 2011;68:951–964. doi: 10.1007/s00018-010-0576-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 12.Sleeman M, Fraser J, McDonald M, Yuan S, White D, et al. Identification of a new fibroblast growth factor receptor, FGFR5. Gene. 2001;271:171–182. doi: 10.1016/s0378-1119(01)00518-2. [DOI] [PubMed] [Google Scholar]
- 13.Trueb B, Zhuang L, Taeschler S, Wiedemann M. Characterization of FGFRL1, a novel fibroblast growth factor (FGF) receptor preferentially expressed in skeletal tissues. J Biol Chem. 2003;278:33857–33865. doi: 10.1074/jbc.M300281200. [DOI] [PubMed] [Google Scholar]
- 14.Steinberg F, Zhuang L, Beyeler M, Kälin RE, Mullis PE, et al. The FGFRL1 receptor is shed from cell membranes, binds fibroblast growth factors (FGFs), and antagonizes FGF signaling in Xenopus embryos. J Biol Chem. 2010;285:2193–2202. doi: 10.1074/jbc.M109.058248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhuang L, Villiger PM, Trueb B. Interaction of the receptor FGFRL1 with the negative regulator Spred1. Cell Signal. 2011;23:1496–1504. doi: 10.1016/j.cellsig.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Baertschi S, Zhuang L, Trueb B. Mice with a targeted disruption of the Fgfrl1 gene die at birth due to alterations in the diaphragm. FEBS J. 2007;274:6241–6253. doi: 10.1111/j.1742-4658.2007.06143.x. [DOI] [PubMed] [Google Scholar]
- 17.Catela C, Bilbao-Cortes D, Slonimsky E, Kratsios P, Rosenthal N, et al. Multiple congenital malformations of Wolf-Hirschhorn syndrome are recapitulated in Fgfrl1 null mice. Dis Model Mech. 2009;2:283–294. doi: 10.1242/dmm.002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, et al. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller U, Wang D, Denda S, Meneses JJ, Pedersen RA, et al. Integrin alpha8beta1 is critically important for epithelial-mesenchymal interactions during kidney morphogenesis. Cell. 1997;88:603–613. doi: 10.1016/s0092-8674(00)81903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ron D, Fuchs Y, Chorev DS. Know thy Sef: a novel class of feedback antagonists of receptor tyrosine kinase signaling. Int J Biochem Cell Biol. 2008;40:2040–2052. doi: 10.1016/j.biocel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Niehrs C, Meinhardt H. Modular feedback. Nature. 2002;417:35–36. doi: 10.1038/417035a. [DOI] [PubMed] [Google Scholar]
- 23.Guy GR, Jackson RA, Yusoff P, Chow SY. Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol. 2009;203:191–202. doi: 10.1677/JOE-09-0110. [DOI] [PubMed] [Google Scholar]
- 24.Poladia DP, Kish K, Kutay B, Hains D, Kegg H, et al. Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol. 2006;291:325–339. doi: 10.1016/j.ydbio.2005.12.034. [DOI] [PubMed] [Google Scholar]
- 25.Katoh M, Katoh M. Cross-talk of WNT and FGF signaling pathways at GSK3beta to regulate beta-catenin and SNAIL signaling cascades. Cancer Biol Ther. 2006;5:1059–1064. doi: 10.4161/cbt.5.9.3151. [DOI] [PubMed] [Google Scholar]
- 26.Valerius MT, McMahon AP. Transcriptional profiling of Wnt4 mutant mouse kidneys identifies genes expressed during nephron formation. Gene Expr Patterns. 2008;8:297–306. doi: 10.1016/j.gep.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmidt-Ott KM, Masckauchan TN, Chen X, Hirsh BJ, Sarkar A, et al. Beta-catenin/TCF/Lef controls a differentiation-associated transcriptional program in renal epithelial progenitors. Development. 2007;134:3177–3190. doi: 10.1242/dev.006544. [DOI] [PubMed] [Google Scholar]
- 28.Fujimura N, Vacik T, Machon O, Vlcek C, Scalabrin S, et al. Wnt-mediated down-regulation of Sp1 target genes by a transcriptional repressor Sp5. J Biol Chem. 2007;282:1225–1237. doi: 10.1074/jbc.M605851200. [DOI] [PubMed] [Google Scholar]
- 29.Cain JE, Hartwig S, Bertram JF, Rosenblum ND. Bone morphogenetic protein signaling in the developing kidney: present and future. Differentiation. 2008;76:831–842. doi: 10.1111/j.1432-0436.2008.00265.x. [DOI] [PubMed] [Google Scholar]
- 30.Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, et al. Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol. 2009;332:273–286. doi: 10.1016/j.ydbio.2009.05.578. [DOI] [PubMed] [Google Scholar]
- 31.Kearns AE, Donohue MM, Sanyal B, Demay MB. Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J Biol Chem. 2001;276:42213–42218. doi: 10.1074/jbc.M106163200. [DOI] [PubMed] [Google Scholar]
- 32.McCright B. Notch signaling in kidney development. Curr Opin Nephrol Hypertens. 2003;12:5–10. doi: 10.1097/00041552-200301000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Shimizu K, Chiba S, Kumano K, Hosoya N, Takahashi T, et al. Mouse jagged1 physically interacts with notch2 and other notch receptors. Assessment by quantitative methods. J Biol Chem. 1999;274:32961–32969. doi: 10.1074/jbc.274.46.32961. [DOI] [PubMed] [Google Scholar]
- 34.Kuure S, Sainio K, Vuolteenaho R, Ilves M, Wartiovaara K, et al. Crosstalk between Jagged1 and GDNF/Ret/GFRalpha1 signalling regulates ureteric budding and branching. Mech Dev. 2005;122:765–780. doi: 10.1016/j.mod.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Wawersik S, Maas RL. Vertebrate eye development as modeled in Drosophila. Hum Mol Genet. 2000;9:917–925. doi: 10.1093/hmg/9.6.917. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Oghi KA, Zhang J, Krones A, Bush KT, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 37.Davis RJ, Shen W, Sandler YI, Amoui M, Purcell P, et al. Dach1 mutant mice bear no gross abnormalities in eye, limb, and brain development and exhibit postnatal lethality. Mol Cell Biol. 2001;21:1484–1490. doi: 10.1128/MCB.21.5.1484-1490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu PX, Adams J, Peters H, Brown MC, Heaney S, et al. Eya1-deficient mice lack ears and kidneys and show abnormal apoptosis of organ primordia. Nat Genet. 1999;23:113–117. doi: 10.1038/12722. [DOI] [PubMed] [Google Scholar]
- 39.Laclef C, Souil E, Demignon J, Maire P. Thymus, kidney and craniofacial abnormalities in Six 1 deficient mice. Mech Dev. 2003;120:669–679. doi: 10.1016/s0925-4773(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 40.Laclef C, Hamard G, Demignon J, Souil E, Houbron C, et al. Altered myogenesis in Six1-deficient mice. Development. 2003;130:2239–2252. doi: 10.1242/dev.00440. [DOI] [PubMed] [Google Scholar]
- 41.Ayres JA, Shum L, Akarsu AN, Dashner R, Takahashi K, et al. DACH: genomic characterization, evaluation as a candidate for postaxial polydactyly type A2, and developmental expression pattern of the mouse homologue. Genomics. 2001;77:18–26. doi: 10.1006/geno.2001.6618. [DOI] [PubMed] [Google Scholar]
- 42.Brunskill EW, Aronow BJ, Georgas K, Rumballe B, Valerius MT, et al. Atlas of gene expression in the developing kidney at microanatomic resolution. Dev Cell. 2008;15:781–791. doi: 10.1016/j.devcel.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koch M, Bohrmann B, Matthison M, Hagios C, Trueb B, et al. Large and small splice variants of collagen XII: differential expression and ligand binding. J Cell Biol. 1995;130:1005–1014. doi: 10.1083/jcb.130.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used for RT-PCR.
(DOC)
Primers used for WISH. Underlined nucleotides indicate restriction sites used for subcloning.
(DOC)