Abstract
EhADH112 is an Entamoeba histolytica Bro1 domain-containing protein, structurally related to mammalian ALIX and yeast BRO1, both involved in the Endosomal Sorting Complexes Required for Transport (ESCRT)-mediated multivesicular bodies (MVB) biogenesis. Here, we investigated an alternative role for EhADH112 in the MVB protein trafficking pathway by overexpressing 166 amino acids of its N-terminal Bro1 domain in trophozoites. Trophozoites displayed diminished phagocytosis rates and accumulated exogenous Bro1 at cytoplasmic vesicles which aggregated into aberrant complexes at late stages of phagocytosis, probably preventing EhADH112 function. Additionally, the existence of a putative E. histolytica ESCRT-III subunit (EhVps32) presumably interacting with EhADH112, led us to perform pull-down experiments with GST-EhVps32 and [35S]-labeled EhADH112 or EhADH112 derivatives, confirming EhVps32 binding to EhADH112 through its Bro1 domain. Our overall results define EhADH112 as a novel member of ESCRT-accessory proteins transiently present at cellular surface and endosomal compartments, probably contributing to MVB formation during phagocytosis.
1. Introduction
Entamoeba histolytica, the causative agent of human amoebiasis provokes the second worldwide highest rates of morbidity and mortality due to protozoa [1]. E. histolytica trophozoites obtain host nutrients from a very active uptake and efficient engulfment of bacteria, red blood cells (RBC), and cell debris [2], which makes them to be considered as professional phagocytes. Since E. histolytica phagocytosis-deficient mutants have a diminished virulence in vitro and in vivo [3, 4], and nonvirulent E. histolytica strains exhibit reduced rates of phagocytosis [5], this cellular event has been defined as a key virulence factor.
EhCPADH, an E. histolytica protein complex formed by the EhADH112 adhesin and the EhCP112 cysteine protease, has been widely involved in adherence to, phagocytosis, and destruction of target cells [6]. Bioinformatics analysis revealed that EhADH112 is structurally related to mammalian ALIX [7], an evolutionarily conserved, ubiquitously expressed and multifunction scaffold protein, originally identified by its association with proapoptotic signaling partners [8, 9]. Additional evidence has established that ALIX modulates other cellular mechanisms, including receptor downregulation [10, 11], endosomal protein sorting [12–14], integrin-mediated cell adhesion and extracellular matrix assembly [15], actin-based cytoskeleton remodeling [16, 17], and membrane invagination and abscission in cytokinesis and retroviral budding [18].
ALIX is an abundant cytoplasmic protein with a multimodular architecture containing an N-terminal “banana”-shaped Bro1 domain [19], a middle “V”-shaped domain [20] and a C-terminal proline-rich region [21]. This tripartite domain organization occurs in the majority of ALIX orthologues and provides them multiple protein-binding sites for specific roles in several cellular processes, and the possibility of linking proteins into distinct networks, thus acting as scaffold proteins [22].
Much of what is known about ALIX has stemmed from the characterization of its closest orthologue, yeast BRO1, a crucial component of the Endosomal Sorting Complexes Required for Transport (ESCRT) pathway [23, 24]. The ESCRT machinery comprises a set of protein complexes (ESCRT-0,-I, -II, -III, and -associated proteins) most of them constituted by the so-called Vacuolar protein sorting (Vps) factors. The assembly of the ESCRT apparatus at the endosomal surface is required to selectively transport ubiquitinated receptors and other cargo proteins into late endosomes known as multivesicular bodies (MVB), towards final degradation into the vacuole or lysosome [25, 26]. In this process, human ALIX or yeast BRO1 promotes endosomal membrane scission for intraluminal vesicles formation of MVB, drived by the direct association of their N-terminal Bro1 domains to the human CHMP4 or yeast Vps32 (also named Snf7) ESCRT-III subunits, respectively [19, 20].
Despite significant advances in the understanding of EhADH112 functions related to parasite virulence [6], its structural relationship with ALIX and BRO1 proteins [7] and recent evidence regarding the existence of most ESCRT components [27] and MVB-like organelles in E. histolytica [28], the potential role of EhADH112 in ESCRT-dependent protein sorting and trafficking along the MVB pathway had not yet been explored. The Bro1 domain occurs in a wide group of eukaryotic proteins that serve as scaffold for linking different cellular networks, including MVB formation dependent on ESCRT. In fact, a well-known hallmark for Bro1 domain functionality is its ability to bind to ESCRT-III subunits. Here, we initiated the characterization of the N-terminal residues comprising the small EhADH112 Bro1 domain originally defined by the Pfam protein domain database [29], but later extended by crystallographic experiments [19]. The tertiary structure modeling of EhADH112, presented in this study, predicted the spatial conformation needed for putative interaction with ESCRT-III subunits via the Bro1 domain. To explore if EhADH112 could be involved in MVB formation during phagocytosis, we generated a trophozoite population (ANeoBro1) overexpressing the first 166 amino acids of the EhADH112 Bro1 domain. ANeoBro1 trophozoites dramatically diminished their rates of phagocytosis, possibly due to an impairment of EhADH112 functions, seemingly produced by exogenous Bro1 accumulation in cytoplasmic vesicles and aberrant complexes, and its absence in plasma membrane and phagosomes, where EhADH112 exerts its role for target cell adherence and phagocytosis. Electron immunolocation of EhADH112 in structures resembling MVB, together with its finding in both soluble and insoluble subcellular fractions, suggested its participation in MVB formation. Moreover, EhADH112 in vitro binding to a protein homologous to Vps32 (EhVps32), strengthened our hypothesis regarding the EhADH112 contribution to ESCRT-mediated protein sorting along the MVB pathway by virtue of its Bro1 domain. Altogether, our results define EhADH112 as a novel member of Bro1 domain-containing proteins present at cellular surface and endosomal compartments with a potential role in the MVB pathway.
2. Materials and Methods
2.1. Tertiary (3D) Protein Modeling
The EhADH112 primary sequence was submitted to the Phyre Server (http://www.sbg.bio.ic.ac.uk/~phyre/) and validated by the Swiss Model Database. EhADH112 3D modeling was performed with human ALIX (2oev) and yeast BRO1 (1z1b) crystallized sequences as templates. Results were documented and analyzed through the DeepView-Swiss-Pdb Viewer software.
2.2. E. histolytica Cultures
Trophozoites of clone A (strain HM1: IMSS) were axenically cultured in TYI-S-33 medium at 37°C [30]. Medium for transfected trophozoites (ANeo, ANeoADH112 and ANeoBro1 populations) was supplemented with 40 μg/mL geneticin (G418) (Life Technologies, Gaithesburg, MD). Trophozoites were harvested in logarithmic growth phase for all experiments and cell viability was monitored by microscopy using Trypan blue dye exclusion tests.
2.3. PCR Amplification of the Bro1-FLAG Encoding Fragment
A 498 bp fragment from the 5′ end of the EhAdh112 gene, corresponding to the first 166 amino acids of the EhADH112-Bro1 domain was PCR-amplified, using the sense (1–24 EhAdh112 nt) 5′-GGGGTACCATGAATAGACAATTCATTCCTGAA-3′ and the antisense (477–497 EhAdh112 nt) 5′- CGGGATCCTTACTTATCGTCGTCATCCTTGTAATCACTACCTGCTGCACATAGTTGG-3′ oligonucleotides, 10 mM dNTPs, 100 ng of E. histolytica genomic DNA as template and 2.5 U of Taq DNA polymerase (Gibco). PCR was carried out for 30 cycles comprising 1 min at 94°C, 30 sec at 59°C, and 40 sec at 72°C. The sense oligonucleotide contained a KpnI restriction site, whereas the antisense oligonucleotide contained the FLAG (DYKDDDDK) tag-encoding sequence (underlined) [31] and a BamH1 restriction site. Oligonucleotides used in this work specifically recognize sequences present in the EhAdh112 gene but not those present in the previously reported EhAdh112-like genes [7].
2.4. Generation of ANeoBro1 Trophozoites by Transfection with the pNeoBro1FLAG Plasmid
The PCR-amplified product (Bro1FLAG) was cloned into the BamH1 and KpnI sites of the pExEhNeo (pNeo) plasmid, which contains E. histolytica-specific transcription signals and the G418 resistance (NeoR) conferring gene as selectable marker [32], producing the pNeoBro1FLAG construct. Escherichia coli DH5α bacteria were transformed with the pNeoBro1FLAG or pNeo plasmids. Both plasmids were purified using the QIAGEN Maxi kit (Chatsworth, CA) and automatically sequenced. Plasmids (200 μg) were transfected by electroporation as previously described [32] into exponentially growing trophozoites of clone A, generating the ANeo and ANeoBro1 trophozoite populations.
2.5. RT-PCR Experiments
cDNAs were synthesized using 1 μg of DNase-treated total RNA, 10 mM dNTPs, 200 U AMV reverse transcriptase (Gibco) and 0.5 μg of oligo dT (Gibco) in a final volume of 10 μL, for 1 h at 42°C. PCR amplifications were carried out using 4 μL (~330 ng) of cDNA, 1 U of Taq polymerase, 2 mM dNTPs, and 100 ng of the sense (1–24 nt) and the antisense (477–497 nt) oligonucleotides from the EhAdh112 gene. Additionally, the sense (1–17 nt) 5′-ATGATTGAACAAGATGG-3′ and the antisense (780–794 nt) 5′-TTAGAAGAACTCGTC-3′ primers were used to amplify a 794 bp fragment of the NeoR gene. Each PCR was performed as described above. Amplified products were separated by 1% agarose gel electrophoresis, ethidium bromide-stained and visualized in a Gel Doc 1000 apparatus (BioRad).
2.6. Immunofluorescence Assays
Trophozoites grown on coverslips were fixed with 4% paraformaldehyde (PFA) (Sigma) at 37°C for 1 h, permeabilized with 0.5% Triton X-100 in PBS (PBS-Triton) for 30 min and incubated with 1% bovine serum albumin (BSA) for 40 min at 37°C. Trophozoites were incubated with mouse monoclonal antibodies against EhADH112 (mαEhADH112) (1 : 10) or rabbit polyclonal anti-FLAG (pαFLAG) (USBiological) antibodies (1 : 500), overnight (ON) at 4°C, followed by incubation with fluorescein-isothiocyanate (FITC)-labeled anti-mouse or anti-rabbit secondary antibodies (1 : 100) (Zymed), respectively, for 1 h at 37°C.
For colocalization experiments, PFA-fixed trophozoites were incubated ON at 4°C with mαEhADH112 and pαFLAG antibodies, followed (1 h at 37°C) by FITC-labeled anti-mouse IgM and tetramethyl-rhodamine-isothiocyanate-(TRITC)-labeled anti-rabbit IgG secondary antibodies. For some experiments, trophozoites were first incubated with fresh RBC (1 : 40) for different times at 37°C, fixed with PFA, contrasted with 3 mM diaminobenzidine (DAB) and treated for immunofluorescence assays as above. All preparations were preserved using the antifade reagent Vectashield (Vector) and examined through a Nikon inverted microscope attached to a laser confocal scanning system (Leica).
2.7. Adherence and Phagocytosis Kinetics
Trophozoites were incubated for 5, 10, and 15 min with freshly obtained human RBC (108 cells/mL) (1 : 100 ratio) at 4°C for adherence, or at 37°C for phagocytosis experiments [33]. RBC were contrasted with DAB and counted at random from three independent experiments to determine the number of RBC adhered to or ingested by 100 trophozoites.
2.8. Transmission Electron Microscopy (TEM) Assays
Fast-freeze fixation followed by cryosubstitution was used for ultrastructural location of EhADH112 and the FLAG-tagged Bro1 recombinant polypeptide. Transfected trophozoites were pelleted and placed into the hole of a 7 mm diameter antiadhesive plastic ring positioned on a foam rubber support and frozen on a copper mirror precooled to liquid nitrogen temperature using a Reichert KF 80 unit. Freeze substitution was achieved with a Reichert CS autosystem in acetone containing 4% osmium tetroxide for 48 h at −80°C. Afterwards, samples were brought to room temperature at a rate of 4°C/h and embedded in epoxy resins. Ultrathin sections were obtained and stained with uranyl acetate and lead citrate. For immunogold labeling, thin sections were placed on formvar-coated nickel grids and incubated for 15 min in 0.1 M ammonium chloride. Then, sections were washed twice with PBS (5 min each), blocked with 1% BSA for 15 min, and incubated with polyclonal rabbit anti-EhADH112 (pαEhADH112) or monoclonal mouse anti-FLAG (mαFLAG) (Sigma) antibodies at 1 : 50 or 1 : 20 dilutions, respectively, for 60 min. Then, sections were washed and incubated for 1 h with goat anti-rabbit or anti-mouse IgG (1 : 20) antibodies conjugated to 15 nm colloidal gold (BBI International, Cardiff, UK). Finally, sections were washed and treated as above to be observed through a Zeiss EM 910 electron microscope. For phagocytosis experiments, trophozoites were first incubated with fresh RBC (1 : 40 ratio) at 37°C for 15 min and treated for TEM as described.
2.9. Isolation of E. histolytica Membranes
E. histolytica cellular fractions were obtained as described by Aley et al. [34]. Briefly, trophozoites from wild-type clone A (40 × 106) were harvested, washed twice with 19 mM potassium phosphate buffer, pH 7.2, and 0.27 M NaCl (PD solution) and pooled. Cell pellet was resuspended to 2 × 107 cells/mL PD solution containing 10 mM MgCl2 and rapidly mixed with an equal volume of 1 mg/mL concanavalin A in the same buffer. After 5 min, cells were spun at 50 ×g for 1 min to remove the excess of concanavalin A. The supernatant was discarded and cell pellet was resuspended in 12 mL of 10 mM Tris-HCl buffer, pH 7.5, containing 2 mM phenylmethylsulfonyl fluoride (PMSF) and 1 mM MgCl2. After 10 min swelling in hypotonic buffer, cells were homogenized by 18–20 strokes of a glass Dounce homogenizer with a tight-fitting pestle (Wheaton Scientific Div.). Cell lysis and membrane sheets formation were verified by phase-contrast microscopy. The homogenate was layered over a two-step gradient consisting of 8 mL of 0.5 M mannitol over 4 mL of 0.58 M sucrose, both in Tris buffer, and spun at 250 ×g for 30 min. Material remaining at the top of the 0.5 M mannitol (SN1) was centrifuged at 40 000 ×g for 1 h to separate soluble molecules (SN2) from small membrane fragments and vesicles (P2). Large plasma membrane fragments and other heavy debris formed a tight pellet at the bottom of the gradient (P1). This pellet was resuspended in 1 mL Tris buffer containing 1 M α-methyl mannoside and left on ice for 40 min with occasional mixing. Plasma membranes free of concanavalin A were diluted into three volumes of Tris buffer, homogenized by 80 strokes with a glass Dounce homogenizer, layered on a 20% sucrose Tris gradient and spun for 30 min at 250 ×g. Vesiculated plasma membranes floating above the initial sucrose layer (SN3) were collected and then concentrated by centrifugation at 40 000 ×g for 1 h. The pellet (P4), enriched in plasma membranes, was resuspended in Tris buffer. All steps were performed at 4°C. Samples (50 μg) were analyzed by SDS-PAGE (10%) and transferred onto nitrocellulose membranes for Western blot assays using rabbit polyclonal antibodies against the last C-terminal 243 amino acids of EhADH112 (pαEhADH243) and peroxidase-labeled anti-rabbit IgG secondary antibodies, at 1 : 300 and 1 : 10 000 dilutions, respectively. As a control, mouse monoclonal anti-E. histolytica actin and peroxidase-labeled anti-IgG corresponding secondary antibodies were used at 1 : 1 500 and 1 : 10 000, respectively.
2.10. In Vitro Binding Assays
A plasmid encoding glutathione S-transferase (GST)-EhVps32 was constructed by inserting a PCR fragment containing the EhVps32 coding sequence into the BamH1 site of pGEX-5X-1 (Pharmacia). Plasmids used for in vitro synthesis of EhAdh112 and EhAdh112-truncated derivatives were constructed by inserting PCR-amplified or restriction fragments containing the corresponding EhADH112 coding sequences in the polylinker of pGBKT7 (Clontech). GST-EhVps32 and GST alone were expressed in E. coli BL21 bacteria. Cultures (50 mL) were induced at 30°C after addition of 0.1 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) and incubated for additional 2.5 h before proceeding with the GST fusion protein purification [35]. [35S]-EhADH112 and [35S]-EhADH112 truncated derivatives were synthesized in vitro using the Promega TNT coupled transcription-translation system in the presence of [35S] methionine (1 000 Ci/mmol). Labeling reaction (2 μl) was added to glutathione beads loaded with GST-EhVps32 or GST proteins and incubated at 4°C for 1 h in 500 mL of a 10 mM Tris HCl, pH 8.0, 1 mM EDTA and 150 mM NaCl solution (STE) with 1% (v/v) Triton X-100. After five washes with STE/1% Triton X-100, beads were boiled in sample buffer and proteins were separated by 10% polyacrylamide SDS-PAGE. Bound proteins were detected by autoradiography or Coomassie blue staining of gels.
3. Results and Discussion
3.1. Tertiary Structure Modeling of EhADH112 Predicts Conformational Similarities to ALIX
Bro1 domain-containing proteins are highly conserved among eukaryotes and exhibit distinct functions in several cellular processes (Figure 1(a)), depending on the partners they interact with [22]. EhADH112 displays 40% homology and 20% identity to human ALIX [7] and its primary sequence aligns with the portion of ALIX corresponding to the N-terminal Bro1 and the middle “V” domains. More divergent relatives of ALIX only contain the Bro1 domain but otherwise, bear little resemblance to the remaining structure [36, 37]. Although EhADH112 lacks the proline-rich C-terminal tract that harbors the majority of protein-binding sites linking ALIX to various functions [7, 22], instead, this protein has a target cell adherence domain, which binds to a 97 kDa protein in epithelial cells [38]. Besides EhADH112, the E. histolytica genome predicts two EhAdh112-related genes encoding the EhADH112-like1 (898 amino acids) and EhADH112-like2 (919 amino acids) proteins [7], both displaying a putative Bro1 domain at their N-terminus (Figure 1(a)). However, the existence of these proteins has not yet been confirmed in trophozoites.
Figure 1.
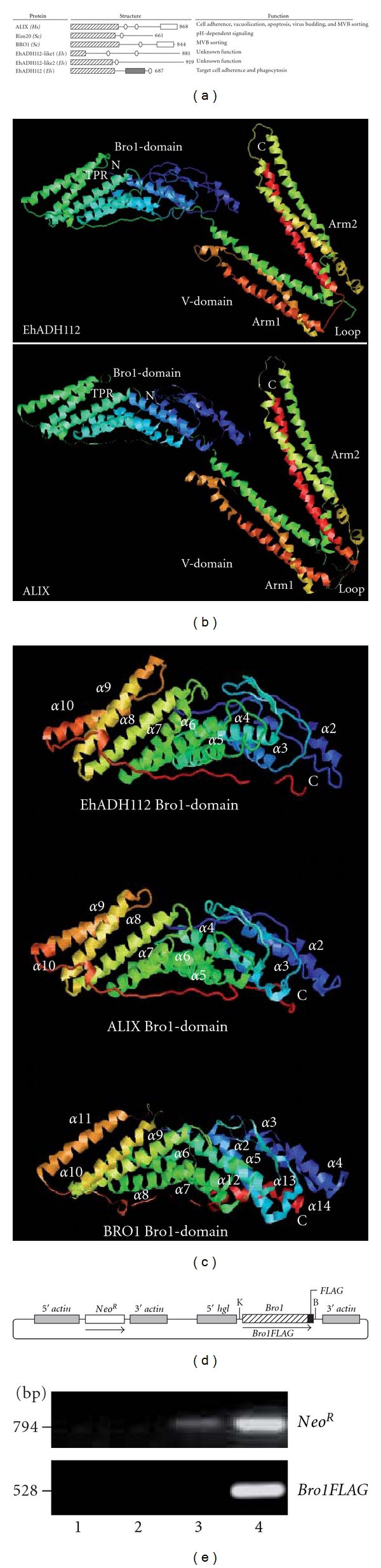
Structural features of EhADH112 and generation of ANeoBro1 trophozoites. (a) Structural features of representative Bro1 domain-containing proteins and EhADH112- and EhADH112-like proteins: Bro1 domain (squares containing diagonal lines), coiled coil regions (ellipses), proline-rich tracts (white squares), and adherence region (gray square). Hs: Homo sapiens; Sc: Saccharomyces cerevisiae; Eh: Entamoeba histolytica. Numbers: amino acid number of each protein. (b-c) Spatial conformation of EhADH112 and its Bro1 domain. (b) Ribbon representation for predicted EhADH112 tertiary and human ALIX crystallized structures and (c) Bro1 domains from EhADH112, ALIX and yeast BRO1 proteins. N: amino terminus. C: carboxy terminus. TPR: tetratricopeptide repeat. (d) Schematic depiction of the pNeoBro1FLAG plasmid. K: KpnI restriction site. B: BamH1 restriction site. (e) Transcripts obtained by RT-PCR assays using oligonucleotides for the NeoR gene or Bro1FLAG sequence and cDNAs synthesized from nontransfected clones A (lane 2), ANeo (lane 3), and ANeoBro1 (lane 4). Lane 1 corresponds to the reaction mixture without reverse transcriptase, using total RNA from ANeoBro1.
Our current predictions for EhADH112 tertiary structure resulted in protein overlapping to ALIX (Figure 1(b)). Models determined the boomerang or “banana”-shaped spatial conformation for the N-terminal EhADH112 Bro1 domain, mostly made of α-helices forming a solenoid, with helices 6 through 11 arranged in a tetratricopeptide repeat-like structure [19], and a central core arranged in two extended three-helix bundles forming elongated arms that fold back into a “V” (Figure 1(b)). Arms conformation suggests that the “V” domain may act as a hinge, changing in response to ligand binding, as described for ESCRT and viral proteins interaction with ALIX [18]. Apparently, EhADH112 conserves the two hydrophobic patches [19] required for ALIX or BRO1 endosomal membrane targeting and association, via direct binding to ESCRT-III CHMP4 or Vps32 subunits, respectively (Figures 1(b) and 1(c)), or Src-tyrosine kinase docking. Since the conserved interaction of ALIX or BRO1 with ESCRT-III is necessary for membrane inward budding and MVB biogenesis during protein sorting and trafficking [39], it is possible that EhADH112 could associate to putative E. histolytica ESCRT-III components via its Bro1 domain.
3.2. Generation of Trophozoites Overexpressing the Bro1 Recombinant Polypeptide
To initiate the characterization of the EhADH112 Bro1 domain, trophozoites of clone A (HM1:IMSS) were transfected with the pNeoBro1FLAG plasmid (Figure 1(d)) driving the expression of the recombinant Bro1 polypeptide and generating the ANeoBro1 population. The presence and expression of plasmids in trophozoites was confirmed by RT-PCR amplification of the NeoR gene. NeoR was amplified from ANeo (transfected with empty pNeo) and ANeoBro1 trophozoites (Figure 1(e), lane 3), whereas a transcript corresponding to the Bro1FLAG fragment, was only detected in the ANeoBro1 population (Figure 1(e), lane 4), as expected. No amplification was detected in the absence of reverse-transcriptase in reaction mixtures or using total RNA from wild type clone A trophozoites (Figure 1(e), lanes 1 and 2).
3.3. ANeoBro1 Trophozoites Localize the Exogenous Bro1 Recombinant Polypeptide in Cytoplasmic Compartments
As part of the EhCPADH complex, an E. histolytica surface heterodimer involved in target cell adherence, phagocytosis, and destruction, EhADH112 is located at trophozoite plasma membrane and cytoplasmic vacuoles [6]. To determine the location of the Bro1 polypeptide overexpressed by ANeoBro1 trophozoites and to distinguish it from the Bro1 domain present in endogenous EhADH112, immunofluorescence experiments were carried out using polyclonal antibodies against the FLAG tag (pαFLAG) and monoclonal antibodies (mαEhADH112) against the EhADH112 carboxy terminus adherence epitope (444–601 amino acids). ANeo trophozoites, as well as a trophozoite population named ANeoADH112 that overexpresses the EhADH112 full length protein fused to a FLAG tag (EhADH112-FLAG), were used here as additional controls [7].
By confocal microscopy, mαEhADH112 antibodies revealed the presence of EhADH112 at the plasma membrane of permeabilized ANeo, ANeoBro1, and ANeoADH112 trophozoites (Figure 2(a)), although fluorescence was higher in ANeoADH112 population, since these trophozoites express both, endogenous EhADH112 and exogenous EhADH112-FLAG.
Figure 2.
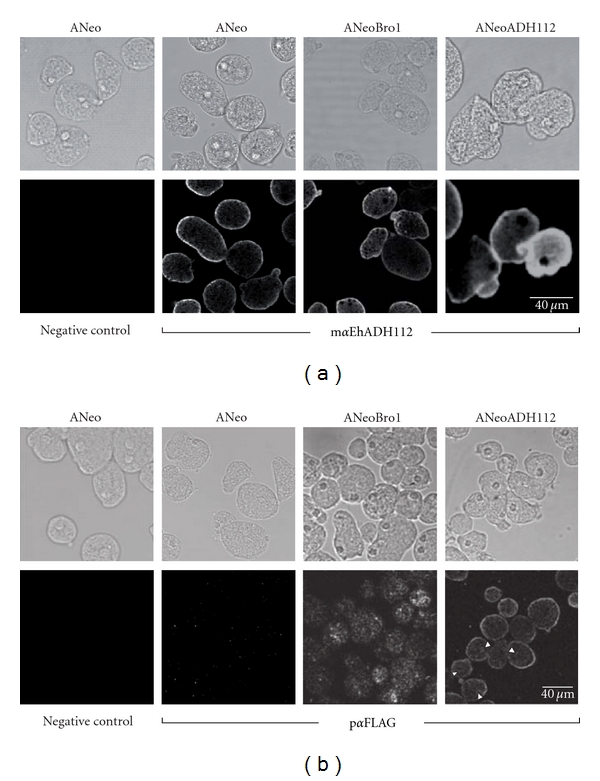
Cellular location of EhADH112 and the Bro1 recombinant polypeptide in transfected trophozoites. Confocal microscopy images of permeabilized ANeo, ANeoBro1, or ANeoADH112 trophozoites incubated with (a) mαEhADH112 or (b) pαFLAG antibodies. Negative controls correspond to trophozoite preparations only incubated with secondary antibodies. Top: phase contrast images. Bottom: corresponding confocal sections. Arrowheads: small cytoplasmic vesicles.
As expected, pαFLAG antibodies gave no reaction with ANeo trophozoites (Figure 2(b)), but traced FLAG-tagged Bro1 as punctuated structures and patches of different sizes in the cytoplasm of ANeoBro1 trophozoites. Interestingly, no signals were detected at plasma membrane (Figure 2(b)), suggesting that the EhADH112 carboxy end, absent in the exogenous Bro1 recombinant polypeptide, could be participating in EhADH112 targeting to the trophozoite surface. Since EhADH112-FLAG appeared at the plasma membrane (Figure 2(b)) and some cytoplasmic vacuoles (Figure 2(b), arrowheads) of ANeoADH112 trophozoites, in a similar pattern to the one exhibited by endogenous EhADH112 in all parasite populations analyzed (Figure 2(a)), we rule out that the FLAG tag could be causing the defective targeting of recombinant Bro1 to the membrane. Negative results were likewise obtained with nontransfected or transfected trophozoites treated only with secondary antibodies. Here, we show results obtained by assaying ANeo trophozoites (Figures 2(a) and 2(b)).
From these experiments, we can conclude that the Bro1 recombinant polypeptide expressed by ANeoBro1 trophozoites is located at a different site than EhADH112, and that transfection procedures or expression of FLAG-tagged proteins did not affect the location of endogenous EhADH112 in trophozoites.
3.4. Expression of the Exogenous Bro1 Recombinant Polypeptide by ANeoBro1 Trophozoites Diminishes Erythrophagocytosis but Not Target Cell Adherence
EhADH112 has been previously characterized by the properties conferred by its C-terminus for target cell primary contact, internalization, and phagocytosis [6, 38]. To further understand the Bro1 domain contributions to EhADH112 functions in trophozoites, we carried out RBC adherence and erythrophagocytosis assays. Adherence efficiency and erythrophagocytosis rates exhibited by ANeo trophozoites at 15 min were taken as 100% in all experiments. Again, as an additional control we used ANeoADH112 trophozoites.
After 15 min of RBC incubation at 4°C, ANeo and ANeoBro1 trophozoites adhered a mean of 5.7 ± 3 and 6.6 ± 2 RBC per trophozoite, respectively (Figures 3(a) and 3(b)), meanwhile at 10 and 5 min they adhered 4.78 ± 1 and 4.56 ± 2, and 2.73 ± 2 and 2.8 ± 2 RBC per trophozoite, respectively. In contrast, ANeoADH112 showed an increased avidity to attach RBC, by adhering above twice more RBC than ANeo and ANeoBro1 at 15, 10, and 5 min (12.5 ± 2, 8.7 ± 2 and 7 ± 2 RBC per trophozoite, respectively) (Figures 3(a) and 3(b)). These results indicate that exogenous expression of the Bro1 recombinant polypeptide did not affect the adherence function of trophozoites and suggest that the EhADH112 N-terminus may not be promoting target cell binding.
Figure 3.
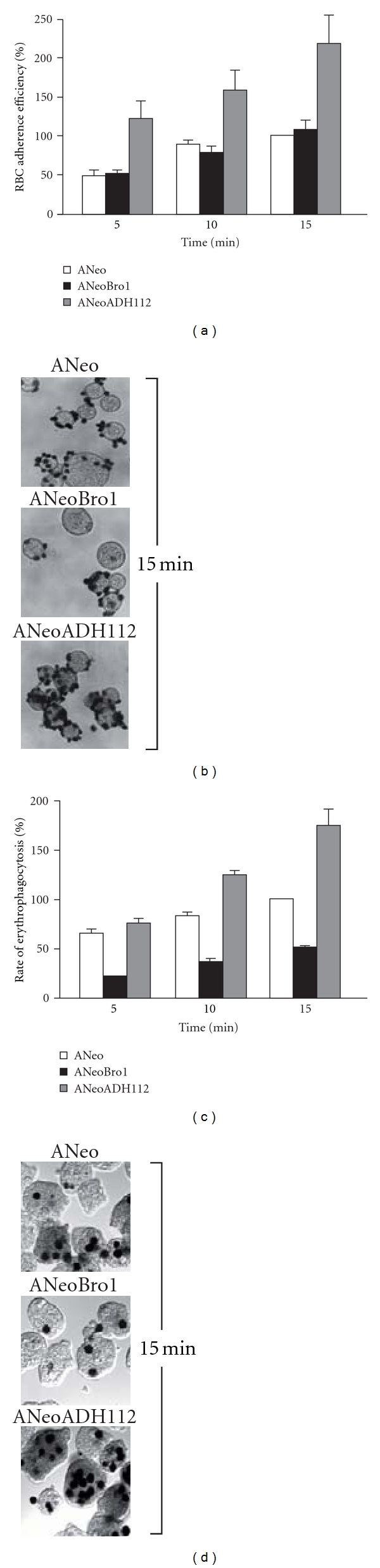
RBC adherence efficiency and rates of erythrophagocytosis exhibited by ANeo, ANeoBro1, and ANeoADH112 trophozoites. (a-b) Trophozoites were incubated with RBC (1 : 100) at 4°C for adherence and (c-d) at 37°C for phagocytosis, for different times. RBC were contrasted with DAB and counted in 100 randomly selected trophozoites under the light microscope to obtain the mean of RBC adhered or ingested by trophozoite. Bars represent the mean ± standard error of at least three independent experiments performed by duplicate.
Interestingly, noteworthy differences were found in assays determining phagocytosis activity of transfected trophozoites. ANeo trophozoites ingested 14.3 ± 2 RBC per parasite at 15 min, 11.6 ± 2 RBC at 10 min and 9.4 ± 2 RBC at 5 min (Figure 3(c)), whereas ANeoBro1 trophozoites only ingested 7.1 ± 2, 5 ± 2 and 2.8 ± 2 RBC per trophozoite at 15, 10 and 5 min, respectively (Figures 3(c) and 3(d)). As previously reported [7], ANeoADH112 trophozoites exhibited augmented phagocytosis rates, by ingesting 76% more RBC than ANeo trophozoites at 15 min of erythrophagocytosis (Figures 3(c) and 3(d)), and 40% and 10% more, at 10 and 5 min, respectively (Figure 3(c)). These latter experiments indicate that overexpression of Bro1 results in a dominant negative effect on phagocytosis. We hypothesized that this phenomenon may be due to recruitment and association of proteins probably involved in target cell internalization and phagocytosis by the truncated EhADH112 protein instead of the endogenous one, thus producing a competition for protein binding sites and reducing trophozoites rates of ingestion, despite efficient primary cell contact. Otherwise, the Bro1 recombinant polypeptide per se, could be producing a conformational change in EhADH112 or preventing EhADH112 accessibility for the interaction with its counterparts, therefore causing a functional impairment.
Transfection procedures did not modify EhADH112 location and function, since ANeo trophozoites displayed similar results to the ones presented by nontransfected clone A trophozoites (data not shown).
3.5. The Bro1 Recombinant Polypeptide Exhibits a Different Cellular Location to Endogenous EhADH112 during Phagocytosis
Previous work using TEM and immunofluorescence experiments determined that the EhADH112 C-terminus mediates target cell adherence but also contributes to phagocytosis activity of E. histolytica trophozoites [6, 38]. The diminished phagocytosis rates exhibited by ANeoBro1 trophozoites led us to precise the location of EhADH112 and the overexpressed Bro1 polypeptide after 5 min of erythrophagocytosis.
Through confocal microscopy, and in agreement to preceding findings, EhADH112 was detected at plasma membrane, target cell contact sites, membrane extensions, and phagosomes of nontransfected clone A trophozoites (data not shown). A similar location was determined for EhADH112 in ANeo trophozoites (data not shown), which adhere to and phagocyte RBC in the same way than wild-type trophozoites do. Regarding ANeoBro1 trophozoites (Figure 4(a)), we observed EhADH112 at the trophozoite plasma membrane, cytoplasmic vacuoles and phagosomes (Figure 4(a), top). Besides, small phagosome-neighbouring vesicles that may correspond to endosomes or lysosomes were detected (Figure 4(a), asterisks). Otherwise, the Bro1 recombinant polypeptide was found in cytoplasmic punctuated and vesicular structures and, significantly, it was accumulated in vacuolar compartments (Figure 4(a), arrows) that did not overlap to RBC location (Figure 4(a)). Images also evidenced that exogenous Bro1 did not reach trophozoites plasma membrane at early stages of phagocytosis and was absent in RBC-containing phagosomes. The different location of EhADH112 and recombinant Bro1 in trophozoites under basal culture conditions, even during phagocytosis, together with the impairment of ANeoBro1 erythrophagocytosis rates and the presence of aggregates containing exogenous Bro1, suggest that this polypeptide could be interfering with the function of trophozoite proteins present at the plasma membrane, probably participating in membrane remodeling and RBC internalization into phagosomes. Control experiments, omitting primary antibodies gave no signals in trophozoites (Figure 4(a), bottom).
Figure 4.
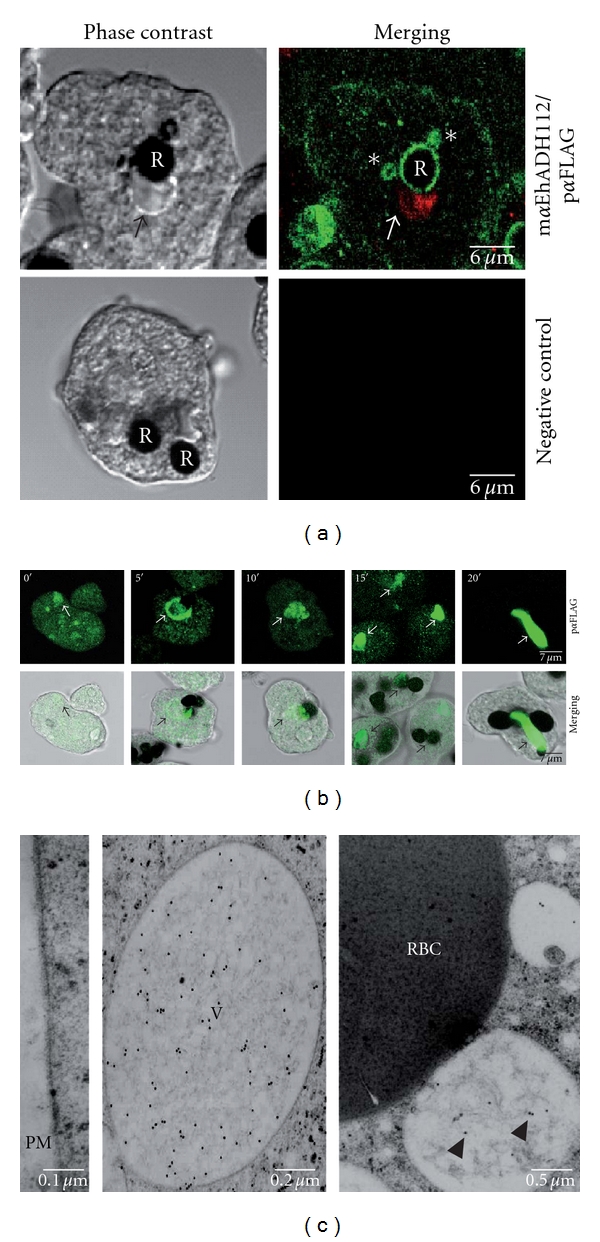
Differential location of endogenous EhADH112 and the Bro1 recombinant polypeptide in ANeoBro1 trophozoites during phagocytosis. (a) Cellular immunolocation of endogenous EhADH112 and exogenous Bro1 in ANeoBro1 trophozoites after RBC ingestion. Trophozoites were incubated with RBC for 5 min and permeabilized. Top: after RBC contrasting with DAB, EhADH112 was detected by mαEhADH112 antibodies and FITC-labeled anti-mouse secondary antibodies. The FLAG-tagged Bro1 recombinant polypeptide was detected by pαFLAG antibodies and TRITC-labeled anti-rabbit secondary antibodies. Bottom: trophozoites incubated with FITC-labeled anti-mouse IgM and TRITC-labeled anti-rabbit IgG secondary antibodies. Preparations were examined through laser confocal microscopy. R: RBC. Arrows: recombinant Bro1 containing vacuoles. Asterisks: EhADH112 present in endosomal or lysosomal-like compartments. (b) Immunolocation of exogenous Bro1 in ANeoBro1 trophozoites at different times of erythrophagocytosis. Trophozoites were incubated with fresh RBC and treated as described above. The Bro1 recombinant polypeptide was detected by pαFLAG and FITC-labeled anti-rabbit secondary antibodies. Preparations were examined through a laser confocal microscope. Top: confocal sections. Bottom: merging of phase contrast images and laser confocal corresponding sections. Arrows: vesicles and large vacuoles containing exogenous Bro1. (c) Ultrastructural location of exogenous Bro1 in ANeoBro1 trophozoites after RBC ingestion. Ultrathin sections of ANeoBro1 trophozoites were processed for immunogold labeling and TEM as described above. The Bro1 recombinant polypeptide was detected by mαFLAG antibodies and gold-labeled secondary antibodies. Left: plasma membrane (PM). Middle: a huge vacuole. Right: vacuoles in the proximity of RBC after 15 min phagocytosis. V: vacuole. Arrowheads: Bro1 gold-labeled particles.
3.6. Exogenous Bro1 Accumulates in Aberrant Compartments at Late Stages of Phagocytosis
It has been previously shown that endogenous EhADH112 protein changes its location within trophozoites during RBC phagocytosis. After target cell contact, this adhesin is translocated from the trophozoite plasma membrane to the phagocytic vacuoles. As the ingestion process advances, EhADH112 is found in RBC and after 30 min, it comes back to the plasma membrane [6]. To elucidate whether exogenous Bro1 localizes at some point to the plasma membrane or to phagosomes and to better understand its fate along the erythrophagocytosis process, we followed this recombinant polypeptide in ANeoBro1 trophozoites at different times of RBC ingestion. Immediately after RBC interaction (0 min), exogenous Bro1 appeared in randomly distributed cytoplasmic vesicles and patches of distinct sizes and morphologies (Figure 4(b)). As RBC ingestion progressed (5 min), the exogenous Bro1 recombinant polypeptide accumulated in a ring-like structure surrounding RBC-containing compartments probably corresponding to phagosomes or phagolysosomes (Figure 4(b)). This Bro1-enriched large structure achieved a closer proximity to RBC at 10 min of phagocytosis, although no RBC overlapping was observed (Figure 4(b)). At late phagocytosis stages (15 and 20 min), the Bro1 recombinant polypeptide appeared in huge tubular structures (Figure 4(b)). In yeast, deletion or inactivation of several Vps factors, which assemble into the ESCRT machinery during protein sorting and trafficking through endosomal compartments, induces the formation of enlarged vacuolated and tubulated organelles that fail to mature into MVB [40, 41]. Hence, the large vacuoles observed in ANeoBro1 trophozoites may correspond to aberrant endosomal compartments where the exogenous Bro1 polypeptide is stuck, affecting the dynamics of RBC internalization from the membrane to the phagosomes. Work in progress in our laboratory, using specific biochemical markers, will allow us to determine the identity of these structures.
Since immunoelectron microscopy is a key technique to place macromolecular functions within a cellular context, we addressed the ultrastructural location of exogenous Bro1 in ANeoBro1 trophozoites under basal culture conditions and at 15 min of erythrophagocytosis. For these experiments we used mouse monoclonal antibodies against the FLAG tag (mαFLAG), which resulted to be more sensitive for specific recognition of the FLAG-tagged Bro1 recombinant polypeptide than the pαFLAG ones.
In agreement with the immunofluorescence results showed here (Figures 2, 4(a) and 4(b)), mαFLAG antibodies did not reveal any signal at the plasma membrane of ANeoBro1 trophozoites in culture (data not shown), and neither at 15 min of RBC ingestion (Figure 4(c), left panel). Numerous gold particles associated to fibrillar material were observed within huge vesicles (Figure 4(c), middle panel) that might correspond to the large vacuoles detected in Figures 4(a) and 4(b). Moreover, mαFLAG antibodies recognized some vesicles close to RBC (Figure 4(c), right panel), confirming that recombinant Bro1 remains in vacuolar compartments of different sizes, but not in RBC or phagosomes (Figure 4(c), right panel). Taken together, our immunolocation results revealed that under culture conditions, exogenous Bro1 is profusely distributed in different-sized cytoplasmic vesicles of ANeoBro1 trophozoites. Similarly, at initial steps of target cell contact, the Bro1 recombinant polypeptide remains in the cytoplasm of ANeoBro1 trophozoites, which preserve their ability to attach to target cells, but significantly decreased their phagocytosis rates. Location of exogenous Bro1 in small structures that gradually come together to transform into exaggerated vacuolar compartments at late stages of phagocytosis suggests that at these sites, recombinant Bro1 could also be retaining proteins involved in RBC internalization, targeting, and phagocytosis, some of them even affecting the function of EhADH112, or that this huge protein complexes may block the recycling of proteins back to the trophozoite membrane, where tentatively they would contribute to the membrane remodeling and protein assembly processes required for phagosome formation.
3.7. EhADH112 Localizes in Structures Resembling MVB in ANeoBro1 Trophozoites
According to our immunofluorescence assays, the location of endogenous EhADH112 at the plasma membrane of ANeoBro1 trophozoites was not affected by the overexpression of recombinant Bro1 under basal conditions. In fact, this resulted in RBC adherence efficiencies similar to that displayed by wild-type and ANeo trophozoites. Since EhADH112 was also found in cytoplasmic vacuoles that could differ or not from that in which recombinant Bro1 accumulated, we assessed the ultrastructural location of EhADH112 in ANeoBro1 trophozoites by immunogold labeling experiments, using polyclonal rabbit antibodies against EhADH112 (pαEhADH112) (Figure 5), which gave a better reactivity on frozen ultrathin sections than mαEhADH112 ones. Of note, pαEhADH112 antibodies do not recognize the EhADH112-like protein sequences previously reported by our group, since Western blot assays specifically detect the band corresponding to the predicted molecular weight of EhADH112 (data not shown).
Figure 5.
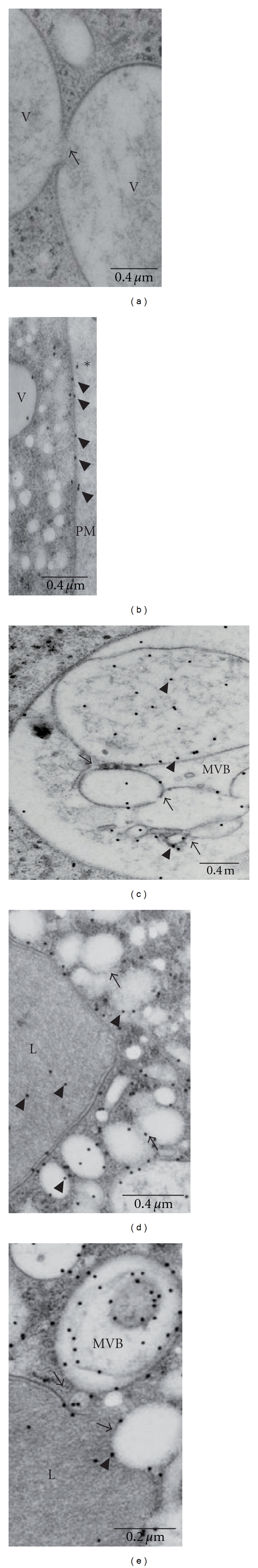
Ultrastructural location of EhADH112 in ANeoBro1 trophozoites. Ultrathin sections of ANeo or ANeoBro1 trophozoites were first prepared by cryosubstitution and incubated with pαEhADH112 and gold-labeled secondary antibodies, and then contrasted and analyzed by TEM. (a) ANeoBro1 trophozoites only incubated with gold-labeled secondary antibodies. (b–e) EhADH112-immunogold labeling in (b) plasma membrane (PM), (c) a structure resembling a MVB, (d) a lysosome-like organelle, (e) MVB and vesicles fusing to a lysosome. V: vacuole. MVB: multivesicular bodies. Arrows: vesicular fusion areas. Arrowheads: EhADH112 gold-labeled molecules.
Through TEM, pαEhADH112 antibodies revealed EhADH112 at the plasma membrane of both, ANeo (data not shown) and ANeoBro1 trophozoites (Figure 5(b)). Gold labeling was also observed at external and internal faces of vesicle membranes and inside vesicles, frequently associated to fibrillar and membranous material (Figures 5(c)–5(e)). Interestingly, EhADH112 was abundant within large vacuoles containing several tubular and vesicular structures. Huge organelles, containing intraluminal vesicles of different sizes and shapes, the majority of them immunolabeled, appeared in many trophozoites (Figure 5(c)). By their morphology, these compartments may correspond to MVB, which showed a high similarity to the ones described in mammals by Denzer et al. [42]. We also detected numerous whitish EhADH112-carrying vesicles outside electrodense structures with lysosome appearance, which also exhibited EhADH112 signals (Figure 5(d)). These lysosome-like structures presented a delimiting double membrane labeled with EhADH112 (Figures 5(d) and 5(e)). Strikingly, several vacuolar structures, one contained inside the other, which suppose membrane inward budding of vesicles (Figure 5(e), arrows), bordered lysosome-like organelles and carried EhADH112. Recurrent and profuse docking of EhADH112 to the membrane of different-sized vesicles, suggest its possible participation in protein sorting along endosomal compartments and vesicle fusion processes (Figures 5(d) and 5(e)).
3.8. EhADH112 Appeared on RBC and Phagosomes during Phagocytosis
At the beginning of erythrophagocytosis, trophozoite membrane proteins make contact with RBC. Immediately, not yet well-understood signaling processes occur, allowing the recruitment of molecules with different roles in the uptake and digestion of target cells. In this paper, our immunofluorescence experiments evidenced that overexpression of the Bro1 recombinant polypeptide in ANeoBro1 trophozoites did not affect EhADH112 location at target cell contact sites and phagosomes (Figure 4(a)). Further, TEM ultrastructural observations, located EhADH112 near to and at invaginating membranes surrounding RBC in trophozoites of ANeo (data not shown) and ANeoBro1 populations (Figure 4(c)). Inside trophozoites, in the vicinity of delimiting membranes surrounding RBC, several small whitish vesicles were seen (Figure 6(b)), sometimes as if they were being released from huge vesicles. By their appearance [42], these vesicles may correspond to MVB (Figure 6(b), asterisks). As phagocytosis advanced, more abundant gold particles were found on RBC (Figures 6(b) and 6(c)). Furthermore, around phagosomes containing RBC, we observed whitish vesicles in arrangements apparently organized, some of them labeled with gold particles (Figure 6(c)).
Figure 6.
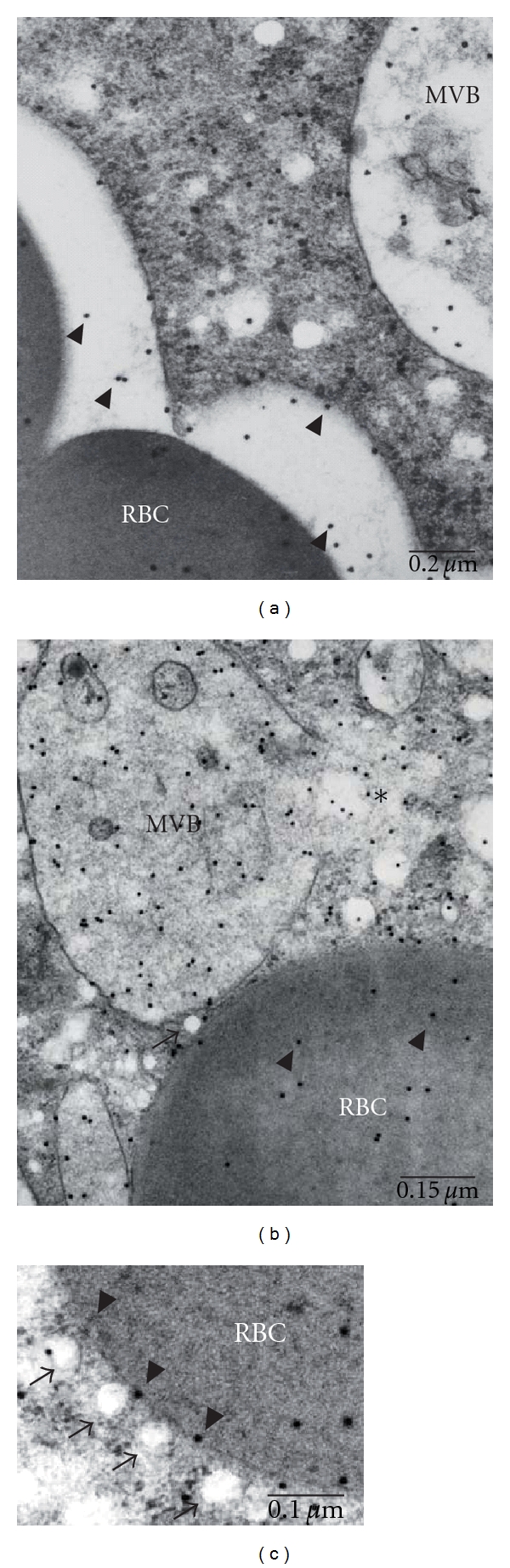
Ultrastructural location of EhADH112 in ANeoBro1 trophozoites after RBC ingestion. After 15 min of interaction with RBC, ANeoBro1 trophozoites were processed for immunogold labeling using pαEhADH112 and secondary antibodies, and contrasted and analyzed by TEM as described above. (a) Section from a trophozoite containing a RBC into phagosomal compartments, displaying a cytoplasmic MVB. (b) MVB and RBC neighboring vesicles; (c) RBC surrounded by small vesicles. MVB: multivesicular bodies. RBC: red blood cells. Arrowheads: EhADH112 gold-labeled molecules. Asterisk: disruption of a larger vacuole and release of vesicles.
Mammalian ALIX and yeast BRO1 are cytoplasmic proteins that associate with endosomal compartments to function in concert with components of the ESCRT machinery during MVB formation [41, 43]. In the endocytic pathway, MVB are formed from early endosomes and then, they fuse to late endosomes or lysosomes [12, 44]. Here, our findings strengthen the role of EhADH112 not only as an adhesin at the trophozoite surface, but also as a protein whose similarity to other Bro1 domain-containing proteins such as ALIX, could assign it an alternative function in endosome, phagosome, and MVB formation. The presence of gold labeling at the surface of small vesicles at the proximity of larger vacuoles could mean that EhADH112 is being targeted to these sites to perform a putative function in vesicle formation or that it is per se a conveyor protein which carries other molecules involved in this process. Moreover, the possibility of EhADH112 participation in vesicle biogenesis implies that it could be translocated from the plasma membrane to the delimiting membranes of internal vesicles. Hence, EhADH112 could be a soluble or insoluble membrane-associated protein, depending on its function and the protein or proteins it binds to.
3.9. EhADH112 Is Mostly Present in Membrane Subcellular Fractions
To investigate whether native EhADH112 remains as a soluble or insoluble membrane-associated protein in wild-type trophozoites, we carried out the procedure described by Aley et al. [34] followed by Western blot assays, using rabbit polyclonal antibodies against the last 243 amino acids of EhADH112 (pαEhADH243), which detect the carboxy-terminus of the adhesin. pαEhDH243 antibodies recognized the expected 78 kDa band corresponding to the EhADH112 molecular weight in crude extracts (TP) obtained after disruption of trophozoites incubated with concanavalin A in the presence of protease inhibitors (Figure 7(a), left panel, lane 1). After centrifugation at 250 ×g for 30 min on a mannitol/sucrose gradient, EhADH112 appeared in the supernatant (SN1), which contains vesicles, small membrane fragments, and soluble proteins (Figure 7(a), left panel, lane 2). We also detected a weak band in the pellet (P1), which contains large fragments of plasma membranes and cell debris (Figure 7(a), left panel, lane 3). As a control, we used mouse monoclonal antibodies against actin (mαactin), which reacted with the corresponding 43 kDa protein in all fractions (Figure 7(a), left panel, lanes 1 to 3).
Figure 7.
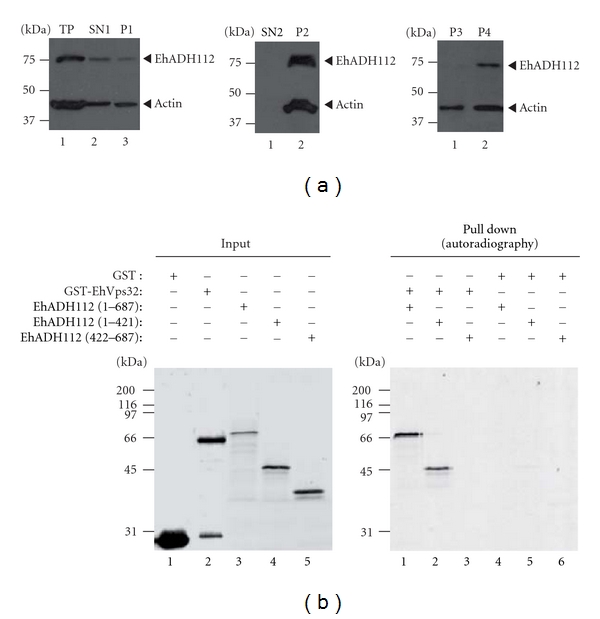
EhADH112 major presence in membrane subcellular fractions and EhADH112 interaction with EhVps32. (a) Location of EhADH112 in wild-type trophozoites subcellular fractions. Proteins (50 μg) from different fractions of trophozoite extracts were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. Western blot assays were performed using pαEhADH243 and mαactin antibodies and corresponding peroxidase-labeled anti-rabbit or anti-mouse IgG secondary antibodies. Membranes were revealed by chemiluminescence. Left: SN1 and P1 fractions resulting after total proteins (TP) centrifugation at 250 ×g for 30 min in a mannitol/sucrose gradient. Middle: SN2 and P2 fractions resulting after SN1 ultracentrifugation at 40 000 ×g for 60 min. Right: P3 and P4 fractions obtained after ultracentrifugation of solubilized P1 at 40 000 ×g for 60 min. (b) Binding of EhADH112 to EhVps32 through the Bro1 domain. GST or GST-EhVps32 proteins were immobilized on glutathione-Sepharose beads and incubated with in vitro synthesized [35S]-EhADH112 or [35S]-EhADH112 derivatives. Left: purified GST and GST-EhVps32 fusion proteins or [35S]-EhADH112 and [35S]-truncated derivatives (autoradiography, lanes 3, 4 and 5) used for binding experiments (8% of the total reaction mixture). Right: pulled-down proteins electrophoresed on 10% SDS-polyacrylamide and detected by autoradiography.
Then, we ultracentrifuged (40 000 ×g) the SN1 fraction to obtain the supernatant (SN2), where soluble components remain, and the pellet (P2), containing internal vesicles and small membrane fragments. EhADH112 was only found in P2 (Figure 7(a), middle panel, lane 2), suggesting that it could be mostly a membrane-associated protein. Actin was also absent in the SN2 fraction and strongly detected in P2 (Figure 7(a), middle panel, lane 2).
Next, we processed the P1 fraction, that according to Aley et al. [34], contains vesiculated and nonvesiculated plasma membrane fragments. The P1 fraction (Figure 7(a), left panel, lane 3) was homogenized with α-methyl mannoside and centrifuged at 250 ×g on a sucrose cushion to separate vesiculated (P4) from nonvesiculated membranes and debris (P3). P3 did not react with pαEhADH243 antibodies (Figure 7(a), right panel, lane 1), indicating that EhADH112 is poorly present in nonvesiculated membranes. Then, we concentrated the supernatant (SN3) by ultracentrifugation at 40 000 ×g to obtain vesiculated membrane fragments (P4). pαEhADH243 antibodies revealed the presence of EhADH112 in P4 (Figure 7(a), right panel, lane 2), thus, confirming that this protein is associated to vesiculated plasma membrane fragments. Meanwhile, actin was localized in P3 and P4, attached to nonvesiculated and vesiculated plasma membranes (Figure 7(a), right panel, lanes 1 and 2).
These data showed the presence of EhADH112 in membrane vesicles, supporting results obtained by microscopy experiments. They also evidence that EhADH112 could be associated to plasma membrane during the active vesicular traffic exhibited by trophozoites, probably inside small vesicles that eventually fuse to plasma membrane.
Most Bro1 domain-containing proteins are cytosolic, and it has been noticed that ALIX is transiently recruited to the plasma membrane to promote membrane fission during budding and release of viral particles, in association to late-acting ESCRT proteins [45]. EhADH112 was originally described as a component of the heterodimeric EhCPADH complex, present at trophozoite plasma membrane and cytoplasmic vacuoles under culture conditions. Interestingly, during erythrophagocytosis, this complex is found at different locations. First, the protein is found at the trophozoite plasma membrane, particularly, at target cell contact sites. Then, this protein is detected together to internalized erythrocytes, in phagosomes and phagolysosomes. At late stages, the EhCPADH complex is detected again at the trophozoite plasma membrane, thus suggesting protein recycling or even, a putative participation of EhADH112 in membrane remodeling. Additional evidence has consistently confirmed the presence of EhADH112 in both, trophozoite plasma membrane and cytoplasmic vesicles. According to our current subcellular fractioning experiments, EhADH112 is mostly present in membrane fractions. However, as it was shown in Figure 7(a), EhADH112 is also found in fractions containing soluble proteins. Therefore, we cannot discard the possibility that EhADH112 exists in soluble form under particular conditions not yet explored. Since the precise protein location is largely determined by its function in the cell, it must be considered that Bro1 domain containing proteins such as ALIX are ubiquitous in order to serve as scaffold proteins connecting several biological processes. Therefore, additional functions should be investigated for EhADH112 to better understand its subcellular location in trophozoites. Particularly, the presence of EhADH112 in different membrane compartments, strongly supports the hypothesis that EhADH112 could perform a role in the endocytic pathway, although it remains to be established if the previously reported E. histolytica ESCRT machinery is indeed participating in this process.
3.10. EhVps32 Binds to the N-Terminus of EhADH112
One conserved feature of the majority of Bro1 domain-containing proteins is their ability to bind the ESCRT-III component Vps32 or CHMP4. This interaction allows BRO1 or ALIX targeting to endosomes during MVB formation and virus budding [13, 19]. Here, we investigated whether EhADH112 interacts with an E. histolytica protein homologous to yeast Vps32. First, we found in the E. histolytica genome database a protein sequence (EhVps32) with an e-value of 2.5 e-12, displaying 48% homology and 25% identity to yeast Vps32. According to multiple sequence analysis and Pfam database predictions, EhVps32 contains a Snf7 domain, present in all members of the Snf7 family. Additionally, the predicted EhVps32 secondary structure using the Jpred program, suggested that EhVps32 conserves the characteristic five α-helices present in the Snf7 family protein. To confirm the predicted interaction between EhADH112 and EhVps32 proteins, pull down experiments were performed. Thus, we expressed a GST-EhVps32 fusion protein in bacteria. GST alone or purified GST-EhVps32 were immobilized on glutathione-sepharose beads (Figure 7(b), left panel, lanes 1 and 2, resp.) and incubated with [35S]-labeled EhADH112 (Figure 7(b), left panel, lane 3) or [35S]-EhADH112 (1-421 amino acids) (Figure 7(b), left panel, lane 4) and [35S]-EhADH112 (422–687 amino acids) derivatives (Figure 7(b), left panel, lane 5), previously synthesized by a coupled transcription-translation system, as described in Section 2.10. GST-EhVps32 beads retained EhADH112 (Figure 7(b), right panel, lane 1), and the EhADH112 (1–421) derivative (Figure 7(b), right panel, lane 2), but not the EhADH112 (422–687 amino acids) polypeptide (Figure 7(b), right panel, lane 3). As expected, GST alone was unable to bind EhADH112 and EhADH112 derivatives (Figure 7(b), right panel, lanes 4 to 6). Proteins present in pull-down reaction mixtures were Coomassie blue stained as an additional control (data not shown). Together, these results strongly suggest that EhADH112 binds EhVps32 through the Bro1 domain, as it has been reported for other Bro1 domain-containing proteins [19, 20].
Although we do not know yet the identity of other proteins associated to or transported by EhADH112 in live trophozoites, based on these results, we hypothesized that EhADH112 may interact with ESCRT proteins in vivo, as it has been reported for ALIX and BRO1 [19, 39, 46, 47]. Interestingly, EhADH112 seems to be a novel member of a subfamily of Bro1 domain-containing proteins present at cellular surface [48] that alternatively could regulate the assembly of proteins at endosomal membranes for MVB biogenesis. Translocation of EhADH112 from the plasma membrane to internal vesicles, endosomes, or phagosomes and back to the surface, could also be related to a scaffold function, as it has been described for its homologues in yeast and mammals [22]. However, protein partnerships in different cellular networks should be addressed further.
4. Conclusions
In this work, we reported for the first time the functional characterization of the N-terminus of EhADH112, an E. histolytica Bro1 domain-containing protein involved in parasite virulence. A dramatic decrease of phagocytosis rates displayed by trophozoites overexpressing an EhADH112 Bro1 recombinant polypeptide, together with an exaggerated accumulation of this protein in aberrant compartments, suggested that the Bro1 domain recruits proteins participating in phagocytosis. Moreover, EhADH112 localization at trophozoite plasma membrane, MVB and phagosomes and in both soluble and insoluble subcellular fractions, provided additional support for an alternative role for this protein in the endosomal MVB pathway. This function is conserved among Bro1 domain-containing proteins, which interact with ESCRT components to associate to endosomes. Here, we also showed the in vitro association of EhADH112 with an E. histolytica protein homologous to the ESCRT-III Vps32 subunit, as a putative hallmark for EhADH112 Bro1 domain function in endosomal protein sorting. Additional efforts should be made to better understand the role of EhADH112 in ESCRT-mediated MVB biogenesis and other functions also assigned to Bro1 domain-containing proteins.
Acknowledgments
Authors thank Abigail Betanzos for providing comments on the paper, Silvia Castellanos for pαEhADH243 antibodies and Alfredo Padilla for his assistance in the artwork. This research was mainly supported by the European Community (Phagoamoeba Project) and the National Council for Science and Technology (CONACyT-México). Authors are also grateful for the Spanish CICYT Grant BFU2005-01970 and PFPI studentship supporting O. Vincent and S. Herranz, respectively.
References
- 1.Stanley SL., Jr. Amoebiasis. The Lancet. 2003;361(9362):1025–1034. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 2.Laughlin RC, Temesvari LA. Cellular and molecular mechanisms that underlie Entamoeba histolytica pathogenesis: prospects for intervention. Expert Reviews in Molecular Medicine. 2005;7(13):1–19. doi: 10.1017/S1462399405009622. [DOI] [PubMed] [Google Scholar]
- 3.Orozco E, Guarneros G, Martínez-Palomo A, Sánchez T. Entamoeba histolytica. Phagocytosis as a virulence factor. Journal of Experimental Medicine. 1983;158(5):1511–1521. doi: 10.1084/jem.158.5.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodríguez MA, Orozco E. Isolation and characterization of phagocytosis- and virulence-deficient mutants of Entamoeba histolytica. Journal of Infectious Diseases. 1986;154(1):27–32. doi: 10.1093/infdis/154.1.27. [DOI] [PubMed] [Google Scholar]
- 5.Pimenta PFP, Diamond LS, Mirelman D. Entamoeba histolytica Schaudinn, 1903 and Entamoeba dispar Brumpt, 1925: differences in their cell surfaces and in the bacteria-containing vacuoles. Journal of Eukaryotic Microbiology. 2002;49(3):209–219. doi: 10.1111/j.1550-7408.2002.tb00525.x. [DOI] [PubMed] [Google Scholar]
- 6.García-Rivera G, Rodríguez MA, Ocádiz R, et al. Entamoeba histolytica: a novel cysteine protease and an adhesin form the 112 kDa surface protein. Molecular Microbiology. 1999;33(3):556–568. doi: 10.1046/j.1365-2958.1999.01500.x. [DOI] [PubMed] [Google Scholar]
- 7.Bañuelos C, García-Rivera G, López-Reyes I, Orozco E. Functional characterization of EhADH112: an Entamoeba histolytica Bro1 domain-containing protein. Experimental Parasitology. 2005;110(3):292–297. doi: 10.1016/j.exppara.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Vito P, Pellegrini L, Guiet C, D’Adamio L. Cloning of AIP1, a novel protein that associates with the apoptosis- linked gene ALG-2 in a Ca2+-dependent reaction. Journal of Biological Chemistry. 1999;274(3):1533–1540. doi: 10.1074/jbc.274.3.1533. [DOI] [PubMed] [Google Scholar]
- 9.Missotten M, Nichols A, Rieger K, Sadoul R. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death and Differentiation. 1999;6(2):124–129. doi: 10.1038/sj.cdd.4400456. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt MHH, Hoeller D, Yu J, et al. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Molecular and Cellular Biology. 2004;24(20):8981–8993. doi: 10.1128/MCB.24.20.8981-8993.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt MHH, Dikic I, Bögler O. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. Journal of Biological Chemistry. 2005;280(5):3414–3425. doi: 10.1074/jbc.M409839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nature Reviews Molecular Cell Biology. 2002;3(12):893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 13.Katoh K, Shibata H, Suzuki H, et al. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. Journal of Biological Chemistry. 2003;278(40):39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 14.Matsuo H, Chevallier J, Mayran N, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303(5657):531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 15.Pan S, Wang R, Zhou X, et al. Extracellular Alix regulates integrin-mediated cell adhesions and extracellular matrix assembly. EMBO Journal. 2008;27(15):2077–2090. doi: 10.1038/emboj.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. Journal of Cell Science. 2005;118(12):2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- 17.Pan S, Wang R, Zhou X, et al. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. Journal of Biological Chemistry. 2006;281(45):34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, Si J, Corvera J, Gallick GE, Kuang J. Decoding the intrinsic mechanism that prohibits ALIX interaction with ESCRT and viral proteins. Biochemical Journal. 2010;432(3):525–534. doi: 10.1042/BJ20100862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, Sitaraman S, Hierro A, Beach BM, Odorizzi G, Hurley JH. Structural basis for endosomal targeting by the Bro1 domain. Developmental Cell. 2005;8(6):937–947. doi: 10.1016/j.devcel.2005.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher RD, Chung HY, Zhai Q, Robinson H, Sundquist WI, Hill CP. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 2007;128(5):841–852. doi: 10.1016/j.cell.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 21.DeJournett RE, Kobayashi R, Pan S, et al. Phosphorylation of the proline-rich domain of Xp95 modulates Xp95 interaction with partner proteins. Biochemical Journal. 2007;401(2):521–531. doi: 10.1042/BJ20061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odorizzi G. The multiple personalities of Alix. Journal of Cell Science. 2006;119(15):3025–3032. doi: 10.1242/jcs.03072. [DOI] [PubMed] [Google Scholar]
- 23.Rothman JH, Howald I, Stevens TH. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO Journal. 1989;8(7):2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nickas ME, Yaffe MP. BRO1, a novel gene that interacts with components of the Pkc1p-mitogen-activated protein kinase pathway in Saccharomyces cerevisiae. Molecular and Cellular Biology. 1996;16(6):2585–2593. doi: 10.1128/mcb.16.6.2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Current Opinion in Cell Biology. 2009;21(4):568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roxrud I, Stenmark H, Malerød L. ESCRT & Co. Biology of the Cell. 2010;102(5):293–318. doi: 10.1042/BC20090161. [DOI] [PubMed] [Google Scholar]
- 27.López-Reyes I, García-Rivera G, Bañuelos C, et al. Detection of the endosomal sorting complex required for transport in Entamoeba histolytica and characterization of the EhVps4 protein. Journal of Biomedicine and Biotechnology. 2010;2010 doi: 10.1155/2010/890674. Article ID 890674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada M, Huston CD, Oue M, et al. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Molecular and Biochemical Parasitology. 2006;145(2):171–183. doi: 10.1016/j.molbiopara.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 29.Bateman A, Birney E, Cerruti L, et al. The pfam protein families database. Nucleic Acids Research. 2002;30(1):276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Transactions of the Royal Society of Tropical Medicine and Hygiene. 1978;72(4):431–432. doi: 10.1016/0035-9203(78)90144-x. [DOI] [PubMed] [Google Scholar]
- 31.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proceedings of the National Academy of Sciences of the United States of America. 1981;78(6 I):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamann L, Nickel R, Tannich E. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proceedings of the National Academy of Sciences of the United States of America. 1995;92(19):8975–8979. doi: 10.1073/pnas.92.19.8975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.García-Rivera G, Sánchez T, Orozco E, Guarneros G. Isolation of Entamoeba histolytica clones deficient in adhision to human erythrocytes. Archivos de Investigacion Medica. 1982;13(3):129–136. [PubMed] [Google Scholar]
- 34.Aley SB, Scott WA, Cohn ZA. Plasma membrane of Entamoeba histolytica. Journal of Experimental Medicine. 1980;152(2):391–404. doi: 10.1084/jem.152.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent O, Rainbow L, Tilburn J, Arst HN, Peñalva MA. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Molecular and Cellular Biology. 2003;23(5):1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu W, Smith FJ, Jr., Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Molecular Biology of the Cell. 2004;15(12):5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichioka F, Kobayashi R, Katoh K, Shibata H, Maki M. Brox, a novel farnesylated Bro1 domain-containing protein that associates with charged multivesicular body protein 4 (CHMP4) FEBS Journal. 2008;275(4):682–692. doi: 10.1111/j.1742-4658.2007.06230.x. [DOI] [PubMed] [Google Scholar]
- 38.Martínez-López C, Orozco E, Sánchez T, García-Pérez RM, Hernández-Hernández F, Rodríguez MA. The EhADH112 recombinant polypeptide inhibits cell destruction and liver abscess formation by Entamoeba histolytica trophozoites. Cellular Microbiology. 2004;6(4):367–376. doi: 10.1111/j.1462-5822.2004.00363.x. [DOI] [PubMed] [Google Scholar]
- 39.Babst M. A protein’s final ESCRT. Traffic. 2005;6(1):2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 40.Bishop N, Woodman P. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Molecular Biology of the Cell. 2000;11(1):227–239. doi: 10.1091/mbc.11.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luhtala N, Odorizzi G. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. Journal of Cell Biology. 2004;166(5):717–729. doi: 10.1083/jcb.200403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Denzer K, Kleijmeer MJ, Heijnen HFG, Stoorvogel W, Geuze HJ. Exosome: from internal vesicle of the multivesicular body to intercellular signaling device. Journal of Cell Science. 2000;113(19):3365–3374. doi: 10.1242/jcs.113.19.3365. [DOI] [PubMed] [Google Scholar]
- 43.Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. Journal of Cell Science. 2003;116(10):1893–1903. doi: 10.1242/jcs.00395. [DOI] [PubMed] [Google Scholar]
- 44.Gruenberg J, Stenmark H. The biogenesis of multivesicular endosomes. Nature Reviews Molecular Cell Biology. 2004;5(4):317–323. doi: 10.1038/nrm1360. [DOI] [PubMed] [Google Scholar]
- 45.Von Schwedler UK, Stuchell M, Müller B, et al. The protein network of HIV budding. Cell. 2003;114(6):701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 46.Boysen JH, Mitchell AP. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Molecular Biology of the Cell. 2006;17(3):1344–1353. doi: 10.1091/mbc.E05-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCullough J, Fisher RD, Whitby FG, Sundquist WI, Hill CP. ALIX-CHMP4 interactions in the human ESCRT pathway. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(22):7687–7691. doi: 10.1073/pnas.0801567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Irie T, Nagata N, Yoshida T, Sakaguchi T. Recruitment of Alix/AIP1 to the plasma membrane by Sendai virus C protein facilitates budding of virus-like particles. Virology. 2008;371(1):108–120. doi: 10.1016/j.virol.2007.09.020. [DOI] [PubMed] [Google Scholar]


