Abstract
Selective neuronal nitric oxide synthase (nNOS) inhibitors have therapeutic applications in the treatment of numerous neurodegenerative diseases. Here we report the synthesis and evaluation of a series of inhibitors designed to have increased cell membrane permeability via intramolecular hydrogen bonding. Their potencies were examined in both purified enzyme and cell-based assays; a comparison of these results demonstrates that two of the new inhibitors display significantly increased membrane permeability over previous analogs. NMR spectroscopy provides evidence of intramolecular hydrogen bonding under physiological conditions in two of the inhibitors. Crystal structures of the inhibitors in the nNOS active site confirm the predicted non-intramolecular hydrogen bonded binding mode. Intramolecular hydrogen bonding may be an effective approach for increasing cell membrane permeability without affecting target protein binding.
Keywords: neuronal nitric oxide synthase, enzyme inhibitors, intramolecular hydrogen bond
1. Introduction
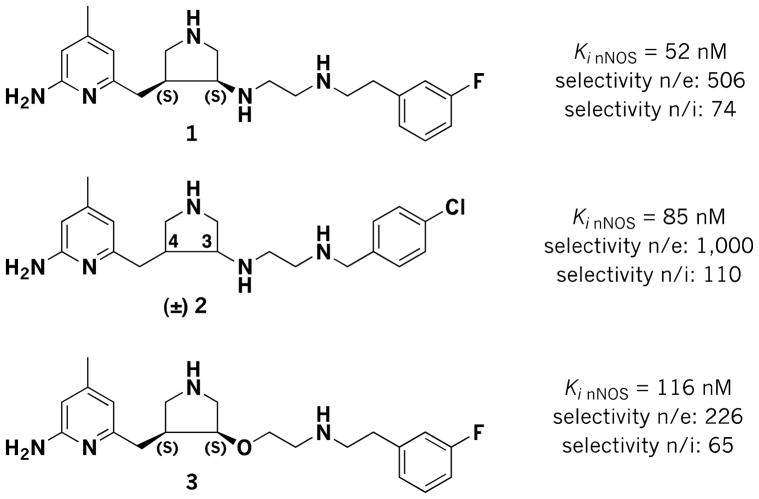
Neurodegeneration, characterized by progressive loss of neurons in isolated areas of the central nervous system (CNS), has become a rapidly spreading health crisis in the world. It affects the lives of millions of people, causing problems with motor skills and memory. Although significant research advances have been made, effective treatments for neurodegenerative disorders have remained elusive. An abnormally high concentration of cerebral nitric oxide (NO), the metabolite of the neuronal isoform of nitric oxide synthases (nNOS), has been observed as a common pathogenic phenomenon shared by various neurodegenerative conditions, including Alzheimer’s, [1] Parkinson’s, [2] Huntington’s,[3] migraine headaches,[4] as well as neuronal damage in stroke.[5] High NO concentrations provide a rationale for the therapeutic use of nNOS inhibitors as a potential simultaneous treatment for different types of neurodegeneration. In the past two decades, a large number of nNOS inhibitors have been reported.[6–9,9] However, none of these compounds has been approved for medical use. Recently, we reported a family of syn-3,4-substituted pyrrolidine inhibitors (e.g., 1–3, Figure 1) that were discovered using structure-based drug design.[10–13] These inhibitors showed remarkable potency for nNOS and excellent isoform selectivity over endothelial NOS (eNOS) and inducible NOS (iNOS).We have previously found that the (3S,4S)-isomers of 1 and 3 bind to nNOS in the predicted mode with the 3-fluorophenyl group bound in the hydrophobic cleft comprised of M336 and L337 and the aminopyridine interacting with E592, but unexpectedly the (3R,4R)-isomers bind with 180° rotation with the 3-fluorophenyl group -stacked over the active site heme and the aminopyridine electrostatically interacting with a carboxylate of heme propionate.[11] Also surprising, the (3R,4R)-isomers are more potent than the (3S,4S)-isomers. However, results from animal tests indicated that racemic mixtures of 1 and 3 (and their corresponding 3R,4R stereoisomers) show minimal blood-brain barrier (BBB) penetration;[13] the BBB is a unique barrier formed by brain capillary endothelial cells that molecules must be able to penetrate to be effective in the CNS. This result limits further investigation of these lead compounds as oral therapeutics. Given the chemical structures of the inhibitors, we reasoned that the multiple positive charges and hydrogen bond donor properties of 1–3 at physiological pH, derived from the amino groups, limit them from penetrating the BBB by passive diffusion.[13] Lead compound 3was designed to eliminate one of the hydrogen bond donors of 1, the amino group attached to the pyrrolidine ring, by replacing it with a hydrogen bond acceptor ether linkage; hydrogen bond donors are believed to lower the ability to cross the BBB more than hydrogen bond acceptors. Supporting our rationale, BBB penetration was slightly improved from the design of 1 to 3, but there is still much room for improvement in the bioavailability of these selective inhibitors.[13]
Figure 1.
Chemical structures and inhibitory activities of inhibitors (3S,4S)-1 and (3S,4S)-3 and (±)-2.
In previous efforts we pursued different strategies to modify the structure of compounds in this class.[13–17] For example, the high pKa amino groups have been replaced by neutral functionalities such as ethers and amides.[13] We have also inserted electron-withdrawing groups, such as ether, monofluoromethylene, difluoromethylene, and cyclopropyl, to inductively decrease the pKa of the amino group.[14–17] Despite the improved membrane permeability of the new inhibitors, the successes of these modified structures were compromised by a significant drop in the in vitro potency and isoform selectivity.
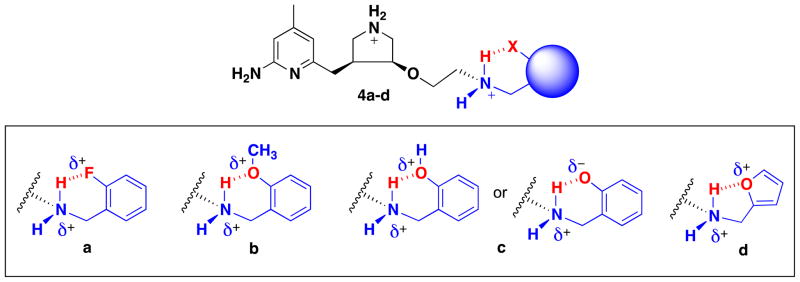
Here we describe the design and synthesis of a new series of nNOS inhibitors (4a–d, Figure 2) to diffuse the overall charge of 3 by incorporation of an intramolecular hydrogen bond, a known strategy in the design of novel inhibitors for a variety of enzymes, [18–23] sometimes used to improve BBB penetration.[24] In the highly hydrophobic environment of the BBB, the inhibitor is hypothesized to adopt a “closed” conformation by forming an intramolecular H-bond, which decreases the overall polarity of the compound to improve BBB permeability. On the other hand, when the inhibitor reaches and binds to nNOS, it may encounter various intermolecular interactions and stabilize an “open” (not intramolecular hydrogen bonded) conformation. In this conformation, the pharmacologically critical amino group may be released from the intramolecular H-bond. Compounds 4a–d have benzyl-like aryl substituents like 2[25] rather than phenylethyl-like aryl substituents like 1[26] and 3, [26] so that 6-membered instead of 7-membered intramolecular hydrogen bonds would form. Therefore, we chose to make the (3S,4S)-isomers that bind in the normal mode, despite the generally lower potency in the phenylethyl series; computer modeling suggested that with a one-carbon shorter side chain compounds would have better binding, although the (3R,4R) compounds have not been made to confirm. Six-membered intramolecular H-bonds are most common, the majority of which are linked by sp2-hybridized atoms and are therefore part of planar conjugated systems; five-membered intramolecular hydrogen bonds also have precedence.[18]
Figure 2.
Chemical structures of (S.S)-4a–d. The desired hydrogen bond is shown in red. Inhibitor 4c may take on a conformation in which the −OH acts as both a H-bond acceptor and donor, delocalizing the positively charged nitrogen.
2. Chemistry
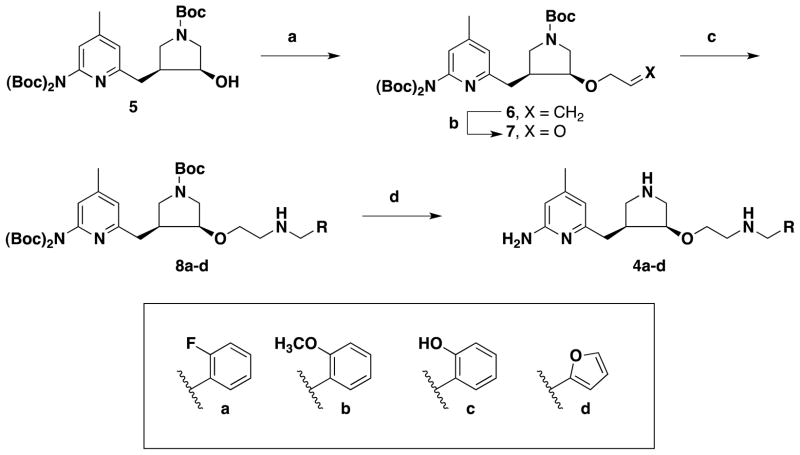
The synthesis of inhibitors 4a–d began with chiral precursor 5.[27] Allylation of 5 using gave alkene 6 in good yields.[28] Ozonolysis allyl methyl carbonate in the presence of Pd(PPh3)4 of 6 yielded aldehyde 7, which was subjected to reductive amination with amines to provide 8a–d. Global deprotection of the three Boc-protecting groups with TFA generated inhibitors 4a–d in quantitative yields.
3. Results and Discussion
3.1 Assay with purified nNOS
The potencies of these compounds have been tested both in a purified enzyme assay and in a cell-based assay. The purified enzyme assay results demonstrate that 4a–d have lower potencies for nNOS and lower selectivities over eNOS and iNOS in comparison with our previous inhibitors 1–3 (Table 1), [25,26] but compounds 4a–d are proof-of-principle compounds to test their ability to permeate a cell membrane.
Table 1.
Ki a values of 1–3 and 4a–4d with all three NOS isoforms.
3.2 Assay in HEK293t cells
We have employed a simple, colorimetric, cell-based assay using HEK293t cells stably transfected with and expressing nNOS to assess the potencies of these compounds in a cellular environment. [29] Although other processes may be influencing nNOS inhibition in these assays, cell permeability can be predicted from the cellular potencies of these compounds as measured by the inhibition of the production of nitric oxide in the HEK293t cells (Table 2). Comparing the IC50 values of these compounds in the purified enzyme assay versus the cellular assay clearly indicates that membrane permeability is a major factor that limits the effective concentration of these inhibitors within cells. Therefore, the Relative Permeability Index (RPI) is a measure of how similar the IC50 value is for a compound as measured in the cell-based assay relative to the purified enzyme assay. If a compound freely passed through the cell membrane, then those IC50 values should be the same (cell based IC50/purified enzyme IC50 = 1.0). The lower the RPI, the more cell membrane-penetrable is the compound. Therefore, 4a and 4b have the highest cell penetrability compared to 1–3, and 4c is comparable in penetrability relative to 3, despite the poorer inhibitory properties of these compounds relative to 1–3. As depicted in Figure 2, however, 4c had the greatest theoretical potential to neutralize the charge on the ammonium ion, yet it did not exhibit high cell permeability. That may suggest that the phenolic hydroxyl group does not act as a hydrogen bond donor. Compound 4d, with the highest IC50 of all compounds in the table, is nonetheless more cell-penetrable than 1 and 2. We also tested just the aminopyridine fragment of inhibitors 1–4 (2-amino-4,6-dimethylpyridine hydrochloride), which is a potent, nonselective inhibitor of purified nNOS (50 nM); however, its RPI is much higher than any of our inhibitors, indicating that it poorly crosses the cell membrane despite having a neutral pKa. Although L-N-nitroarginine (L-NNA) is a relatively poor inhibitor of purified nNOS, it appears to penetrate the cell membrane freely, having the same IC50 value in the cell-based assay as the isolated enzyme assay and a RPI of 1.
Table 2.
IC50 valuesa of NOS inhibitors in purified enzyme assay and cell-based assay. See methods for assay details.
| Compound | Purified IC50 (μM) | Cell-based IC50 (μM) | Relative Permeability Index (RPI)b (Cell based IC50/Purified IC50) |
|---|---|---|---|
| 1 | 0.45 | 19 | 42 |
| 2 | 0.74 | 42 | 57 |
| 3 | 1.0 | 13 | 13 |
| 4a | 3.6 | 11 | 3.1 |
| 4b | 7.1 | 22 | 3.1 |
| 4c | 3.5 | 43 | 12 |
| 4d | 3.4 | 100 | 29 |
| aminopyridinec | 0.05 | 25 | 500 |
| L-NNAd | 5.0 | 5.0 | 1 |
See methods. IC50 values represent at least triplicate measurements with standard deviations of less than10%.
The smaller the number, the closer the difference in IC50 for the cell-based assay and purified enzyme assay, which suggests improved permeability into cells.
aminopyridine = 2-amino-4,6-dimethylpyridine hydrochloride.
L-NNA = L-N-nitroarginine.
It is interesting to note that the relative permeability index has little correlation with partition coefficients for the inhibitors. As calculated by Chem Axon [30], the cLogP values are as follows: 2.18 (1); 2.35 (2); 2.49(3); 2.2 (4a); 1.9 (4b); −0.09 (4c); 1.12 (4d). For this series, cLogP seems to be a poor predictor of permeability as measured by the NOS cell-based assay.
3.3 Crystallography of 4a–d bound to nNOS
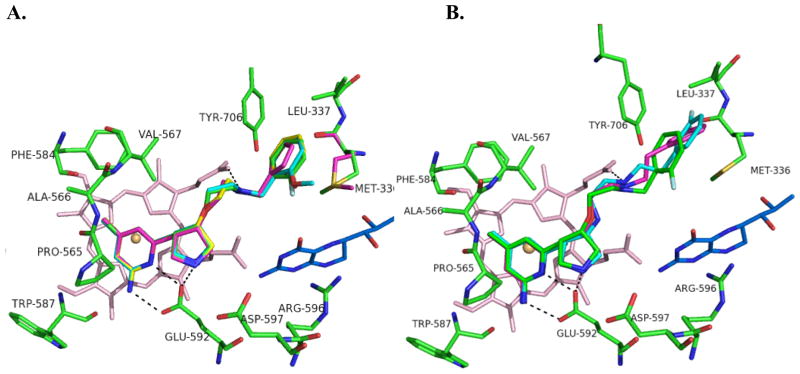
Crystal structures of all four compounds bound to nNOS were obtained, showing that when bound to nNOS, they all adopt the open, non-intramolecular hydrogen bonded form (Figure 3). Inhibitors 4a–4d (like 1–3), bear a (3S,4S) configuration at the pyrrolidine ring, and all bind to the nNOS active site in the normal binding mode, with the 2-amino-4-methylpyridine stacked over the heme plane and hydrogen bonding with Glu592.[11] The pyrrolidine nitrogen hydrogen bonds with Glu592 and interacts through a water-bridged hydrogen bond with Asp597, the residue that can be exploited for selectivity between nNOS and eNOS (Asn368 in eNOS). The amino group that was designed to form an intramolecular hydrogen bond with the substituent from the tail aromatic ring is exposed in the crystal structures and hydrogen bonds with heme propionate D in all four inhibitors. Compounds 4a–c have a substituted phenyl tail, but 4d contains a furan in place of the phenyl ring. The substituted phenyls and the furan ring occupy the pocket lined with hydrophobic residues Met336, Leu337, and Tyr706. The furan is smaller than the phenyl substituents, so it is able to accommodate a different rotamer of Met336 in which it rotates in toward the inhibitor molecule. The slightly higher Ki of 4b, the methoxyl-substituted inhibitor, may result from a slight steric clash with Trp306 (subunit B) in this pocket.
Figure 3.
A. Binding conformations of 4a–4d overlaid: 4a (green), 4b (cyan), 4c (yellow), 4d (magenta); heme is shown in light pink; H4B (tetrahydrobiopterin) is blue. The enzyme conformation is highly conserved between all four structures; therefore only the active site residues from the crystal structure of 4a are shown. Met336 of the 4d crystal structure is also shown (magenta) because of the difference in its configuration. B. Binding conformations of 1 (3JWS, cyan), 3 (3NLK, magenta) and 4a (green), overlaid. Residues overlay almost exactly; therefore only those corresponding to the crystal structure of 4a are shown. Hydrogen bonds are depicted with dashed lines. PDB codes: nNOS-4a, 3TYL; nNOS-4b, 3TYM; nNOS-4c, 3TYN; nNOS-4d, 3TYO.
Figure 3B shows a comparison of the binding mode of 4a–d with that of previously designed (3S,4S) inhibitors 1–3 using an overlay of 1 and 3 with 4a. Inhibitor 1 (2 as well) contains a nitrogen atom in place of the oxygen atom adjacent to the pyrrolidine in 3 and 4a–d. It is this amino nitrogen in 1 (and 2) that makes a hydrogen bond with a different heme propionate (of pyrrole A). The ether oxygen in 3 and 4a–d does not have a proton to make this hydrogen bond. Instead, 3 and 4a–d all curl so that the other amino group next to the aromatic tail group can hydrogen bond with heme propionate D. The inhibitors with an amine next to the pyrrolidine (1 and 2), generally bind tighter to nNOS (Figure 1) than the ones bearing an ether oxygen at the same position (3 and 4a–d, Table 1). However, the ether inhibitors show better bioavailability.[14,17] Another noticeable difference in the crystal structure between inhibitors 1 and 3 and series 4a–d occurs because of one less carbon unit in the chain to the phenyl substituent in the latter. The hydrophobic pocket is large enough to accommodate either chain length. However, the inhibitors with a longer chain length make tighter van der Waals contacts with Met336, Leu337, and Tyr706, thus showing better binding affinity than the shorter tailed 4a–d. Inhibitor 2 has the same chain length as 4a–d but 2 has better binding affinity than 4a–d. This is because the amino group next to the pyrrolidine in 2 hydrogen bonds to heme propionate A, so that the rest of its fluorophenyl tail adopts an extended conformation rather the curled one seen in 4a–d, allowing tighter hydrophobic interactions with the surrounding protein.
3.4 NMR spectroscopy to determine extent of hydrogen bonding
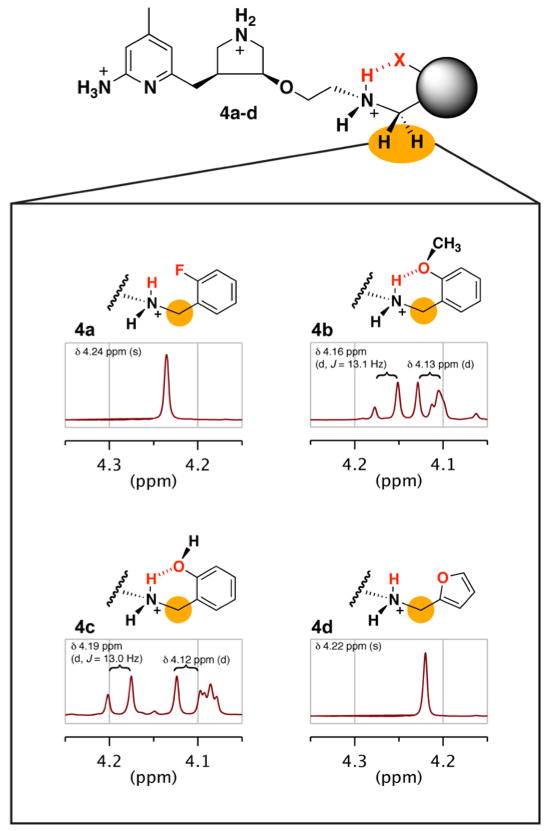
Inspection of the 1H NMR spectra of these compounds in D2O at room temperature shows a striking difference in the splitting pattern of the methylene group (4.2 ppm) in the arylalkylside chain (Figure 4). When the intramolecular H-bond forms, the interaction of the phenyl ortho-substituent differentiates the two protons of the ammonium and makes the ammonium nitrogen chiral. This anisotropy [32,34] is evident as a second-order effect in the 1H NMR spectrum because the side chain methylene protons are diastereotopic. In the 1H NMR spectra of 4b and 4c, these protons exhibit an AB pattern, two separate but coupled doublets, instead of the singlet expected for these identical protons. The average shift of the AB quartet is centered at the chemical shift of the expected singlet, and the doublets for the individual protons are found shifted upfield or downfield.
Figure 4.
1H NMR spectra of 4a–4d taken in D2O at 25 °C. Doublets appear in place of the singlet usually seen with compounds of this scaffold for the methylene protons adjacent to the H-bondedamine in 4b and 4c; this is evidence for the H-bond. In both cases, the doublet further upfield overlaps with the triplet from one of the pyrrolidine hydrogens; a coupling value cannot be reported for the doublet further upfield but is presumed to be the same.
In contrast, the observed signal in the NMR spectra for the methylene protons of 4a and 4d (Figure 4) is the expected singlet at approximately 4.2 ppm. In these two compounds the methylene protons are also diastereotopic (as in 4b and 4c), but the lack of a stable interaction to differentiate the protons of the ammonium gives rise to a singlet for the methylene protons. Therefore, there is likely no significant intramolecular hydrogen bonding interaction occurring in these compounds at room temperature in an aqueous environment, and more importantly, not under physiological conditions (37 °C). Fluorine-mediated hydrogen bonds are weak (if observable at all), and these results (4a) are consistent with this observation. A five-membered intramolecular hydrogen bond (4d) is known to be the least likely to form.[18]
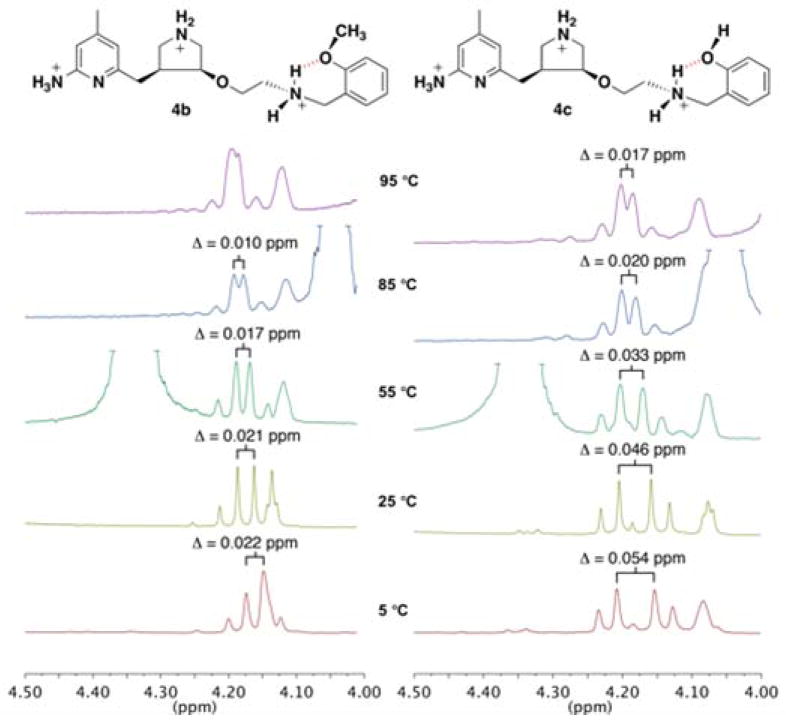
An examination of the NMR spectra of 4b and 4c in D2O at various temperatures provides further evidence of the hydrogen bond character. The two doublets shift at higher temperatures, as seen in Figure 5. Unfortunately the doublets of 4b are slightly obscured by the shift of the pyrrolidine proton (at 25 °C, 4.12 ppm) and spectral resolution decreases at 95 °C; nonetheless, Figure 5 illustrates that the chemical shifts of the doublets are moving closer together as the temperature increases. In both inhibitors, hydrogen bonding is observed in water at physiological pH, and the temperature at which doublets would be expected to collapse into a singlet is well above the physiological temperature of 37 °C. This suggests that the intramolecular hydrogen bonds of 4b and 4c are stable in solution at high temperatures and that this approach would be amenable to enhancing membrane permeability in vivo.
Figure 5.
1H NMR spectra of 4b and 4c in D2O at 95, 85, 55, 25, and 5 °C. The AB doublets shift toward one another at higher temperatures. In the spectra of 4b, the doublet further upfield is obscured by the shift of a pyrrolidine proton (4.12 at 25 °C). The J value within the doublets remains the same, J = ~13 Hz. The difference between the chemical shifts ( ) is reported to better illustrate the changes in the chemical shifts, as instrument resolution is lost at higher temperatures.
Interestingly, 4a and 4b demonstrate the highest relative permeability (Table 2), although it is 4b and 4c that display NMR spectral evidence of a H-bond. The o-fluorophenyl group of permeable inhibitor 4a is the most lipophilic side chain group of the series and also the most electron withdrawing; the increased electron withdrawing character lowers the pKa of the ammonium group, making it less basic. Both of these properties might account for the enhanced permeability of 4a. This is consistent with the findings of Ballet et al. who concluded that lipophilicity rather than side chain flexibility is the key determinant for BBB penetration.[35] Compound 4c, despite its ability to intramolecular hydrogen bond has a polar phenolic hydroxyl group, which would hinder cell permeability.
4. Conclusion
New nNOS inhibitors 4a–d have been synthesized derived from chiral pyrrolidine lead 2. Crystal structures confirm that these compounds bind to nNOS in the “open” conformation, while 1H NMR spectra provide evidence of intramolecular hydrogen bonding, the “closed” conformation, in two of the compounds (4b and 4c). It appears, however, that incorporation of an intramolecular hydrogen bond is not sufficient to enhance cell membrane permeability; lipophilicity seems to play a larger role, as evidenced by the good permeability of 4a and the poor permeability of 4c. Inhibitors 4a and 4b demonstrate greatly improved cell permeability when compared to previous inhibitors 1–3. For inhibitor 4b, its improved permeability likely results from intramolecular hydrogen bonding to diffuse the positive charge on the vicinal ammonium ion. Once the compounds enter the active site of nNOS, however, intermolecular binding interactions overcome the intramolecular hydrogen bonds, resulting in the predicted open-conformation binding mode. This suggests that intramolecular hydrogen bonding can accomplish the desired decreased polarity for enhanced cell membrane permeability, but not interfere with desired binding interactions in the target protein. The purpose of these studies was to demonstrate the effect of hydrogen bonding on cell permeability; the next step in the process will be to modify the compounds for better binding potency without loss of cell membrane permeability.
5. Experimental Section
5. 1 General Methods
All experiments were conducted under anhydrous conditions in an atmosphere of argon, using flame-dried apparatus and employing standard techniques in handling air-sensitive materials. All solvents were distilled and stored under an argon or nitrogen atmosphere before using. All reagents were used as received. Aqueous solutions of sodium bicarbonate, sodium chloride (brine), and ammonium chloride were saturated. Analytical thin layer chromatography was visualized by ultraviolet light. Flash column chromatography was carried out under a positive pressure of nitrogen. 1H NMR spectra were recorded on 500 MHz spectrometers. Data are presented as follows: chemical shift (in ppm on the δ scale relative to δ = 0.00 ppm for the protons in tetramethylsilane (TMS)), integration, multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad), coupling constant (J/Hz). Coupling constants were taken directly from the spectra and are uncorrected. 13C NMR spectra were recorded at 125 MHz, and all chemical shift values are reported in ppm on the δ scale with an internal reference of δ 77.0 or 49.0 for CDCl3 or MeOD, respectively. High-resolution mass spectra were measured by liquid chromatography/time-of-flight mass spectrometry (LC-TOF). The purity of the final compounds was determined by HPLC analysis to be ≥95%. The column used was a Higgins Analytical, TARGA®C18, 5 μm, 4.6 × 250 mm (p/n: TS-2546-C185, The Nest Group, Inc., http://www.nestgrp.com). The column was thoroughly equilibrated at 100% solvent A, minimum 5 volumes. The compounds were eluted with a gradient from solvent A (water, 0.1% TFA) to solvent B (acetonitrile, 0.1% TFA), 0–5 minutes, 0–25% B, 5–10, 25–35% B, 10–20 35% isocratic. The compounds had the following retention times, 4a, 11.2 min, 4b, 11.6 min, 4c, 11.4 min, 4d, 10.5 min.
5.2 Preparation and Characterization of New Compounds
5.2.1 (3S,4S)-tert-Butyl-3-(allyloxy)-4-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)pyrrolidine-1-carboxylate (6)
To a solution of 5 (1.15 g, 2.0 mmol) and Pd(Ph3P)4 (235 mg, 0.2 mmol) in dry THF (50 mL) was added allyl methyl carbonate (700 μL, 6.0 mmol). The reaction mixture was allowed to stir at 45 °C for 5 h and then concentrated. The resulting material was purified by flash column chromatography (silica gel, EtOAc/hexanes, 1:2, Rf = 0.40) to yield 6 (675 mg, 66%) as a colorless oil: 1H NMR (500 MHz, CDCl3) 1.40–1.50 (m, 27H), 2.25–2.27 (m, 3H), 2.60–2.75 (m, 1H), 2.78–2.85 (dd, J = 9.0, 13.5 Hz, 1H), 2.98–3.05 (dd, J = 9.0, 13.5 Hz, 1H), 3.10–3.21 (m, 1H), 3.25–3.29 (dd, J = 4.0, 12.5 Hz, 1H), 3.40–3.62 (m, 2H), 3.75–3.85 (m, 2H), 4.00–4.10 (td, J = 5.5, 13.0 Hz, 1H), 5.15–5.17 (d, J = 10.5 Hz, 1H), 5.25–5.29 (d, J = 17.0 Hz, 1H), 5.84–5.91 (ddd, J = 5.0, 10.5, 17.0 Hz, 1H), 6.85–6.95 (m, 2H); 13C NMR (125 MHz, CDCl3) 20.9, 27.9, 28.4, 28.5, 34.7, 34.8, 42.7, 43.3, 48.9, 49.2, 50.4, 51.0, 70.2, 70.3, 77.8, 78.6, 79.1, 79.2, 82.8, 116.7, 116.9, 119.6, 122.9, 134.6, 134.7, 149.50, 149.52, 151.4, 151.5, 151.8, 154.5, 154.8, 159.2, 159.3; LC-TOF (M+H+) calcd for C29H46N3O7 548.3336, found 548.3339.
5.2.2 (3S,4S)-tert-Butyl-3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-oxoethoxy)pyrrolidine-1-carboxylate (7)
A solution of 6 (100 mg, 0.19 mmol) in CH2Cl2 (10 mL) was cooled to −78 °C, to which O3 was charged until the reaction solution turned purple (~10 min). The O3 flow was stopped, and the reaction was allowed to stir at the same temperature for 30 min. To the resulting solution was added Me2S (150 μL). The reaction mixture was then warmed to room temperature and kept stirring at room temperature for an additional 2 h. The solvent was removed by rotary evaporation and the resulting crude product was purified by flash column chromatography (EtOAc/hexanes, 1:1, Rf = 0.2) to yield 6 (87 mg, 87%) as a colorless oil: 1H NMR (500 MHz, CDCl3) 1.45 (s, 27H), 2.22 (s, 3H), 2.70–2.85 (m, 1H), 2.85–2.95 (m, 1H), 3.05–3.15 (m, 1H), 3.16–3.25 (m, 1H), 3.30–3.37 (m, 1H), 3.45–3.70 (m, 2H), 3.85–3.95 (m, 2H), 4.05–4.20 (t, J = 10.0 Hz, 1H), 6.90–6.93 (m, 2H), 9.66 (s, 1H); 13C NMR (125 MHz, CDCl3) 20.9, 24.7, 27.9, 28.5, 29.7, 34.4, 42.5, 43.2, 48.8, 49.1, 50.3, 51.0, 74.6, 74.9, 79.4, 79.5, 80.4, 83.0, 119.66, 119.73, 122.8, 149.7, 149.8, 151.57, 151.60, 151.8, 154.4, 154.8, 159.87, 158.94, 200.2, 200.6; LC-TOF (M+H+) calcd for C28H44N3O8 550.3128, found 550.3130.
5.2.3 (3S,4S)-tert-Butyl-3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-(2-fluorobenzylamino)ethoxy)pyrrolidine-1-carboxylate (8a)
To a solution of aldehyde 7 (100 mg, 0.18 mmol) in DCM (4 mL) was added 2,2-difluoro-2-(3-fluorophenyl)ethanamine hydrochloride (83 mg, 0.36 mmol), triethylamine (11 μL, 0.36 mmol), followed by NaHB(OAc)3 (100 mg, 0.45 mmol). The reaction mixture was stirred for an additional 3 h and then concentrated. The crude product was purified by flash column chromatography (EtOAc/hexanes, 1:1, Rf = 0.15) to yield 8a (115 mg, 91%) as a colorless oil: 1H NMR (500 MHz, CDCl3) 1.43–1.44 (s, 27H), 2.29–2.30 (m, 3H), 2.60–2.75 (m, 1H), 2.76–2.85 (m, 2H), 2.92–3.00 (m, 1H), 3.05–3.14 (m, 1H), 3.20–3.50 (m, 5H), 3.52–3.70 (m, 2H), 3.72–3.80 (m, 1H), 3.91 (s, 2H), 6.84–6.85 (d, J = 8.0 Hz, 1H), 6.89–6.91 (d, J = 10.0 Hz, 1H), 7.02–7.06 (dd, J = 9.0, 9.5 Hz, 1H), 7.10–7.13 (dd, J = 7.0, 7.5 Hz, 1H), 7.20–7.30 (m, 1H), 7.35–7.40 (m, 1H); 13C NMR (125 MHz, CDCl3) 20.9, 24.7, 27.9, 28.47, 28.50, 29.7, 34.6, 34.7, 36.6, 42.6, 43.3, 46.8, 48.2, 48.8, 49.1, 50.3, 50.8, 68.0, 68.2, 78.7, 79.2, 79.3, 79.4, 82.8, 115.2, 115.3, 115.4, 115.5, 119.5, 119.6, 122.8, 124.17, 124.19, 128.89, 128.92, 128.95, 128.99, 130.46, 130.51, 130.55, 149.6, 151.4, 151.5, 151.8, 154.5, 154.8, 159.08, 159.14, 160.3, 162.2; LC-TOF (M+H+) calcd for C35H52FN4O7 659.3820, found 659.3818.
5.2.4 (3S,4S)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-(2-methoxybenzylamino)ethoxy)pyrrolidine-1-carboxylate (8b)
Compound 8b was synthesized using a similar procedure to that of 8a (88%): 1H NMR (500 MHz, CDCl3) 1.42–1.43 (s, 27H), 2.31–2.33 (m, 3H), 2.60–2.70 (m, 1H), 2.70–2.80 (m, 1H), 2.90–3.10 (m, 4H), 3.28–3.32 (m, 1H), 3.35–3.52 (m, 3H), 3.65–3.75 (m, 1H), 3.79–3.81 (m, 2H), 3.85–3.87 (m, 3H), 4.01–4.20 (m, 2H), 6.75–7.00 (m, 4H), 7.26–7.30 (m, 1H), 7.35–7.40 (m, 1H); 13C NMR (125 MHz, CDCl3) 20.9, 21.9, 27.9, 28.3, 28.4, 28.5, 29.7, 34.4, 34.5, 42.5, 43.2, 46.1, 47.5, 47.7, 48.7, 48.9, 50.3, 50.8, 55.4, 55.5, 55.6, 60.4, 65.2, 65.3, 79.3, 79.4, 79.8, 82.92, 82.94, 110.5, 110.6, 119.7, 119.8, 121.0, 121.2, 121.3, 122.7, 122.8, 130.50, 130.52, 131.4, 149.9, 151.47, 151.51, 151.77, 151.79, 154.5, 154.7, 157.7, 158.89, 158.91, 176.2; LC-TOF (M+H+) calcd for C36H55N4O8 671.4020, found 671.4016.
5.2.5 (3S,4S)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-(2-hydroxybenzylamino)ethoxy)pyrrolidine-1-carboxylate (8c)
Compound 8c was synthesized using a similar procedure to that of 8a (55%): 1H NMR (500 MHz, CDCl3) 1.45–1.46 (s, 27H), 2.34 (s, 3H), 2.60–2.75 (m, 2H), 2.75–2.85 (m, 1H), 2.85–3.00 (m, 2H), 3.00–3.07 (m, 1H), 3.07–3.20 (m, 1H), 3.20–3.33 (m, 1H), 3.33–3.51 (m, 3H), 3.51–3.65 (m, 1H), 3.65–3.74 (m, 2H), 3.74–3.90 (m, 1H), 4.00–4.18 (m, 2H), 6.75–6.85 (m, 2H), 6.93 (s, 1H), 7.02–7.10 (m, 1H), 7.15–7.20 (m, 1H); 13C NMR (125 MHz, CDCl3) 20.9, 21.1, 27.9, 28.5, 29.7, 34.1, 42.4, 43.1, 46.8, 47.0, 48.4, 48.9, 50.1, 50.3, 50.7, 50.8, 53.4, 60.3, 66.3, 66.4, 78.6, 79.3, 79.5, 79.6, 82.9, 83.2, 83.3, 116.1, 116.3, 118.9, 119.4, 119.7, 120.3, 120.5, 120.9, 122.9, 123.0, 123.2, 128.8, 128.9, 129.4, 129.6, 129.7, 150.3, 150.6, 151.4, 151.6, 151.7, 154.7, 154.9, 157.2, 157.3, 159.1, 159.2; LC-TOF (M+H+) calcd for C35H53N4O8 657.3863, found 657.3874.
5.2.6 (3S,4S)-tert-Butyl 3-((6-(bis(tert-butoxycarbonyl)amino)-4-methylpyridin-2-yl)methyl)-4-(2-(furan-2-ylmethylamino)ethoxy)pyrrolidine-1-carboxylate(8d)
Compound 8b was synthesized using a similar procedure to that of 8a (90%): 1H NMR (500 MHz, CDCl3) 1.43–1.44 (s, 27H), 2.30–2.32 (m, 3H), 2.50–2.60 (m, 1H), 2.75–2.83 (m, 2H), 2.92–3.17 (m, 3H), 3.20–3.50 (m, 5H), 3.52–3.70 (m, 2H), 3.77–3.80 (m, 1H), 3.81 (s, 2H), 6.20–6.21 (d, J = 3.0 Hz, 1H), 6.32 (s, 1H), 6.86–6.92 (m, 2H), 7.37 (s, 1H); 13C NMR (125 MHz, CDCl3) 19.1, 20.9, 21.0, 23.4, 24.7, 27.9, 28.39, 28.48, 28.51, 29.7, 30.6, 34.6, 34.7, 36.6, 42.6, 43.2, 45.8, 45.9, 48.2, 48.3, 48.8, 49.1, 50.3, 50.8, 64.4, 68.2, 68.4, 78.7, 79.2, 79.3, 79.4, 82.85, 82.86, 107.0, 107.2, 110.16, 110.24, 119.57, 119.61, 122.8, 141.90, 141.94, 141.97, 149.6, 151.45, 151.50, 151.8, 153.4, 154.5, 154.7, 159.1, 159.2, 171.3; LC-TOF (M+H+) calcd for C33H51N4O8 631.3703, found 631.3703.
5.2.7 6-(((3S,4S)-4-(2-(2-Fluorobenzylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (4a)
To a solution of 8a (70 mg, 0.10 mmol) in MeOH (2 mL) was added 6 N HCl (4 mL) at room temperature. The mixture was stirred for 12 h and then concentrated. The crude product was purified by recrystallization (EtOH/H2O) to give inhibitor 4a(38 mg, 97%): 1H NMR (500 MHz, D2O) 2.18 (s, 3H), 2.68–2.73 (m, 1H), 2.76–2.82 (dd, J = 7.0, 15.5 Hz, 1H), 2.85–2.90 (dd, J = 8.0, 15.0 Hz, 1H), 3.04–3.09 (t, J = 11.5 Hz, 1H), 3.15–3.25 (m, 2H), 3.39–3.43 (dd, J = 8.5, 11.5 Hz, 1H), 3.50–3.53 (d, J = 13.5 Hz, 1H), 3.53–3.60 (m, 1H), 3.73–3.80 (m, 1H), 4.05–4.10 (m, 1H), 4.22 (s, 2H), 6.47 (s, 1H), 6.53 (s, 1H), 7.12–7.14 (dd, J = 1.0, 8.5 Hz, 1H), 7.16–7.18 (dd, J = 1.0, 8.0 Hz, 1H), 7.35–7.40 (m, 1H); 13C NMR (125 MHz, D2O) 21.0, 28.8, 41.3, 44.67, 44.70, 46.4, 47.1, 49.3, 63.9, 78.0, 110.3, 114.0, 115.8, 115.9, 117.4, 117.5, 125.0, 125.1, 132.1, 132.2, 132.3, 132.4, 145.7, 153.8, 158.1, 160.1, 162.0; LC-TOF (M+H+) calcd for C20H28FN4O 359.2247, found 359.2253.
5.2.8 6-(((3S,4S)-4-(2-(2-Methoxybenzylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (4b)
Compound 4b was synthesized using a similar procedure to that of 4a (96%): 1H NMR (500 MHz, CDCl3) 2.18 (s, 3H), 2.65–2.85 (m, 3H), 3.03–3.10 (t, J = 11.5 Hz, 1H), 3.17–3.32 (m, 3H), 3.40–3.47 (dd, J = 8.5, 12.0 Hz, 1H), 3.50–3.57 (m, 2H), 3.72–3.80 (m, 4H), 4.05–4.17 (m, 3H), 6.43 (s, 1H), 6.49 (s, 1H), 6.91–6.94 (dd, J = 7.0, 7.5 Hz, 1H), 6.97–6.99 (d, J = 8.5 Hz, 1H), 7.23–7.25 (dd, J = 1.0, 7.5 Hz, 1H), 7.34–7.38 (ddd, J = 1.5, 8.5, 9.0 Hz, 1H); 13C NMR (125 MHz, CDCl3) 21.0, 38.7, 41.0, 46.2, 46.9, 47.1, 49.1, 55.4, 64.0, 77.8, 110.3, 111.2, 113.8, 118.2, 121.0, 131.6, 131.8, 145.6, 153.8, 157.6, 158.0; LC-TOF (M+H+) calcd for C21H31N4O2 371.2447, found 371.2450.
5.2.9 2-((2-((3S,4S)-4-((6-Amino-4-methylpyridin-2-yl)methyl)pyrrolidin-3-yloxy)ethylamino)methyl)phenol (4c)
Compound 4c was synthesized using a similar procedure to that of 4a (92%): 1H NMR (500 MHz, CDCl3) 2.15 (s, 3H), 2.60–2.70 (m, 1H), 2.77–2.82 (m, 2H), 3.00–3.10 (t, J = 11.5 Hz, 1H), 3.18–3.22 (m, 3H), 3.42–3.51 (dd, J = 8.5, 11.5 Hz, 1H), 3.51–3.56 (m, 2H), 3.73–3.80 (m, 1H), 4.00–4.25 (m, 3H), 6.38 (s, 1H), 6.50 (s, 1H), 6.83–6.88 (m, 2H), 7.21–7.25 (m, 2H); 13C NMR (125 MHz, CDCl3) 21.0, 28.8, 41.2, 46.0, 47.0, 47.1, 49.2, 63.7, 77.8, 110.3, 114.0, 115.4, 117.0, 120.6, 131.6, 131.7, 145.6, 153.8, 155.0, 158.0; LC-TOF (M+H+) calcd for C20H29N4O2 357.2291, found 357.2277.
5.2.10 6-(((3S,4S)-4-(2-(Furan-2-ylmethylamino)ethoxy)pyrrolidin-3-yl)methyl)-4-methylpyridin-2-amine (4d)
Compound 4b was synthesized using a similar procedure to that of 4a (96%): 1H NMR (500 MHz, CDCl3) 2.32 (s, 3H), 2.65–2.73 (m, 1H), 2.76–2.83 (dd, J = 7.5, 15.0 Hz, 1H), 2.88–2.93 (dd, J = 7.5, 14.5 Hz, 1H), 3.04–3.09 (t, J =11.5 Hz, 1H), 3.17–3.24 (m, 3H), 3.39–3.43 (dd, J = 9.0, 11.5 Hz, 1H), 3.48–3.51 (d, J = 13.5 Hz, 2H), 3.51–3.56 (m, 1H), 3.70–3.75 (m, 1H), 4.05–4.10 (m, 1H), 4.22 (s, 2H), 6.38–6.40 (dd, J = 2.0, 5.0 Hz, 1H), 5.48 (s, 1H), 6.52–6.53 (d, J = 3.5 Hz, 1H), 6.55 (s, 1H),; 13C NMR (125 MHz, CDCl3) 21.0, 28.8, 41.4, 43.1, 46.0, 47.0, 49.4, 64.0, 78.1, 110.3, 110.8, 111.0, 113.0, 114.0, 144.1, 144.3, 144.9, 145.8, 153.9, 158.1; LC-TOF (M+H+) calcd for C18H27N4O2 331.2134, found 331.2136.
5.3 NOS Purified Enzyme Assays
The three NOS isoforms, rat nNOS, murine iNOS, and bovine eNOS were obtained as recombinant enzymes overexpressed in and purified from E. coli as previously reported.[36–39] The hemoglobin capture assay was used to measure nitric oxide production.[40] Briefly, the assay was run at 37 °C in 100 mM HEPES buffer (10% glycerol; pH 7.4) in the presence of 10 μM L-arginine. The following were also included in the assay: 100 μM NADPH, 10 μM tetrahydrobiopterin, 1 mM CaCl2, 11.6 μg/mL calmodulin and 3.0 μM oxyhemoglobin. For iNOS, calmodulin and CaCl2 were omitted because iNOS is calcium independent; CaM is bound tightly. All NOS isozymes were used at a concentration of approximately 100 nM. The assay was run in a 96 well plate, using the Synergy 4 by BioTek, at the Northwestern University High Throughput Analysis Facility. The assay was run in triplicate; Forty-eight 100 μL reactions were performed at once. The addition of hemoglobin and NOS were automated with a maximum of a 30 second delay before the reactions could be recorded at 401 nm. The absorbance increase at 401 nm is due to the formation of NO via the conversion of oxyhemoglobin to methemoglobin, The IC50 values were obtained using non-linear regression in GraphPad Prism5 software. Subsequent Ki values were determined using the Cheng-Prusoff relationship:[41]
The following known Km values were used: rat nNOS 1.3 μM; murine iNOS 8.3 μM; bovine eNOS 1.7 μM.
5.4 nNOS Cell-Based Assay
HEK293t cells stably transfected with rat nNOS were cultured as previously described.28 The nNOS inhibition assay was performed as previously reported,28 with the following modifications: assays were performed in 96-well plates with a total volume of 100 μL, and 10 μM A23187 (Sigma Aldrich, St Louis, MO, USA) (added as 100x stock in 50% DMSO) was used in place of 5 μM. A23187 is a calcium ionophore; it transports calcium into the cells and activates nNOS. Nine concentrations of each inhibitor were tested, in at least triplicate wells. All inhibitors were assayed within the same experiment to assure consistencies in cell concentration and passage number. The entire assay was repeated at least three times for each inhibitor, and the IC50 values averaged. Cells were plated in the 96-well plates 24 hours before activation. After 6 hours of activation (in the presence or absence of inhibitor, which was added 30 minutes before activation) 50 μL aliquots of the media were removed and nitrite production was quantified using Griess reagent.35
5.5 Crystal Preparation, X-ray Diffraction Data Collection, and Structure Refinement
The nNOS heme domain protein used for crystallization was generated by limited trypsin digest from the full length nNOS and further purified through a Superdex 200 gel filtration column (GE Healthcare) as described previously.[42] The nNOS heme domain at 7–9 mg/mL containing 20 mM histidine was used for the sitting drop vapor diffusion crystallization setup under the conditions reported [42] with minor modification by addition of 10% (v/v) ethylene glycol in well solution. Fresh crystals were first passed stepwise through cryo-protectant solutions as described [42] and then soaked with 10 mM inhibitor for 4–6 h at 4 °C before being flash cooled by plunging into liquid nitrogen.
The cryogenic (100K) X-ray diffraction data were collected remotely at beamline 7-1 at Stanford Synchrotron Radiation Light source through the data collection control software Blu-Ice [43] and the crystal mounting robot. Raw data frames were indexed, integrated, and scaled using HKL2000.[44] The binding of inhibitors was detected by the initial difference Fourier maps calculated with REFMAC.[45] The inhibitor molecules were then modeled in COOT [46] and refined using REFMAC. Water molecules were added in REFMAC and checked by COOT. The TLS [47] protocol was implemented in the final stage of refinements with each subunit as one TLS group. The refined structures were validated in COOT before deposition to RCSB protein data bank. The crystallographic data collection and structure refinement statistics are summarized in Table 3 with PDB accession codes included.
Table 3.
Crystallographic data collection and refinement statistics
| Data seta | nNOS-4a | nNOS-4b | nNOS-4c | nNOS-4d |
|---|---|---|---|---|
| PDB code | 3TYL | 3TYM | 3TYN | 3TYO |
| Data collection | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell dimensions | ||||
| a, b, c (Å) | 51.9, 110.8, 164.3 | 51.8, 110.7, 164.2 | 51.8, 110.8, 164.2 | 51.9, 110.9, 164.3 |
| Resolution (Å) | 1.90 (1.93-1.90) | 2.00 (2.03-2.00) | 1.97 (2.00-1.97) | 1.93 (1.96-1.93) |
| Rsym or Rmerge | 0.055 (0.629) | 0.056 (0.590) | 0.046 (0.564) | 0.047 (0.605) |
| I/σ | 29.2 (2.1) | 15.6 (2.0) | 31.0 (2.5) | 32.1 (2.5) |
| No. unique reflections | 75,516 | 64,550 | 67,186 | 72,491 |
| Completeness (%) | 99.9 (100.0) | 99.9 (97.8) | 98.8 (98.0) | 99.8 (99.9) |
| Redundancy | 4.1 (4.1) | 4.1 (4.1) | 4.1 (4.1) | 4.1 (4.0) |
| Refinement | ||||
| Resolution (Å) | 1.90 | 2.00 | 1.97 | 1.93 |
| No. reflections | 71,520 | 61,119 | 63,641 | 68,646 |
| Rwork/Rfree2 | 0.179/0.212 | 0.186/0.227 | 0.185/0.223 | 0.180/0.215 |
| No. atoms | ||||
| Protein | 6709 | 6671 | 6679 | 6671 |
| Ligand/ion | 185 | 183 | 181 | 177 |
| Water | 377 | 283 | 321 | 366 |
| Mean B-factor | 45.16 | 49.50 | 48.17 | 47.50 |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.014 | 0.013 | 0.013 | 0.012 |
| Bond angles (°) | 1.411 | 1.386 | 1.403 | 1.324 |
See Figure 1 for chemical formula of inhibitors.
Supplementary Material
Scheme 1.
Synthesis of 4a–d.
Reagents and conditions: (a) allyl methyl carbonate, Pd(PPh3)4, 45 °C, 5 h, 66%; (b) O3, −78 °C, 30 min, (ii) Me2S, −78 °C to rt, 2 h, 87%; (c) amines, NaHB(OAc)3, rt, 3 h, 91%; (d) 6 N HCl in MeOH (2:1), rt, 12 h, 99%.
Acknowledgments
We are grateful to the National Institutes of Health, Grants GM049725 to RBS and GM057353 to TLP. We thank Dr. Bettie Sue Siler Masters (NIH grant GM52419, with whose laboratory P.M. and L.J.R. are affiliated). B.S.S.M. also is grateful to the Welch Foundation for a Robert A. Welch Distinguished Professorship in Chemistry (AQ0012). P.M. is supported by grant 0021620849from MSMT of the Czech Republic. We thank the SSRL beamline staff for their support during remote X-ray diffraction data collection. We thank Drs. Josh Kurutz and Yuyang Wu of the Northwestern University IMSERC facility for valuable assistance and discussions concerning the NMR experiments.
Abbreviations
- CNS
central nervous system
- NO
nitric oxide
- nNOS
neuronal nitric oxide synthase
- eNOS
endothelial nitric oxide synthase
- iNOS
inducible nitric oxide synthase
- BBB
blood brain barrier
- H-bond
hydrogen bond
- L-NNA
L -N-nitroarginine
- H4B
tetrahydrobiopterin
Footnotes
PDB accession codes: The PDB accession codes for compounds 4a–d with nNOS, shown in Figure 3 are as follows: nNOS-4a, 3TYL; nNOS-4b, 3TYM; nNOS-4c, 3TYN; nNOS-4d, 3TYO.
Supplementary data associated with this article can be found in the online version of the article. These data include full1H and 13 C NMR spectra of 4a–4d.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dorheim MA, Tracey WR, Pollock JS, Grammas P. Biochem Biophys Res Commun. 1994;205:659–665. doi: 10.1006/bbrc.1994.2716. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Dawson VL, Dawson TM. Pharmacol Ther. 2006;109:33–41. doi: 10.1016/j.pharmthera.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 3.Norris PJ, Waldvogel HJ, Faull RLM, Love DR, Emson PC. Neuroscience. 1996;72:1037–1047. doi: 10.1016/0306-4522(95)00596-x. [DOI] [PubMed] [Google Scholar]
- 4.Ashina M. Expert Opin Pharmacother. 2002;3:395–399. doi: 10.1517/14656566.3.4.395. [DOI] [PubMed] [Google Scholar]
- 5.Sims NR, Anderson MF. Neurochem Int. 2002;40:511–526. doi: 10.1016/s0197-0186(01)00122-x. [DOI] [PubMed] [Google Scholar]
- 6.Southan GJ, Szabó C. Biochem Pharmacol. 1996;51:383–394. doi: 10.1016/0006-2952(95)02099-3. [DOI] [PubMed] [Google Scholar]
- 7.Babu BR, Griffith OW. Curr Opin Chem Biol. 1998;2:491–500. doi: 10.1016/s1367-5931(98)80125-7. [DOI] [PubMed] [Google Scholar]
- 8.Alderton WK, Cooper CE, Knowles RG. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji H, Erdal E, Litzinger EA, Seo J, Zhu Y, Xue F, Fang J, Huang J, Silverman RB. In: Frontiers in Medicinal Chemistry. 5. Reitz AB, Choudhary MI, Atta-ur-Rahman, editors. Bentham Science Publishers; 2009. pp. 842–882. [Google Scholar]
- 10.Silverman RB. Acc Chem Res. 2009;42:439–451. doi: 10.1021/ar800201v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delker SL, Ji H, Li H, Jamal J, Fang J, Xue F, Silverman RB, Poulos TL. J Am Chem Soc. 2010;132:5437–5442. doi: 10.1021/ja910228a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji H, Tan S, Igarashi J, Li H, Derrick M, Martásek P, Roman LJ, Vásquez-Vivar J, Poulos TL, Silverman RB. Ann Neurol. 2009;65:209–217. doi: 10.1002/ana.21555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawton GR, Ralay Ranaivo H, Chico LK, Ji H, Xue F, Martásek P, Roman LJ, Watterson DM, Silverman RB. Bioorg Med Chem. 2009;17:2371–2380. doi: 10.1016/j.bmc.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue F, Fang J, Lewis WW, Martásek P, Roman LJ, Silverman RB. Bioorg Med Chem Lett. 2010;20:554–557. doi: 10.1016/j.bmcl.2009.11.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xue F, Huang J, Ji H, Fang J, Li H, Martásek P, Roman LJ, Poulos TL, Silverman RB. Bioorg Med Chem. 2010;18:6526–6537. doi: 10.1016/j.bmc.2010.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xue F, Li H, Fang J, Roman LJ, Martásek P, Poulos TL, Silverman RB. Bioorg Med Chem Lett. 2010;20:6258–6261. doi: 10.1016/j.bmcl.2010.08.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue F, Li H, Delker SL, Fang J, Martásek P, Roman LJ, Poulos TL, Silverman RB. J Am Chem Soc. 2010;132:14229–14238. doi: 10.1021/ja106175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn B, Mohr P, Stahl M. J Med Chem. 2010;53:2601–2611. doi: 10.1021/jm100087s. [DOI] [PubMed] [Google Scholar]
- 19.Laurence C, Brameld KA, Graton J, Le Questel JY, Renault E. J Med Chem. 2009;52:4073–4086. doi: 10.1021/jm801331y. [DOI] [PubMed] [Google Scholar]
- 20.Jansma A, Zhang Q, Li B, Ding Q, Uno T, Bursulaya B, Liu Y, Furet P, Gray NS, Geierstanger BH. J Med Chem. 2007;50:5875–5877. doi: 10.1021/jm700983a. [DOI] [PubMed] [Google Scholar]
- 21.McDonagh AF, Lightner DA. J Med Chem. 2007;50:480–488. doi: 10.1021/jm0609521. [DOI] [PubMed] [Google Scholar]
- 22.Sasaki S, Cho N, Nara Y, Harada M, Endo S, Suzuki N, Furuya S, Fujino M. J Med Chem. 2003;46:113–124. doi: 10.1021/jm020180i. [DOI] [PubMed] [Google Scholar]
- 23.Alex A, Millan DS, Perez M, Wakenhut F, Whitlock GA. Med Chem Commun. 2011;2:669–674. [Google Scholar]
- 24.Ashwood VA, Field MJ, Horwell DC, Julien-Larose C, Lewthwaite RA, McCleary S, Pritchard MC, Raphy J, Singh L. J Med Chem. 2001;44:2276–2285. doi: 10.1021/jm010825z. [DOI] [PubMed] [Google Scholar]
- 25.Ji H, Li H, Martásek P, Roman LJ, Poulos TL, Silverman RB. J Med Chem. 2009;52:779–797. doi: 10.1021/jm801220a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji H, Delker SL, Li H, Martásek P, Roman LJ, Poulos TL, Silverman RB. J Med Chem. 2010;53:7804–7824. doi: 10.1021/jm100947x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue F, Kraus JM, Labby KJ, Ji H, Mataka J, Xia G, Li H, Delker SL, Roman LJ, Martásek P, Poulos TL, Silverman RB. J Med Chem. 2011;54:6399–6403. doi: 10.1021/jm200411j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haight AR, Stoner EJ, Peterson MJ, Grover VK. J Org Chem. 2003;68:8092–8096. doi: 10.1021/jo0301907. [DOI] [PubMed] [Google Scholar]
- 29.Fang J, Silverman RB. Anal Biochem. 2009;390:74–78. doi: 10.1016/j.ab.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viswanadhan VN, Ghose AK, Revankar GR, Robins RK. J Che Inf Comput Sci. 1989;29:163–172. [Google Scholar]
- 31.Oki M. Top Stereochem. 1983;14:1–81. [Google Scholar]
- 32.Clayden J, Moran WJ, Edwards PJ, LaPlante SR. Angew Chem Int Ed. 2009;48:6398–6401. doi: 10.1002/anie.200901719. [DOI] [PubMed] [Google Scholar]
- 33.Kraus JM, Tatipaka HB, McGuffin SA, Chennamaneni NK, Karimi M, Arif J, Verlinde CLMJ, Buckner FS, Gelb MH. J Med Chem. 2010;53:3887–3898. doi: 10.1021/jm9013136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson JC, Munro SLA, Craik DJ. Magn Reson Chem. 1995;33:367–374. [Google Scholar]
- 35.Ballet S, Misicka A, Kosson P, Lemieux C, Chung NN, Schiller PW, Lipkowski AW, Tourwé D. J Med Chem. 2008;51:2571–2574. doi: 10.1021/jm701404s. [DOI] [PubMed] [Google Scholar]
- 36.Hevel JM, White KA, Marletta MA. J Biol Chem. 1991;266:22789–22791. [PubMed] [Google Scholar]
- 37.Roman LJ, Sheta EA, Martasek P, Gross SS, Liu Q, Masters BS. Proc Natl Acad Sci USa. 1995;92:8428–8432. doi: 10.1073/pnas.92.18.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerber NC, Ortiz de Montellano PR. J Biol Chem. 1995;270:17791–17796. doi: 10.1074/jbc.270.30.17791. [DOI] [PubMed] [Google Scholar]
- 39.Martasek P, Liu Q, Liu J, Roman L, Gross S. Biochem Biophys Res Comm. 1996 doi: 10.1006/bbrc.1996.0238. [DOI] [PubMed] [Google Scholar]
- 40.Hevel JM, Marletta MA. Methods Enzymol. 1994;233:250–258. doi: 10.1016/s0076-6879(94)33028-x. [DOI] [PubMed] [Google Scholar]
- 41.Cheng YC, Prusoff WH. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 42.Li H, Shimizu H, Flinspach M, Jamal J, Yang W, Xian M, Cai T, Wen EZ, Jia Q, Wang PG, Poulos TL. Biochemistry. 2002;41:13868–13875. doi: 10.1021/bi020417c. [DOI] [PubMed] [Google Scholar]
- 43.McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. J Synchrotron Radiat. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, Isupov MN, Murshudov GN. Acta Crystallogr, Sect D: Biol Crystallogr. 2001;D57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.