Abstract
Objective
We set out to test the hypothesis that pharmacometabolomic data could be efficiently merged with pharmacogenomic data by SNP imputation of metabolomic-derived pathway data on a “scaffolding” of genome-wide association (GWA) SNP data to broaden and accelerate “pharmacometabolomics-informed pharmacogenomic” studies by eliminating the need for initial genotyping and by making broader SNP association testing possible.
Methods
We previously genotyped 131 tag SNPs for six genes encoding enzymes in the glycine synthesis and degradation pathway using DNA from 529 depressed patients treated with citalopram/escitalopram to pursue a glycine metabolomics “signal” associated with selective serotonine reuptake inhibitor response. We identified a significant SNP in the glycine dehydrogenase gene. Subsequently, GWAS SNP data were generated for the same patients. In this study, we compared SNP imputation within 200 kb of these same six genes with results of the previous tag SNP strategy as a rapid strategy for merging pharmacometabolomic and pharmacogenomic data.
Results
Imputed genotype data provided greater coverage and higher resolution than did tag SNP genotyping, with a higher average genotype concordance between genotyped and imputed SNP data for “1000 Genomes” (96.4%) than HapMap 2 (93.2%) imputation. Many low p-value SNPs with novel locations within genes were observed for imputed compared with tag SNPs, thus altering the focus for subsequent functional genomic studies.
Conclusions
These results indicate that the use of GWAS data to impute SNPs for genes in pathways identified by other “omics” approaches makes it possible to rapidly and economically identify SNP markers to “broaden” and accelerate pharmacogenomic studies.
Keywords: Pharmacometabolomics, pharmacogenomics, imputation, tag SNPs, 1000 Genomes, HapMap, selective serotonin reuptake inhibitors, SSRIs, major depressive disorder, MDD
Introduction
A major challenge in the current era of “systems pharmacology” [1] is that of merging a variety of high throughput datasets from differing platforms to generate and test hypotheses with regard to mechanisms for variation in drug response. Pharmacogenomics represents only one of the “omics” disciplines that can be applied to understand and, eventually, predict variation in drug response phenotypes. The development of “pharmacometabolomics” – the use of a series of high throughput metabolomic platforms to detect and quantify hundreds or thousands of small molecule “metabolites” [2] – could, in theory, compliment and guide subsequent genomic studies. However, merging pharmacometabolomic data with pharmacogenomics, and doing so rapidly and efficiently, remains a significant challenge.
We recently utilized both metabolomic and genomic data to study selective serotonin reuptake inhibitor (SSRI) treatment outcomes in patients with major depressive disorder (MDD) by use of a “pharmacometabolomic-informed” pharmacogenomic research strategy [3]. That approach began with the identification of glycine as a marker for SSRI treatment outcomes by using a GC-MS metabolomic platform, an observation that raised the possibility that sequence variation in genes encoding glycine synthesis and degradation enzymes might contribute to the metabolomic findings. As the next step, we applied a “traditional” tag SNP approach, genotyped the six genes in the glycine “synthesis-degradation” pathway and identified and replicated a SNP in the glycine dehydrogenase (GLDC) gene that was associated with SSRI treatment outcomes in MDD patients. That process required eight months to complete with an expense, just for genotyping, of over $20,000. Subsequently, we obtained GWAS SNP data for the same 529 MDD patients used in the initial study, and – at nearly the same time – pilot “1000 Genomes” data were published [4]. These two developments, following soon after the completion of our tag SNP genomic studies in pursuit of the glycine pharmacometabolomic signal, made it possible to directly compare the tag SNP approach with the use of imputation based on GWAS SNP data to merge candidate pathway data from another “omic” discipline with pharmacogenomics – and to do so quickly, in a cost-effective fashion and with broader coverage of individual genes than was practically possible with tag SNPs.
In the present study we have directly compared genotype data and association results for SNPs that were genotyped as tag SNPs in our previous study with SNPs imputed using HapMap and “1000 Genomes” data. The efficacy of imputation for “1000 Genomes” data was marginally improved compared to HapMap data but, in both cases, we found that using GWAS data to impute across our pharmacometabolomic candidate pathway provided data that correlated well with that obtained by tag SNP genotyping. However, the use of imputation was much faster, much more economical and – of greatest importance – it identified many additional candidates for validation by genotyping and follow-up functional genomic studies. As additional pharmacometabolomic platforms (e.g. LC-MS/MS, GC-MS/MS, Coulomb array, etc.) are applied that could detect different sets of metabolites, this approach can be applied in an iterative fashion to the novel candidates / pathways identified, all using the same GWAS data as genetic background for imputation, allowing these two independent omics disciplines to guide and inform each other.
Methods
Study participants
Patient enrollment, patient characteristics, and treatment procedures for the Mayo PGRN SSRI trial have been described elsewhere [3]. Briefly, DNA samples were obtained from 529 MDD patients enrolled in an ongoing clinical trial in which the patients are treated with the SSRIs citalopram/escitalopram for 8 weeks. Baseline as well as week 4 and 8 blood samples were obtained. Citalopram/escitalopram treatment outcomes were established using the “Quick Inventory of Depressive Symptomatology – Clinician rated” (QIDS-C) [5]. The “remission” phenotype was defined as a post-treatment QIDS-C score ≤5 and “response” was defined as a QIDS-C reduction of ≥50%. All patients provided written informed consent for participation in the trial, and the Mayo Clinic Institutional Review Board reviewed and approved the study protocol.
Genotyping
In our original tag SNP study, we genotyped a panel of 131 tag SNPs for genes encoding glycine synthesis (SHMT1 and SHMT2) and glycine degradation enzymes (AMT, DLD, GCSH and GLDC) using all 529 of the Mayo PGRN SSRI clinical trial DNA samples [3]. When we completed the present study, four of the SNPs that we imputed were selected for validation and replication, and three were successfully genotyped utilizing Applied Biosystems TaqMan technology (Carlsbad CA). Of the three SNPs that were successfully genotyped, rs11172135 was a pre-designed assay, while the remaining two (rs2108227, and rs12371684) were custom TaqMan SNP genotyping assays designed with Applied Biosystems’ online custom TaqMan assay design tool (www.appliedbiosystems.com). Primer and probe sequences for these assays are available upon request. PCR protocols were followed according to the manufacturer’s guidelines for the 384 well format. PCR products were amplified with a Thermo Fisher Scientific Hybrid thermal cycler (Waltham MA), and were analyzed on an Applied Biosystems 7900HT. Genome-wide SNP data for the same 529 DNA samples from patients enrolled in the Mayo PGRN SSRI clinical trial were generated by the RIKEN Center for Genomic Medicine, Yokohama, Japan, using the Illumina Human610-Quad BeadChip platform.
Imputation
For the six candidate genes in the glycine synthesis and degradation pathway (SHMT1, SHMT2, AMT, DLD, GCSH and GLDC), untyped SNP genotypes were imputed with the software package MaCH 1.0 [6]. We selected MaCH 1.0 for SNP imputation based on previous reports of its performance and practical usage [7, 8]. This imputation method estimates genotypes for untyped SNPs in a set of samples with typed data using a reference set of genotype data. We used the CEPH HapMap (phase 2, release 22) and “1000 Genomes” (pilot phase release October, 2010) as the reference sets, and the genome-wide SNP data for the SSRI samples as the genetic background on which we imputed. Variants within 200 kb of each gene were identified using a two-step imputation procedure. In the first step, 250 randomly selected subjects were used to calibrate the model parameters, and genotype imputation was performed during the second step with all subjects using the learned parameters. Imputation quality estimates were established by masking 10% of the genotypes at random and imputing the masked genotypes to compare the original and imputed masked genotypes. The MaCH “Rsq” (designated imputation-Rsq hereafter) value was used as a quality control measure after imputation, with the recommend thresholds of 0.3 and 0.5 used for HapMap and “1000 Genomes” data, respectively [6]. The imputation-Rsq value is an estimate of the squared correlation between imputed and true genotypes. For imputed genotypes, the estimated allelic dosage values were used to perform the association analyses.
Quality control and single SNP associations
Our GWAS SNP data were processed with genotyping quality control measures that included examining sample and SNP call rates, discordance of duplicate samples, departures from Hardy-Weinberg Equilibrium (HWE) and the distribution of minor allele frequency (MAF) values. SNPs were removed from the analysis because of low MAFs (≤0.01), genotyping failure, low call rates (≤0.95), or significant departures from HWE (p≤0.000001). Study subjects were excluded if their DNA samples failed during genotyping or had low sample call rates (≤0.98). The analyses reported here were limited to self-reported Non-Hispanic White subjects who were enrolled in the Mayo-PGRN SSRI study, but as detailed subsequently, we also corrected for possible population stratification.
The software package STRUCTURE [9] was used to infer ancestry of the subjects with a subset of 4,855 SNPs in linkage equilibrium with each other. Genetic ancestry for subjects was also determined by comparison with genetic profiles for DNA samples from 287 lymphoblastoid cell lines in the “Human Variation Panel” that included three ethnic groups which were obtained from the Coriell Institute (Camden, NJ). For each study subject, probabilities of membership in each of the three known ancestral groups of the cell lines were calculated. Those analyses verified race for the 509 subjects with a self-reported white non-Hispanic (WNH) race and identified five additional subjects with a self-reported non-specific race as white non-Hispanic. Sample population stratification was determined by calculating eigenvectors with the EIGENSOFT software EIGENSTRAT (smartpca) [10] based on a set of genome-wide SNPs in low or no LD with each other (data not presented). A Tracy-Widom test determined the use of four eigenvectors to adjust analyses for population stratification. Thus, 514 WNH subjects were identified for use in the analyses.
To make it possible to compare our data with the results reported in our previous tag SNP study [3], the same statistical modeling methods from our previous study were used here for single SNP association analyses. A logistic regression model, assuming a log-additive allele effect, was used with the SSRI treatment outcome remission and response phenotypes (QIDS-C≤5) at week 8 or week 4 if week 8 status was not available.
Results
SNP imputation
Genomewide SNP genotyping of the DNA samples from SSRI-treated patients that we had studied previously using a tag SNP strategy made it possible for us to impute SNPs for genomic regions containing the six genes of interest for the glycine synthesis and degradation pathway to analyze genetic variation using HapMap and “1000 Genomes” imputation. As shown subsequently, imputation provided a much higher resolution and much more comprehensive coverage of sequence variation than did the tag SNP study, with a total of 1578 SNPs from HapMap and 11154 SNPs from “1000 Genomes” data for the six candidate genes. The estimated average per genotype error rate – established by masking 10% of the genotypes at random, imputing them and comparing with the masked original – was 4.6% and 4.3% for HapMap and “1000 Genomes” imputed data, respectively. After removal of poorly imputed SNPs based on their imputation-Rsq values, 1506 and 5349 imputed SNPs from HapMap and “1000 Genomes”, respectively, remained for analysis (Table 1). A majority of the imputed SNPs were common (MAF > 0.05) (Supplemental Figure 1).
Table 1.
The “tag SNPs” were those used to genotype these same genes in our original study [3]; “GWAS SNPs” were obtained by GWAS genotyping of these DNA samples after the tag SNP study was completed; and the “HapMap” imputed and “1000 Genomes” imputed columns list the number of SNPs imputed from those two reference sets after removal of poorly imputed SNPs based on imputation-Rsq values.
| Gene | Tag SNPs | GWAS SNPs | HapMap Imputed |
“1000 Genomes” Imputed |
|---|---|---|---|---|
| GLDC | 76 | 105 | 346 | 1466 |
| AMT | 2 | 17 | 107 | 351 |
| DLD | 12 | 103 | 333 | 765 |
| GCSH | 15 | 117 | 441 | 1802 |
| SHMT1 | 19 | 38 | 167 | 561 |
| SHMT2 | 7 | 38 | 112 | 404 |
| Total | 131 | 418 | 1506 | 5349 |
Gene-specific Manhattan plots
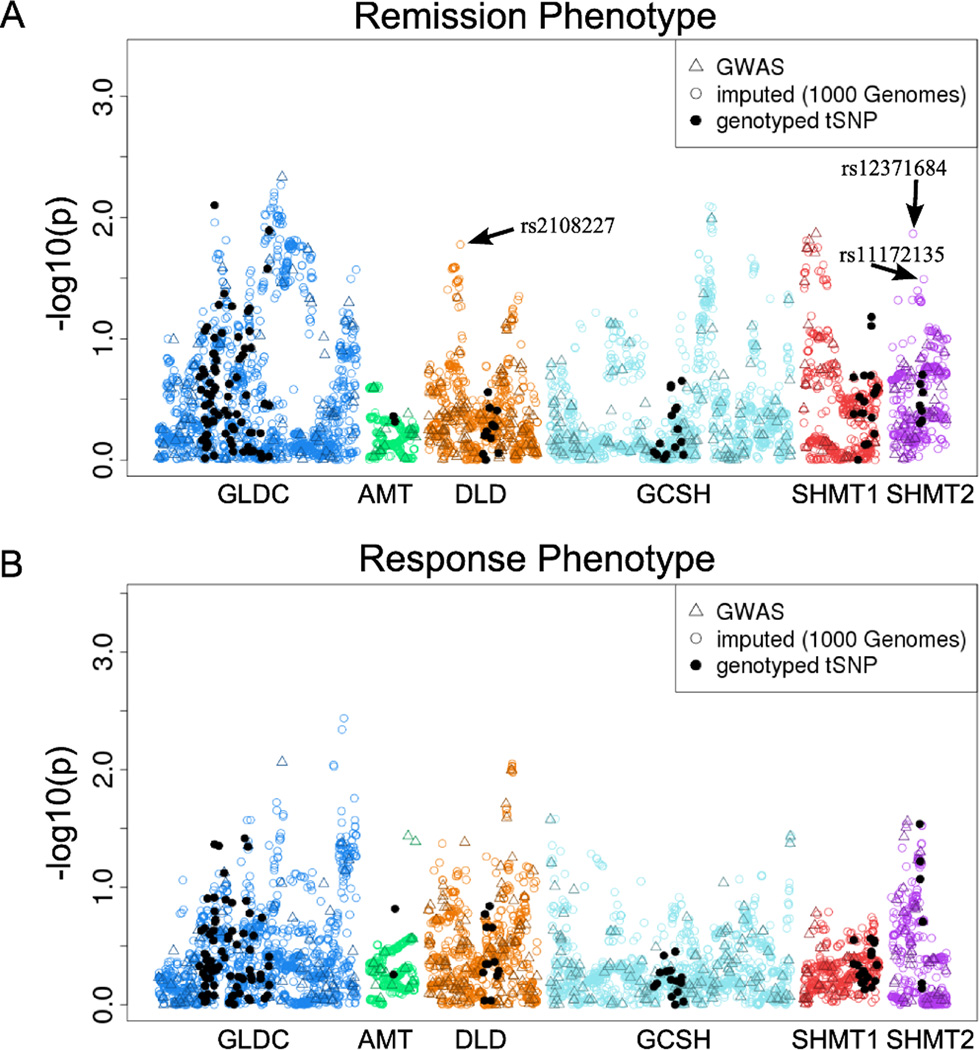
A practical advantage of the use of imputation is the ability to comprehensively study genetic variation within a region of interest without additional genotyping time or expense, whereas the ability to study variation with traditional tag SNP approaches is limited by cost and requires significant time. The potential benefit from the increase in coverage and resolution by imputation using “1000 Genomes” data is illustrated in a striking fashion by the plots of log transformed p-values for association with the remission and response phenotypes for SSRI therapy versus SNP location that are shown in Figure 1. These plots represent miniature Manhattan plots for each of 6 genes in the glycine synthesis-degradation pathway. Among the most striking features of the plots are the relatively “narrow” areas covered by the tag SNP approach (black dots) and the fact that a number of novel association signals (i.e. low p-values) were observed using the imputed SNPs that had greater significance than those seen for the previously analyzed tag SNP data. The imputation results continued to indicate that GLDC was the most important “glycine synthesis and degradation pathway” gene of the six candidate genes analyzed for association with SSRI outcomes in patients with MDD, as reported in our original tag SNP study, but they also focused attention on additional areas of these genes for validation and functional pursuit. These plots illustrate, in a striking fashion, the value of the use of this approach as well as the relatively “narrow” view of genetic architecture provided, primarily because of cost constraints, of the tag SNP approach. For example, for the imputed HapMap data there were 5 SNPs with minimum p-values < 0.01 (4 upstream of GLDC), while the “1000 Genomes” imputation data had 20 SNPs with p-values < 0.01 (18 upstream of GLDC).
Figure 1.
Log transformed p-values for SSRI remission (A) and response (B) phenotypes using GWAS, “1000 Genomes” imputed, and genotyped tag SNPs (tSNPs) for the six candidate genes. The arrows indicate SNPs in the DLD and SHMT2 genes that were selected for genotyping to validate the imputation associations.
Genotype verification
To validate the imputation data, we focused on novel “remission” phenotype signals for DLD and SHMT2 and selected the top two imputed SNPs from each of these genes for validation by genotyping (see Figure 1A). One SNP from DLD failed the Applied Biosystems TagMan assay design process. We observed associations with the remission phenotype were very similar after genotyping for the three successfully genotyped SNPs (rs2108227, rs12371684, rs11172135) (Table 2). We next determined: 1) imputation quality for the SNPs that were previously genotyped as tag SNPs and were also imputed using HapMap or “1000 Genomes” data, and 2) whether we could achieve association results with the imputed data similar to those obtained using tag SNPs.
Table 2.
Remission association results using “1000 Genomes” imputation data and actual genotype data for the three imputed SNPs with the lowest p-values in DLD and SHMT2 (see Figure 1A) that were successfully genotyped for validation.
| Genotyped |
“1000 Genomes” Imputation |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP | chr | gene | bp | MAF | OR (95% CI) | pvalue | MaCH Rsq | MAF | OR (95% CI) | pvalue |
| rs2108227 | 7 | DLD | 107465340 | 0.239 | 1.50(1.1,2.06) | 0.0110 | 0.9158 | 0.2346 | 1.47(1.07,2.01) | 0.0167 |
| rs12371684 | 12 | SHMT2 | 57564478 | 0.1028 | 1.55(1.04,2.31) | 0.0325 | 0.653 | 0.1183 | 1.78(1.12,2.81) | 0.0136 |
| rs11172135 | 12 | SHMT2 | 57647374 | 0.088 | 1.63(1.06,2.53) | 0.0280 | 0.694 | 0.1017 | 1.68(1.04,2.72) | 0.0323 |
chr = chromosome; MAF = minor allele frequency; OR = Odds ratio; CI = confidence interval;
Comparisons between imputed and genotyped tag SNPs and association analyses
To assess the quality of imputation with HapMap and “1000 Genomes” data, we directly compared results from our previous study [3] with results obtained by imputation. A total of 131 tag SNPs were successfully genotyped and analyzed out of the 144 tag SNPs selected by Ji et al. (2011) during the tag SNP study. Those tag SNPs had been selected to cover +/− 10 kb of flanking sequence for each gene with tag SNPs with a linkage disequilibrium (LD) r2 (designated LD-r2 hereafter) threshold of 0.9 and a minimum MAF of 0.05 [3]. Of the 131 genotyped tag SNPs, 42 were included among the GWAS SNPs while 65 and 86 of the 131 tag SNPS were imputed with the HapMap and “1000 Genomes” data, respectively (Supplemental Table 1). For tag SNPs that were not imputed or genotyped on the GWAS panel, there were 15 and three imputed SNPs from HapMap and “1000 Genomes”, respectively, that were in high LD (LD-r2 >= 0.8) with those SNPs. No imputed HapMap SNPs among the 65 were removed due to low imputation-Rsq, while nine of the 86 imputed “1000 Genomes” SNPs were removed for that reason. Supplemental Table 2 shows the MAF distributions of the imputed, GWAS, and genotyped tag SNPs.
We next performed a series of comparisons to assess the quality of the imputed data. First, direct comparison of the most likely imputed genotypes with the observed tag SNP genotypes showed high concordance rates for both imputed HapMap (93.2%) and “1000 Genomes” data (97%). Similar results were observed by correlating the estimated allele dosages after imputation with the observed genotyped dosage values (HapMap, Spearman rank correlation coefficient = 0.94, “1000 Genomes”, Spearman rank correlation coefficient = 0.96). The imputed SNP MAFs were also strongly correlated with the observed tag SNP MAFs for both imputed HapMap and “1000 Genomes” values (intraclass correlation=0.99, Spearman rank correlation=0.99).
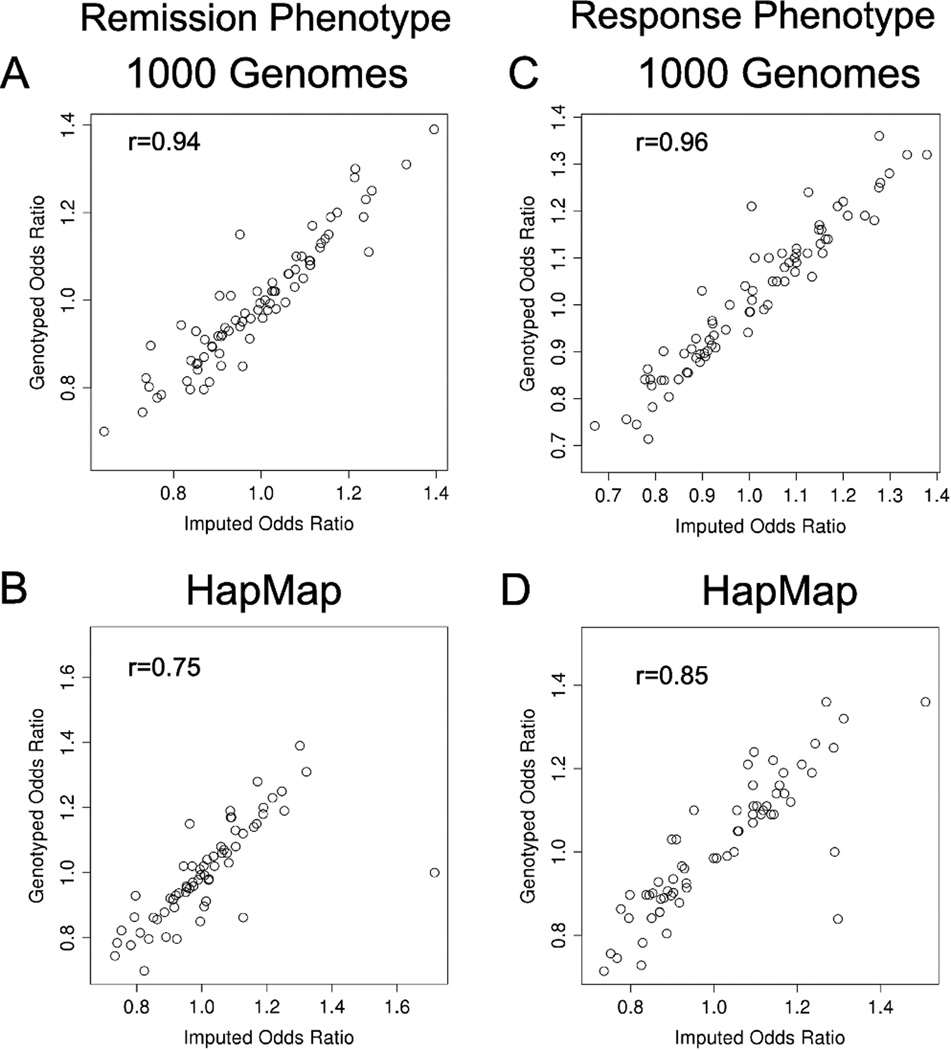
We also determined that highly correlated imputation data translated to highly correlated association results. For the imputed SNPs with reliable imputation-Rsq values, there were high correlations between association results for SSRI treatment “remission” and “response” phenotypes for imputed values with the genotyped tag SNP data. Figure 2 shows the high correlations observed between odds ratios (ORs) for both phenotypes for imputed and genotyped SNPs. Tests for differences between p-values for imputed and genotyped results for both phenotypes were not significant.
Figure 2.
Scatter plots of genotyped and imputed odds ratios for the remission (A,B) and response (C,D) phenotypes using “1000 Genomes” (A,C) and HapMap (B,D) imputation data compared to genotyped tag SNP data. r = Spearman rank correlation coefficient.
We also wanted to determine if the results that we had obtained with the tag SNPs [3] could be achieved using the imputed data. Table 3 shows a comparison of the genotyped and imputation results for the SSRI treatment outcome associated variants with p-values < 0.05 that were identified during our original tag SNP study [3]. Of the nine SNPs with p-values < 0.05 for remission and response, five were genotyped on the GWAS panel while three and four were imputed with HapMap and “1000 Genomes” data, respectively. Three of the previously reported SNPs, including the replicated genotyped rs10975461 SNP, had nominal p-values < 0.05 using the “1000 Genomes” imputed data, while none had nominal p-values < 0.05 using HapMap imputed data. If the same region previously tagged for variation were to be analyzed with the “1000 Genomes” imputed data, rs10975461 would have been the third most significant SNP associated with SSRI treatment remission behind two imputed SNPs that were in high LD with each other. Therefore, although imputation may result in missed findings, these results show that imputation utilizing “1000 Genomes” data would have resulted in conclusions very similar to those that we reported for our tag SNP study [3] and, of course, candidate SNPs identified by imputation still have to be verified by genotyping, as described subsequently, but the number of SNPs genotyped is much smaller than in a tag SNP study and the areas of the gene studied can be much larger.
Table 3.
Genotyped and imputed - by “1000 Genomes” or HapMap data - association results for the remission and response phenotypes for the top associated SNPs identified by Ji et al. (2011) as a result of tag SNP genotyping [3].
| Genotyped |
“1000 Genomes” |
HapMap |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs# | chr | gene | MAF | OR (95% CI) | pvalue | MaCH Rsq | OR (95% CI) | pvalue | MaCH Rsq | OR (95% CI) | pvalue |
|
“Remission” SSRI Outcome Phenotype | |||||||||||
| rs10975641 | 9 | GLDC | 0.39 | 0.70 (0.53,0.91) | 0.008 | 0.5 | 0.62 (0.42,0.89) | 0.011 | 0.72 | 0.82 (0.61,1.12) | 0.21 |
|
“Response” SSRI Outcome Phenotype | |||||||||||
| rs11612037 | 12 | SHMT2 | 0.03 | 0.46 (0.23,0.92) | 0.029 | 0.26 | 0.88 (0.36,2.14) | 0.78 | ___________________ | ||
| rs1755615 | 9 | GLDC | 0.24 | 0.71 (0.50,1.0) | 0.039 | 0.83 | 0.84 (0.61,1.16) | 0.28 | 0.86 | 0.74 (0.51,1.06) | 0.1 |
| rs12004478 | 9 | GLDC | 0.3 | 0.75 (0.55,1.0) | 0.045 | 0.96 | 0.73 (0.56,0.96) | 0.022 | 0.99 | 0.77 (0.57,1.04) | 0.085 |
| rs10975641 | 9 | GLDC | 0.4 | 0.73 (0.53,0.99) | 0.043 | 0.5 | 0.62 (0.42,0.89) | 0.01 | 0.72 | 0.83 (0.58,1.17) | 0.28 |
chr = chromosome; MAF = minor allele frequency; OR = Odds ratio; CI = confidence interval;
Discussion
This study has directly tested an approach designed to facilitate the merger of pharmacometabolomic and pharmacogenomic data by determining the genetic variation within genes in pathways identified during metabolomic studies by genotype imputation rather than by traditional tag SNP genotyping. Central to this strategy is the availability of GWAS SNPs for the sample set being studied and reference genomic data to make it possible to perform genotype imputation for the candidate genes and/or pathways. Our hypothesis was that the use of pathway imputation might both accelerate and broaden the scope of the analysis of pharmacogenomic candidate genes and/or pathways by making it possible to survey more widely and drastically reduce the need to genotype prior to replication. We tested that hypothesis directly by using a recent “pharmacometabolomics-informed pharmacogenomics” study that we had performed which involved six genes encoding enzymes that catalyze the synthesis and degradation of glycine, and compared SNP imputation with our recent LD-based tag SNP genotype study [3]. Obviously, imputation of genomic data has been widely applied – but our purpose was to highlight its use to rapidly merge data from other “omics” disciplines with pharmacogenomic studies, and to allow these “omics” disciplines to inform each other.
We found that imputation using both HapMap and “1000 Genomes” datasets provided similar estimated genotype data for a majority of the SNPs genotyped with the LD tagging approach but they also identified many more SNPs within and surrounding sequences for the six genes studied, significantly altering subsequent functional genomic experiments (see Figure 1). Comparisons among the imputed and genotyped SNP data and the related association results supported imputation as a reliable fine-mapping approach, but genotyping still remains a step that must be taken to validate SNPs selected from imputed data for functional studies. While a number of studies have assessed the quality of SNP imputation to perform fine-mapping in a candidate gene study [11, 12], the use of imputation to facilitate merging of “omics” data from divergent platforms to perform pharmacogenomic follow-up studies has not been tested directly. Many more common and rare variants were imputed from “1000 Genomes” than HapMap phase 2 data, as anticipated, and comparisons with genotyped data and results were similar for the two imputation datasets. The differences between HapMap and “1000 Genomes” imputed data and results for our study are most likely due to several factors, including reference sample size, reference markers set, and reference genetic background. Overall, based on these differences, “1000 Genomes” should probably be used to perform future studies of this type. In addition, our study provides conservative estimates for the value of imputation using “1000 Genomes” data, since future releases of “1000 Genomes” reference data will continue both to increase sample sizes and improve data quality.
A great advantage of imputation is that it can effectively and rapidly focus on appropriate variants in or around a gene for possible functional pursuit (Figure 1). This advantage was highlighted in our study by the association peaks observed with the imputed “1000 Genomes” data that were in gene locations that the tag SNP approach was not able to cover during the original study. We used genotyping to follow up three of the novel top associated SNPs identified using “1000 Genomes” imputation for the SSRI remission phenotype with subsequent genotyping to validate the imputed results. While two of three SNP p-values increased slightly when they were genotyped, the three SNP p-values all remained nominally significant and similar to the imputed values (Table 2). These observations illustrate the need to validate results produced by “1000 Genomes” imputation, especially for SNPs with lower imputation-Rsq values and MAFs that are likely to have variable association estimates. Nonetheless, imputation can direct the focus of subsequent studies to putatively associated SNPs that can be rapidly pursued by very limited genotyping and functional characterization, followed by replication. While there is the large initial cost for acquiring GWAS genotyping data, those data are becoming increasingly available for sample sets with drug response phenotypes. When GWAS data are available and new “omics” data become available, utilizing the GWAS data to perform imputation to quickly analyze genes and/or pathways of interest dramatically reduces downstream cost and time compared to the use of a tag SNP approach.
It is clear that imputation can quickly provide more comprehensive coverage of genetic variation for candidate genes than can a tag SNP approach, but there is an analytical trade-off as well as the limitation of analyzing a much larger set of SNPs, requiring correction for multiple testing. However, this situation also occurs in DNA sequencing studies, in which additional analytical methods will be required to identify association signals with more power than for single marker analysis. The imputed SNPs that we verified in DLD and SHMT2 were not significant based on a conservative Bonferroni correction, but these SNPs can now be pursued in replication and functional genomic studies.
In summary, the strategy proposed here represents a useful approach for merging other “omics” data with pharmacogenomics. Our results demonstrate that the use of GWAS data to impute SNPs with “1000 Genomes” data can improve pharmacogenomic candidate gene/pathway studies by accelerating and broadening the analysis. Our results also highlight the need to validate signals identified by the use of imputation by limited genotyping before moving on to replication, but we have also demonstrated the gains that can occur in terms of cost and time savings by using imputation as compared to traditional tag SNP selection and genotyping. In the context of “pharmacometabolomics-informed pharmacogenomics”, this approach might become increasingly important to identify findings undetected by GWAS and/or candidate gene strategies.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health grants R01 GM28157, R01 CA132780, CA140879, U19 GM61388 (The Pharmacogenomics Research Network), R24 GM78233 (the Metabolomic Research Network for Drug Response Phenotypes); a PhRMA Foundation Center of Excellence in Clinical Pharmacology Award; and the NIH Pharmacogenomics Research Network–RIKEN Center for Genomic Medicine Global Alliance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sorger PK, Allerheiligen SRB, Abernethy DR, Altman RB, Brouwer KLR, Califano A, et al. Quantitative and Systems Pharmacology in the Post-genomic Era: New Approaches to Discovering Drugs and Understanding Therapeutic Mechanisms. In: Ward R, editor. QSP Workshop Group. 2011. [ http://www.nigms.nih.gov/NR/rdonlyres/8ECB1F7C-BE3B-431F-89E6-A43411811AB1/0/SystemsPharmaWPSorger2011.pdf]. [Google Scholar]
- 2.Kaddurah-Daouk R, Kristal BS, Weinshilboum RM. Metabolomics: a global biochemical approach to drug response and disease. Annu Rev Pharmacol Toxicol. 2008;48:653–683. doi: 10.1146/annurev.pharmtox.48.113006.094715. [DOI] [PubMed] [Google Scholar]
- 3.Ji Y, Hebbring S, Zhu H, Jenkins GD, Biernacka J, Snyder K, et al. Glycine and a glycine dehydrogenase (GLDC) SNP as citalopram/escitalopram response biomarkers in depression: pharmacometabolomics-informed pharmacogenomics. Clinical pharmacology and therapeutics. 2011;89:97–104. doi: 10.1038/clpt.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durbin RM, Altshuler DL, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rush AJ, Trivedi MH, Ibrahim HM, Carmody TJ, Arnow B, Klein DN, et al. The 16-Item Quick Inventory of Depressive Symptomatology (QIDS), clinician rating (QIDS-C), and self-report (QIDS-SR): a psychometric evaluation in patients with chronic major depression. Biological psychiatry. 2003;54:573–583. doi: 10.1016/s0006-3223(02)01866-8. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biernacka JM, Tang R, Li J, McDonnell SK, Rabe KG, Sinnwell JP, et al. Assessment of genotype imputation methods. BMC proceedings. 2009;3 Suppl 7:S5. doi: 10.1186/1753-6561-3-s7-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nothnagel M, Ellinghaus D, Schreiber S, Krawczak M, Franke A. A comprehensive evaluation of SNP genotype imputation. Human genetics. 2009;125:163–171. doi: 10.1007/s00439-008-0606-5. [DOI] [PubMed] [Google Scholar]
- 9.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 11.Michel S, Liang L, Depner M, Klopp N, Ruether A, Kumar A, et al. Unifying Candidate Gene and GWAS Approaches in Asthma. PLoS ONE. 2010;5:e13894. doi: 10.1371/journal.pone.0013894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, Roos C, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57:3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.