Abstract
Genome-wide association studies (GWASs) have primarily focused on marginal effects for individual markers and have incorporated external functional information only after identifying robust statistical associations. We applied a new approach combining the genetics of gene expression and functional classification of genes to the GWAS of basal cell carcinoma (BCC) to identify potential biological pathways associated with BCC. We first identified 322,324 expression-associated single-nucleotide polymorphisms (eSNPs) from two existing GWASs of global gene expression in lymphoblastoid cell lines (n=995), and evaluated the association of these functionally annotated SNPs with BCC among 2,045 BCC cases and 6,013 controls in Caucasians. We then grouped them into 99 KEGG pathways for pathway analysis and identified two pathways associated with BCC with p-value < 0.05 and false discovery rate (FDR) < 0.5: the autoimmune thyroid disease pathway (mainly HLA class I and II antigens, p < 0.001, FDR = 0.24) and JAK-STAT signaling pathway (p = 0.02, FDR = 0.49). Seventy nine (25.7%) out of 307 eSNPs in the JAK-STAT pathway were associated with BCC risk (p < 0.05) in an independent replication set of 278 BCC cases and 1,262 controls. In addition, the association of JAK-STAT signaling pathway was marginally validated by using 16,691 eSNPs identified from 110 normal skin samples (p = 0.08). Based on the evidence of biological functions of the JAK-STAT pathway on oncogenesis, it is plausible that this pathway is involved in BCC pathogenesis.
Keywords: Pathway analysis, Basal cell carcinoma, GWAS, JAK-STAT
Introduction
Basal cell carcinoma (BCC) is the most common cancer in Caucasian populations, accounting for approximately 80 percent of all non-melanoma skin cancers, and its incidence is increasing (de Vries et al. 2004). Although the mortality attributable to BCC is not high, the disease is responsible for considerable morbidity, imposing a growing burden on healthcare services. Besides the known risk factors for BCC, including exposure to ultraviolet radiation and host factors such as fair complexion, red or blond hair, and light eye color (Gallagher et al. 1995; Lear et al. 1997; Rubin et al. 2005), germline polymorphic variants have been implicated as playing a role in disease development (Lear et al. 2005). More recently, genome-wide association studies (GWAS) have identified several new loci as associated with BCC (Stacey et al. 2008; Stacey et al. 2009). However, the GWAS studies have focused only on the most significant SNPs (those that fulfilled a stringent statistical “genome-wide” significance criterion), while paying little attention to the rest. Thus, new methods are emerging to utilize the remaining genetic information in the GWAS data.
Given that gene-gene interactions may contribute to complex diseases, combining the modest association signals in the GWAS data with information on biological pathways and networks can help to detect the joint effects of multiple genes (Pedroso 2010). Some have argued that a large proportion of disease susceptibility genes will be found to be functionally related and/or interact with one another in biological pathways, and that only a limited number of biological pathways contribute to the etiology of complex traits (Carlborg and Haley 2004). Thus, pathway-based approaches have been applied to the GWAS of several complex diseases in determining whether the cumulative contribution of genes with a common biological denominator is greater than expected by chance (Baranzini et al. 2009; Elbers et al. 2009; Menashe et al. ; Wang et al. 2007; Wang et al. 2009). In these studies, the representative SNPs for the genes were chosen from all the SNPs physically located in the gene region; however, SNPs in a gene region might not represent functional variants of the gene, and a true disease-associated gene may have multiple independent functional variants. Furthermore, a gene may be regulated in trans by genetic variants that are physically distant from the structural gene (Schadt et al. 2008). It has been demonstrated that SNPs associated with gene expression in liver and adipose tissues are enriched for association with type 2 diabetes (Zhong et al. 2010a). By applying this method to the GWAS of type 2 diabetes, several well-known pathways as well as some novel ones have been identified in the etiology of this disease (Zhong et al. 2010b). Therefore, a new approach integrating the genetics of gene expression and GWAS pathway analysis is appealing for its ability to improve the power of detecting the association (Zhong et al. 2010b).
In this study, we adapted this approach to integrate pathway information and two existing databases of gene expression quantitative trait loci (eQTL) for the GWAS on BCC. We used the expression-associated SNPs (eSNPs) discovered in two GWAS of global gene expression in Epstein-Barr virus transformed lymphoblastoid cell lines (LCL) (Dixon et al., 2007 and unpublished data, for details see method), and sought to uncover the important biological pathways associated with BCC risk that might help provide insight into the etiology of BCC.
Materials and Methods
eSNP Processing and Analysis
Global gene expression data were analyzed by two techniques in two independent samples. The first sample (referred to as MRCA) contains 405 children of British descent (Dixon et al. 2007), organized into 206 sibships, including 297 sib pairs and 11 half-sib pairs. The families were identified through a proband with childhood asthma, and siblings were included regardless of disease status. Global gene expression in LCLs was measured using Affymetrix HG-U133 Plus 2.0 chips. All 405 children and their parents were genotyped using the Illumina Sentrix Human-1 Genotyping BeadChip (ILMN100K, including 105,713 autosomal SNPs), and 378 children were also genotyped using Illumina Sentrix HumanHap300 BeadChip (ILMN300K, including 307,981 autosomal SNPs) according to the manufacturer's instructions (Dixon et al. 2007; Moffatt et al. 2007).
The second sample (referred to as MRCE, data unpublished) of 950 individuals from 320 families of British descent was genotyped using the Illumina Sentrix HumanHap300 Genotyping Beadchip. The genotyped sample consisted of 347 subjects with asthma and 487 subjects with atopic dermatitis (259 subjects with both diseases). Expression arrays using Illumina Human 6 BeadChips were available on 550 children. Expression values were estimated using BeadStudio (Illumina, San Diego), and bead summary data were used for downstream analysis. From a total of 47,293 probes, we excluded 30,806 probes deemed “absent” (detection score <0.95) in more than 80% arrays to eliminate noise. We retained 16,487 probes representing 15,576 genes for analysis. The data were then normalized using quantile normalization (Bolstad et al. 2003).
We performed genotype imputation in the MRCA and MRCE samples independently. The ILMN300K genotypes were used to impute the polymorphic SNPs in the Phase II HapMap. Imputation was carried out using a hidden Markov model programmed in MACH (Li et al. 2009).
We estimated non-genetic contributions in gene expression measures using principal component analysis (Leek and Storey 2007; Stegle et al. 2008). Top principal components were included in the eQTL regression model as covariates. The number of principal components used was chosen to maximize the number of genome-wide significant cis eQTLs. Association analysis was applied with the FASTASSOC option implemented in MERLIN (Abecasis et al. 2002; Chen and Abecasis 2007).
Finally, we obtained 367,232 eSNPs associated with at least one gene in cis with LOD > 4 (corresponding to false discovery rate, FDR < 0.3%, accounting for all cis SNP-gene pairs within 1Mb) or one gene in trans with FDR < 5%. In total, 18,988 genes were found to be associated with at least one of these eSNPs.
The skin eQTL analysis was conducted among 110 normal skin tissues (Ellinghaus et al. ; Nair et al. 2009). We identified 16,697 cis SNPs as skin eQTLs, which were associated with the expression of at least one gene in the skin with LOD > 4.
BCC GWAS
Description of Study Populations
The BCC GWAS comprised 2,045 BCC cases and 6,013 controls from five GWAS sets (Hunter et al. 2007; Qi et al. 2010; Rimm et al. 1991), which reflects the postmenopausal invasive breast cancer case-control study (controls only) nested within the Nurses’ Health Study (BC_NHS, BCC cases=248, BCC controls=816), the type 2 diabetes case-control study nested within the NHS (T2D_NHS, BCC cases=665, BCC controls=2,162), the type 2 diabetes case-control study nested within the Health Professionals Follow-up Study (T2D_HPFS, BCC cases=597, BCC controls=1,555), the coronary heart disease case-control study nested within the NHS (CHD_NHS, BCC cases=253, BCC controls=765), and the coronary heart disease case-control study nested within the HPFS (CHD_HPFS, BCC cases=282, BCC controls=715). All the cases are self-reported BCC patients, and we excluded those who carried diagnosis of other common cancers before diagnosis of BCC based on common cancers reported by National Cancer Institute and American Cancer Society. For BCC controls, we also excluded those individuals who carried diagnosis of BCC or other common cancers. Those common cancers included melanoma, SCC, breast cancer, endometrial cancer, ovarian cancer, colorectal cancer, bladder cancer, lung cancer, pancreatic cancer, kidney (renal cell) carcinoma, leukemia, Non-Hodgkin Lymphoma, thyroid cancer, and oral cancer. The overlapping samples among the five GWAS sets were removed. All the individuals in our study are of European ancestry and live in the United States. The study protocol was approved by the Institutional Review Board of Brigham and Women's Hospital and Harvard School of Public Health. Detailed descriptions of the study populations and genotyping platforms for each study have been reported previously (Hunter et al. 2007; Qi et al. 2010; Rimm et al. 1991).
Genotyping and Genotype Imputation
We used the Illumina HumanHap550 array for the genotyping in BC_NHS (Hunter et al. 2007) and used the Affymetrix 6.0 array for the other four GWAS sets (T2D_NHS, T2D_HPFS, CHD_NHS, and CHD_HPFS) (Qi et al. ; Rimm et al. 1991). Within each of the five studies, we used the MACH program (Li et al. 2009) to impute genotypes for more than 2.5 million SNPs based on the genotyped SNPs and haplotype information in the HapMap phase II data build 36 (CEU) (Li et al. 2009; Marchini et al. 2005). eSNPs with imputation R2 > 0.4 and minor allele frequency (MAF) > 2.0% in all of the five sets were included in further analysis. Finally, 322,324 out of 367,232 eSNPs in the LCL eQTL database were used for the pathway enrichment analysis. In the replication stage, 16,691 out of the 16,697 skin eSNPs with imputation R2 > 0.4 and MAF > 2.0% in the BC_NHS set were used. Due to the very limited number of the identified skin eQTLs, we conducted the replication analysis in the BC_NHS set, which had the largest number of eQTLs passing quality control.
Association analysis
Associations between each SNP and BCC risk were determined using a multivariable logistic regression model; the first three principal components of genetic variation were adjusted in the model within each of the five sets. These principal components were calculated for all individuals on the basis of ca. 10,000 unlinked markers using the EIGENSTRAT software (Price et al. 2006). For each SNP, within-cohort association results were combined in an inverse variance–weighted meta-analysis by using software METAL (http://www.sph.umich.edu/csg/abecasis/Metal/index.html).
Pathway data construction
We collected pathway data from the Kyoto Encyclopedia of Genes and Genomes (KEGG). Ninety-nine pathways that contain at least 20 but at most 200 genes represented by eSNPs were tested. The cutoff of 20–200 genes was selected as a good balance for gene sets to reduce the multiple-testing issue and to avoid testing overly narrow or broad functional categories, as suggested by Wang et. al.(Wang et al. 2007)
Statistical Method of eSNP Enrichment Analysis
We applied the method of Zhong et al. (Zhong et al. 2010b) to integrate the eSNP information into the pathway-based GWAS analysis. Specifically, for each gene, we evaluated the association between BCC risk and all the eSNPs which were associated with the expression of this gene, and then assigned the eSNP which was most significantly associated with BCC risk into each gene as its representative. Then we assigned these genes into the pathways. We used the Kolmogorov-Smirnov test statistic to evaluate the association between each pathway and BCC risk. For each pathway, we calculated the Enrichment Score (ES), which reflects the overrepresentation of genes within this pathway at the top of the entire ranked list of genes in the genome. A permutation procedure (1,000 permutations, permuting the case-control status and re-calculating the statistic value) was used to assess the significance of the ES. A normalized enrichment score (NES) was calculated for each pathway in the observed and permutated data to allow direct comparison of pathways of different sizes. A false discovery rate (FDR) was calculated to estimate the proportion of false positive findings by using NES (Reiner et al. 2003). The detailed algorithm was described by Zhong et al. (Zhong et al. 2010b) and Wang et al. (Wang et al. 2007). We set the significance level for the pathway analysis as p value < 0.05 and FDR < 0.5.
Replication set
We included the individuals in the kidney stone case-control study (KS) nested in the NHS, the Nurses’ Health Study II (NHS2) and the HPFS as the replication set. NHS2 was established in 1989, when 116,678 female registered nurses aged 25-42 and residing in the United States at the time of enrollment responded to an initial questionnaire on their medical histories and baseline health-related exposures. Using the same criteria of case identification as used for the BCC GWAS set, we identified 278 BCC cases and 1,262 controls in the KS set. Illumina Human610 array was used for genotyping and the same imputation procedure was conducted as for the BCC GWAS set.
Result
From two existing eQTL databases based on LCL (Dixon et al., 2007 and unpublished data, for details see method), we identified 367,232 eSNPs associated with gene expression in LCLs. Among them, 322,324 eSNPs had MAF > 2.0% and imputation R2 > 0.4 in our BCC GWAS, and were used for further analysis. A total of 11,478 genes with eSNPs significantly associated with their abundance of transcripts in LCL were assigned to the KEGG pathways. Finally, 99 KEGG pathways that contained between 20 and 200 eligible genes with qualified eSNPs associated with their expression were tested for association with BCC risk in our GWAS data.
From the pathway enrichment analysis, eight (8.08%) out of the 99 KEGG pathways reached a nominal p value < 0.05, which was 1.63 times as high as the number expected by chance (0.05 × 99 = 4.95, a conservative estimate, for that pathways may be correlated due to overlapping genes and the effective number should be smaller than 99). Two of these eight pathways reached a FDR of less than 0.5 (Table 1; complete association results for all the 99 KEGG pathways are listed in Table S1). The two pathways with significant enrichment in the BCC GWAS were the autoimmune thyroid disease pathway (p < 0.001, FDR = 0.24) and the Janus kinase-signal transducer and activator of transcription (JAK-STAT) signaling pathway (p = 0.02, FDR = 0.49). In addition, interleukin 6 signal transducer (IL6ST, smallest p-value for BCC association, PBCC = 0.01) and interleukin 6 receptor (IL-6R, smallest PBCC = 0.03) in the JAK-STAT signaling pathway were associated with BCC risk. According to the pathway definition in the KEGG database, there is some overlap between the two pathways: 32.7% of the genes in the autoimmune thyroid disease pathway are also in the JAK-STAT signaling pathway.
Table 1.
Pathways with significant enrichment in the BCC GWAS.
| Pathway1 | Gene Count2 | Genes with eSNP (%) | FDR3 | Pathway Enrichment p-value | Genes with Pbcc < 0.014 | eSNP5 | eSNP Pbcc | Chr | Position | Distance (bp) |
|---|---|---|---|---|---|---|---|---|---|---|
| Autoimmune thyroid disease | 55 | 25(45.5) | 0.24 | <0.001 | HLA-B | rs3093562 | 1.21E-04 | 6 | 31659516 | 226,548 |
| HLA-G | rs805262 | 5.15E-04 | 6 | 31736712 | 1,829,834 | |||||
| HLA-DQB1 | rs2736171 | 7.53E-04 | 6 | 31703466 | 1,032,168 | |||||
| HLA-DRB36 | rs2856683 | 8.19E-04 | 6 | 32763196 | 97,656 | |||||
| HLA-DRB1 | rs7536479 | 1.92E-03 | 1 | 99838156 | NA7 | |||||
| HLA-DOB | rs10947342 | 1.92E-03 | 6 | 32828758 | 59,759 | |||||
| IFNA14 | rs9299397 | 4.09E-03 | 9 | 93514692 | 72,284,714 | |||||
| HLA-DPA1 | rs13215763 | 4.11E-03 | 6 | 33104578 | 36,193 | |||||
| HLA-DRB4 | rs7775228 | 4.72E-03 | 6 | 32766057 | 285,383 | |||||
| HLA-C | rs2854050 | 5.10E-03 | 6 | 32293583 | 945,749 | |||||
| HLA-A | rs9461216 | 5.38E-03 | 6 | 25916722 | 4,101,587 | |||||
| HLA-DMA | rs2857211 | 5.54E-03 | 6 | 32848420 | 175,952 | |||||
| HLA-DPB1 | rs3130171 | 6.26E-03 | 6 | 33106707 | 45,030 | |||||
| HLA-DOA | rs6920606 | 7.29E-03 | 6 | 33105652 | 20,285 | |||||
| JAK-STAT signaling pathway | 155 | 60(38.7) | 0.49 | 0.02 | IL21R | rs4787976 | 2.93E-03 | 16 | 27513198 | 143,582 |
| STAM2 | rs10930939 | 2.96E-03 | 2 | 152688179 | 6,619 | |||||
| IFNA14 | rs9299397 | 4.09E-03 | 9 | 93514692 | 72,284,714 | |||||
| IL15RA | rs8177600 | 7.43E-03 | 10 | 6065069 | 4,921 | |||||
| CSF2RB | rs2284027 | 9.25E-03 | 22 | 35595025 | 44,595 | |||||
| IL6ST | rs17730330 | 9.47E-03 | 12 | 91522645 | NA7 | |||||
| PIK3CB | rs6439828 | 9.68E-03 | 3 | 140113284 | 152,409 | |||||
Pathways are from the Kyoto Encyclopedia of Genes and Genomes (KEGG)
The number of genes in the pathway according to the KEGG database
Based on 1000 permutations and 99 tested pathway in total
PBCC represents p values of eSNPs for BCC association. Only the genes whose representative eSNPs reached PBCC < 0.01 are listed here and the information of all enriched genes in the two pathways are listed in Table S2
Denotes genes with the smallest PBCC
The four genes HLA-DRB3, HLA-DRB5, HLA-DQA1, and HLA-DQA2 have the same representative eSNP: rs2856683
This is a trans eQTL. The eSNP is not on the same chromosome as the associated gene.
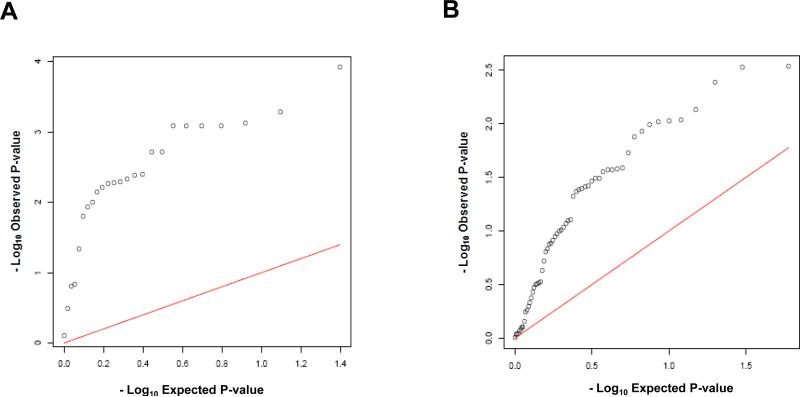
There were 25 genes with eSNPs in the autoimmune thyroid disease pathway, among which 21 (84.0%) genes had eSNPs with PBCC less than 0.05. Twenty-five (41.7%) out of 60 genes with eSNPs in the JAK-STAT signaling pathway had eSNPs with PBCC less than 0.05. The representative eSNPs of the genes in the two pathways that had PBCC < 0.01 are listed in Table 1, and all the representative eSNPs in the two pathways are listed in Table S2. The quantile quantile (QQ) plots of all the representative eSNPs for the two pathways are shown in Figure 1.
Figure 1.
Quantile Quantile Plots of the representative eSNP p values for two pathways: A) autoimmune thyroid disease pathway and B) Jak-STAT signaling pathway.
We attempted to validate the findings of the two pathways with BCC using the skin eQTLs generated from 110 normal skin tissues (Ding et al. 2010). A total of 16,697 skin eQTLs were identified and 16,691 of them with MAF > 2.0% and imputation R2 > 0.4 in the BCC GWAS were used in the final analysis. There were 619 (77%) out of the 803 genes in skin eQTL data overlapping with those from the LCL eQTL data. As a result, the association for JAK-STAT pathway was marginal significant (p-value = 0.08). The association between the autoimmune thyroid disease pathway and BCC risk was not significant using the skin eQTL data (p-value = 0.56).
We further conducted a replication analysis in an additional set of 278 BCC cases and 1,262 controls for the JAK-STAT pathway. We found that 79 (25.7%) out of 307 LCL eSNPs within this pathway were associated with BCC in the replication set (p < 0.05), which were 5.3-folder more than by chance (307×0.05=15). This finding validated the enrichment of association signals for the eSNPs within this pathway.
Discussion
GWASs have primarily focused on marginal effects for individual markers and functional pathways were investigated only after robust statistical associations were identified. In this study, we integrated pathway analysis and functional annotation of SNPs from existing eQTL databases for a GWAS on BCC risk. This approach improved power to detect relevant functional pathways that are directly regulated by BCC-associated genetic variants. This method has recently shown its potential strength in the context of type 2 diabetes GWAS (Zhong et al. 2010b); however, application to cancer has not been reported. Using this method, we integrated the LCL eQTL information and identified two pathways that might be implicated in the etiology of BCC: the autoimmune thyroid disease pathway and the JAK-STAT signaling pathway. The latter was also found to be marginal significant using the skin eQTL information based on a small dataset.
Ideally, the skin eQTLs are more relevant to the study on BCC. However, the available eQTLs from skin tissues were based on a small sample (n=110 for skin tissue samples vs. n= 995 for LCL samples), which limits the power to identify eQTLs that are specific to the skin. After quality control, there were only 16,691 skin eQTLs identified for the association with the expressions of 803 genes compared to 322,324 LCL eQTLs identified for 11,487 genes, which would substantially reduce the statistical power of the pathway analysis and limit the number of genes within each pathway. Although patterns of gene expression may vary across different tissue types, eQTLs could be substantially shared between certain tissues. Hence, eQTLs identified from one tissue type could be a useful surrogate to study the genetic of gene expression in another tissue (Cookson et al. 2009; Ding et al. 2010). The eQTLs identified in the LCL (include the eQTLs used in our paper and other LCL data such as the HapMap samples) have helped interpret findings from GWAS where the LCL is not the directly relevant tissue, including human height, body mass index, waist-hip ratio, osteoporosis-related traits, childhood asthma and Crohn disease among many others (Czarnecki et al. ; Dixon et al. 2007; Heid et al. ; Hsu et al. ; Lango Allen et al. ; Libioulle et al. 2007). In a recent study, it was estimated that ~70% of cis-eQTLs in LCLs are shared with skin (Ding et al. 2010). In addition, the study on psoriasis reported a substantial overlap (>95%) of cis-eQTLs among three types of skin tissues (normal skin in healthy controls, lesional psoriatic skin and uninvolved skin in psoriasis patients) (Ding et al. 2010). Therefore, we used the LCL eQTLs in the discovery stage, and the skin eQTLs for validation. As a result, we marginally validated the finding of one pathway but not the other. It is possible that some important eSNPs for genes in the other pathway were missed using the current skin eQTL data due to insufficient power (n=110 skin expression data vs n=995 in LCL expression data). Nevertheless, we do not rule out the possibility that some eQTLs were specific to lymphocytes and not observed in the skin eQTL dataset.
Signal transducer and activator of transcription (STAT) has been implicated in programming gene expression in biological events such as embryonic development, programmed cell death, organogenesis, innate immunity, adaptive immunity and cell growth regulation in many organisms. The JAK-STAT pathway is the principal signaling mechanism for a wide array of cytokines and growth factors including IL-6, -21, -23 and IFN, and is well known to contribute to oncogenesis (Boudny and Kovarik 2002). In particular, IL-6 signaling via STAT3 promotes the Th-17 phenotype and IFNgamma signaling via STAT4 promoted Th-1 phenotype, both important T cell subtypes for cancer surveillance. Changes with signaling via JAK/STAT, either aberrant function resulting in impaired surveillance or enhanced activation can result in susceptibility to cancer. Persistent activation of STAT signaling has been shown in an increasing number of human cancers, including a wide variety of solid tumors such as breast and prostate cancer, as well as many leukemias and lymphomas (Boudny and Kovarik 2002; Buettner et al. 2002; Yu and Jove 2004). As for BCC, it has been reported that the microenvironment of BCC is dominated by several cytokines including IL-6 (Elamin et al. 2008), which enhances tumorigenic potential and anti-apoptotic activity in human BCC (Jee et al. 2001). The JAK-STAT pathway has been reported to transmit IL-6 signals and induce cell survival and angiogenic activity in BCC cell lines (Jee et al. 2002; Jee et al. 2004), and over-expression of IL-6 has been reported in the tissue of BCC (Gambichler et al. 2006). In addition, an IL-6 polymorphism has been associated with BCC risk in a population study (Wilkening et al. 2006). Consistent with these reports, we found that IL6ST (p = 0.01) and IL-6R (p = 0.03) were associated with BCC risk in the JAK-STAT signaling pathway, which further suggests the involvement of IL-6 in disease development via JAK-STAT signaling.
As for the autoimmune thyroid disease pathway, it includes HLA class I and class II antigens, which play an important role in tumor immune surveillance by presenting tumor antigens to T lymphocytes (Streilein 1991). The association between various HLA antigens and BCC risk has been reported by a number of publications (Bavinck et al. 2000; Bouwes Bavinck et al. 1997; Bouwes Bavinck et al. 1991; Cerimele et al. 1988; Czarnecki et al. 1991; Myskowski et al. 1985). However, the association of this pathway was not validated using the skin eQTLs and is upon further replication.
Although none of the eSNPs of the genes in the identified pathway reached genome-wide significance (0.05/2,500,000 = 2×10-8) in our BCC GWAS, we tested for the enrichment of BCC risk in predefined pathways by incorporating prior functional information available from existing eQTL databases. This approach could improve power to detect functionally relevant groups of genes. There are still some limitations in this study. First, we used the LCL eQTLs to surrogate those of the skin and it is possible that some important genes that are specifically expressed in skin tissue and play critical roles in BCC etiology may be missed. Second, we focused only on the eQTLs that affect gene expression. Hence, other potential functional SNPs (e.g. methylation SNPs, splicing SNPs and missense/nonsense SNPs) may be missed. This approach also highly depends on the accuracy and completeness of the existing eQTL database. As the sample size increases and newer technology covers more depth of gene expression, more eSNPs are likely to be identified. Third, the FDR of the identified pathway is not particularly low. Further replication using additional samples and functional investigation would be required to illustrate the true effect of this pathway.
In this study, we identified the JAK-STAT pathway with known biological functions in oncogenesis that may be involved in BCC development. The role of the JAK/STAT pathway in the host Th1/Th17 response may be associated with risk for BCC. Further studies are warranted to verify our findings and additional functional characterization of this pathway is needed to elucidate its biological mechanism in the etiology of BCC. It is noteworthy that this approach, which integrates the pathways and eSNPs, may help identify potential biological mechanisms underlying other diseases using GWAS data.
Supplementary Material
Acknowledgements
We thank Dr. David Hunter, Dr. Frank B. Hu, and Dr. Eric B. Rimm for their work on the five GWAS sets (BC_NHS, T2D_NHS, T2D_HPFS CHD_NHS and CHD_HPFS). We thank Dr. Hongmei Nan for her work in BCC GWAS dataset construction and Dr. Mousheng Xu for his technical support in programming. We thank the participants in the Nurses’ Health Study and the Health Professionals Follow-Up Study for their dedication and commitment. We also thank the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Grant support:
The NHS and HPFS cohorts are supported by NIH grants CA87969, CA055075, and CA49449.
Footnotes
Conflict of Interest
The authors state no conflict of interest.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin--rapid analysis of dense genetic maps using sparse gene flow trees. Nature Genetics. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BM, Kappos L, Polman CH, Matthews PM, Hauser SL, Gibson RA, Oksenberg JR, Barnes MR. Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18:2078–90. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavinck JN, Bastiaens MT, Marugg ME, Beckers RC, Westendorp RG, Vermeer BJ, Claas FH. Further evidence for an association of HLA-DR7 with basal cell carcinoma on the tropical island of Saba. Arch Dermatol. 2000;136:1019–22. doi: 10.1001/archderm.136.8.1019. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Boudny V, Kovarik J. JAK/STAT signaling pathways and cancer. Janus kinases/signal transducers and activators of transcription. Neoplasma. 2002;49:349–55. [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Claas FH, Hardie DR, Green A, Vermeer BJ, Hardie IR. Relation between HLA antigens and skin cancer in renal transplant recipients in Queensland, Australia. J Invest Dermatol. 1997;108:708–11. doi: 10.1111/1523-1747.ep12292086. [DOI] [PubMed] [Google Scholar]
- Bouwes Bavinck JN, Vermeer BJ, van der Woude FJ, Vandenbroucke JP, Schreuder GM, Thorogood J, Persijn GG, Claas FH. Relation between skin cancer and HLA antigens in renal-transplant recipients. N Engl J Med. 1991;325:843–8. doi: 10.1056/NEJM199109193251203. [DOI] [PubMed] [Google Scholar]
- Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–54. [PubMed] [Google Scholar]
- Carlborg O, Haley CS. Epistasis: too often neglected in complex trait studies? Nat Rev Genet. 2004;5:618–25. doi: 10.1038/nrg1407. [DOI] [PubMed] [Google Scholar]
- Cerimele D, Contu L, Carcassi C, Costa G, La Nasa G, Sanna E, Campus GV. HLA and multiple skin carcinomas. Dermatologica. 1988;176:176–81. doi: 10.1159/000248700. [DOI] [PubMed] [Google Scholar]
- Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. American Journal of Human Genetics. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czarnecki D, Lewis A, Nicholson I, Tait B. Multiple basal cell carcinomas and HLA frequencies in southern Australia. J Am Acad Dermatol. 1991;24:559–61. doi: 10.1016/0190-9622(91)70082-d. [DOI] [PubMed] [Google Scholar]
- de Vries E, Louwman M, Bastiaens M, de Gruijl F, Coebergh JW. Rapid and continuous increases in incidence rates of basal cell carcinoma in the southeast Netherlands since 1973. J Invest Dermatol. 2004;123:634–8. doi: 10.1111/j.0022-202X.2004.23306.x. [DOI] [PubMed] [Google Scholar]
- Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, Nair RP, Elder JT, Abecasis GR. Gene expression in skin and lymphoblastoid cells: Refined statistical method reveals extensive overlap in cis-eQTL signals. Am J Hum Genet. 2010;87:779–89. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Elamin I, Zecevic RD, Vojvodic D, Medenica L, Pavlovic MD. Cytokine concentrations in basal cell carcinomas of different histological types and localization. Acta Dermatovenerol Alp Panonica Adriat. 2008;17:55–9. [PubMed] [Google Scholar]
- Elbers CC, van Eijk KR, Franke L, Mulder F, van der Schouw YT, Wijmenga C, Onland-Moret NC. Using genome-wide pathway analysis to unravel the etiology of complex diseases. Genet Epidemiol. 2009;33:419–31. doi: 10.1002/gepi.20395. [DOI] [PubMed] [Google Scholar]
- Ellinghaus E, Ellinghaus D, Stuart PE, Nair RP, Debrus S, Raelson JV, Belouchi M, Fournier H, Reinhard C, Ding J, Li Y, Tejasvi T, Gudjonsson J, Stoll SW, Voorhees JJ, Lambert S, Weidinger S, Eberlein B, Kunz M, Rahman P, Gladman DD, Gieger C, Wichmann HE, Karlsen TH, Mayr G, Albrecht M, Kabelitz D, Mrowietz U, Abecasis GR, Elder JT, Schreiber S, Weichenthal M. Franke A Genome-wide association study identifies a psoriasis susceptibility locus at TRAF3IP2. Nat Genet. 42:991–5. doi: 10.1038/ng.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher RP, Hill GB, Bajdik CD, Fincham S, Coldman AJ, McLean DI, Threlfall WJ. Sunlight exposure, pigmentary factors, and risk of nonmelanocytic skin cancer. I. Basal cell carcinoma. Arch Dermatol. 1995;131:157–63. [PubMed] [Google Scholar]
- Gambichler T, Skrygan M, Hyun J, Bechara F, Tomi NS, Altmeyer P, Kreuter A. Cytokine mRNA expression in basal cell carcinoma. Arch Dermatol Res. 2006;298:139–41. doi: 10.1007/s00403-006-0673-1. [DOI] [PubMed] [Google Scholar]
- Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpelainen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Allen HL, Weyant RJ, Wheeler E, Wood AR, Estrada K, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard-Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti-Proenca C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K, Tammesoo ML, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YH, Zillikens MC, Wilson SG, Farber CR, Demissie S, Soranzo N, Bianchi EN, Grundberg E, Liang L, Richards JB, Estrada K, Zhou Y, van Nas A, Moffatt MF, Zhai G, Hofman A, van Meurs JB, Pols HA, Price RI, Nilsson O, Pastinen T, Cupples LA, Lusis AJ, Schadt EE, Ferrari S, Uitterlinden AG, Rivadeneira F, Spector TD, Karasik D, Kiel DP. An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits. PLoS Genet. 6:e1000977. doi: 10.1371/journal.pgen.1000977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, Hankinson SE, Wacholder S, Wang Z, Welch R, Hutchinson A, Wang J, Yu K, Chatterjee N, Orr N, Willett WC, Colditz GA, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Hayes RB, Tucker M, Gerhard DS, Fraumeni JF, Jr., Hoover RN, Thomas G, Chanock SJ. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jee SH, Chiu HC, Tsai TF, Tsai WL, Liao YH, Chu CY, Kuo ML. The phosphotidyl inositol 3-kinase/Akt signal pathway is involved in interleukin-6-mediated Mcl-1 upregulation and anti-apoptosis activity in basal cell carcinoma cells. J Invest Dermatol. 2002;119:1121–7. doi: 10.1046/j.1523-1747.2002.19503.x. [DOI] [PubMed] [Google Scholar]
- Jee SH, Chu CY, Chiu HC, Huang YL, Tsai WL, Liao YH, Kuo ML. Interleukin-6 induced basic fibroblast growth factor-dependent angiogenesis in basal cell carcinoma cell line via JAK/STAT3 and PI3-kinase/Akt pathways. J Invest Dermatol. 2004;123:1169–75. doi: 10.1111/j.0022-202X.2004.23497.x. [DOI] [PubMed] [Google Scholar]
- Jee SH, Shen SC, Chiu HC, Tsai WL, Kuo ML. Overexpression of interleukin-6 in human basal cell carcinoma cell lines increases anti-apoptotic activity and tumorigenic potency. Oncogene. 2001;20:198–208. doi: 10.1038/sj.onc.1204076. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 467:832–8. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear JT, Hoban P, Strange RC, Fryer AA. Basal cell carcinoma: from host response and polymorphic variants to tumour suppressor genes. Clin Exp Dermatol. 2005;30:49–55. doi: 10.1111/j.1365-2230.2004.01669.x. [DOI] [PubMed] [Google Scholar]
- Lear JT, Tan BB, Smith AG, Bowers W, Jones PW, Heagerty AH, Strange RC, Fryer AA. Risk factors for basal cell carcinoma in the UK: case-control study in 806 patients. J R Soc Med. 1997;90:371–4. doi: 10.1177/014107689709000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libioulle C, Louis E, Hansoul S, Sandor C, Farnir F, Franchimont D, Vermeire S, Dewit O, de Vos M, Dixon A, Demarche B, Gut I, Heath S, Foglio M, Liang L, Laukens D, Mni M, Zelenika D, Van Gossum A, Rutgeerts P, Belaiche J, Lathrop M, Georges M. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–7. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- Menashe I, Maeder D, Garcia-Closas M, Figueroa JD, Bhattacharjee S, Rotunno M, Kraft P, Hunter DJ, Chanock SJ, Rosenberg PS, Chatterjee N. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 70:4453–9. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, Heinzmann A, Simma B, Frischer T, Willis-Owen SA, Wong KC, Illig T, Vogelberg C, Weiland SK, von Mutius E, Abecasis GR, Farrall M, Gut IG, Lathrop GM, Cookson WO. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- Myskowski PL, Pollack MS, Schorr E, Dupont B, Safai B. Human leukocyte antigen associations in basal cell carcinoma. J Am Acad Dermatol. 1985;12:997–1000. doi: 10.1016/s0190-9622(85)70127-2. [DOI] [PubMed] [Google Scholar]
- Nair RP, Duffin KC, Helms C, Ding J, Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, Ruether A, Schreiber S, Weichenthal M, Gladman D, Rahman P, Schrodi SJ, Prahalad S, Guthery SL, Fischer J, Liao W, Kwok PY, Menter A, Lathrop GM, Wise CA, Begovich AB, Voorhees JJ, Elder JT, Krueger GG, Bowcock AM, Abecasis GR. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso I. Gaining a pathway insight into genetic association data. Methods Mol Biol. 2010;628:373–82. doi: 10.1007/978-1-60327-367-1_20. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Qi L, Cornelis MC, Kraft P, Stanya KJ, Linda Kao WH, Pankow JS, Dupuis J, Florez JC, Fox CS, Pare G, Sun Q, Girman CJ, Laurie CC, Mirel DB, Manolio TA, Chasman DI, Boerwinkle E, Ridker PM, Hunter DJ, Meigs JB, Lee CH, Hu FB, van Dam RM. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19:2706–15. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–75. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Rimm EB, Giovannucci EL, Willett WC, Colditz GA, Ascherio A, Rosner B, Stampfer MJ. Prospective study of alcohol consumption and risk of coronary disease in men. Lancet. 1991;338:464–8. doi: 10.1016/0140-6736(91)90542-w. [DOI] [PubMed] [Google Scholar]
- Rubin AI, Chen EH, Ratner D. Basal-cell carcinoma. N Engl J Med. 2005;353:2262–9. doi: 10.1056/NEJMra044151. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, Zhu J, Millstein J, Sieberts S, Lamb J, GuhaThakurta D, Derry J, Storey JD, Avila-Campillo I, Kruger MJ, Johnson JM, Rohl CA, van Nas A, Mehrabian M, Drake TA, Lusis AJ, Smith RC, Guengerich FP, Strom SC, Schuetz E, Rushmore TH, Ulrich R. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey SN, Gudbjartsson DF, Sulem P, Bergthorsson JT, Kumar R, Thorleifsson G, Sigurdsson A, Jakobsdottir M, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Scherer D, Rudnai P, Gurzau E, Koppova K, Hoiom V, Botella-Estrada R, Soriano V, Juberias P, Grasa M, Carapeto FJ, Tabuenca P, Gilaberte Y, Gudmundsson J, Thorlacius S, Helgason A, Thorlacius T, Jonasdottir A, Blondal T, Gudjonsson SA, Jonsson GF, Saemundsdottir J, Kristjansson K, Bjornsdottir G, Sveinsdottir SG, Mouy M, Geller F, Nagore E, Mayordomo JI, Hansson J, Rafnar T, Kong A, Olafsson JH, Thorsteinsdottir U, Stefansson K. Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet. 2008;40:1313–8. doi: 10.1038/ng.234. [DOI] [PubMed] [Google Scholar]
- Stacey SN, Sulem P, Masson G, Gudjonsson SA, Thorleifsson G, Jakobsdottir M, Sigurdsson A, Gudbjartsson DF, Sigurgeirsson B, Benediktsdottir KR, Thorisdottir K, Ragnarsson R, Scherer D, Hemminki K, Rudnai P, Gurzau E, Koppova K, Botella-Estrada R, Soriano V, Juberias P, Saez B, Gilaberte Y, Fuentelsaz V, Corredera C, Grasa M, Hoiom V, Lindblom A, Bonenkamp JJ, van Rossum MM, Aben KK, de Vries E, Santinami M, Di Mauro MG, Maurichi A, Wendt J, Hochleitner P, Pehamberger H, Gudmundsson J, Magnusdottir DN, Gretarsdottir S, Holm H, Steinthorsdottir V, Frigge ML, Blondal T, Saemundsdottir J, Bjarnason H, Kristjansson K, Bjornsdottir G, Okamoto I, Rivoltini L, Rodolfo M, Kiemeney LA, Hansson J, Nagore E, Mayordomo JI, Kumar R, Karagas MR, Nelson HH, Gulcher JR, Rafnar T, Thorsteinsdottir U, Olafsson JH, Kong A, Stefansson K. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–14. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegle O, Kannan A, Durbin R, Winn J. Accounting for Non-genetic Factors Improves the Power of eQTL Studies. 2008. pp. 411–422.
- Streilein JW. Immunogenetic factors in skin cancer. N Engl J Med. 1991;325:884–7. doi: 10.1056/NEJM199109193251210. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Bucan M. Pathway-Based Approaches for Analysis of Genomewide Association Studies. Am J Hum Genet. 2007;81 doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Zhang H, Kugathasan S, Annese V, Bradfield JP, Russell RK, Sleiman PM, Imielinski M, Glessner J, Hou C, Wilson DC, Walters T, Kim C, Frackelton EC, Lionetti P, Barabino A, Van Limbergen J, Guthery S, Denson L, Piccoli D, Li M, Dubinsky M, Silverberg M, Griffiths A, Grant SF, Satsangi J, Baldassano R, Hakonarson H. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S, Hemminki K, Rudnai P, Gurzau E, Koppova K, Kumar R, Forsti A. Case-control study in basal cell carcinoma of the skin: single nucleotide polymorphisms in three interleukin promoters pre-analysed in pooled DNA. Br J Dermatol. 2006;155:1139–44. doi: 10.1111/j.1365-2133.2006.07440.x. [DOI] [PubMed] [Google Scholar]
- Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- Zhong H, Beaulaurier J, Lum PY, Molony C, Yang X, Macneil DJ, Weingarth DT, Zhang B, Greenawalt D, Dobrin R, Hao K, Woo S, Fabre-Suver C, Qian S, Tota MR, Keller MP, Kendziorski CM, Yandell BS, Castro V, Attie AD, Kaplan LM, Schadt EE. Liver and adipose expression associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 2010a;6:e1000932. doi: 10.1371/journal.pgen.1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010b;86:581–91. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.