Abstract
Many pathogenic Gram-negative bacteria utilize type III secretion systems (T3SS) to alter the normal functions of target cells. Shigella flexneri uses its T3SS to invade human intestinal cells to cause bacillary dysentery (shigellosis) which is responsible for over one million deaths per year. The Shigella type III secretion apparatus (T3SA) is comprised of a basal body spanning both bacterial membranes and an exposed oligomeric needle. Host altering effectors are secreted through this energized unidirectional conduit to promote bacterial invasion. The active needle tip complex of S. flexneri is composed of a tip protein, IpaD, and two pore-forming translocators, IpaB and IpaC. While the atomic structure of IpaD has been elucidated and studied, structural data on the hydrophobic translocators from the T3SS family remain elusive. We present here the crystal structures of a protease-stable fragment identified within the N-terminal regions of IpaB from S. flexneri and SipB from Salmonella enterica serovar Typhimurium determined at 2.1 Å and 2.8 Å limiting resolution, respectively. These newly identified domains are comprised of extended length (114 Å in IpaB and 71 Å in SipB) coiled-coil motifs that display a high degree of structural homology to one another despite the fact that they share only 21% sequence identity. Further structural comparisons also reveal substantial similarity to the coiled-coil regions of pore-forming proteins from other Gram-negative pathogens, notably colicin Ia. This suggests that these mechanistically-separate and functionally-distinct membrane-targeting proteins may have diverged from a common ancestor during the course of pathogen-specific evolutionary events.
Keywords: Bacterial Pathogenesis, Type Three Secretion, Translocator, Structural Biology, Colicins
Shigella and Salmonella spp. are leading causes of gastroenteritis and severe diarrhea. Of the 1.1 million deaths that are caused by Shigella each year, nearly two thirds are children under five years of age 1. Salmonella enterica serovar Typhimurium is the leading cause of hospitalization and death due to food-borne gastroenteritis in the U.S. The pathogenesis of these enterics involves the invasion of epithelial cells of the gastrointestinal tract, which requires the use of a type III secretion system (T3SS).
The T3SS is a common virulence factor among Gram-negative pathogens. It is used to deliver bacterial effector proteins to the membrane and cytoplasm of target cells where they subvert normal cellular functions for the benefit of the pathogen 2; 3. Much research has been focused on the diverse activities of the effector proteins injected into the target cytoplasm via the T3SS. Likewise, the type III secretion apparatus (T3SA) of S. flexneri and S. Typhimurium have been extensively studied with respect to structure, function, and assembly. The T3SA resembles a molecular needle and syringe and it serves as a nanomachine that ultimately forms a unidirectional, energized conduit from the bacterial cytoplasm to the host cell membrane. In toto, the T3SA injectisome is comprised of a bulb within the bacterial cytoplasm, a basal body that localizes to the inner and outer membranes of the pathogen 4, and an external needle that creates a hollow channel extending from the base, formed by the polymerization of a single needle protein (MxiH in Shigella and PrgI in Salmonella) 5. The needle has an inner diameter of 2.5-3.0 nm and extends 40-80 nm, or just beyond the lipopolysaccharide layer of the bacterial outer membrane 6.
At the outermost tip of the T3SA needle resides the hydrophilic translocator or ‘needle tip protein’, IpaD in Shigella spp. and SipD in Salmonella spp., which is required for proper control of type III secretion 7. The needle tip proteins form a putative pentamer at the exposed end of the needle where they serve as environmental sensors for controlling the delivery of T3SA secretion substrates 8. In the presence of certain small molecules in the extracellular milieu, IpaD undergoes a conformational change that promotes the mobilization of IpaB, hereafter described as the first translocator protein due to its initial presence at the needle tip complex prior to the second translocator IpaC, to a position at the end of the needle distal to where IpaD is anchored 9; 10; 11. Though apparently less pronounced, SipD also undergoes a conformational change upon binding small molecules 12; 13; however, it is not known whether this serves as a trigger for mobilization of SipB to the Salmonella needle tip. In Shigella, newly exposed IpaB detects host cell contact via host membrane components that induce recruitment of IpaC, the second and final translocator protein, to the bacterial surface where it works in concert with IpaB to form a pore in the host cell membrane 14. This allows subsequently secreted effector proteins to pass through the unidirectional conduit and into the target cell membrane. As is the case in all T3SSs, the tip protein and both translocators are essential to bacterial pathogenesis 15.
While the atomic structures of several T3SA needle 16; 17; 18; 19; 20 and tip proteins 8; 12; 21 have been determined, solving the structures of the translocators has met with little success. This is due in part to the apolar physical nature of these membrane-penetrating proteins. Lack of success in this area continues to hamper developing the full mechanistic understanding of the T3SA and the role of type III secretion in causing highly debilitating disease. Because the tip protein (IpaD) and the first hydrophobic translocator protein (IpaB) are surface exposed prior to pathogen invasion and control type III secretion, they represent attractive targets for the development of prophylactic therapeutics. Thus, any information derived from their structure-function relationships would provide the foundation for subunit vaccines toward the prevention of shigellosis and salmonellosis. While the use of live, attenuated Shigella strains as viable vaccine candidates has met with little success 22, preliminary data indicate that IpaB is indeed a protective antigen. Identification of distinct structural domains within the surface-exposed translocator would be expected to provide clues to discrete regions that are responsible for this protein’s protective capacity. In this study, we present the structure for the N-terminal region of IpaB and its homolog SipB. From these structures, a common structural theme appears to be emerging as the T3SA of Shigella and Salmonella is built up from the needle into the maturing tip complex and possibly into the formation of a translocon pore. We also examine the potential relationship of this structural theme to that of certain bacterial toxins, especially the pore forming toxins within the colicin family.
Identification of a Soluble N-terminal Region within the T3SS First Translocators IpaB and SipB
Given the limited structural information for the T3SS translocators and the instability of full length IpaB in the absence of its cognate chaperone IpgC 23; 24; 25; 26, we sought to identify a soluble fragment of the translocator that would be more amenable to crystallographic analysis (see Supplementary Material). Both IpaB and SipB contain a conserved central hydrophobic region that is predicted to form a single α-helical transmembrane hairpin 24; 25; 27; 28. It has also been shown that the region C-terminal to the hydrophobic domain contains membrane-binding properties 23; 25. Thus, it seemed likely that removal of this entire C-terminal region might yield a soluble, stable translocator fragment.
When purified, recombinant translocator/chaperone complexes were subjected to limited subtilisin digestion, yielding protease resistant translocator fragments for both the IpaB/IpgC and SipB/SicA complexes (SFig. 1a, b). Following separation by SDS-PAGE, the resulting fragments were characterized by LC-MS/MS analysis of their respective tryptic peptides 29. One such fragment corresponded to residues 28-226 of S. flexneri IpaB (S3-103-4 in SFig. 1a), while a similarly sized fragment was also observed for S. Typhimurium SipB (S4-103-4 in SFig. 1b). The fact that both fragments were derived from a closely overlapping area of distinct proteins supports our hypothesis that a largely stable and otherwise soluble domain is a conserved structural feature within the N-terminal region of these translocators. Indeed, recombinant forms of both IpaB28.226 and SipB30.237 were readily expressed in the absence of chaperone, and were found to be monodisperse in minimal buffers (as judged by both analytical gel-filtration chromatography and dynamic light scattering) even at concentrations greater than 5 mg/ml protein.
Crystal Structures of the IpaB and SipB Fragments Reveal a Structurally-conserved Intramolecular Coiled-Coil
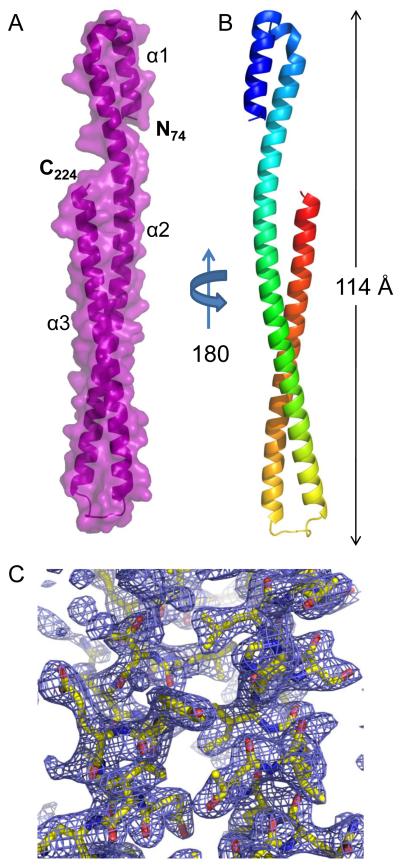
Despite the encouraging physical properties and the apparent stability of IpaB28.226, initial crystallization screening was unsuccessful. However, subsequent adventitious proteolytic degradation within the N-terminal region of IpaB28.226 was observed, which yielded a readily crystallizable core consisting of residues 74-224 (see Supplementary material). The structure of S. flexneri IpaB74.224 was solved by MAD using a platinum derivatized crystal exposed to synchrotron X-rays at two wavelengths corresponding to Pt peak and remote energies (Table 1), and the final structure was refined to 2.1 Å limiting resolution in space group P21. The final model consists of two complete polypeptides (SFig. 2a) that are comprised of residues 74-224 (Fig. 1).
Table 1. Diffraction Data Collection and Structure Refinement Statistics.
| Data Collectiona | ||||||
|---|---|---|---|---|---|---|
| Crystal | IpaB28.226 | IpaB28.226 | IpaB28.226 | SipB30.237 | SipB30.237 | |
| Native | Pt Remote | Pt Peak | SeMet Remote | SeMet Peak | ||
| Beamline | APS 22-BM | APS 22-BM | APS 22-BM | APS 22-BM | APS 22-BM | |
| Space Group | P21 | P21 | P21 | P21212 | P21212 | |
| Unit Cell Dimensions | ||||||
| a (Å) | 52.391 | 52.300 | 52.300 | 51.144 | 51.052 | |
| b (Å) | 28.372 | 28.060 | 28.060 | 84.646 | 84.715 | |
| c (Å) | 104.804 | 104.760 | 104.760 | 159.155 | 159.336 | |
| β(°) | 95.966 | 95.920 | 95.920 | |||
| Wavelength (Å) | 1.0000 | 1.0000 | 1.0719 | 0.9724 | 0.9793 | |
| Resolution (Å) | 50-2.1 | 50-2.5 | 50-2.5 | 50-2.8 | 50-3.0 | |
| Completeness (%) | 91.7 (59.3) | 95.7 (72.0) | 97.2 (80.5) | 98.0 (87.2) | 99.2 (95.5) | |
| Reflections (total) | 61,832 | 68,016 | 71,247 | 209,952 | 86,789 | |
| Reflections (unique) | 18,186 | 10,464 | 10,634 | 17,478 | 14,590 | |
| Redundancy (fold) | 3.4x | 6.5x | 6.7x | 12.0x | 5.9x | |
| <I>/<σI> | 13.4 (2.0) | 18.6 (2.82) | 19.4 (3.6) | 18.8 (2.55) | 14.3 (2.05) | |
| Rmerge (%)b | 8.5 (42.5) | 8.4 (35.0) | 8.2 (27.8) | 10.0 (67.1) | 11.5 (61.1) | |
| Refinement | ||||||
| RCSB Accession Code | 3U0C | 3TUL | ||||
| Protein Molecules/AU | 2 | 4 | ||||
| Rwork/Rfree (%)c | 24.53/29.45 | 29.73/31.67 | ||||
| Number of Atoms | ||||||
| Protein | 2426 | 3877 | ||||
| Solvent | 88 | 43 | ||||
| Ramachandran Plot (%) | ||||||
| Favored | 97.7 | 95.7 | ||||
| Allowed | 0.6 | 4.0 | ||||
| Outliers | 1.7 | 0.2 | ||||
| RMSD | ||||||
| Bond | ||||||
| Lengths (Å) | 0.008 | 0.009 | ||||
| Bond | ||||||
| Angles (°) | 1.066 | 1.240 | ||||
| B factor (Å2) | ||||||
| Protein | 44.76 | 52.56 | ||||
| Solvent | 50.60 | 58.10 |
Numbers in parentheses are for the highest-resolution no.
Rmerge = ΣhΣi|Ii(h)-<I(h)>|/ΣhΣiIi(h), where Ii(h) is the ith measurement of reflection h and <I(h)> is a weighted mean of all measurements of h.
R = Σh|Fobs(h)-Fcalc(h)|/Σh|Fobs|. Rcryst and Rfree were calculated from the working and test reflection sets, respectively. The test set constituted 5% of the total reflections not used in refinement.
Procedures detailing crystallization and data collection of IpaB28.226 and SipB30.237 are detailed in Supplementary Material. Briefly, crystals of both proteins were obtained by the hanging drop vapor diffusion method. Following data collection, individual reflections were indexed, integrated, and scaled using HKL2000 49. Both structures were determined by two-wavelength MAD phasing using the program AutoSol within the PHENIX suite 50. The experimental models were iteratively improved by manual building in Coot 51; 52, and refined using phenix.refine within the PHENIX suite 50.
Fig. 1. Crystal Structure of IpaB74.224 at 2.1 Å Resolution.
A, Crystal structure of S. flexneri IpaB (residues 74-224) shown in cartoon ribbon format surrounded by surface representation (colored purple). Two copies of each polypeptide are found within the asymmetric unit (single copy shown for clarity). B, Crystal structure rotated 180° about the long axis, colored blue (N-terminus) to red (C-terminus). C, Representative model-to-map correlation for IpaB74.224; 2Fo-Fc weighted electron density (contoured at 2.0 σ) is drawn as a blue cage around a region of the coiled-coil. Representations of all structures were generated using PyMol 46.
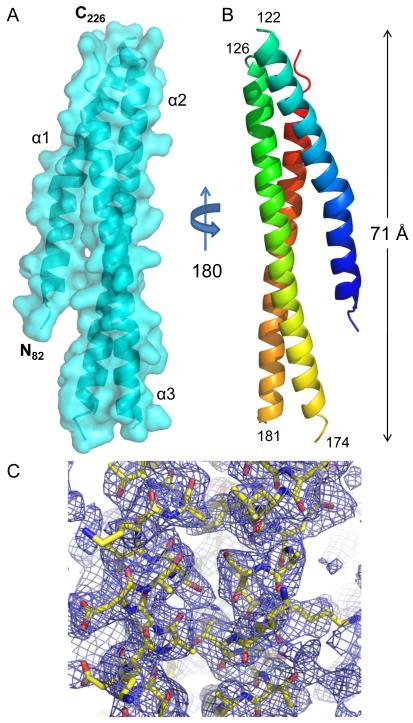
Similarly, the structure of S. Typhimurium SipB82.226 was solved by MAD using crystals of Seleno-L-Methionine-labeled protein exposed to synchrotron X-rays at two wavelengths corresponding to Se peak and remote energies (see Supplementary material). The final model was refined to a 2.8 Å limiting resolution in the space group P21212 and consisted of four polypeptides (SFig. 2b). While high-quality model/map correlation was observed for residues 82-122, 126-174 and 182-226 (Fig. 2), the electron density map corresponding to solvent exposed loop regions (i.e. residues 123-125 and 175-181) was too weak to model accurately. Interestingly, two of the four SipB polypeptides in the asymmetric unit exhibit different conformations within the 20 N-terminal residues of helix α1 (SFig. 2c). This region is characterized by a high degree of conformational flexibility and culminates in a 16.8 Å shift as measured from the carbonyl of Gly83 within chains A and B. Such intrinsic flexibility is apparently absent from the IpaB crystal structure.
Fig. 2. Crystal Structure of SipB82.226 at 2.8 Å Resolution.
A, Crystal structure of S. Typhimurium SipB (residues 82-226) shown in cartoon ribbon format surrounded by surface representation (colored cyan). The electron density map corresponding to solvent exposed loop regions (i.e. residues 123-125 and 175-181) was too weak to model accurately. The start and end of each missing loop region is labeled in panel B. Four copies of SipB are found within the asymmetric unit (single copy shown for clarity). B, Crystal structure rotated 180° about the long axis; colored blue (N-terminus) to red (C-terminus). C, Representative model-to-map correlation for SipB82.226; 2Fo-Fc weighted electron density (contoured at 1.5 σ) is drawn as a blue cage around a region of the coiled-coil. Representations of all structures were generated using PyMol 46.
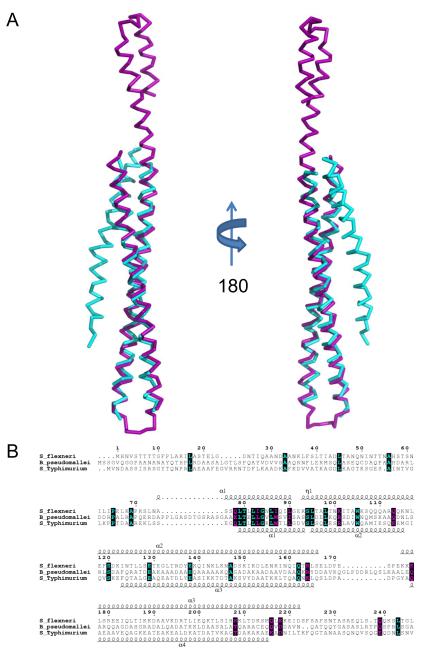
Both IpaB74.224 and SipB82.226 are almost entirely α-helical and are comprised of three anti-parallel helices compressed into an intramolecular coiled-coil tertiary structure (Figs. 1 and 2). It is interesting to note that previous structure-function analysis of IpaB and SipB predicted that the N-terminal regions of both IpaB (110-170) and SipB (180-216) would contain a coiled-coil domain 25; 30; 31; however, the predicted region is much smaller, and covers only a portion of the entire tertiary structure that is reported here. Remarkably, the latter part of the second and third helices of each structure (residues 104-224 and 126-226 for IpaB and SipB, respectively) display very strong structural identity with one another. Overall 93 of 94 Cα positions superimpose within 5.0 Å with an RMSD of 1.42 Å (Fig. 3a), indicating that this coiled-coil is a conserved feature of these T3SS translocators. This occurs despite the fact that IpaB shares relatively low sequence identity (21%) to the corresponding residues 1-240 of SipB (Fig. 3b).
Fig. 3. Structural Superposition of Translocator Coiled-coils.
A, Ribbon diagram of a structural alignment of the coiled-coils from IpaB (residues 120-224, purple) and SipB (residues 126-226, cyan) with an RMSD of 1.42 Å over 93/94 Cα atoms within 5.0 Å, rotated 180° about the long axis. Although the overall topology of both structures is similar, there are differences within the N-terminal region spanning the first helix (α1) and turn as well as the length of the second helix (α2). Such differences within the N-terminus of the structures reported here could be reflective of the apparent instability of the chaperone binding domains (CBD) in the absence of their cognate chaperones. B, Limited structure-based sequence alignment of type III secretion first translocators (residues 1-240) colored according to residue conservation (cyan=absolute and purple=similar) as judged by the BLOSUM62 matrix. Alignment was generated using ClustalW and rendered with ESPRIPT. Numbers above the sequences correspond to S. flexneri IpaB. Secondary structure elements of IpaB and SipB are shown above and below the alignment, respectively. Representations of all structures were generated using PyMol 46. Three-dimensional structures were superimposed using the Local-Global Alignment method (LGA) 47. Sequence alignments were carried out using CLUSTALW 48 and aligned with secondary structure elements using ESPRIPT 49.
Conservation of the Intramolecular Coiled-coil Motif within Type Three Secretion Proteins
The small protein monomers that comprise the T3SA needle of diverse organisms themselves consist of a helix-turn-helix motif that is essentially a short intramolecular coiled-coil 16; 17; 18; 19; 20. These monomers self-assemble into a superhelical bundle that forms the hollow, yet extended T3SA needle characteristic of these various pathogens 16. Similarly, a longer intramolecular coiled-coil is a highly conserved feature among the T3SA needle tip proteins from several families of pathogens 32, where it appears to constitute a centralized structural scaffold. The fact that IpaD’s presence at the T3SA needle tip is also required for stable maintenance of IpaB following its initial recruitment 11 suggests that these two proteins are likely to physically interact within the context of the T3SA needle assembly. Conceivably this could be accomplished most readily through a nearly parallel alignment of the tip protein and translocator coiled-coils in a manner similar to what has been previously described for the T3SA needle protein monomers 16. In this regard, it is likely significant that the coiled-coil domains of IpaB and SipB (residues 104-224 and 126-226, respectively) share a high degree of structural identity with the central coiled-coil region of their respective tip proteins, IpaD (RMSD of 2.47 Å over 101/121 Cα atoms within 5.0 Å) and SipD (RMSD of 2.39 Å over 88/94 Cα atoms within 5.0 Å) (SFig. 3a, b).
The structural homology shared between the needle monomers, tip proteins, and the N-terminal translocator fragments presented here further underscores the general importance of intramolecular coiled-coils as structural scaffolds within T3S proteins 33. While much work clearly remains to be done (particularly in terms of the remaining regions of IpaB and SipB), these initial structures provide an attractive starting point for understanding the mechanism of matured translocon formation. We believe that the increasing lengths of the coiled-coil structures in MxiH/PrgI, IpaD/SipD and the extreme length of the coiled-coils presented here (114 Å in Shigella and 71 Å in Salmonella) may allow the putative pore-forming hydrophobic domain of the first membrane-penetrating translocators to properly oligomerize above the IpaD/SipD-based tip complex. In this position, the assembled translocator multimer can sense host contact via interactions with lipids. Such an interaction would be responsible for concomitant recruitment of the second translocator, IpaC, to the T3SA needle tip, and thus facilitate needle insertion into the host cell membrane 14. It is interesting to note, then, that IpaC is also predicted to contain a coiled-coil 33; 34; by analogy, this domain might interact directly with the coiled-coil region indentified here within the first translocator, IpaB. Once the mature tip complex (IpaD-IpaB-IpaC in Shigella, SipD-SipB-SipC in Salmonella) has thus been assembled, full-scale translocation of effectors can then commence, which signals the beginning of T3SS-dependent cellular invasion.
The IpaB/SipB Coiled-coil Domain is Related to Those of Pore-Forming Colicins
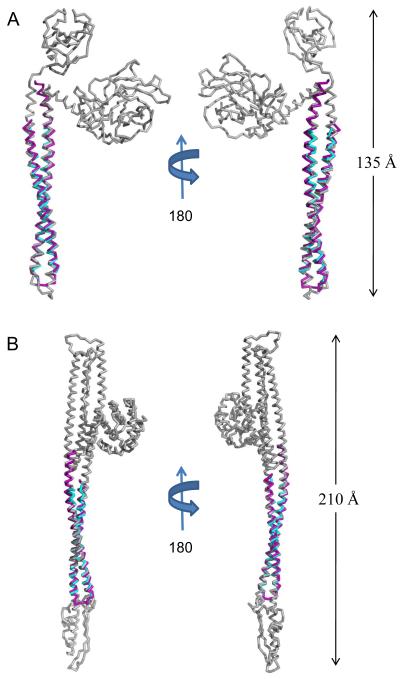
Despite their limited sequence conservation, the coiled-coil regions of IpaB (residues 104-224) and SipB (residues 126-226) share a high level of structural identity with one another. Furthermore, these intramolecular coiled-coils are reminiscent of the centralized helical scaffold within T3SA needle tip proteins. Together, these observations raised questions as to whether this structurally-conserved region of the T3SS first translocators might also be evolutionarily related to other proteins. To this end, the refined first translocator coiled-coils (comprised of IpaB104.224 and SipB126.226) were used to search for structurally-related motifs within other proteins via the DALI server 35 with the top 10 unique hits listed in Table 2. Whereas two significant similarities were detected between the translocator coiled-coils and distinct bacterial proteins involved in flagellum biogenesis (which itself is a variant of the T3SS), six additional of the highest-scoring matches were intriguingly derived from proteins involved in membrane targeting and/or insertion/pore-formation events 36; 37; 38; 39. In particular, colicin E3 (IpaB, Z-score = 9.7, RMSD of 2.03 Å over 118/121 Cα atoms; SipB, Z-score = 8.0, RMSD of 1.19 Å over 92/94 Cα atoms) and colicin Ia (IpaB, Z-score = 10.2, RMSD of 1.73 Å over 121/121 Cα atoms; SipB, Z-score = 10.1, RMSD of 1.21 Å over 93/94 Cα atoms) were among the highest scoring matches emanating from this search (Fig. 4a, b). Both of these colicins contain extended-length coiled-coil motifs (160 Å for colicin Ia and 100 Å for colicin E3) within their receptor domains 39. In all of these cases, the coiled-coil domains are responsible for spanning long distances with the purpose of mediating a contact dependent function, notably obtaining access to or transversing a membrane barrier 36; 37; 38; 40. At a fundamental level, this appears to be directly in line with a critical function already ascribed to T3SS first translocator proteins.
Table 2. First Translocator Coiled-coil DALI Search Statistics.
| Protein name | PDB code | Z-scorea | RMSD | Ca rangeb | %IDc | |
|---|---|---|---|---|---|---|
| IpaB104.224 | HP0958 | 3NA7 | 11.8 | 2.9 | 119/237 | 13 |
| Phosphatidylinositol 3-kinase regulatory subunit beta | 3L4Q | 10.3 | 2 | 119/163 | 8 | |
| Colicin E3 | 2B5U | 10.3 | 2.5 | 120/470 | 7 | |
| Adhesin A | 3ETX | 10.0 | 3.6 | 107/107 | 9 | |
| Colicin E2 | 2YSU | 9.5 | 2.8 | 114/123 | 9 | |
| ATP Synthase Gamma chain | 1JNV | 9.5 | 2.9 | 108/273 | 0 | |
| Lysine-specific Histone Demethylase 1 | 2X0L | 9.5 | 2.7 | 112/670 | 13 | |
| Chemotaxis receptor domain | 1QU7 | 9.2 | 2.3 | 109/227 | 6 | |
| HIV-1 envelope glycoprotein GP41 | 3P30 | 9.0 | 2 | 84/84 | 7 | |
| Colicin Ia | 1CII | 8.9 | 2.4 | 120/602 | 8 | |
|
| ||||||
| SipB126.226 | Colicin E2 | 2YSU | 10.6 | 1.4 | 93/132 | 19 |
| Macrolide-specific efflux protein MacA | 3FPP | 10.5 | 1.8 | 91/266 | 9 | |
| Colicin Ia | 1CII | 10.1 | 1.7 | 93/602 | 8 | |
| Flagellar FliJ Type III exportin protein | 3AJW | 9.6 | 1.2 | 90/134 | 12 | |
| Nucleocapsid protein | 2IC6 | 9.2 | 1.6 | 75/76 | 7 | |
| Prefoldin chaperone beta subunit | 2ZQM | 9.1 | 1.7 | 92/114 | 13 | |
| CT670 Chlamydia trachomatis YscO homolog | 3K29 | 9.0 | 1.6 | 92/162 | 11 | |
| Colicin E3 | 1UJW | 8.9 | 2.2 | 92/115 | 11 | |
| Chemotaxis receptor domain | 1QU7 | 8.7 | 2.4 | 93/227 | 4 | |
| Arfaptin 2 | 1I49 | 8.6 | 2.2 | 91/201 | 12 | |
Similarity score representing a function that evaluates the overall level of similarity between two structures. Z-scores higher than 8.0 indicate the two structures are most likely homologous 35.
Denotes the number of residues from the query structure that superimpose within an explicit distance cut-off of an equivalent position in the aligned structure.
Denotes the percent sequence identity across the region of structural homology.
Fig. 4. Translocator Coiled-coils Share Structural Homology with Pore-forming Proteins.
A, Ribbon diagram of IpaB (residues 104-224, purple) and SipB (residues 126-226, cyan) superimposed upon E. coli Colicin E3 (colored grey; PDB code= 1JCH) with an RMSD of 2.03 Å over 118/121 Cα atoms for IpaB and an RMSD of 1.19 Å over 92/94 Cα atoms for SipB. Crystal structures are rotated 180° about their long axis. B, Ribbon diagram of IpaB (residues 104-224, purple) and SipB (residues 126-226, cyan) superimposed over E. coli Colicin Ia (colored grey; PDB code= 1CII) with an RMSD of 1.73 Å over 121/121 Cα atoms for IpaB and an RMSD of 1.21 Å over 93/94 Cα atoms for SipB. Crystal structures are rotated 180° about their long axis. The DALI server was used to query available structures within the PDB for structural homology 35. Representations of all structures were generated using PyMol 46. Three-dimensional structures were superimposed using the Local-Global Alignment method (LGA) 47.
Colicins comprise a diverse series of bacteriocidal toxins produced by certain strains of Escherichia coli that exert their various activities targeting other E. coli cells through an array of distinct mechanisms, such as formation of transmembrane, voltage-gated pores, DNA and/or RNA cleavage through a potent intrinsic nuclease activity and even cell wall degradation 38; 39; 41. Despite the significant differences in their precise bacteriocidal activities, the initial movement of colicins across the target bacterial cell membrane systems appears to be accomplished by a largely similar mechanism, regardless of the method of killing, involving interaction with outer membrane (OM) receptor proteins 38; 41. For colicin Ia, which possesses a region that was structurally among the most similar to the coiled-coil region of both IpaB and SipB within the DALI search (Table 2), recognition of an OM receptor by a region at one end of its long coiled-coil allows a translocation domain to promote access of the protein to the periplasmic space of the target bacterium 37; 41; 42. While still anchored to the outer membrane receptor, the coiled-coil of colicin Ia allows the protein to span the periplasmic space to promote the insertion of a hydrophobic domain into the bacterial inner (cytoplasmic) membrane where it ultimately forms an ion channel. Considering their structural homology to members of the colicin family, we can thus begin to make larger-scale proposals relevant to T3SS first translocator function. In this regard, it seems very likely that the extended N-terminal coiled-coil domains of IpaB and SipB serve a similar role in both anchoring the first hydrophobic translocators to the IpaD/SipD-based T3SA needle tip complex and placing their respective hydrophobic domains in an extended position where recognition of and penetration into a target host cell plasma membrane can occur. This would not only maintain IpaB/SipB as a stable component of the maturing T3SA needle tip complex, but also ensure that the unidirectional, energized conduit remains contiguous.
The structures of the first translocator N-terminal fragments presented here cover approximately 25% of these proteins’ sequence. However, they do not provide direct information on the C-terminal half of these proteins, which contribute to their critical pore-forming structures 27; 28. The Fold and Function Assignment Server (FFAS) utilizes comparisons between sequences of interest and proteins of known function to then make structural predictions for the sequences of interest 43. When we used FFAS to query the PDB with sequences that correspond to the putative α-helical hairpin regions, the most conserved section of the first translocators within the Inv-Mxi-Spa family (SFig. 4), spanning residues 310-370 of IpaB 25; 30 and residues 320-380 of SipB 23; 24; 25, a region within the pore-forming domain of colicin Ia (residues 579-611) was identified. The pore-forming domain of colicin Ia is comprised of its C-terminal 176 residues, and consists of 10 α-helices arranged in a bundle-type structure (SFig. 5) that is highly similar to other colicin pore-forming domains 37; 40. Perhaps most significantly, the precise region identified by FFAS also corresponds to the hydrophobic helical hairpin that is responsible for anchoring the colicin pore within the host lipid bilayer 39. Thus, while the structural identity between the IpaB and SipB coiled-coils and members of the colicin family is clear, bioinformatic tools suggest that there may be even further relationships between these two groups of proteins.
Conclusions and Future Directions
S. flexneri and S. Typhimurium use the T3SS to promote entry into both macrophages and epithelial cells 44. Through the investigation of the Shigella T3SA we have been able to demonstrate that the mature needle tip complex is assembled in a stepwise manner. Initially, IpaD localizes to the needle tip where it controls secretion and represents completion of the assembly of the nascent T3SA needle tip complex 45. Interaction of IpaD with environmental small molecules such as host bile salts results in recruitment of IpaB to the needle tip, representing a maturation of the tip complex into a form that detects host cell contact 10. Finally, IpaB interacts with liposomes, preferentially those containing cholesterol and sphingomyelin, which mimic the host cell membrane. This ultimately leads to the recruitment of IpaC to the distal end of the needle complex 14. Yet despite this substantial level of mechanistic detail, our understanding of this process at a molecular level has been hampered by the lack of structural information on the translocators, IpaB and IpaC.
In the past, approaches based around biochemical characterization of gene deleted strains and comparative analysis of operon structures have proven instrumental in the identification of novel T3SSs from diverse gram-negative bacteria. Yet despite their many similarities at a functional level, analyses based upon sequence conservation alone have not played as significant a role in fostering understanding of various T3SSs - particularly for the Inv-Mxi-Spa T3SS family that is shared between Shigella, Salmonella, and Burkholderia spp. Within this context, the IpaB and SipB translocators of the Inv-Mxi-Spa T3SS family possess a relatively low level of sequence conservation within their N-terminal 240 residues (SFig. 4); however, as we have shown here, there exists a telling level of structural identity within this region, but one that required a biochemical mapping approach to identify. Thus, even though regions of these T3SS components may be diverging rapidly at the sequence level, it seems that there is strong selective pressure to maintain the coiled-coil structure within the N-terminal region of the first translocators. Intriguingly, this level of structural conservation appears to be shared between T3SS first translocators and members of the colicin family that likewise appear to have related membrane targeting and/or penetrating functions 39. At a fundamental level, this “shared structure, but low sequence homology” relationship appears to suggest an unexpected functional link between T3SSs and bacteriocins. However, the possibility that it may have even broader implications for creating and understanding phylogenies among secreted bacterial proteins is hard to ignore.
Because of the essential role of first translocators in type III secretion, the data presented here represent an important step forward which will allow detailed mechanistic dissection of the delivery of effectors to host cells. As importantly, because IpaB, and presumably SipB, is surface exposed prior to host cell contact, they represent potentially valuable targets for vaccine development. Since current vaccines against these pathogens are serotype specific, the development of IpaB and SipB (or defined regions thereof) into subunit-based vaccines could provide highly sought after heterologous protection among various serovars. As a result, the structures presented here provide valuable information on not only the mechanism of the first translocator’s role in T3SS-related disease, but may also suggest a plausible and attractive route to prevention of these diseases in the first place.
Supplementary Material
Acknowledgements
The authors acknowledge generous technical assistance of Drs. Rod Salazar and Andy Howard during X-ray diffraction data collection. Members of the Picking laboratories are also acknowledged for their critical reading of the manuscript. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) beamlines at the Advanced Photon Source, Argonne National Laboratory. A list of supporting member institutions may be found at www.ser-cat.org/members.html. This work was supported by National Institutes of Health Grants AI071028, AI067858, AI090149 to B.V.G., W.D.P., and W.L.P, and AI084203 to N.E.D. Additional support was provided by the Oklahoma Center for the Advancement of Science and Technology (HR10-128S).
ABBREVIATIONS
- T3SS
type III secretion system
- T3SA
type III secretion apparatus
- IpaB
invasion plasmid antigen B
- SipB
Salmonella invasion protein B
- IpaD
invasion plasmid antigen D
- SipD
Salmonella invasion protein D
- IpgC
invasion plasmid gene C
- SicA
Salmonella invasion chaperone A
Footnotes
Accession Numbers
The refined coordinates and structure factors (codes 3U0C and 3TUL) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rscb.org/). Translocator sequences used in alignment, along with their respective GenBank accession numbers, were as follows: Shigella flexneri (GenBank ID: 281603883), Burkholderia pseudomallei (GenBank ID: 127950892), and Salmonella enterica serovar Typhimurium (GenBank ID: 975295). Diffraction data and refinement statistics for all crystals discussed here are provided in Table 1.
REFERENCES
- 1.Diarrheal Diseases: Shigellosis. Vol. 2009. World Health Organization; 2009. http://www.who.int/vaccine_research/diseases/diarrhoeal/en/index6.html. [Google Scholar]
- 2.Mueller C, Broz P, Cornelis G. The type III secretion system tip complex and translocon. Mol Microbiol. 2008;68:1085–95. doi: 10.1111/j.1365-2958.2008.06237.x. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis GR. The type III secretion injectisome, a complex nanomachine for intracellular ‘toxin’ delivery. Biol Chem. 2010;391:745–51. doi: 10.1515/BC.2010.079. [DOI] [PubMed] [Google Scholar]
- 4.Blocker A, Jouihri N, Larquet E, Gounon P, Ebel F, Parsot C, Sansonetti P, Allaoui A. Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol Microbiol. 2001;39:652–63. doi: 10.1046/j.1365-2958.2001.02200.x. [DOI] [PubMed] [Google Scholar]
- 5.Darboe N, Kenjale R, Picking WL, Picking WD, Middaugh CR. Physical characterization of MxiH and PrgI, the needle component of the type III secretion apparatus from Shigella and Salmonella. Protein Sci. 2006;15:543–52. doi: 10.1110/ps.051733506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–2. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picking WL, Nishioka H, Hearn PD, Baxter MA, Harrington AT, Blocker A, Picking WD. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73:1432–40. doi: 10.1128/IAI.73.3.1432-1440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Self-chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–44. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. Conformational Changes in IpaD from Shigella flexneri upon Binding Bile Salts Provide Insight into the Second Step of Type III Secretion. Biochemistry. 2010 doi: 10.1021/bi101365f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stensrud KF, Adam PR, La Mar CD, Olive AJ, Lushington GH, Sudharsan R, Shelton NL, Givens RS, Picking WL, Picking WD. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem. 2008;283:18646–54. doi: 10.1074/jbc.M802799200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun. 2007;75:2626–9. doi: 10.1128/IAI.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell S, De Guzman RN. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 2011;20:75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lunelli M, Hurwitz R, Lambers J, Kolbe M. Crystal structure of PrgI-SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. doi: 10.1371/journal.ppat.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Epler CR, Dickenson NE, Olive AJ, Picking WL, Picking WD. Liposomes recruit IpaC to the Shigella flexneri type III secretion apparatus needle as a final step in secretion induction. Infect Immun. 2009;77:2754–61. doi: 10.1128/IAI.00190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ménard R, Sansonetti PJ, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WD, Picking WL, Blocker AJ, Lea SM. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci U S A. 2006;103:12529–33. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol. 2006;359:322–30. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Ouellette AN, Egan CW, Rathinavelan T, Im W, De Guzman RN. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J Mol Biol. 2007;371:1304–14. doi: 10.1016/j.jmb.2007.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol. 2008;377:819–30. doi: 10.1016/j.jmb.2007.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinaud M, Plé S, Job V, Contreras-Martel C, Simorre JP, Attree I, Dessen A. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci U S A. 2007;104:7803–8. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derewenda U, Mateja A, Devedjiev Y, Routzahn KM, Evdokimov AG, Derewenda ZS, Waugh DS. The structure of Yersinia pestis V-antigen, an essential virulence factor and mediator of immunity against plague. Structure. 2004;12:301–6. doi: 10.1016/j.str.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Levine MM, Kotloff KL, Barry EM, Pasetti MF, Sztein MB. Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat Rev Microbiol. 2007;5:540–53. doi: 10.1038/nrmicro1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hayward RD, Hume PJ, McGhie EJ, Koronakis V. A Salmonella SipB-derived polypeptide blocks the ‘trigger’ mechanism of bacterial entry into eukaryotic cells. Mol Microbiol. 2002;45:1715–27. doi: 10.1046/j.1365-2958.2002.03124.x. [DOI] [PubMed] [Google Scholar]
- 24.McGhie EJ, Hume PJ, Hayward RD, Torres J, Koronakis V. Topology of the Salmonella invasion protein SipB in a model bilayer. Mol Microbiol. 2002;44:1309–21. doi: 10.1046/j.1365-2958.2002.02958.x. [DOI] [PubMed] [Google Scholar]
- 25.Hume PJ, McGhie EJ, Hayward RD, Koronakis V. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol Microbiol. 2003;49:425–39. doi: 10.1046/j.1365-2958.2003.03559.x. [DOI] [PubMed] [Google Scholar]
- 26.Ménard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;79:515–25. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 27.Kaniga K, Tucker S, Trollinger D, Galán JE. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–71. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baudry B, Kaczorek M, Sansonetti PJ. Nucleotide sequence of the invasion plasmid antigen B and C genes (ipaB and ipaC) of Shigella flexneri. Microb Pathog. 1988;4:345–57. doi: 10.1016/0882-4010(88)90062-9. [DOI] [PubMed] [Google Scholar]
- 29.Kinter M, Serman NE. Protein sequencing and identification using tandem mass spectrometry. Wiley-Interscience; New York: 2000. [Google Scholar]
- 30.Guichon A, Hersh D, Smith MR, Zychlinsky A. Structure-function analysis of the Shigella virulence factor IpaB. J Bacteriol. 2001;183:1269–76. doi: 10.1128/JB.183.4.1269-1276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use of pairwise residue correlations. Proc Natl Acad Sci U S A. 1995;92:8259–63. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Espina M, Ausar SF, Middaugh CR, Baxter MA, Picking WD, Picking WL. Conformational stability and differential structural analysis of LcrV, PcrV, BipD, and SipD from type III secretion systems. Protein Sci. 2007;16:704–14. doi: 10.1110/ps.062645007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delahay RM, Frankel G. Coiled-coil proteins associated with type III secretion systems: a versatile domain revisited. Mol Microbiol. 2002;45:905–16. doi: 10.1046/j.1365-2958.2002.03083.x. [DOI] [PubMed] [Google Scholar]
- 34.Kueltzo LA, Osiecki J, Barker J, Picking WL, Ersoy B, Picking WD, Middaugh CR. Structure-function analysis of invasion plasmid antigen C (IpaC) from Shigella flexneri. J Biol Chem. 2003;278:2792–8. doi: 10.1074/jbc.M208383200. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soelaiman S, Jakes K, Wu N, Li C, Shoham M. Crystal structure of colicin E3: implications for cell entry and ribosome inactivation. Mol Cell. 2001;8:1053–62. doi: 10.1016/s1097-2765(01)00396-3. [DOI] [PubMed] [Google Scholar]
- 37.Wiener M, Freymann D, Ghosh P, Stroud RM. Crystal structure of colicin Ia. Nature. 1997;385:461–4. doi: 10.1038/385461a0. [DOI] [PubMed] [Google Scholar]
- 38.Kleanthous C. Swimming against the tide: progress and challenges in our understanding of colicin translocation. Nat Rev Microbiol. 2010;8:843–8. doi: 10.1038/nrmicro2454. [DOI] [PubMed] [Google Scholar]
- 39.Zakharov SD, Kotova EA, Antonenko YN, Cramer WA. On the role of lipid in colicin pore formation. Biochim Biophys Acta. 2004;1666:239–49. doi: 10.1016/j.bbamem.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 40.Hilsenbeck JL, Park H, Chen G, Youn B, Postle K, Kang C. Crystal structure of the cytotoxic bacterial protein colicin B at 2.5 A resolution. Mol Microbiol. 2004;51:711–20. doi: 10.1111/j.1365-2958.2003.03884.x. [DOI] [PubMed] [Google Scholar]
- 41.Cao Z, Klebba PE. Mechanisms of colicin binding and transport through outer membrane porins. Biochimie. 2002;84:399–412. doi: 10.1016/s0300-9084(02)01455-4. [DOI] [PubMed] [Google Scholar]
- 42.Ghosh P, Mel SF, Stroud RM. The domain structure of the ion channel-forming protein colicin Ia. Nat Struct Biol. 1994;1:597–604. doi: 10.1038/nsb0994-597. [DOI] [PubMed] [Google Scholar]
- 43.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–8. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder G, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21:134–56. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Espina M, Olive AJ, Kenjale R, Moore DS, Ausar SF, Kaminski RW, Oaks EV, Middaugh CR, Picking WD, Picking WL. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri. Infect Immun. 2006;74:4391–400. doi: 10.1128/IAI.00440-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. 2009 2002. http://www.pymol.org.
- 47.Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003;31:3370–4. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J, Higgins D, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gouet P, Courcelle E, Stuart D, Métoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–8. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.