Abstract
With its unparalleled ability to safely generate high-contrast images of soft tissues, magnetic resonance imaging (MRI) has remained at the forefront of diagnostic clinical medicine. Unfortunately due to resolution limitations, clinical scans are most useful for detecting macroscopic structural changes associated with a small number of pathologies. Moreover, due to a longstanding inability to directly observe magnetic resonance (MR) signal behavior at the cellular level, such information is poorly characterized and generally must be inferred. With the advent of the MR microscope in 1986 came the ability to measure MR signal properties of theretofore unobservable tissue structures. Recently, further improvements in hardware technology have made possible the ability to visualize mammalian cellular structure. In the current study, we expand upon previous work by imaging the neuronal cell bodies and processes of human and porcine α-motor neurons. Complimentary imaging studies are conducted in pig tissue in order to demonstrate qualitative similarities to human samples. Also, apparent diffusion coefficient (ADC) maps were generated inside porcine α-motor neuron cell bodies and portions of their largest processes (mean = 1.7±0.5 μm2/ms based on 53 pixels) as well as in areas containing a mixture of extracellular space, microvasculature, and neuropil (0.59±0.37 μm2/ms based on 33 pixels). Three-dimensional reconstruction of MR images containing α-motor neurons shows the spatial arrangement of neuronal projections between adjacent cells. Such advancements in imaging portend the ability to construct accurate models of MR signal behavior based on direct observation and measurement of the components which comprise functional tissues. These tools would not only be useful for improving our interpretation of macroscopic MRI performed in the clinic, but they could potentially be used to develop new methods of differential diagnosis to aid in the early detection of a multitude of neuropathologies.
Keywords: MRM, diffusion, neuron, human, axon, dendrite
Introduction
Owing to the fact that the technique does not rely on endoscopy or ionizing radiation, MRI is now arguably the most dominant non-invasive, non-destructive imaging modality of biological systems, with utility in both medical and material sciences. Despite finding widespread use in the clinic for identifying macroscopic features of a limited number of pathologies—e.g. tumors (Quadery and Okamoto, 2003), ischemic stroke infarct (Moseley et al., 1990)—MRI's utility for differential diagnosis is limited by a lack of specificity and fundamental understanding regarding the nature of how pathologies alter the MR contrast of microscopic tissue structures. Moreover, because the hallmarks of many diseases are often first detectable at the cellular level, understanding the characteristics of these changes at resolutions sufficient to observe them using MR is integral to creating new methods for detecting diseases at their earliest stages of development. Thus, to investigate such tissue contrast in the microscopic regime, we propose using magnetic resonance microscopy (MRM) as a means of characterizing MR contrast properties in cellular elements of human and porcine central nervous system tissue.
Although there were early concerns regarding its feasibility (Mansfield and Morris, 1982), MRM has evolved into a bona fide imaging regime of MR in the same sense that optical microscopy is a regime of optical imaging (Callaghan, 1991). As predicated in the literature, MR microscopy is loosely referred to as imaging (two-dimensional mapping) conducted at resolutions below 100μm in at least one of the two spatial dimensions (Benveniste and Blackband, 2005; Tyszka et al., 2005). True three-dimensional MRM may have isotropic resolution, but most often the in-plane dimensions are equal, while the third, through-plane dimension is larger. Such resolutions are employed to take advantage of certain samples' inherent symmetry which, when oriented so as to achieve structural homogeneity in the through-plane dimension, allows for larger slice geometries and increases the signal to noise ratio (SNR) without loss of structural delineation due to volume averaging.
MRM offers unique challenges as an imaging modality. The SNR is, in the majority of cases, the single greatest factor to consider in terms of attaining the highest spatial resolution. Increased spectral line widths associated with employing higher magnetic field strengths are another important consideration for microscopy studies as they have the potential to limit resolution (Cho et al., 1990; Ciobanu et al., 2003). Moreover, for the spatial resolution implied by the voxel size to be real, said voxel size must dominate potential blurring imposed by both the intrinsic transverse relaxation time (T2) and diffusivity (D) of the nuclear species being imaged. In summation, generating accurate MRM data requires sufficient SNR and strong gradients, limiting most experiments to those imaging the hydrogen contained in water of relatively small samples at high magnetic field strengths.
The first such works published on MRM (Aguayo et al., 1986; Eccles and Callaghan, 1986; Johnson et al., 1986) appeared in 1986: over a decade after the first publication on MRI (Lauterbur, 1973). The Eccles and Callaghan study employed a plant stem, achieving an in-plane resolution of 20μm, and—taking advantage of the sample's symmetry—a relatively large (1.5mm) slice thickness. The study by Johnson et al. was the first published work conducted on animal tissue, achieving a resolution of 50 × 50 × 1000μm in the rat brain. Experiments conducted by Aguayo et al. were the first to visualize single animal cells—frog ova, ≈ 1mm diameter—which was achieved using an image resolution of 10 × 13 × 250μm. Later studies explored the utility of Aplysia californica's L7 neuron as an alternative model for cellular imaging (Hsu et al., 1997). Such aquatic animal models became popular because their cell size allowed for excellent accessibility; however, the diameters of such cells are two to ten times greater than even the largest examples of mammalian cells (100-150μm) and orders of magnitude greater than the majority of mammalian cells (5-15μm). This size disparity was not trivial regarding the early limits of cellular visualization using MRM because, while the cells of certain aquatic organisms could be visualized using relatively thick slice geometries, the through-plane volume averaging associated with such scans precluded the detection of mammalian cellular structures by these means.
Most recently however, improvements in MR hardware technology—higher fields, micro surface-coils (Badilita et al., 2010; Ehrmann et al., 2007; Massin et al., 2003; Weber et al., 2011), and strong, rapidly-switching imaging gradients (Weiger et al., 2008)—have enabled the first reported instance of mammalian cell imaging using MRM (Flint et al., 2009). Perikarya of α-motor neurons in the ventral horn of the rat spinal cord were observed (7.8 × 7.8 × 25μm) using diffusion-weighted microimaging methods. Identification of the specific microstructural elements within such MRM images can be achieved using light-microscopy based correlative histology techniques (Meadowcroft et al., 2007; Nabuurs et al., 2011). In the present study, we extend these experiments to include the first MRM images of individual human cells. Cell bodies and neural processes of human α-motor neurons are visualized with native MR contrast in excised portions of intact, fixed spinal cord tissue. MRM studies on porcine tissue are also reported as a means of demonstrating its similarity in morphological and MR signal characteristics to human tissue.
Methods
Tissue Preparation
All procedures involving procurement, imaging, and disposal of human tissue specimen were reviewed and approved by the University of Florida's Internal Review Board (IRB # 129-2010). Immersion-fixed (4% Formaldehyde) spinal cord sections of 1.5cm length were excised by gross dissection from both cervical and lumbar enlargements. Histological cross sections (50μm) were cut in an ice-cold bath of phosphate-buffered saline (PBS) (137mM NaCl, 2.7mM KCl, 10mM Na2HPO4, and 1.8mM KH2PO4: pH 7.4) using a vibratome (Ted Pella, Lancer series 1000). Prior to imaging, slices were washed overnight in PBS to reduce fixative levels in the samples.
MRM
Imaging of Human α-Motor Neuron Perykaria
All MR imaging was performed using a 600 MHz Oxford spectrometer interfaced to a Bruker Biospin console. An area of the spinal cord's ventral horn (3mm × 3mm × 50μm) containing α-motor neurons was identified and excised from each spinal cord cross section (n = 3) prior to being secured in the tissue well of a 500μm diameter micro surface-coil (Bruker Biospin, B6370) as described previously (Flint et al., 2010). Briefly, ventral horn tissue was excised from intact slices by hand and secured over the micro surface-coil using a nylon mesh insert of 50μm pore size (Small Parts, CMN-0053-C) held in place by a retention ring of 300μm thickness fashioned from a nylon washer (Small Parts, WN-M02-C). Sample selection and placement were achieved with the aid of a dissecting microscope (Zeiss, OPMI 1-FC). After securing the sample, additional PBS was added to the microcoil's tissue well and a piece (1cm × 1cm) of polymerase chain reaction (PCR) film (ABgene, AB-0558) was used to seal the well and prevent sample dehydration. Diffusion-weighted, 2D images (TR/TE = 2000ms/23.5ms, res = 7.8μm × 7.8μm × 50 μm, temp = 23°C, Δ = 8.36ms, δ = 2ms, b = 2000s/mm2, Avg = 40, scan time = 5h40min) were employed to visualize cell bodies of human α-motor neurons. A linear smoothing algorithm (Bruker Biospin, ParaVision 3.0.2) helped define the boundaries of cell bodies present in our MR images.
Imaging of Human and Pig α-Motor Neuron Processes
Spinal cord samples (n = 3) were prepared and secured as described above. In experiments targeting processes, a micro surface-coil of 100μm diameter was employed (Bruker Biospin, B6372) and three dimensional spin-echo images (TR/TE = 2000ms/12.75ms, res = 6.25μm isotropic, Avg = 14, scan time = 63h43min) were collected. One such dataset was analyzed using an automated segmentation tool (Amira 3.1.1) to visualize cellular structures from multiple vantage points and display the close spatial arrangement of processes extending from adjacent neurons. In order to investigate intracellular diffusivity, three dimensional diffusion-weighted images (TR/TE = 2000ms/6.2ms, res = 6.25μm isotropic, temp = 23°C, δ = 0.31ms, Δ = 2ms, Avg = 14, scan time = 63h43min) were collected at two b values (b = 300s/mm2; b = 600s/mm2) and used to map the apparent diffusion coefficient (ADC) in the sample.
Correlative Histology
Nissl Stain for Neuronal Perikarya
Following diffusion-weighted, 2D MRM in the ventral horn of the human spinal cord, samples were removed from the micro surface-coil and stained for neuronal cell bodies. Samples were immersed in a solution of Nissl stain (0.5% cresyl violet acetate, 0.3% glacial acetic acid, 99.2% ddH2O) for 2min prior to receiving quick rinses in baths of destain solution (0.3% glacial acetic acid, 99.7% ddH2O) followed by PBS. Stained samples were then adhered to precleaned microscope slides (Fisher, 12-550-13) using Histomount (National Diagnostics, HS-103) mounting medium. False-color, bright field microscopy images were collected (Zeiss, Axioplan 2) using a 20× objective lens and captured with a digital camera (QImaging, Retiga 4000R Fast 1394 Color). Color adjustment in histology photographs was achieved using imaging software (QCapture Pro 6.0) and was performed uniformly over the entire image for the purpose of aiding in visualization of the labeled cell bodies.
Bielschowsky 's Silver Stain for Neuronal Processes
After collecting T2- and diffusion-weighted 3D MRM datasets, human and porcine spinal cord samples (100μm thick) were treated with a modified Bielschowsky stain (Stanely and Bielschowsky, 1925) in order to visualize neuronal processes in the tissue. Samples were exposed to an aqueous, 10% silver nitrate solution at 37°C for a period of 30min followed by 3×3min washes in ddH2O. Ammonium hydroxide (800μl, 1.0%) was added to the silver staining bath and the slices were immersed for a period of 15min. Next, slices were placed in serial baths (3×2min) of 1.0% ammonium hydroxide at ambient temperature. Reducing agent (200μl 37% formaldehyde, 3.57μl 70% nitric acid, 0.05g citric acid, 12ml ddH20) was added (70μl) to the heated silver staining bath and the slices were immersed in this solution for 20min. After this exposure, tissue was again placed in serial baths (3×2min) of 1.0% ammonium hydroxide followed by 3×2min washes in ddH2O. Lastly, tissue was toned in 0.2% gold chloride solution for 2min prior to being mounted and photographed as described above.
Results
Figure 1 shows three MRM images containing human cells and their accompanying correlative histology. Cell bodies of α-motor neurons stand out with hypointense signal contrast compared to the surrounding tissue in the ventral horn. In each case, the spatial locations of Nissl-stained cell bodies in our histological data match those patterns of cell bodies seen in the corresponding MRM. Interestingly, areas of the histological slices which do not contain parenchyma, but are devoid of tissue (i.e. microscopic tissue tears and holes, Figure 1, yellow arrow) also manifest as hypointense areas on our diffusion-weighted images. Figure 2 shows a sequence of representative 2D MR images selected from our T2-weighted, 3D datasets. Adjacent panels are consecutive in the 3D data and represent sections of tissue 6.25μm thick. Signal-to-noise ratio (SNR) values for hyperintense regions surrounding the cell bodies and roots of large processes range from approximately 8 to 21 for these scans and contrast to noise (CNR) between this tissue and cell bodies range from 5 to 11. Details of this analysis are provided in the figure text. Physical projections which possess equivalent signal to and are continuous with the cell bodies of α-motor neurons are visualized extending through multiple panels of the 3D MRI datasets. Tissue structures described in Figure 2 are positively identified through correlative histology in Figure 3. Here we see a representative sample of consecutive MR image slices taken from a diffusion-weighted (b = 600s/mm2), 3D dataset which contain components from three distinct porcine α-motor neurons. The cellular components visualized in our MRM data (two perikarya and portions of three processes) can be identified in accompanying light-microscopy photographs of the tissue following silver staining. The fact that all of the cellular components are not readily visible in a single frame of our MR dataset is indicative of slight differences in their through-plane spatial positions. Such differences are not apparent in the corresponding bright field microscopy image which projects through the entirety of the 100μm-thick spinal cord section. In Figure 4, two diffusion-weighted, 3D datasets have been used to generate an ADC map which illustrates differences in apparent diffusion between a cell—the perikaryon of an α-motor neuron and the proximal region of one of its processes—and the neural tissue of the ventral horn surrounding these structures. In this case, two representative images from the first dataset (left column; b = 300s/mm2) and their corresponding images from the second dataset (middle column; b = 600s/mm2) are shown with calculated ADC maps (right column). The apparent diffusivity inside this cell (soma and visible process) was estimated to be 1.7±0.5 μm2/ms (based on 53 pixels) suggesting higher diffusion rates in these regions compared to the remaining parenchyma where the ADC was estimated to be 0.59±0.37 μm2/ms (based on 33 pixels). Signal-to-noise ratio (SNR) values for regions adjacent to α-motor neurons range from approximately 11 to 17 for these scans and contrast to noise (CNR) between these tissues and cell bodies range from 0.4 to 3.9. Details of this analysis are provided in the figure text. Figure 5 shows a segmentation reconstruction of the tissue from one of our 3D, porcine MRM datasets (Figure 2, A). Continuity of the neural projections and their respective cell bodies is evident in the reconstructed 3D data which can be viewed as a movie file (Movie S5_3D reconstruction of porcine alpha motor neuron cell bodies and processes.mpg). online as part of this study's supplementary materials (Movie S5_3D reconstruction of porcine alpha motor neuron cell bodies and processes.mpg).
Figure 1. Human cell bodies of α-motor neurons detected using magnetic resonance microscopy and confirmed through correlative histology.
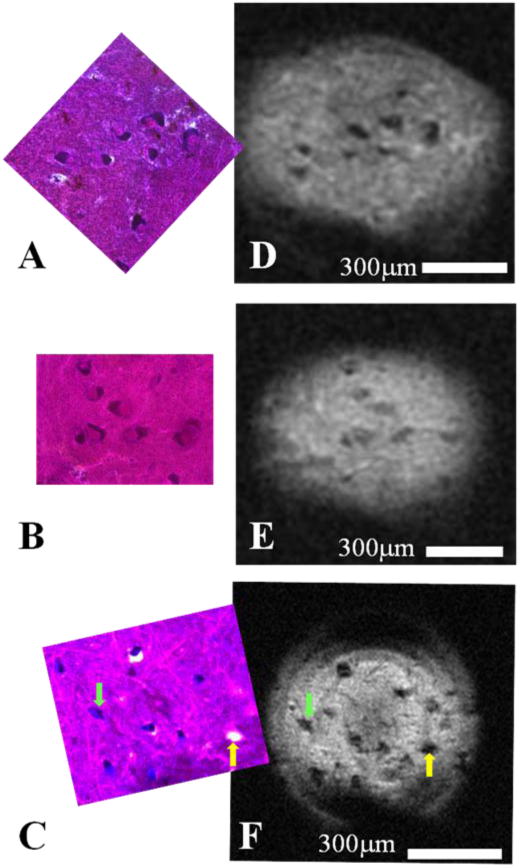
Diffusion-weighted (b = 2000s/mm2) MRM (n = 3) taken in the ventral horn of human spinal cord enlargements using a 500μm micro surface-coil and their corresponding correlative histology. MR images (DEF) show perikarya of human α-motor neurons which appear dark in contrast to the surrounding tissue of the ventral horn's gray matter. Nissl-stained histology images (ABC) contain the same cell bodies whose cytoplasmic compartments have been darkly stained due to the presence of Nissl bodies. The nuclear compartment is visible in a fraction of cells present in the histology and can be identified by its relative lack of Nissl staining and the presence of a nucleolus; however, subcellular compartments have not been visualized in the corresponding MRM. Interestingly, as is seen in panels C and F, cell bodies (green arrow) can share morphological and contrast characteristics with microscopic tissue tears (yellow arrow) at the resolutions employed.
Figure 2. Neuronal processes in the human and pig visualized using magnetic resonance microscopy.
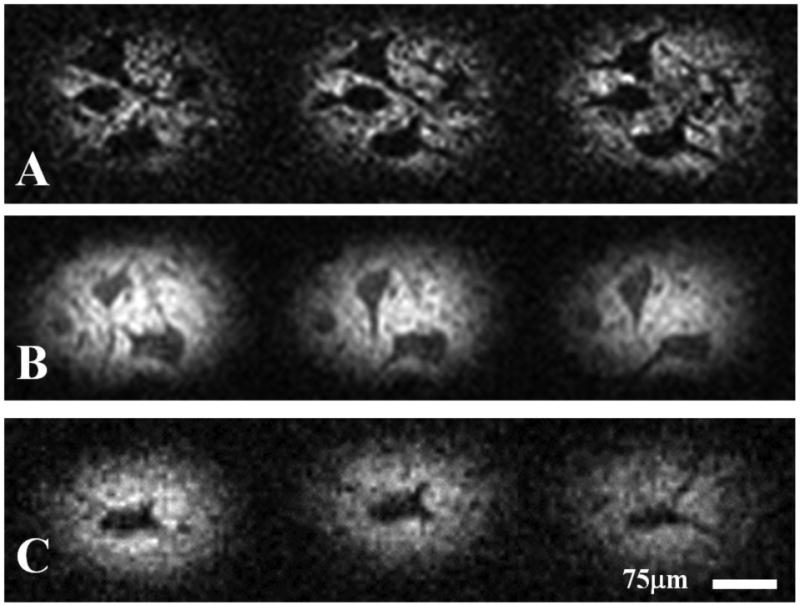
Representative MRM images (6.25μm isotropic resolution) taken from three-dimensional T2-weighted datasets (A & B: porcine; C: human) illustrating the hypointense projections emanating from cell bodies of α-motor neurons. These projections are continuous with the cells' perikarya and extend through multiple frames of the 3D datasets. Adjacent frames located in rows of the figure represent consecutive, voxel-thick sections of the tissue slice. SNR in tissue surrounding the α-motor neurons from top to bottom row: 7.85, 21.1 and 7.77. CNR (between α-motor neuron and surrounding tissues) from top to bottom row: 6.13, 11.0 and 5.01.
Figure 3. Correlative histology identifying porcine neuronal processes with Bielschowsky's silver stain.
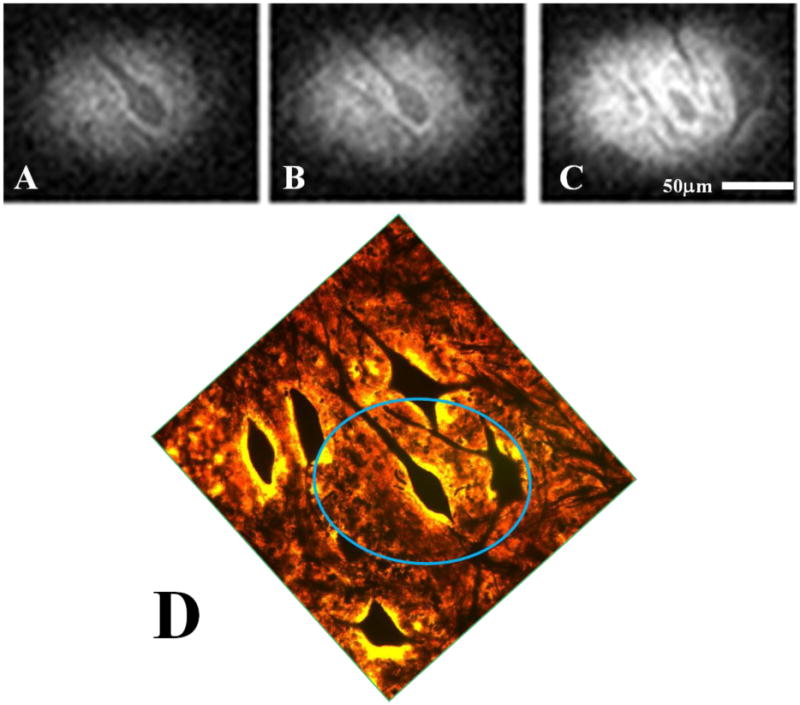
Three consecutive images (ABC) taken from our 3D, diffusion-weighted dataset (6.25μm isotropic) and the corresponding correlative histology (D) which positively identifies the structures seen in our MRM as cell bodies and their neuronal processes. The highlighted region in the histology image (blue oval) roughly approximates the portion of the tissue slice captured in our MR data.
Figure 4. Representative scans from two diffusion-weighted, 3D datasets and their corresponding ADC maps.
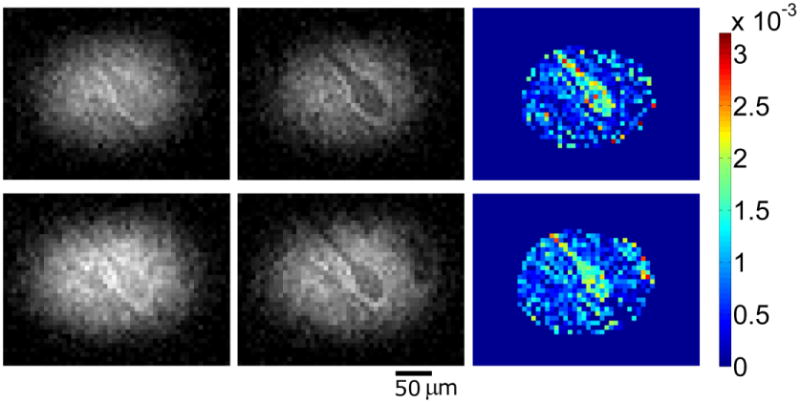
Left to right by row (b = 300s/mm2; b = 600s/mm2; ADC map). Differences in apparent diffusion between visible structures belonging to an α-motor neuron and the remainder of gray matter tissue in the ventral horn which surrounds it are clearly demonstrated in raw data and the calculated ADC maps. Most notably, the ADC maps illustrate high apparent diffusivity inside the perikaryon and proximal axon of a porcine α-motor neuron relative to adjacent tissues. Colorbar units = mm2/s. SNR values (region surrounding motor neuron) top row: (left) 12.8 and (middle) 10.8; bottom row: (left) 17.0 and (middle) 13.9. CNR (between cell interior and surrounding tissues) top row: (left) 0.38 and (middle) 3.39; bottom row: (left) 1.42 and (middle) 3.86.
Figure 5. Three-dimensional reconstruction of segmented MRM data illustrating that closely spaced processes from adjacent porcine α-motor neurons can be resolved.

Reconstruction of eight voxel-thick slices (8 × 6.25μm = 50μm) from one of our 3D porcine datasets. Individual frames were segmented into volumes containing visualized portions of α-motor neurons (filled, pink) and surrounding tissues of the ventral horn (transparent, green) automatically using commercial imaging software (Amira 3.1.1) Voxels located in the space occupied by noise were removed manually. Following segmentation, the individual frames were compiled, and the ‘surface gen’ tool was used to visualize the 3D reconstruction.
Discussion and Conclusions
To our knowledge, this work represents the first reported direct cellular imaging in human and pig tissue using MRM methods. More important than our ability to observe these structures with MRM—as other microscopy techniques offer greater specificity, higher resolution, and cheaper operating costs—is what it can reveal to us regarding the signal content of microscopic MR images. MRM makes it possible to distinguish microscopic tissue components based on MR properties and contrast specific to each component. It is the only microscopy method capable of generating MR contrast, and is thus the only means of studying the MR signal properties of tissues at the microscopic level. Such contrast is an inherent property of tissues and does not require the introduction of dyes, photobleaching, or free-radical inducing laser-based excitation methods common to other microscopy techniques. If such analyses are required, MRM may be employed prior to these techniques with no reduction in the quality of histology data. Because MRI shares the distinction with positron emission tomography (PET) and computed tomography (CT) as being one of the methods used on humans for diagnostic imaging in the clinic, increasing our understanding of MR image content—in particular, how and to what extent it is altered by pathology—offers a means to improve one of the most widely used and effective imaging tools for disease diagnosis ever devised. This is especially important for improving our capability to detect diseases in central nervous system tissues which are seldom taken for biopsy: particularly from asymptomatic individuals who may nonetheless be succumbing to the earliest stages of neurological disease. Also, unlike PET or CT, MRI does not employ ionizing radiation, which means that—in addition to being the least invasive human imaging modality—there are few limitations when taking MRM techniques developed in vitro and moving them to in vivo or even longitudinal studies. The ability to perform MRM is integral to understanding the microscopic origins of MR signal, which is in turn integral to the formulation of tissue models relevant to improving MRI's specificity. Furthermore, with the ability to make direct, quantitative comparison between MRM data and histology, MRM methods can serve as tools to validate and improve existing MR methods (Hansen et al., 2011). It is our hope that this work will help reconcile disparities between inferred diffusion behavior at microscopic resolutions and observations made in the macroscopic regime. Interestingly, cell bodies of human α-motor neurons appear hypointense in diffusion-weighted scans when compared to adjacent tissues suggesting higher diffusion rates in the perikarya. Quantitative ADC measurements later confirmed that the observed contrast was, in fact, diffusion based. Using modeling techniques, the works of Niendorf et al. attributed fast (80%) and slow (20%) diffusing components to the water signal, which correspond to the approximate volume fractions of the intra and extracellular compartments, respectively (Niendorf et al., 1996). Other modeling techniques employed ex vivo or in vitro phantoms (Latour et al., 1993, 1994) as well as synthetic geometries (Stanisz et al., 1997; Szafer et al., 1995) intended to describe different aspects of the water diffusion behavior observed in functional tissues. The reader is referred to the review article by Norris for a detailed synopsis of these methods (Norris, 2001). More recently, alternative interpretations to compartment-based contrast—e.g. restricted water motion near the cell membrane as proposed in the review by Le Bihan—have been offered (Le Bihan, 2007). Measurements taken in the L7 neuron of aplysia were reported to exhibit relatively low, hindered diffusion in the cytoplasm compared to higher ADCs in the nucleus (Hsu et al., 1997). Results in the current study show that certain subregions of the intracellular compartment in mammals—specifically the perikarya and large-diameter portions of neuronal processes from α-motor neurons—exhibit relatively high apparent diffusion. It is not yet clear why these cells possess such disparate diffusion characteristics, but such results should engender caution, especially when attempting to generalize measurements obtained in a specific animal model or tissue type to another organism or tissue type. Also, it is important to note that ADC values reported in the current study are expected to vary from measurements collected in ex vivo or in vivo conditions (Shepherd et al., 2009). Because it is unknown whether the diffusion characteristics will be represented across a multitude of tissue and cell types or to what degree fixation will alter these measurements, characterizing such properties in ex vivo models of living tissue will form the subject of future studies.
Cell bodies observed in the earliest, projection-style DWI images taken at anisotropic resolutions are similar in appearance and diffusion contrast to microscopic tissue tears as seen in panels C and F of figure 2. Such tears were identified in our light microscopy images and can be recognized by their saturated appearance resulting from the uninterrupted bright-field light source. Although not within the scope of the present study, additional quantitation will be needed to develop reliable methods for distinguishing between cell bodies and tissue tears in MR images taken at resolutions which prevent such distinctions to be made by comparative morphology.
Because processes were not visualized in our first series of diffusion experiments, questions remained as to whether these structures possessed diffusion characteristics similar or dissimilar to cell bodies. It was possible that these theretofore unseen processes may have exhibited higher diffusion signal due to increasing restrictive properties or equivalent diffusion signal which was rendered undetectable in the lower resolution images due to volume averaging. While our highest-resolution diffusion-weighted datasets and their corresponding ADC map clearly show higher apparent diffusivity in the visualized portions of these structures—those which are contiguous with and immediately adjacent to their cell bodies—it is important not to assume such contrast is maintained to the farthest reaches of the dendritic arbor. Although this is among the most appealing possibilities since it would allow for complete, diffusion-based compartmentation of intra- and extracellular tissue components, the MR signal characteristics exhibited by these narrowest of neuronal structures remains to be seen. This, of course, is a nontrivial issue because cell bodies themselves account for only a small volume fraction of total brain tissue: Sherwood et al. estimated the volume fraction taken up by glial and neuronal cell bodies in primary motor cortex to be approximately 15-16% in macaques and only 11-12% in humans (Sherwood et al., 2004). The remaining 84-89% of tissue is divided between the extracellular space (with a volume fraction of approximately 20%) (Nicholson, 2001), neuropil (dendrites, axons, synapses, and glial cell processes; 60-65% volume fraction), and blood vessels (3-4% volume fraction) (Norris, 2001; Pannese, 1994; Schlaug et al., 1993). Hence, these subcellular structures which are yet-to-be observed using MRM make up more than half of our cortical tissue by volume, and understanding their signal characteristics is of great importance to improving interpretation of macroscopic MR imaging employed in the clinic. Furthermore, some of these microscopic brain structures are subject to structural change on the timescale of minutes (Dailey and Smith, 1996) or even seconds (Fischer et al., 1998). This plasticity forms the basis of the brain's ability to perform functions such as adapting to environmental change and storing memories: i.e. it is the basis of many aspects of normal brain function. On the other hand, when disease alters or disrupts nervous tissue structure, function is affected, often with severe consequences for the patient. Examples of such effects are found among major neurological diseases with high mortality (e.g. Alzheimer's, amyotrophic lateral sclerosis, multiple sclerosis, and Parkinson's disease) but also occur in conditions such as stress, post-traumatic stress disorder (PTSD), depression, anxiety and schizophrenia. The development of imaging techniques that enable investigation of brain tissue microstructure is therefore important for expanding our understanding of the normal and diseased brain alike.
Neural processes in the current study possessed physical characteristics consistent with motor neuron morphology reported in both in vivo and in vitro experiments (Van Damme et al., 2007). While processes were visualized in human and porcine tissue using diffusion weighting, T2-weighted spin-echo data taken in pig samples resulted in images with higher contrast which aided in the delineation of structural boundaries. Intracellular T2 of the α-motor neurons appears to be shorter than that measured in the tissues surrounding them in the ventral horn. Short intracellular T2 was also observed in the only other animal cells studied to date (frog ova and Aplysia neurons). The short T2 has been presumed to arise due to interactions with neighboring spins in the relatively dense cytoplasmic environment, but elucidation of its exact origins requires further quantitative studies. This is especially interesting as a function of tissue perturbation where Aplyisa neurons undergoing a 20% hypotonic perturbation, were found to have T2 increased by 24% while D stayed constant: consistent with the behaviour expected from a perfect osmometer (Hsu et al., 1996). We plan to perform similar perturbation studies in mammalian tissues. Diffusion contrast was qualitatively similar between porcine and human tissue in that both cell bodies and processes manifested as hypointense signal regions as compared to the mixture of intracellular and extracellular tissues comprising the hyperintense signal regions. Our human dataset in which processes were visualized (Figure 2, C) exhibited reduced SNR as a result of increased transverse magnetization spoiling. Histological images confirm dysmorphic characteristics in the human cells most likely resulting from protracted postmortem periods prior to tissue fixation. This variable is particularly difficult to control and virtually impossible to eliminate in the case of human studies as its length is dictated by cultural practices surrounding death. However, we have previously obtained pathological human brain tissue with relatively short postmortem intervals (Shepherd et al., 2003), and thus this issue may be addressed in those cases.
In conclusion, we have performed the first direct MR microscopy of individual neuronal cell bodies and their proximal processes in human and porcine tissue. ADC maps calculated from scans in which porcine neurons were resolved show higher rates of apparent water diffusion in the α-motor cell bodies and their adjacent processes relative to the tissue which surrounds them. Thus the origins of MR signals at resolutions needed to visualize human cells may be explored. Methods that will allow for future quantitative studies on living tissues are currently in development. Once equipped with sufficient quantitative information, realistic models of MR signals in tissue may be developed that promise to aid in the interpretation and improve the diagnostic potential of clinical MRI.
Supplementary Material
Acknowledgments
We would like to thank members of the AMRIS staff at the University of Florida's McKnight Brain Institute for technical support. We would like to thank Dr. Gerry Shaw for his help in obtaining porcine tissue. We would also like to thank ScienceCare for their help in obtaining human tissue. Funded by the NIH (1R01EB012874), the NSF through the National High Magnetic Field Laboratory, and the Danish National Research Foundation (95093538-2458, project 100297).
Footnotes
Author Contributions: JF, BH, and SB wrote the paper, JF and SP performed MR acquisition, JF performed histology, BH performed quantitative data analysis, SP performed pulse sequence programming, CHL performed segmentation of 3D data, MF and FV performed micro surface-coil development and application, SB and PV provided MRM expertise, funding, and performed manuscript editing with JF, BH, and SP.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguayo JB, Blackband SJ, Schoeniger J, Mattingly MA, Hintermann M. Nuclear magnetic resonance imaging of a single cell. Nature. 1986;322:190–191. doi: 10.1038/322190a0. [DOI] [PubMed] [Google Scholar]
- Badilita V, Kratt K, Baxan N, Mohmmadzadeh M, Burger T, Weber H, Elverfeldt Dv, Hennig J, Korvink JG, Wallrabe U. On-chip three dimensional microcoils for MRI at the microscale. Lab Chip. 2010;10:1387–1390. doi: 10.1039/c000840k. [DOI] [PubMed] [Google Scholar]
- Benveniste H, Blackband SJ. Translational neuroscience and MR microscopy. Lancet Neurol. 2005;5(6):536–544. doi: 10.1016/S1474-4422(06)70472-0. [DOI] [PubMed] [Google Scholar]
- Callaghan PT. Principles of Nuclear Magnetic Resonance Microscopy. New York: Oxford University Press; 1991. [Google Scholar]
- Cho ZH, Ahn CB, Juh SC, Jo JM, Friedenberg RM, Fraser SE, Jacobs RE. Recent progress in NMR microscopy towards cellular imaging. Phil Trans R Soc Lond. 1990;A333(1632):469–475. [Google Scholar]
- Ciobanu L, Webb AG, Pennington CH. Magnetic resonance imaging of biological cells. Prog Nucl Magn Reson Spectrosc. 2003;42:69–93. [Google Scholar]
- Dailey ME, Smith SJ. The dynamics of dendritic structure in developing hippocampal slices. J Neurosci. 1996;16(9):2983–2994. doi: 10.1523/JNEUROSCI.16-09-02983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccles CD, Callaghan PT. High resolution imaging: the NMR microscope. J Magn Reson. 1986;68:393–398. [Google Scholar]
- Ehrmann K, Saillen N, Vincent F, Stettler M, Jordan M, Wurm FM, Besse PA, Popovic R. Microfabricated solenoids and Helmholtz coils for NMR spectroscopy of mammalian cells. Lab Chip. 2007;7(3):373–380. doi: 10.1039/b614044k. [DOI] [PubMed] [Google Scholar]
- Fischer M, Kaech S, Knutti D, Matus A. Rapid actin-based plasticity in dendritic spines. Neuron. 1998;20(5):847–854. doi: 10.1016/s0896-6273(00)80467-5. [DOI] [PubMed] [Google Scholar]
- Flint JJ, Hansen B, Fey M, Schmidig D, King MA, Vestergaard-Poulsen P, Blackband SJ. Cellular-level diffusion tensor microscopy and fiber tracking in mammalian nervous tissue with direct histological correlation. Neuroimage. 2010;52(2):556–561. doi: 10.1016/j.neuroimage.2010.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint JJ, Lee CH, Hansen B, Fey M, Schmidig D, Bui JD, King MA, Vestergaard-Poulsen P, Blackband SJ. Magnetic resonance microscopy of mammalian neurons. Neuroimage. 2009;46(4):1037–1040. doi: 10.1016/j.neuroimage.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen B, Flint JJ, Heon-Lee C, Fey M, Vincent F, King MA, Vestergaard-Poulsen P, Blackband SJ. Diffusion tensor microscopy in human nervous tissue with quantitative correlation based on direct histological comparison. Neuroimage. 2011;57(4):1458–1465. doi: 10.1016/j.neuroimage.2011.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu EW, Aiken NR, Blackband SJ. Nuclear magnetic resonance microscopy of single neurons under hypotonic perturbation. Am J Physiol. 1996;271(6 Pt 1):C1895–C1900. doi: 10.1152/ajpcell.1996.271.6.C1895. [DOI] [PubMed] [Google Scholar]
- Hsu EW, Aiken NR, Blackband SJ. A study of diffusion isotropy in single neurons by using NMR microscopy. Magn Reson Med. 1997;37(4):624–627. doi: 10.1002/mrm.1910370425. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Thompson MB, Gewalt SL, Hayes CE. Nuclear magnetic resonance imaging at microscopic resolution. J Magn Reson. 1986;68(1):129–137. [Google Scholar]
- Latour LL, Mitra PP, Kleinberg RL, Sotak CH. Time-dependent diffusion coefficient of fluids in porous media as a probe of surface-to-volume ratio. J Magn Reson. 1993;101A(3):342–346. [Google Scholar]
- Latour LL, Svoboda K, Mitra PP, Sotak CH. Time-Dependent Diffusion of Water in a Biological Model System. Proc Natl Acad Sci USA. 1994;91(4):1229–1233. doi: 10.1073/pnas.91.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauterbur PC. Image formation by induced local interactions: examples employing nuclear magnetic resonance. Nature. 1973;242:190–191. [PubMed] [Google Scholar]
- Le Bihan D. The ‘wet mind’: water and functional neuroimaging. Phys Med Biol. 2007;52(7):R57–R90. doi: 10.1088/0031-9155/52/7/R02. [DOI] [PubMed] [Google Scholar]
- Mansfield P, Morris PG. NMR imaging in biomedicine Volume 2 of Advances in Magnetic Resonance. New York: Academic Press; 1982. [Google Scholar]
- Massin C, Vincent F, Homsy A, Ehrmann K, Boero G, Besse PA, Daridon A, Verpoorte E, de Rooij NF, Popovic RS. Planar microcoil-based microfluidic NRM probes. J Magn Reson. 2003;164(2):242–255. doi: 10.1016/s1090-7807(03)00151-4. [DOI] [PubMed] [Google Scholar]
- Meadowcroft MD, Zhang S, Liu W, Park BS, Connor JR, Collins CM, Smith MB, Yang QX. Direct magnetic resonance imaging of histological tissue samples at 3.0 T. Magn Reson Med. 2007;57(5):835–841. doi: 10.1002/mrm.21213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley ME, Kucharczyk J, Mintorovitch J, Cohen Y, Kurhanewicz J, Derugin N, Asgari H, Norman D. Diffusion-weighted MR imaging of acute stroke: correlation with T2-weighted and magnetic susceptibility-enhanced MR imaging in cats. AJNR. 1990;11(3):423–429. [PMC free article] [PubMed] [Google Scholar]
- Nabuurs RJ, Hegeman I, Natté R, van Duinen SG, van Buchem MA, van der Weerd L, Webb AG. High-field MRI of single histological slices using an inductively coupled self-resonant microcoil: application of ex vivo samples of patients with Alzheimer's disease. NMR Biomed. 2011;24(4):351–357. doi: 10.1002/nbm.1598. [DOI] [PubMed] [Google Scholar]
- Nicholson C. Diffusion and related transport mechanisms in brain tissue. Rep Prog Phys. 2001;64(7):815–884. [Google Scholar]
- Niendorf T, Dijkhuizen RM, Norris DG, van Lookeren Campagne M, Nicolay K. Biexponential diffusion attenuation in various states of brain tissue: implications for diffusion-weighted imaging. Magn Reson Med. 1996;36(6):847–857. doi: 10.1002/mrm.1910360607. [DOI] [PubMed] [Google Scholar]
- Norris DG. The effects of microscopic tissue parameters on the diffusion weighted magnetic resonance imaging experiment. NMR Biomed. 2001;14(2):77–93. doi: 10.1002/nbm.682. [DOI] [PubMed] [Google Scholar]
- Pannese E. Neurocytology: Fine Structure of Neurons, Nerve Processes, and Neuroglial Cells. New Youk: Thieme Medical Publisher, Inc.; 1994. [Google Scholar]
- Quadery FA, Okamoto K. Diffusion-weighted MRI of haemangioblastomas and other cerebellar tumors. Neuroradiol. 2003;45(4):212–219. doi: 10.1007/s00234-003-0951-y. [DOI] [PubMed] [Google Scholar]
- Schlaug G, Armstrong E, Schleicher A, Zilles K. Layer V pyramidal cells in the adult human cingulate cortex. A quantitative Golgi study. Anat Embryol. 1993;187(6):515–522. doi: 10.1007/BF00214429. [DOI] [PubMed] [Google Scholar]
- Shepherd TM, Thelwall PE, Stanisz GJ, Blackband SJ. Aldehyde fixative solutions alter the water relaxation and diffusion properties of nervous tissue. Magn Reson Med. 2009;62(1):26–34. doi: 10.1002/mrm.21977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd TM, Wirth ED, 3rd, Thelwall PE, Chen HX, Roper SN, Blackband SJ. Water diffusion measurements in perfused human hippocampal slices undergoing tonicity changes. Magn Reson Med. 2003;49(5):856–863. doi: 10.1002/mrm.10456. [DOI] [PubMed] [Google Scholar]
- Sherwood CC, Holloway RL, Erwin JM, Schleicher A, Zilles K, Hof PR. Cortical orofacial motor representation in Old World monkeys, great apes, and humans. I. Quantitative analysis of cytoarchitecture. Brain Behav Evol. 2004;63(2):61–81. doi: 10.1159/000075672. [DOI] [PubMed] [Google Scholar]
- Stanely C, Bielschowsky M. A method for intra-vital staining with silver ammonium oxide solution. Journal für Psychologie und Neurologie. 1925;31:301–304. [Google Scholar]
- Stanisz GJ, Szafer A, Wright GA, Henkelman RM. An analytical model of restricted diffusion in bovine optic nerve. Magn Reson Med. 1997;37(1):103–111. doi: 10.1002/mrm.1910370115. [DOI] [PubMed] [Google Scholar]
- Szafer A, Zhong J, Gore JC. Theoretical model for water diffusion in tissues. Magn Reson Med. 1995;33(5):697–712. doi: 10.1002/mrm.1910330516. [DOI] [PubMed] [Google Scholar]
- Tyszka JM, Fraser SE, Jacobs RE. Magnetic resonance microscopy: recent advances and applications. Curr Opin Biotechnol. 2005;16(1):93–99. doi: 10.1016/j.copbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Van Damme P, Bogaert E, Dewil M, Hersmus N, Kiraly D, Scheveneels W, Bockx I, Braeken D, Verpoorten N, Verhoeven K, Timmerman V, Herijgers P, Callewaert G, Carmeliet P, Van Den Bosch L, Robberecht W. Astrocytes regulate GluR2 expression in motor neurons and their vulnerability to excitotoxicity. PNAS. 2007;104(37):14825–14830. doi: 10.1073/pnas.0705046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Baxan N, Paul D, Maclaren J, Schmidig D, Mohammadzadeh M, Hennig J, Elverfeldt Dv. Microcoil-based MRI: feasibility study and cell culture applications using a conventional animal system. MAGMA. 2011;24(3):137–145. doi: 10.1007/s10334-011-0244-0. [DOI] [PubMed] [Google Scholar]
- Weiger M, Schmidig D, Denoth S, Massin C, Vincent F, Schenkel M, Fey M. NMR-microscopy with isotropic resolution of 3.0 μm using dedicated hardware and optimized methods. Concepts Magn Reson Part B. 2008;33B(2):84–93. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


