Abstract
The cbb3-type cytochrome c oxidases (cbb3-Cox) constitute the second most abundant cytochrome c oxidase (Cox) group after the mitochondrial-like aa3-type Cox. They are present in bacteria only, and are considered to represent a primordial innovation in the domain of Eubacteria due to their phylogenetic distribution and their similarity to nitric oxide (NO) reductases. They are crucial for the onset of many anaerobic biological processes, such as anoxygenic photosynthesis or nitrogen fixation. In addition, they are prevalent in many pathogenic bacteria, and important for colonizing low oxygen tissues. Studies related to cbb3-Cox provide a fascinating paradigm for the biogenesis of sophisticated oligomeric membrane proteins. Complex subunit maturation and assembly machineries, producing the c-type cytochromes and the binuclear heme b3-CuB center, have to be coordinated precisely both temporally and spatially to yield a functional cbb3-Cox enzyme. In this review we summarize our current knowledge on the structure, regulation and assembly of cbb3-Cox, and provide a highly tentative model for cbb3-Cox assembly and formation of its heme b3-CuB binuclear center.
Keywords: cbb3-type cytochrome c oxidase, co- or post-translational cofactor insertion, assembly of membrane proteins, copper acquisition, photosynthesis and respiration, energy transduction
1. Introduction
1.1 Generalities
The emerging ease of whole genome sequencing approaches has revealed the existence of multiple terminal oxidases in bacteria, which allow them to utilize efficiently varying oxygen (O2) concentrations in their environments. Most of the terminal oxidases belong to the heme-Cu: O2 reductase superfamily (heme-Cu: O2 reductases); they reduce O2 to H2O and couple electron transfer with vectorial proton translocation across the membrane [1]. Members of this superfamily are present in eukarya, archaea and eubacteria. They are thought to be of monophyletic origin, possibly related to nitric oxide reductases (NOR) [2]. NOR catalyze reduction of NO to N2O during denitrification. Although they do not reduce O2 and do not generate a proton gradient [3], their structural organization is similar to heme-Cu: O2 reductases.
The heme-Cu: O2 reductases can be divided into different subgroups in respect to their electron donors: those that use a c-type cytochrome (i. e., cytochrome c oxidase, Cox) and those that use a quinol (i. e., quinol oxidase, Qox). Cox are universally conserved oligomeric membrane proteins that terminate the respiratory chains of aerobic and facultative aerobic organisms. The structure and function of Cox enzymes have been studied intensely because of their central role in energy metabolism. Given the complexity of these multi-subunit, multi-cofactor enzymes, their assembly also attracted much attention. Indeed, assembly defects in the human Cox are major causes of mitochondrial disorders, and are crucial for neurodegenerative diseases, cancer and ageing [4–7]. Among the Cox enzymes, the aa3-type Cox (aa3-Cox) of mitochondria and many bacterial species are highly abundant and the best-studied group. However, the heme-Cu: O2 reductases represent a diverse ensemble of enzymes with significantly different subunit compositions and cofactors [8]. In this review, we focus on the structure, subunit maturation and assembly of the cbb3-type Cox (cbb3-Cox) that represent the second most abundant group after the aa3-Cox. Most of the studies on cbb3-Cox biogenesis were performed with the facultative phototrophic α-proteobacterium Rhodobacter capsulatus. This species is used as a model organism for these studies as it contains only one heme-Cu:O2 reductase, cbb3-Cox. R. capsulatus is still capable of aerobic respiration in the absence of it because it also contains an unrelated terminal oxidase (bd-type quinol oxidase) (Fig. 1) [9, 10].
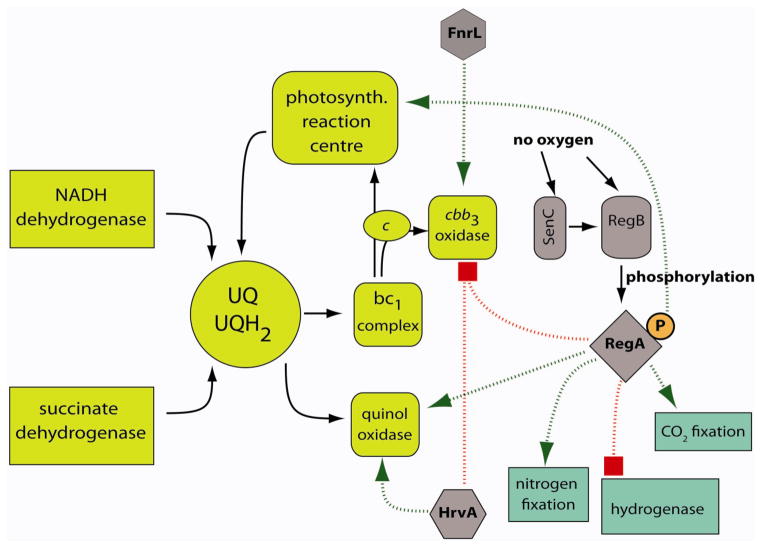
Figure 1.
The respiratory chain of R. capsulatus and its regulation in response to oxygen availability. The protein complexes and electron carrier of the respiratory chain are shown in yellow. UQ and UQH2 correspond to the oxidized and reduced form of quinones. Soluble cytochrome c2 and the membrane-bound cytochrome cy (indicated as c in the figure) are electron donors for the photosynthetic reaction center and cbb3-Cox. Metabolic processes which also consume redox equivalents are shown in blue. Regulatory proteins are shown in grey. RegB is a sensor kinase that phosphorylates the response regulator RegA under low oxygen concentrations. Phosphorylated RegA (RegA-P) inhibits (shown as the red square) expression of ccoNOQP (cbb3-Cox) and hupSLC ([NiFe] hydrogenase), but stimulates the expression of the bd-type quinol oxidase and of genes required for photosynthesis. In addition, RegA-P stimulates (shown as an arrow) the expression of nifHDK (nitrogenase) and the cbb1 and cbb2 operons (CO2 fixation) both directly and indirectly [154]. Under microaerobic and anaerobic conditions HvrA further represses ccoNOQP expression but stimulates bd-type quinol oxidase expression. Finally, FnrL probably activates ccoNOQP expression in the transition phase between aerobic and anaerobic conditions. R. capsulatus SenC and its R. sphaeroides homologue PrrC were suggested to be involved in oxygen-dependent expression of ccoNOQP and photosynthetic genes. FixLJ-K regulatory system discussed in the text is not shown in the figure.
1.2 Classification of heme-Cu: O2 reductases
Members of heme-Cu: O2 reductases are diverse in terms of their subunit composition, heme cofactor content, electron donor, and O2 affinity (Table 1) [8, 11, 12]. All members have a conserved core subunit (Subunit I), containing at least 12 transmembrane helices. This subunit contains a low spin heme (of a- or b-type), a binuclear metal center composed of a high spin heme (of a-, o-, or b-type heme, referred to as a3, o3 or b3)-iron, and a Cu atom (CuB), as well as a tyrosine residue covalently linked to a histidine ligand of CuB [1, 8]. Besides subunit I, which is the catalytic heart where O2 is reduced to H2O, heme-Cu:O2 reductases have at least two other core subunits: Subunit II is the primary electron acceptor, and in some cases, it harbors extra cofactors like a binuclear Cu center (CuA), or is a c-type cytochrome. Subunit III in many cases does not contain any cofactor except in cbb3-Cox where it is a diheme c-type cytochrome (Table 1) [1]. Most bacterial Cox are composed of these three core subunits, but mitochondrial Cox are more complex with 13 subunits of which only three (subunits I, II, and III) are encoded in the mitochondrial genome. An additional fourth subunit with a single transmembrane helix is also present in some bacterial Cox. However, this subunit is unlike any of the ten nuclear encoded mitochondrial subunits [13].
Table 1.
Characteristic features of different Cox and NOR enzymes
| cbb3-Cox | NOR | ba3-Cox | aa3-Cox | |
|---|---|---|---|---|
| Subunit composition |
|
|
|
|
| Catalytic binuclear centre |
|
|
|
|
| Enzymatic Properties | Km~ 7 nM (for O2) Km~ 12 mM (for Km NO) | Km ~ 0,1μM | Km 0.7 - 2 μM | |
| Proton pathway | ‘K’ pathway | Protons from periplasm | K pathway | K and D pathways |
catalytic subunit of the enzyme
A classification, based on common features of the core subunits, and key residues in proton transfer pathways, defines three (A, B and C) types of heme-Cu: O2 reductases (Table 1) (Fig. 2) [1]. Mitochondrial- aa3-Cox (Type A), ba3-Cox of Thermus thermophilus (Type B) and cbb3-Cox of Rhodobacter capsulatus (Type C) are the most representative members of these types.
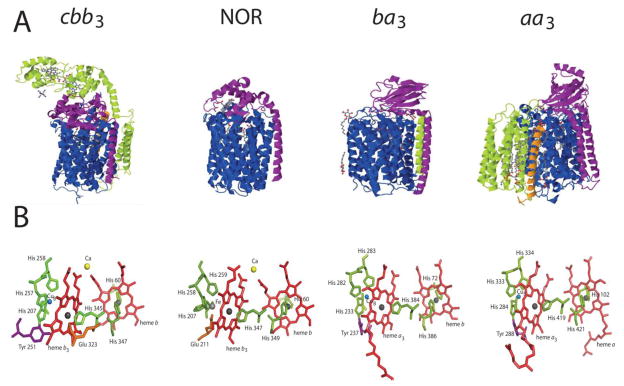
Figure 2.
A. 3D structures of different types of heme-Cu:O2 reductases and NO reductase: Catalytic subunit I blue (cbb3-Cox CcoN), subunit II (cbb3-Cox CcoO) – magenta, subunit III (cbb3-Cox CcoP) – green, additional subunit –orange. B. Architecture of the catalytic binuclear center of the different types of heme-Cu:O2 reductases and NO reductase. Heme – red, His – green, Tyr – violet, Glu – orange. Spheres: copper – blue, heme iron - dark grey, non-heme iron – light grey, calcium – yellow. The structures depicted are taken from protein database (PDB) entries PDB ID: 3MK7 (cbb3-Cox) [14], PDB ID: 3O0R (NOR ) [60], PDB ID: 1M56 (aa3-Cox) [19], PDB ID: 1EHK (ba3-Cox) [17] using Jmol software (http://www.jmol.org/).
Available three-dimensional (3D) structures of the heme-Cu:O2 reductase family members (i. e., the aa3-Cox from P. denitrificans, R. sphaeroides and bovine heart mitochondria; ba3-Cox from T. thermophilus, bo3-Qox from E. coli and the cbb3-Cox from Pseudomonas stutzerii) (Fig. 2A) [14–19] reveal a remarkably conserved Cu and heme cofactors arrangement in these enzymes. Subunit I of type A enzymes contain two proton-transfer pathways referred to as K- and D- channels (Table 1). The four electrons required for O2 reduction at the binuclear heme-CuB center are conveyed sequentially via a non-covalently attached low spin heme, which itself receives electrons from the CuA center in subunit II [15, 19]. Type B enzymes are present only in bacteria and archaea, but not in eukaryotes, and constitute the least abundant group [20]. The catalytic center of type B enzymes is similar to that of type A, except that they lack the D- channel for proton transfer [21]. The prototypes of type C are the cbb3-Cox, which are present only in bacteria, and considered to represent the most distant members of heme-Cu: O2 reductases. They also lack the D-channel for proton transfer [22–24], their subunits II and III are c-type cytochromes (Table 1), and they exhibit higher O2 affinity as compared with other types of Cox enzymes.
2. The cbb3-Cox
2.1 Distribution of cbb3-Cox in bacteria and its role in pathogenesis
The cbb3-Cox enzymes are common to proteobacteria and also found in the cytophaga, flexibacter and bacteriodes (CFB) group [12]. They were first described in the facultative symbiotic N2-fixing rhizobiacaea [25, 26]. In B. japonicum expression of cbb3-Cox is required for symbiotic N2 fixation under very low O2 conditions in soybean root nodules [26]. Due to their importance for symbiotic N2 fixation, the four structural genes of cbb3-Cox were initially termed fixNOQP [26]. Subsequent studies identified these genes and their products also in non-symbiotic bacteria such as R. capsulatus, and referred to them as ccoNOQP [27–29]. A recent bioinformatics study, based on the occurrence of ccoN and ccoO genes, identified the ccoNOQP cluster in all bacterial species, except the Thermotogales, Deinococcales and Firmicutes [12]. So far, no cbb3-Cox coding sequences were identified in archaea [30]. Interestingly, ccoNO genes were found recently in the last mitochondrial ancestor bacterium Midichloria mitochondrii [31].
The cbb3-Cox is present in many microaerophilic pathogenic bacteria and implicated in host colonization (Table 2). In some species including Campylobacter jejuni [32], Helicobacter pylori [33, 34], Neisseria meningitidis and Neisseria gonorrhoea [35] cbb3-Cox is the sole Cox, and it is suggested that its high O2 affinity allows these pathogens to colonize low O2 containing tissues [11]. Correlations between cbb3-Cox and host colonization have so far been examined mainly in C. jejuni, which is a non-fermenting microaerophile that colonizes the small and large intestines of humans and animals [36], causing acute gastroenteritis in humans. C. jejuni has a branched aerobic electron transport chain, with a cbb3-Cox of high O2 affinity (Km = 40 nM) and a quinol oxidase (CioAB) of low (Km = 800 nM) O2 affinity. Gene expression studies have shown that CioAB was mainly expressed at high O2 conditions [37], while cbb3-Cox was induced significantly in C. jejuni cells colonizing the chick caecum, which is a low O2 environment [38]. C. jejuni mutants lacking cbb3-Cox were unable to colonize the chick caecum, whereas mutants lacking CioAB were slightly impaired in colonization [38]. The ceacum being primarily colonized by obligate anaerobic bacteria, C. jejuni is likely to be exposed to anaerobic conditions transiently. Probably, it tolerates anaerobiosis by using alternative electron acceptors like nitrate, nitrite, DMSO or TMAO [39], while its cbb3-Cox would keep cellular O2 concentrations low to protect its crucial O2-labile enzymes and allow establishment of anaerobic respiration [40, 41]. Indeed, C. jejuni mutants lacking nitrate or nitrite reductases are significantly attenuated for colonizing the chick caecum [39]. Thus, anaerobic respiration together with cbb3-Cox seems to allow C. jejuni to adapt successfully to environmental changes at their ecological niches.
Table 2.
Various terminal oxidases and NOR in some pathogenic bacteria.
| cbb3-Cox | NOR | aa3-Cox | bd-type Qox | |
|---|---|---|---|---|
| Campylobacter jejuni | ✓ | ✓ | ✓ | |
| Helicobacter pylori J99 | ✓ | |||
| Neisseria gonorrhoea | ✓ | ✓ | ||
| Neisseria meningitidis | ✓ | ✓ | ||
| Pseudomonas aeruginosa PAO1 | ✓ | ✓ | ✓ | ✓ |
| Vibrio cholera | ✓ | ✓ |
A cbb3-Cox is also present in Brucella suis, which is an intracellular Gram-negative pathogen and the causative agent of brucellosis [42]. Under microaerobic conditions in liquid cultures, B. suis expresses mainly cbb3-Cox. However inside macrophages, it relies almost exclusively on a bd-type quinol oxidase [42], which apparently has a higher O2 affinity than cbb3-Cox in this species. In H. pylori which is responsible for stomach ulcers, cbb3-Cox is the sole respiratory terminal oxidase recognizable in its genome (Table 2). Transposon insertion mutants in ccoN, ccoO and ccoP genes of this species were obtained under microaerobiosis, indicating that cbb3-Cox is not essential under these conditions [43]. Whether H. pylori needs cbb3-Cox to colonize the intestine is unknown. The fact that H. pylori mutants that lack fumarate reductase fail to colonize the mouse stomach [44] indicates that anaerobic respiration is crucial for virulence of this and other microaerophilic pathogens.
Although not yet studied extensively, the role of cbb3-Cox during pathogenesis might provide a potential antibacterial target for therapeutic interventions. Chemicals that interfere with cbb3-Cox might act as powerful specific antibiotics with minimal side effects due to the absence of this type of Cox in mammals [45].
2.2 Regulation of cbb3-Cox expression
In R. capsulatus and most other bacteria the steady-state concentration and activity of cbb3-Cox is dependent on the environmental O2 concentrations. This activity is high under microaerobic, low under fully aerobic and even lower under fully anaerobic growth conditions [46]. Several components, including RegA (or PrrA), RegB (or PrrB), FnrL, HvrA, and FixLJ-K that respond to different O2 and redox conditions, are thought to coordinately regulate this process (Fig. 1) [47]. RegA and RegB constitute a global two-component regulatory system that controls the expression of many genes involved in energy metabolism, including cbb3-Cox, bd-quinol oxidase, and various genes related to photosynthesis, hydrogen utilization, nitrogen fixation and carbon assimilation [46, 48–51]. RegA is a response regulator with a conserved helix-turn-helix DNA binding motif, whereas RegB is a sensor kinase proposed to contain a conserved region that monitors redox changes inside the cells. Under microaerobic and aerobic conditions the dephosphorylated form of RegA activates cbb3-Cox expression. Conversely, under anaerobic conditions RegA is phosphorylated by the sensor kinase RegB, and phosphorylated RegA represses cbb3-Cox and enhances photosynthesis genes expression [47]. In R. sphaeroides, it was proposed that cbb3-Cox monitors electron flow through its CcoN subunit [52]. When this electron flow is high, it sends an inhibitory signal to PrrA and PrrB (homologues of RegA and RegB) to repress photosynthesis genes expression [52]. Thus, in this species abolishing cbb3-Cox results in activation of photosynthetic genes under aerobic conditions [53].
Besides RegA and RegB, some other genes also regulate cbb3-Cox expression. FnrL-like genes are found upstream of ccoNOQP in R. capsulatus [54], and upstream of both ccoNOQP and ccoGHIS (involved in the assembly of cbb3-Cox, see below) in other organisms including R. sphaeroides, P. denitrificans, B. japonicum. Studies performed in E. coli established that FnrL controls switching from aerobic to anaerobic respiration [55]. Inactivation of FnrL in R. capsulatus decreases cbb3-Cox activity by about 80% under anaerobic growth conditions, suggesting that it acts as an activator in the absence of O2 [47]. This effect is counter balanced by the trans-acting regulatory protein HvrA, which functions as a repressor of ccoNOQP under microaerobic and anaerobic conditions (Fig. 1) [47]. Another major set of regulators of microaerophilic growth as well as photosynthesis is the FixLJ-K regulatory system. FixL is a histidine kinase that can bind O2 via a heme b group, whereas FixJ is a transcription factor that undergoes phosphorylation by FixL under microaerophilic conditions. Phosphorylated FixL then activates the expression of FixK, a downstream transcription regulator that binds specifically upstream of ccoN gene to activate its expression [56–58]. The FixLJ-K system regulating expression of cbb3-Cox is also found in many other bacteria, including B. japonicum, Caulobacter crescentus and Novosphingobium aromativorans [59].
2.3 Subunit composition and structure of cbb3-Cox
Usually, cbb3-Cox enzymes are composed of CcoN (subunit I), CcoO (subunit II), CcoQ (subunit IV) and CcoP (subunit III) proteins (Table 1) [12]. Excitingly, the 3D structure of Pseudomonas stutzerii cbb3-Cox was solved recently at a resolution of 3.2 Å [14]. The structure revealed that the overall shape and size of the membrane-embedded part of cbb3-Cox is similar to those of R. sphaeroides aa3-Cox [19] and T. thermophilus ba3-Cox, [17] whereas its periplasmic part, mainly formed by the heme containing domains of CcoO and CcoP, is more surface-exposed than in other Cox enzymes (Fig. 2A). Moreover, highlighting the close evolutionary relationship between the cbb3-Cox and NOR enzymes, the 3D structure of CcoN was found very similar to that of Pseudomonas aeruginosa NOR [60], even though the amino acid sequence conservation between these two proteins is below 40%.
CcoN forms a “clamshell”-like structure, with its the amino (N-) and carboxyl (C-) termini located close to each other on the cytoplasmic side. CcoN also has two β strands that form a hairpin loop between the helices V and VI. Two calcium atoms with a proposed stabilizing role are found in the structure (Fig. 2B). One of these atoms interacts directly with the heme b3-CuB catalytic center, while the second one is located between the loops IV and V at the edge of the structure. The clamshell structure surrounds the catalytic center located near the outer membrane surface and consists of a low spin heme b, a high spin heme b3 and a Cu (CuB) atom. The open edge of the clamshell is close to heme b, whereas heme b3 and CuB are at its distal end. Like in NOR, the hemes b and b3 are linked together by a calcium atom, which is coordinated to the carboxyl groups of pyrrole D rings of both hemes. Like other Cox enzymes, three histidine residues coordinate the CuB atom, and one of them is covalently ligated to a tyrosine residue on helix VII (Fig. 2B). This linkage is absent in NOR, and in aa3-Cox the ligating tyrosine residue is located on helix VI (Table 1) [19]. Like the type B enzymes, cbb3-Cox contains only one proton pathway that is positioned similar to the K-channel in the type A enzymes. In cbb3-Cox, hydrophobic residues block the equivalent of the D-channel, and moreover, none of the residues of the proposed proton pathway of cbb3-Cox (Ser240, Tyr223, His243, Tyr317, Thr215, Tyr251 of CcoN and Glu49 of CcoP) is conserved in type A heme-Cu:O2 reductases. However, Tyr223 and Tyr317 can be found in the predicted proton channel of type B enzyme ba3-Cox of T. thermophilus [17]. Similar proton pathways have not been identified in NOR which does not pump protons, but it is noteworthy that some cbb3-Cox also exhibit low NOR activity (Table 1) [61].
CcoO is a mono-heme c-type cytochrome, and has an N-terminally located transmembrane helix as an anchor (Table 1). It is thought to convey electrons to heme b of CcoN, and makes strong contacts with the CcoN α-helices. In particular, CcoO contacts the calcium atom that coordinates heme b and heme b3 at the catalytic center via its Ser102 residue. Of the two cavities that are visible on the 3D structure of cbb3-Cox, one is close to the periplasmic face of the membrane, between CcoO and CcoN. This cavity is located at a position equivalent to the end of the D-channel of type A enzymes, and is proposed to provide an exit path for protons and water molecules from the catalytic site to the periplasm. The second cavity is membrane-embedded and connected by narrow and hydrophobic channels to the catalytic site, possibly providing an O2 access pathway. An equivalent hydrophobic channel is also present in the NOR structure, and is suggested to allow access of NO to the catalytic site of this enzyme [60].
CcoP is a di-heme c-type cytochrome with both of its heme-groups solvent exposed. In the case of P. stutzerii, CcoP has two transmembrane helices connected via a long linker that makes multiple contacts to the cytoplasmic part of CcoN (Table 1). In some other organisms, including R. capsulatus, CcoP probably contains only one transmembrane helix as its anchor. The distal heme group of CcoP is thought to accept electrons from a soluble or membrane bound electron donor (e. g., cytochrome c2 or cytochrome cy in R. capsulatus) and transfers them to the proximal heme of this subunit. This latter heme then conveys electrons to the heme group of CcoO, which transfers them to the catalytic center in CcoN.
CcoQ is a small subunit formed by a single transmembrane helix, and is not present in all cbb3-Cox. It does not contain any cofactor, and its elimination does not completely abolish cbb3-Cox activity. CcoQ is absent in P. stutzerii 3D structure, which instead contains an unassigned α-helix, located close to helices IX and XI of CcoN. The location of CcoQ in R. capsulatus cbb3-Cox is unknown, but it can be cross-linked to CcoP, suggesting that it is associated with this subunit [62].
3. Assembly of the cbb3-Cox
In general, Cox assembly is an intrinsically complex process because this oligomeric enzyme is membrane embedded and contains multiple cofactors [63]. Membrane insertion and maturation of individual subunits, insertion of cofactors, and assembly of cognate partners have to be coordinated to produce an active enzyme [63, 64]. In eukaryotes, aa3-Cox assembly involves more than 30 factors, which provide timely availability of mitochondrially-encoded subunits, or which associate transiently with various assembly intermediates [13, 65]. Assembly of bacterial aa3-Cox is possibly less complex, but still relies on specific components that mediate heme and Cu insertion into the subunits. Assembly of cbb3-Cox is particularly challenging because the maturation of CcoO and CcoP require the c-type cytochrome maturation (Ccm) machinery, whereas CcoN relies on heme b and Cu atom insertion processes. A set of genes, ccoGHIS are located immediately downstream of ccoNOQP operon in most cbb3-Cox containing bacteria [54, 66–68]. The roles of ccoGHIS products in the assembly of cbb3-Cox are described below.
3.1 An assembly pathway for the subunits of cbb3-Cox and role of CcoH
Assembly of cbb3-Cox has been studied in R. capsulatus membranes using blue-native polyacrylamide gel electrophoresis (BN-PAGE) analyses (Fig. 3A) [62, 67, 69]. This technique allows identification of membrane protein complexes in their native state, and is particularly suitable for examining abundant and stable respiratory chain complexes [70]. In R. capsulatus membranes, BN-PAGE, activity staining and immunodetection analyses showed that cbb3-Cox forms an active complex of 230 kDa, which contains all four (CcoNOQP) structural subunits [67]. However, BN-PAGE does not necessarily reflect the correct molecular mass of a protein complex, as binding of lipids, detergent and Coomassie blue can interfere with the migration on BN-PAGE [71]. Indeed R. capsulatus cbb3-Cox purified after detergent-solubilization migrates as an active 160 kDa complex on BN-PAGE (Pawlik et al., unpublished data).
Figure 3.
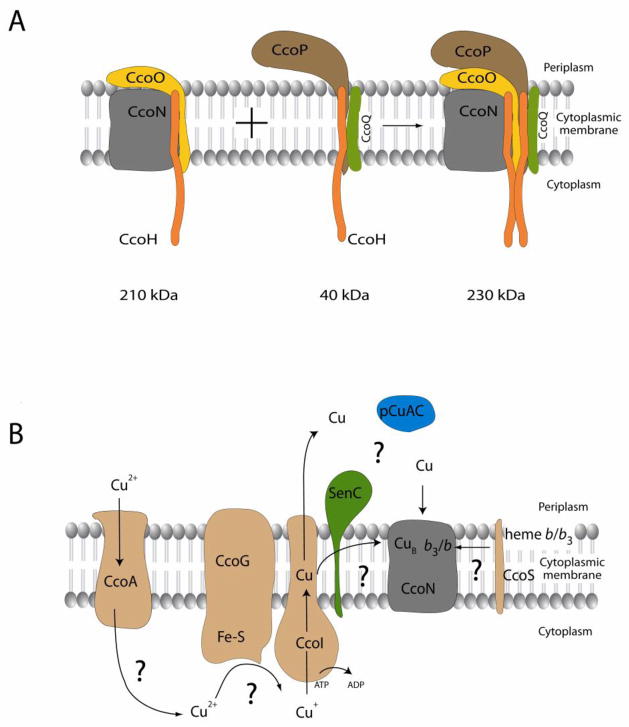
A. An assembly scheme for cbb3-Cox in R. capsulatus. Using BN-PAGE, active cbb3-Cox is detectable in R. capsulatus membranes as a 230 kDa complex. Assembly of the 230 kDa complex proceeds via two inactive assembly intermediates: a 210 kDa subcomplex that contains the catalytic subunit CcoN, the mono-heme cytochrome c subunit CcoO and the single-spanning membrane protein CcoH, and a 40 kDa subcomplex composed of the di-heme cytochrome c subunit CcoP, the small subunit CcoQ and a second copy of CcoH. Full assembly of cbb3-Cox is probably achieved by the dimerization of the cytoplasmic domains of the two CcoH. CcoH was originally considered to function as an assembly factor, but recent data demonstrate that it is a stable component of the fully assembled cbb3-Cox. The c-type cytochromes in both assembly subcomplexes have their heme groups attached indicating that c-type cytochrome maturation preceded the formation of the assembly intermediates.
B. A tentative model for the maturation of CcoN subunit. The maturation of CcoN requires Cu and heme b insertions. Cu is probably imported into the cytoplasm of R. capsulatus by the major facilitator superfamily (MFS) protein CcoA as oxidized Cu2+. On the cytoplasmic face of the membrane Cu2+ is reduced to Cu+ by the ferredoxin-like protein CcoG and transported back to the periplasmic face of the membrane by the PIB-type ATPase CcoI. CcoI then supplies Cu+ to CcoN either directly within the membrane or from the periplasm, or via some putative periplasmic Cu binding proteins such as SenC and PCuAC. The insertion of heme b and heme b3 into CcoN is probably mediated by the assembly factor CcoS.
Later on, it was also found that the putative assembly factor CcoH is a part of the 230 kDa complex (Fig. 3A) [69]. CcoH encodes a single-spanning membrane protein with an extended periplasmic domain that is suggested to serve as a dimerization domain [69]. CcoH was also found in the purified R. capsulatus cbb3-Cox (Pawlik et al., unpublished results). Unlike the mitochondrial aa3-Cox, which forms together with other respiratory complexes a network of supercomplexes [70], no similar large macromolecular assemblies were so far observed in the case of cbb3-Cox in R. capsulatus membranes [67]. In addition to the active 230 kDa complex, using BN-PAGE one large (210 kDa) and one small (~ 40 kDa) inactive assembly intermediates were also detected in R. capsulatus membranes (Fig. 3A). The 210 kDa complex contained CcoN, CcoO and CcoH, but lacked CcoP [67, 69], and the ~ 40 kDa complex had CcoP [67], CcoQ and CcoH [62, 69]. The CcoO subunit in the 210 kDa, and the CcoP subunit in the ~ 40 kDa complexes contained their covalently attached heme groups as indicated by their peroxidase activity [67]. Thus, maturation of the c-type cytochrome subunits occurred prior to their assembly into cbb3-Cox. Genetic data supported these findings, as mutants expressing truncated CcoP derivatives contained only the 210 kDa, and not the 230 kDa complex [67], and mutations abolishing the c-type cytochrome maturation process also prevented cbb3-Cox assembly [72]. Although not essential, CcoQ seemed to improve cbb3-Cox assembly as in its absence active enzymes were produced at reduced amounts. More recent chemical cross-linking data suggest that interactions between CcoQ and CcoP favor assembly of the ~ 40 kDa complex with the 210 kDa complex [62]. The surprising finding that CcoH is associated with the assembly intermediates of cbb3-Cox [69] suggested that this protein behaved more like a bona fide subunit of R. capsulatus enzyme rather than an assembly factor. This is further supported by the observation that the steady-state stability of CcoH is strictly dependent on the presence of CcoNOQP [69]. Available data suggest that cbb3-Cox assembly might occur via a fusion between the CcoNOH and CcoQPH subcomplexes mediated by CcoH (Fig. 3A) [67, 69]. The 3D structure of cbb3-Cox indicates that CcoO is sandwiched between CcoN and CcoP [14]. A similar assembly pathway has also been proposed for B. japonicum cbb3-Cox, although CcoH was not included in this model [73].
3.2 Maturation of cbb3-Cox subunits
3.2.2 CcoN: Roles of CcoS, CcoI and CcoG in hemes b and Cu incorporation
CcoN is an integral membrane protein and is most likely co-translationally inserted into R. capsulatus cytoplasmic membrane [74, 75]. The two heme b and the Cu cofactor are deeply buried within CcoN, and their incorporation into this subunit might be critical for proper folding and stabilization of cbb3-Cox. However, whether these cofactors are inserted co-translationally into CcoN, and if so, in which order, is unknown. A co-translational insertion process for hemes a and Cu cofactors for aa3-Cox has been proposed [76], but not yet established.
In R. capsulatus, CcoS is a small membrane protein of 56 amino acids with no putative heme or Cu binding motifs, which is highly tolerant towards mutations of its conserved residues (Pawlik et al., unpublished results). A mutant lacking CcoS assembles an inactive cbb3-Cox variant of 230 kDa that lacks both of the heme b cofactors of CcoN, but contains properly matured CcoO and CcoP subunits [66]. This finding indicates that c-type cytochrome maturation is not affected, and further supports that the CcoN and c-type cytochrome maturation processes occur independently of each other [66]. Whether the inactive cbb3-Cox variant present in a R. capsulatus ccoS knock out mutants contains CuB is unknown. Clearly, CcoS is required for proper maturation of CcoN, but defining its precise role in cbb3-Cox assembly deserves further studies.
In both mitochondria and bacteria, heme insertion into aa3-Cox is mediated by Surf1 (called Shy1 in yeast), which accepts heme a directly from heme a synthase (CtaA) and is thought to transfer it to subunit I [77]. In the absence of Surf1, aa3-Cox assembly is significantly reduced, but not completely abolished [78–80] implying that Surf1 is important, but not essential for heme a insertion into the subunit I. Moreover, Surf1 homologues are not present in all species that contain heme a containing Cox (e. g. they are missing in B. subtilis and T. thermophilus). In the case of T. thermophilus, a protein called CbaX, which has no homology to Surf1, was implicated in heme a insertion into ba3-Cox [81]. A Surf1 homologue is absent in R. capsulatus genome, and a R. sphaeroides mutant that lacks SurfI is impaired only in the assembly of aa3-but not cbb3-Cox [78]. Available data indicate that Surf1 is dedicated to heme a, and not involved in heme b insertion. In fact, how heme b is inserted into any membrane protein is still enigmatic.
Copper is an essential cofactor of many enzymes such as heme-Cu:O2 reductases, Cu-Zn superoxide dismutase, multicopper oxidase (or laccase) and tyrosinase. Like many metals, free Cu is basically undetectable in cells, minimizing the risk of reactive oxygen species production. Hence, Cu acquisition, trafficking, storage, and delivery to the target sites are strictly controlled processes [82]. Cu can be found in two redox states: the reduced Cu+ is considered to be the transported form while the oxidized Cu+2 is probably the catalytically active form. In eukaryotes, the Cu transporter (Ctr) proteins represent the main Cu importers, and they have been studied intensely [83, 84]. How Cu is imported into the bacterial cytoplasm is not clear. In the Gram-positive Enterococcus hirae a universally conserved P-type ATPase [84], and in B. subtilus the membrane protein YcnJ [85] were suggested to import Cu. In addition, two P-type ATPases are required to import Cu into cyanobacteria (i. e., CtaA and PacS) [86, 87] and into the thylakoids of plant chloroplasts (i. e., PAA1/HMA6 and PAA2/HMA8) [86, 87]. Very recently, CcoA, a novel member of the major facilitator superfamily (MFS) of membrane proteins, was implicated in Cu import in R. capsulatus (Ekici et al., unpublished results, see below) and Schizosaccharomyces pombe [88]. The MFS-type transporters are structurally and functionally distinct from the Ctr-type Cu transporters, and might illustrate a novel Cu uptake system.
Different to Cu import pathways, bacterial Cu efflux pathways are intensely studied, mainly motivated by the toxicity of Cu for all organisms. A major Cu efflux mechanism is provided by P-type ATPases (PIB subgroup). P-type ATPases are integral membrane pumps that hydrolyse ATP for maintaining ion homeostasis, electrochemical gradients and lipid asymmetry [89]. The P-type ATPase superfamily is composed of 11 distinct subgroups of which the PIB is one of the largest and most widespread. PIB-type ATPases use ATP hydrolysis for Cu extrusion across the cytoplasmic membrane [90–93], and contain the characteristic motifs of P-type ATPases, including the ATP binding domain, the phosphorylation domain, and the phosphatase domain [94]. They also contain an N-terminal heavy metal binding domain (HMBD) with the conserved Cu-binding motif (MXCXXC), the membrane embedded ion translocation (CPX)- and the conserved histidine-proline (HP)-motifs [94]. The mechanism of loading cytosolic Cu to Cu-ATPases is not well understood, but HMBD domains are thought to interact with specific metallochaperones to deliver Cu atoms to these transporters [95, 96]. The recent resolution of the first 3D structure of a PIB-type ATPase from Legionella pneumophila revealed a putative docking platform for Cu chaperones [97]. The PIB-type ATPases pump Cu+ out of the bacterial cytoplasm, but their specific roles seem to be determined by their rate of Cu efflux [98]. For example two of them, CopA1 and CopA2 of Pseudomonas aeruginosa, are highly homologous to each other (35% identity and 50% similarity), yet their functions differ. CopA1 has a faster Cu efflux rate and is required for Cu detoxification as its absence renders cells highly sensitive to Cu. CopA2 has a much slower Cu efflux rate and its absence does not affect cellular Cu sensitivity, but leads to the loss of cbb3-Cox activity [99].
CcoI is a homologue of CopA2, and is the product of ccoI gene located in the ccoGHIS cluster in many species [54, 68]. In B. japonicum and R. capsulatus, CcoI appears to be specifically required for cbb3-Cox assembly, but a Rubrivivax gelatinosus mutant lacking the CcoI homologue CtpA seems to be defective in other Cu containing enzymes such as caa3-Cox and N2O reductase as well [100]. In R. capsulatus, absence of CcoI [66] or mutations in its N-terminal HMBD domain or CPC motifs, drastically reduce the steady-state amounts of cbb3-Cox subunits [66] (Pawlik et al., unpublished results). Cu supplementation does not suppress the cbb3-Cox defect of a mutant lacking CcoI, which is also not sensitive to Cu [66]. Whether CuB is inserted into CcoN before or after heme b is not known. In case of aa3-Cox, the availability of enzyme variants containing heme a, but lacking CuB suggests that heme a is likely to be inserted before CuB [78].
The essential role of CcoI for cbb3-Cox assembly suggests a model that involves transport of Cu+ from the cytoplasm to the periplasm, then its concomitant or subsequent insertion into CcoN (Fig. 3B). However it is intriguing that a mutant lacking CcoI is not rescued by exogenous Cu addition, because Cu is thought to be diffused freely across outer membrane into the periplasm. A possibility is that either Cu+ transported by CcoI is inserted into CcoN without its full transport across membrane, or that it is delivered upon transport to specific periplasmic Cu chaperones (perhaps SenC or PCuAC, see below), which then supplies it to CcoN (Fig. 3B). This model would rationalize the cytoplasmic origin of Cu, and suggests that in the absence of CcoI, its loading onto the specific chaperone might be inefficient.
Among the R. capsulatus mutants lacking any one of the ccoGHIS products, those devoid of CcoG exhibited the mildest effect on cbb3-Cox assembly. In the absence of CcoG, all subunits were present at quasi wild type levels, and the enzyme activity was not significantly reduced [66]. CcoG is an integral membrane protein, which has five predicted transmembrane helices and two putative [4Fe-4S] cluster-binding motifs, but whether it contains an iron-sulfur cluster has not been shown experimentally. The exact function of CcoG remains unknown. It has been proposed that it might be involved in intracellular oxidation of Cu+ to Cu2+ [68]. Another possibility is that CcoG might reduce Cu2+ to Cu+ to provide the substrate for CcoI. If so, then CcoG might work together with the putative MFS-type Cu importer CcoA, provided that CcoA imports Cu2+ into the cytoplasm (as suggested by the insensitivity of ccoA mutant to silver which mimics Cu+) (Ekici et al., unpublished results) (Fig. 3B). Clearly, both the structure and function of CcoG are poorly defined and warrant further studies.
3.2.2 CcoO and CcoP: c-type cytochrome maturation (Ccm) process
The CcoO and CcoP subunits of cbb3-Cox are matured by a c-type cytochrome maturation (Ccm) process, which operates independently from CcoN maturation. Ccm is a post-translation and post-translocation process that occurs on the periplasmic face of the cytoplasmic membrane (Fig. 4). During this process, heme b groups are covalently and stereo-specifically attached by thioether bonds formed between their vinyl groups and the cysteine thiols of conserved CXXCH motifs of apocytochromes [101, 102]. Several Ccm systems are encountered in nature, and in α- and γ-proteobacteria and Deinococcus species maturation of the CcoO and CcoP subunits relies on Ccm-system I, which was reviewed in detail recently [103, 104]. Ccm-system I can be divided into three operation modules that are described briefly below.
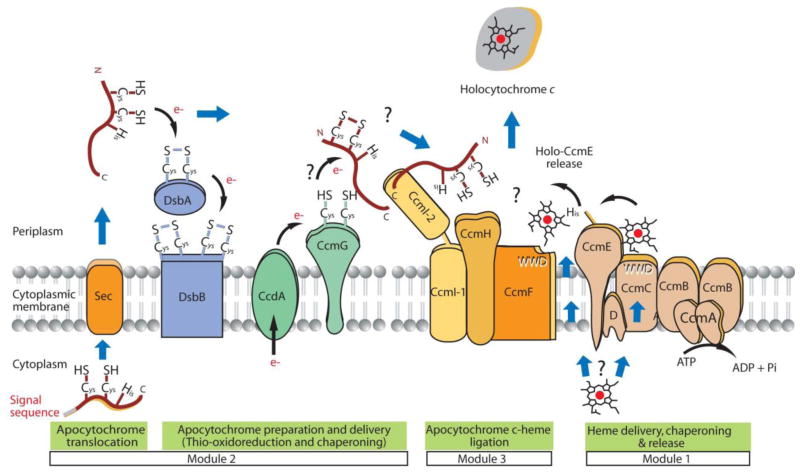
Figure 4.
Cytochrome c maturation (Ccm) system in Rhodobacter capsulatus. The cytochrome c maturation components can be divided into three modules. First module (right) is the transport of heme and its preparation for ligation to apocytochrome that involves CcmABCD proteins which forms a ABC-type transporter that has a role in the delivery of heme to the heme chaperone CcmE before the ligation. It is unknown whether heme is transported via CcmABCD complex or by another unknown protein. CcmA and CcmB are required for the release of heme-bound CcmE from CcmC and CcmD. CcmC and CcmD are involved in the heme attachment to CcmE. Second module (left) involves the apocytochrome thioredox and chaperoning processes. After the translocation of apocytochrome into the periplasm, the cysteine thiols are first oxidized by DsbA-DsbB, then reduced by CcdA, CcmG and/or CcmH. Third module (middle) consists of CcmHIF which is the heme ligation core complex. CcmI binds to C-terminal part of apocytochrome to deliver it to the core complex for the catalysis of the thioether bond formation between reduced apocytochrome and heme vinyl groups forming the mature cytochrome c (adapted from [155])
Module 1: Transport and relay of heme
The function of this module is to translocate cytoplasmically synthesized heme b to the periplasm, and to prepare it for ligation to apo-cytochromes (Fig. 4, right). This module consists of five proteins, named CcmABCDE. CcmABCD is suggested to form an ATP binding cassette (ABC)-type transporter involved in loading heme b to CcmE. CcmE has a single membrane-anchoring helix and a conserved histidine residue that covalently binds heme, which then acts as a heme donor to apocytochromes. CcmA has an ATP binding domain and Walker A and B motifs needed for ATPase activity. CcmB is an integral membrane protein with six transmembrane helices, required for membrane localization of CcmA. CcmC contains a conserved a tryptophan-rich WWD motif and is involved in loading heme to CcmE, and apparently CcmD enhances this process. A recent bioinformatics study grouped CcmC and its homologues as the “heme handling proteins” (HHP) [105]. To what extent CcmAB and CcmCD function together or separately for loading heme onto CcmE and delivering heme-loaded CcmE to the heme ligation complex is unclear. A possibility is that CcmC and CcmD are necessary for attachment of heme to CcmE [106–108], and that CcmAB is required for ATP dependent release of heme-loaded CcmE from CcmCD [109]. On the other hand, whether CcmAB has another, yet to be defined, role in this process is unknown [110, 111].
Module 2: Apocytochrome thio-oxidoreduction and chaperoning
Like most c-type apocytochromes, CcoO and CcoP are thought to be secreted via the SecYEG secretory pathway (Fig. 4, left). Their signal-anchor sequences are not processed, and serve as their N-terminal membrane-anchors. Upon translocation across the cytoplasmic membrane, the DsbA-DsbB dependent oxidative protein-folding pathway [112, 113] is thought to rapidly oxidize the thiol groups of apocytochromes to possibly avoid their proteolytic degradation. The disulfide bonds formed between the cysteines at the conserved heme binding (CXXCH) motifs need to be reduced prior to heme b attachment. Thus, a Ccm-specific thioreduction pathway involving CcdA, CcmG and CcmH proteins has been proposed [114, 115]. CcdA is an integral membrane protein with six transmembrane helices and is responsible for conveying from the cytoplasm to the periplasm reducing equivalents required for this process. CcmG and CcmH are membrane-anchored thioredoxin-like proteins that contain a single CXXC domain facing the periplasm [115], and thought to reduce the disulfide bonds at the heme binding sites. Of the three components, CcdA and CcmG are not required in the absence of DsbA-DsbB dependent thio-oxidation pathway, but CcmH is still required for c-type cytochrome production, suggesting that it has an additional role. Moreover, the 3D structure of CcmG showed that, in addition to its canonical thioredoxin fold, it has a cavity that might bind apocytochromes which is in accordance with a putative holdase role of CcmG as shown in R. capsulatus [116]
Module 3: Apocytochrome and heme b ligation
In R. capsulatus, CcmF, CcmH and CcmI proteins have been proposed to form a heme ligation core complex (Fig. 4, middle), bases on reciprocal co-purification experiments documenting protein-protein interactions between these components [117]. Of these proteins, CcmF is a large integral membrane protein with 11 transmembrane helices, with a trytophan-rich (WWD) signature motif and four conserved histidine residues facing the periplasm proposed to bind heme b before its ligation to apocytochromes [106]. It interacts with heme-loaded CcmE [118], is a heme handling protein (HHP) like CcmC [106]. CcmI is a bipartite protein that contains a membrane embedded N-terminal region with two transmembrane helices and a cytoplasmic leucine-zipper-like motif containing loop (CcmI-1 domain) and a large periplasmic C-terminal extension with tetratricopeptide repeat (TPR)-like motifs (CcmI-2 domain) [119–121]. Unlike most other bacteria, in E. coli the homologue of CcmI-2 domain is fused to the C-terminal end of CcmH, leaving only CcmF and a modified “CcmH” as components of heme ligation core complex [122]. R. capsulatus, mutants lacking CcmI can be suppressed by overproduction of CcmF and CcmH or CcmG. Complementation studies with two distinct domains of CcmI indicated that CcmF and CcmH overproduction relates to the functional role of CcmI-1 domain [123], whereas CcmG overproduction to that of CcmI-2 [120]. These findings suggested that CcmI might be a junction point between the CcdA-CcmG dependent thio-reduction and CcmF-CcmH dependent heme ligation processes. Indeed, CcmI was initially proposed to chaperone apocytochromes to the heme ligation complex [121], and very recently in vitro studies provided strong biochemical evidence to support this proposal (Verissimo et al, unpublished results). Protein-protein interaction studies conducted using purified CcmI and purified apocytochrome c2 indicated that the C-terminal CcmI-2 domain of CcmI recognizes and binds tightly the most C-terminal helix of apocytochrome c2 in the absence of heme b. The folding process of cytochrome c (and cytochrome c2) [124] indicates that their most C-terminal helix interacts with their most N-terminal heme binding helix to form a stable folding intermediate that can trap heme b non-covalently [125]. Altogether, these observations led to the proposal that at least some c-type apocytochromes are first recognized via their C-terminal helices by the periplasmic CcmI-2 domain of the CcmFHI core complex (Fig. 4, middle), and then released from this domain upon transfer of heme b from CcmF to apocytochrome, and subsequently, the thioether bonds are formed. Once the thioether bonds are formed, cytochrome c folds into its final structure.
Recently, another type of post-translational modification was seen in CcoP of pathogenic Neisseria species [126]. The CcoP subunit of N. gonorrhoeae contains a O-linked glycosylation in its periplasmic moiety [126]. The role of this glycosylation, perhaps important for pathogenesis, remains to be determined. It is noteworthy that O-linked glycosylation was also observed for ScoI (see below SenC) and cytochrome CycB, which are involved in cbb3-Cox assembly and electron transfer to cbb3-Cox, respectively [126]. Whether this or other kinds of modification of cbb3-Cox subunits also occurs in other species, and whether this affects biogenesis of this enzyme deserves further studies.
3.2.3 Additional proteins involved in cbb3-Cox biogenesis
Several additional proteins of not yet fully defined roles, including SenC [127], PCuAC [128, 129], DsbA [130] and CcoA (Ekici et al., unpublished results) might be involved in cbb3-Cox biogenesis, and described below.
SenC (ScoI)
SenC is a membrane-anchored protein with a single transmembrane helix, and in its absence, cbb3-Cox biogenesis in R. capsulatus [127] and in P. aeroginosa [131] is drastically decreased and cbb3-Cox activity is regained upon addition of exogenous Cu. Its periplasmic domain contains a thioredoxin fold and a conserved Cu binding motif comprising CxxxC and a His ligand [127]. SenC is a close homologue of the universally conserved ScoI protein, which has been implicated mainly into the assembly of the CuA center of subunit II of aa3-Cox. Whether ScoI acts as a direct donor of Cu to subunit II [132], or is a thiol:disulfide oxidoreductase reducing appropriate cysteines of subunit II for subsequent Cu delivery by another Cu chaperone [129] is not yet clear [133]. B. japonicum mutants lacking SenC were impaired in the assembly of aa3- but not cbb3-Cox and showed reduced symbiotic N2 fixation [134]. Similarly, R. sphaeroides mutants lacking PrrC (a SenC homologue) had no effect on cbb3-Cox assembly although they were defective in photosynthetic growth [52]. Like R. capsulatus senC, R. sphaeroides prrC is located next to prrAB (homologues of R. capsulatus regAB) genes that encode a two component regulatory system controlling energy processes, including photosynthesis [46, 135]. Whether prrC mutations have any polar effect on downstream prrB gene, indirectly interfering with cellular amounts of PrrB, is not known (Fig. 1).
Several studies have shown that the role of prokaryotic ScoI homologues is not restricted to their involvement in aa3-Cox assembly. A role of these proteins in oxidative stress response [136] or photosynthetic gene regulation [52] has been proposed. Bioinformatics based genome surveys indicated that ScoI homologues are also present in many bacterial species, like R. capsulatus, which do not have a CuA containing Cox [133]. R. capsulatus mutants lacking SenC produce very low amounts of cbb3-type Cox [127, 137], a phenotype which was rescued by addition of exogenous Cu [127]. In vitro experiments showed that in a mutant lacking SenC cbb3-Cox assembly proceeded at a significantly reduced level (Lohmeyer et al., unpublished results). Very recent chemical cross-linking studies indicated that SenC interacts with CcoP and CcoH in vivo, suggesting that it is involved directly in cbb3-Cox assembly (Lohmeyer et al., unpublished results) (Fig. 3B).
PCuAC (DR1885)
The periplasmic CuA-chaperone (PCuAC) was first identified in Deinococcus radiodurans and its structural gene is located near ScoI [128]. In this species, PCuAC is thought to provide Cu to the CuA center after ScoI reduces the appropriate disulfide bond at subunit II of aa3-Cox. In the case of T. thermophilus PCuAC is involved in Cu transfer to CuA of ba3-Cox [129]. Homologues of PCuAC are also found in many species that lack aa3-Cox such as R. capsulatus. Unlike D. radiodurans, in R. capsulatus the structural gene of PCuAC is not located near that of SenC, but its product contains similar conserved metal binding motifs. In this species whether PCuAC affects cbb3-Cox CuB center assembly is unknown as no chromosomal PCuAC knockout mutant could be obtained so far (Fig. 3B).
DsbA
R. capsulatus mutants lacking the periplasmic thiol:disulfide oxidoreductase DsbA, primarily involved in oxidative protein folding pathway [112], overproduce the periplasmic protease DegP [138]. They have pleiotropic phenotypes, including temperature sensitivity for growth (35° C), osmosensitivity, filamentation and decreased respiratory capabilities. Even at permissive growth temperature (25° C), they exhibit reduced cbb3-Cox activity. Supplementation of the growth medium with redox active chemicals like cysteine/cystine or Cu2+, restores both temperature sensitive growth and cbb3-Cox defects of mutants lacking DsbA. Remarkably, DsbA knock out mutants revert frequently to regain their growth ability at 35° C without any need for redox active supplements, and concomitantly, they restore their cbb3-Cox production. These revertants acquire mutations in degP that decrease drastically the protease activity of DegP [138]. These findings point to important links between the formation of disulfide bond, degradation of misfolded periplasmic proteins, and production of active cbb3-Cox. In the absence of DsbA, whether exogenously supplied Cu2+ acts exclusively as a source of oxidant, or also eases acquisition of Cu as a missing cofactor for cbb3-Cox production, or both is unknown. Future studies might clarify these emerging links between major cellular processes and cbb3-Cox biogenesis.
CcoA
Very recent studies of R. capsulatus mutants that exhibit “Cu supplement dependent cbb3-Cox production” phenotype yielded a novel component, CcoA involved in cbb3-Cox biogenesis (Ekici et al., unpublished results). These mutants lacked cbb3-Cox activity on Cu-depleted media, but regained it upon exogenous Cu2+ supplementation. Molecular genetic studies established that these mutants were defective in an open reading frame (ORF) of previously unknown function, now named ccoA due to its role in cbb3-Cox biogenesis. CcoA is an integral membrane protein formed by two subdomains of six transmembrane helices each, which are separated by a large cytoplasmic loop. Its sequence is highly homologous to the Major Facilitator Superfamily (MFS) of transporters [139]. It contains the AVYGRR and ARFGRE between its second-third and eight-nine transmembrane helices, respectively. These motifs are highly similar to DRXGRR motifs that characterize members of the MFS family. In addition, unlike many other MFS members, CcoA contains a putative metal (Cu) binding (MXxM) motif as found in Ctr type Cu importers [140], suggesting a role in metal transport. In R. capsulatus mutants lacking CcoA, transcription and translation initiation of ccoNOQP occur, but the steady-state amounts of cbb3-Cox subunits are quasi-absent (Ekici et al., unpublished results). Thus, absence of CcoA perturbs a co- or post-translational step(s) of cbb3-Cox biogenesis. Remarkably, supplementation with Cu2+, but not with other metals like Mn2+, Zn2+ or Fe3+, palliates the cbb3-Cox production defect. CcoA knock out mutants are not sensitive to Cu2+, but their cellular Cu content is lower than normal cells, and they revert very frequently to restore their cbb3-Cox activity. These by-pass suppressors restore normal cellular Cu content and production of cbb3-Cox. Surprisingly, they also become hypersensitive to Cu2+ but not to other metals, including Ag+. These findings suggest that the MFS-type transporter CcoA may be involved in Cu acquisition by a hitherto unknown pathway, which seems needed for normal biogenesis of cbb3-Cox. Very recently, the MFC1 protein of Schizosaccahromyces pombe, which is also a MFS-type transporter with a MXxM motif and homologous to CcoA, was shown to import Cu across the plasma membrane [88]. In S. pombe, the Ctr-type transporters are thought to mediate high affinity Cu uptake, and MFC1 might provide an additional low affinity Cu import route. Clearly, future studies of this novel family of MFS-type putative Cu importers might provide invaluable information about bacterial Cu acquisition pathways and their link to cbb3-Cox biogenesis.
3.2.4 Role of membrane lipids for cbb3-Cox biogenesis
Membrane lipids, especially those that act as “lipochaperones”, are important determinants for membrane protein structure and activity [141]. These lipids bind to specific locations of proteins in stochiometric amounts, and influence their folding, stability, steady-state amounts and activity [142]. Phospholipids were shown to co-crystallize with heme-Cu: O2 reductases from many different species [143–145], and the stability and organization of respiratory chain super complexes were significantly influenced by some phospholipids like cardiolipin [146, 147]. A specific role of lipids on biogenesis and catalytic activity of cbb3-Cox is also likely, as changes in membrane lipid composition seem to affect the amounts of cbb3-Cox found in R. capsulatus membranes [148]. Moreover, different lipids environments were also shown to affect carbonmonoxide binding to CcoP [149]. R. capsulatus mutants that were defective in ornithine lipid (OL) biosynthesis lacked cbb3-Cox [148]. Studies of these mutants defined two genes, olsA and olsB coding for N-acyltransferase and O-acyltransferase, respectively, which are both required for OL biosynthesis [148, 150]. OL is a non-phosphorus membrane lipid that usually accounts only for a small fraction of the total lipids found in bacteria. In some species, OL are synthesized under phosphate limiting conditions, and are used as replacement for phosphate containing lipids [151–153]. In R. capsulatus OL biosynthesis is not regulated by phosphate availability, but absence of OL affects the steady-amounts of a group of membrane proteins, including most of the c-type cytochromes. Mutants unable to synthesize OL have very low amounts of cbb3-Cox and the electron carrier cytochromes c2 and cy. They contain a small amount of cytochrome c1, and consequently, very low ubiquinol: cytochrome c2 oxidoreductase (cytochrome bc1) activity. Interestingly, these defects are both temperature- and growth medium-dependent. Especially on enriched media at regular growth temperature (35° C), mutants lacking OL are photosynthesis-incompetent and have no cbb3-Cox. Pulse-chase studies conducted using cytochrome cy indicated that absence of OL decreased drastically its cellular amount [148], suggesting that in the absence of OL, a group of proteins including cbb3-Cox, may be more prone to misfolding and enhanced degradation.
4. Summary and future prospects
In recent years, enormous amount of progress has been accomplished in studies aimed at understanding the biogenesis of cbb3-Cox. This second most common class of heme-Cu: O2 reductases is becoming attractive for future studies due to the availability of an excellent model system like R. capsulatus providing molecular genetic analyses, a 3D structure at high resolution allowing refined structural and functional inquiries, and exquisite biochemical approaches developed and practiced in various laboratories. Salient recent accomplishments on this topic include an emerging subunit assembly process highlighting the indispensible role of CcoH as a “subcomplex assembler”, the better defined role of CcoI subgroup among the PIB-type ATPases as a Cu exporter to cbb3-Cox, and possibly to a few other Cu containing enzymes. Furthermore, the discovery of a periplasmic Cu chaperone PCuAC, the emerging interactions between the ScoI homologue SenC/PrrC and cbb3-Cox subunits and its assembly factors, as well as the novel MFS-type putative Cu importer CcoA, whose Cu acquisition functions correlate with the production of cbb3-Cox, provide exciting novel insight into the complexity of binuclear center assembly. An attempt was made to organize all known components into a highly speculative working scheme that outlines an emerging path of Cu delivery to the heme-CuB center of cbb3-Cox (Fig. 3B). Indeed, much work is needed to probe experimentally the validity of the various facets of this working model under construction. Currently, we are reaching a stage where the crucial players required for cbb3-Cox assembly are becoming identified, and ongoing molecular genetic and biochemical studies are now initiating research aimed to untangle the sophisticated and unique biogenesis process of cbb3-Cox.
Highlights.
Structure-function, regulation and biogenesis of cbb3-type cytochrome c oxidases are reviewed.
An emerging multistep assembly pathway of the subunits of this membrane enzyme is discussed.
A tentative model for formation of the heme b3-CuB catalytic center of this enzyme is proposed.
Acknowledgments
This work was supported by grants DOE 91ER20052 and NIH GM38239 to F.D., and by grants from the German Science Foundation (DFG-GRK1478) and from the German-French PhD college (DFH/UFA) on “Membranes and Membrane Proteins” to H.G.K.
Abbreviations
- Cox
cytochrome c oxidase
- Qox
quinol oxidase
- Ccm
cytochrome c maturation
- NOR
nitric oxide reductase
- BN-PAGE
blue-native polyacrylamide gel electrophoresis
- Cu
Copper
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pereira MM, Santana M, Teixeira M. A novel scenario for the evolution of haem-copper oxygen reductases. Biochimica et biophysica acta. 2001;1505:185–208. doi: 10.1016/s0005-2728(01)00169-4. [DOI] [PubMed] [Google Scholar]
- 2.Castresana J, Lubben M, Saraste M, Higgins DG. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flock U, Reimann J, Adelroth P. Proton transfer in bacterial nitric oxide reductase. Biochem Soc Trans. 2006;34:188–190. doi: 10.1042/BST0340188. [DOI] [PubMed] [Google Scholar]
- 4.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 5.Won KY, Lim SJ, Kim GY, Kim YW, Han SA, Song JY, Lee DK. Regulatory role of p53 in cancer metabolism via SCO2 and TIGAR in human breast cancer. Hum Pathol. 2011 doi: 10.1016/j.humpath.2011.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Vesela K, Hulkova H, Hansikova H, Zeman J, Elleder M. Structural analysis of tissues affected by cytochrome C oxidase deficiency due to mutations in the SCO2 gene. APMIS. 2008;116:41–49. doi: 10.1111/j.1600-0463.2008.00772.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein C, Facista A, Nguyen H, Zaitlin B, Hassounah N, Loustaunau C, Payne CM, Banerjee B, Goldschmid S, Tsikitis VL, Krouse R, Bernstein H. Cancer and age related colonic crypt deficiencies in cytochrome c oxidase I. World J Gastrointest Oncol. 2010;2:429–442. doi: 10.4251/wjgo.v2.i12.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Horsman JA, Barquera B, Rumbley J, Ma J, Gennis RB. The superfamily of heme-copper respiratory oxidases. J Bacteriol. 1994;176:5587–5600. doi: 10.1128/jb.176.18.5587-5600.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marrs B, Gest H. Genetic mutations affecting the respiratory electron-transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J Bacteriol. 1973;114:1045–1051. doi: 10.1128/jb.114.3.1045-1051.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daldal F, Mandaci S, Winterstein C, Myllykallio H, Duyck K, Zannoni D. Mobile cytochrome c2 and membrane-anchored cytochrome cy are both efficient electron donors to the cbb3- and aa3-type cytochrome c oxidases during respiratory growth of Rhodobacter sphaeroides. J Bacteriol. 2001;183:2013–2024. doi: 10.1128/JB.183.6.2013-2024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitcher RS, Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochimica et biophysica acta. 2004;1655:388–399. doi: 10.1016/j.bbabio.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 12.Ducluzeau AL, Ouchane S, Nitschke W. The cbb3 oxidases are an ancient innovation of the domain bacteria. Mol Biol Evol. 2008;25:1158–1166. doi: 10.1093/molbev/msn062. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann JM, Funes S. Biogenesis of cytochrome oxidase-sophisticated assembly lines in the mitochondrial inner membrane. Gene. 2005;354:43–52. doi: 10.1016/j.gene.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 14.Buschmann S, Warkentin E, Xie H, Langer JD, Ermler U, Michel H. The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science. 2010;329:327–330. doi: 10.1126/science.1187303. [DOI] [PubMed] [Google Scholar]
- 15.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 16.Abramson J, Riistama S, Larsson G, Jasaitis A, Svensson-Ek M, Laakkonen L, Puustinen A, Iwata S, Wikstrom M. The structure of the ubiquinol oxidase from Escherichia coli and its ubiquinone binding site. Nat Struct Biol. 2000;7:910–917. doi: 10.1038/82824. [DOI] [PubMed] [Google Scholar]
- 17.Soulimane T, Buse G, Bourenkov GP, Bartunik HD, Huber R, Than ME. Structure and mechanism of the aberrant ba(3)-cytochrome c oxidase from thermus thermophilus. EMBO J. 2000;19:1766–1776. doi: 10.1093/emboj/19.8.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 19.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann BH, Nitsche CI, Fee JA, Rusnak F, Munck E. Properties of a copper-containing cytochrome ba3: a second terminal oxidase from the extreme thermophile Thermus thermophilus. Proc Natl Acad Sci U S A. 1988;85:5779–5783. doi: 10.1073/pnas.85.16.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang HY, Hemp J, Chen Y, Fee JA, Gennis RB. The cytochrome ba3 oxygen reductase from Thermus thermophilus uses a single input channel for proton delivery to the active site and for proton pumping. Proc Natl Acad Sci U S A. 2009;106:16169–16173. doi: 10.1073/pnas.0905264106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arslan E, Kannt A, Thony-Meyer L, Hennecke H. The symbiotically essential cbb(3)-type oxidase of Bradyrhizobium japonicum is a proton pump. FEBS Lett. 2000;470:7–10. doi: 10.1016/s0014-5793(00)01277-1. [DOI] [PubMed] [Google Scholar]
- 23.Preisig O, Zufferey R, Thony-Meyer L, Appleby CA, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hemp J, Han H, Roh JH, Kaplan S, Martinez TJ, Gennis RB. Comparative genomics and site-directed mutagenesis support the existence of only one input channel for protons in the C-family (cbb3 oxidase) of heme-copper oxygen reductases. Biochemistry. 2007;46:9963–9972. doi: 10.1021/bi700659y. [DOI] [PubMed] [Google Scholar]
- 25.Kahn D, David M, Domergue O, Daveran ML, Ghai J, Hirsch PR, Batut J. Rhizobium meliloti fixGHI sequence predicts involvement of a specific cation pump in symbiotic nitrogen fixation. J Bacteriol. 1989;171:929–939. doi: 10.1128/jb.171.2.929-939.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Preisig O, Anthamatten D, Hennecke H. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc Natl Acad Sci U S A. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thony-Meyer L, Beck C, Preisig O, Hennecke H. The ccoNOQP gene cluster codes for a cb-type cytochrome oxidase that functions in aerobic respiration of Rhodobacter capsulatus. Mol Microbiol. 1994;14:705–716. doi: 10.1111/j.1365-2958.1994.tb01308.x. [DOI] [PubMed] [Google Scholar]
- 28.Gray KA, Grooms M, Myllykallio H, Moomaw C, Slaughter C, Daldal F. Rhodobacter capsulatus contains a novel cb-type cytochrome c oxidase without a CuA center. Biochemistry. 1994;33:3120–3127. doi: 10.1021/bi00176a047. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Horsman JA, Berry E, Shapleigh JP, Alben JO, Gennis RB. A novel cytochrome c oxidase from Rhodobacter sphaeroides that lacks CuA. Biochemistry. 1994;33:3113–3119. doi: 10.1021/bi00176a046. [DOI] [PubMed] [Google Scholar]
- 30.Hemp J, Gennis RB. Diversity of the heme-copper superfamily in archaea: insights from genomics and structural modeling. Results Probl Cell Differ. 2008;45:1–31. doi: 10.1007/400_2007_046. [DOI] [PubMed] [Google Scholar]
- 31.Sassera D, Lo N, Epis S, D’Auria G, Montagna M, Comandatore F, Horner D, Pereto J, Luciano AM, Franciosi F, Ferri E, Crotti E, Bazzocchi C, Daffonchio D, Sacchi L, Moya A, Latorre A, Bandi C. Phylogenomic evidence for the presence of a flagellum and cbb3 oxidase in the free-living mitochondrial ancestor. Mol Biol Evol. 2011 doi: 10.1093/molbev/msr159. [DOI] [PubMed] [Google Scholar]
- 32.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 33.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 34.Alm RA, Ling LS, Moir DT, King BL, Brown ED, Doig PC, Smith DR, Noonan B, Guild BC, deJonge BL, Carmel G, Tummino PJ, Caruso A, Uria-Nickelsen M, Mills DM, Ives C, Gibson R, Merberg D, Mills SD, Jiang Q, Taylor DE, Vovis GF, Trust TJ. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Hopper A, Overton T, Squire DJ, Cole J, Tovell N. Organization of the electron transfer chain to oxygen in the obligate human pathogen Neisseria gonorrhoeae: roles for cytochromes c4 and c5, but not cytochrome c2, in oxygen reduction. J Bacteriol. 2010;192:2395–2406. doi: 10.1128/JB.00002-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Perez GI, Blaser MJ. Campylobacter and Helicobacter. University of Texas Medical Branch at Galveston; Galveston (TX): 1996. 2011/03/18 ed. [PubMed] [Google Scholar]
- 37.Jackson RJ, Elvers KT, Lee LJ, Gidley MD, Wainwright LM, Lightfoot J, Park SF, Poole RK. Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni: the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J Bacteriol. 2007;189:1604–1615. doi: 10.1128/JB.00897-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodall CA, Jones MA, Barrow PA, Hinds J, Marsden GL, Kelly DJ, Dorrell N, Wren BW, Maskell DJ. Campylobacter jejuni gene expression in the chick cecum: evidence for adaptation to a low-oxygen environment. Infect Immun. 2005;73:5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weingarten RA, Grimes JL, Olson JW. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl Environ Microbiol. 2008;74:1367–1375. doi: 10.1128/AEM.02261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones CW, Brice JM, Wright V, Ackrell BA. Respiratory protection of nitrogenase in Azotobacter vinelandii. FEBS Lett. 1973;29:77–81. doi: 10.1016/0014-5793(73)80530-7. [DOI] [PubMed] [Google Scholar]
- 41.Oelze J. Respiratory protection of nitrogenase in Azotobacter species: is a widely held hypothesis unequivocally supported by experimental evidence? FEMS Microbiol Rev. 2000;24:321–333. doi: 10.1111/j.1574-6976.2000.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 42.Loisel-Meyer S, Jimenez de Bagues MP, Kohler S, Liautard JP, Jubier-Maurin V. Differential use of the two high-oxygen-affinity terminal oxidases of Brucella suis for in vitro and intramacrophagic multiplication. Infect Immun. 2005;73:7768–7771. doi: 10.1128/IAI.73.11.7768-7771.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salama NR, Shepherd B, Falkow S. Global transposon mutagenesis and essential gene analysis of Helicobacter pylori. J Bacteriol. 2004;186:7926–7935. doi: 10.1128/JB.186.23.7926-7935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ge Z, Feng Y, Dangler CA, Xu S, Taylor NS, Fox JG. Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb Pathog. 2000;29:279–287. doi: 10.1006/mpat.2000.0391. [DOI] [PubMed] [Google Scholar]
- 45.Smith MA, Finel M, Korolik V, Mendz GL. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch Microbiol. 2000;174:1–10. doi: 10.1007/s002030000174. [DOI] [PubMed] [Google Scholar]
- 46.Swem LR, Elsen S, Bird TH, Swem DL, Koch HG, Myllykallio H, Daldal F, Bauer CE. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J Mol Biol. 2001;309:121–138. doi: 10.1006/jmbi.2001.4652. [DOI] [PubMed] [Google Scholar]
- 47.Swem DL, Bauer CE. Coordination of ubiquinol oxidase and cytochrome cbb(3) oxidase expression by multiple regulators in Rhodobacter capsulatus. J Bacteriol. 2002;184:2815–2820. doi: 10.1128/JB.184.10.2815-2820.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsen S, Dischert W, Colbeau A, Bauer CE. Expression of uptake hydrogenase and molybdenum nitrogenase in Rhodobacter capsulatus is coregulated by the RegB-RegA two-component regulatory system. J Bacteriol. 2000;182:2831–2837. doi: 10.1128/jb.182.10.2831-2837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vichivanives P, Bird TH, Bauer CE, Robert Tabita F. Multiple regulators and their interactions in vivo and in vitro with the cbb regulons of Rhodobacter capsulatus. J Mol Biol. 2000;300:1079–1099. doi: 10.1006/jmbi.2000.3914. [DOI] [PubMed] [Google Scholar]
- 50.Mosley CS, Suzuki JY, Bauer CE. Identification and molecular genetic characterization of a sensor kinase responsible for coordinately regulating light harvesting and reaction center gene expression in response to anaerobiosis. J Bacteriol. 1994;176:7566–7573. doi: 10.1128/jb.176.24.7566-7573.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sganga MW, Bauer CE. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell. 1992;68:945–954. doi: 10.1016/0092-8674(92)90037-d. [DOI] [PubMed] [Google Scholar]
- 52.Eraso JM, Kaplan S. From redox flow to gene regulation: role of the PrrC protein of Rhodobacter sphaeroides 2.4.1. Biochemistry. 2000;39:2052–2062. doi: 10.1021/bi9923858. [DOI] [PubMed] [Google Scholar]
- 53.O’Gara JP, Eraso JM, Kaplan S. A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1998;180:4044–4050. doi: 10.1128/jb.180.16.4044-4050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koch HG, Hwang O, Daldal F. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J Bacteriol. 1998;180:969–978. doi: 10.1128/jb.180.4.969-978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cotter PA, Darie S, Gunsalus RP. The effect of iron limitation on expression of the aerobic and anaerobic electron transport pathway genes in Escherichia coli. FEMS Microbiol Lett. 1992;79:227–232. doi: 10.1111/j.1574-6968.1992.tb14045.x. [DOI] [PubMed] [Google Scholar]
- 56.Rey FE, Harwood CS. FixK, a global regulator of microaerobic growth, controls photosynthesis in Rhodopseudomonas palustris. Mol Microbiol. 2010;75:1007–1020. doi: 10.1111/j.1365-2958.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 57.Fischer HM. Environmental regulation of rhizobial symbiotic nitrogen fixation genes. Trends Microbiol. 1996;4:317–320. doi: 10.1016/0966-842x(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 58.Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99:1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 59.Cosseau C, Batut J. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch Microbiol. 2004;181:89–96. doi: 10.1007/s00203-003-0641-5. [DOI] [PubMed] [Google Scholar]
- 60.Hino T, Matsumoto Y, Nagano S, Sugimoto H, Fukumori Y, Murata T, Iwata S, Shiro Y. Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science. 2010;330:1666–1670. doi: 10.1126/science.1195591. [DOI] [PubMed] [Google Scholar]
- 61.Forte E, Urbani A, Saraste M, Sarti P, Brunori M, Giuffre A. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur J Biochem. 2001;268:6486–6491. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 62.Peters A, Kulajta C, Pawlik G, Daldal F, Koch HG. Stability of the cbb3-type cytochrome oxidase requires specific CcoQ-CcoP interactions. J Bacteriol. 2008;190:5576–5586. doi: 10.1128/JB.00534-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cobine PA, Pierrel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochimica et biophysica acta. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 64.Greiner P, Hannappel A, Werner C, Ludwig B. Biogenesis of cytochrome c oxidase--in vitro approaches to study cofactor insertion into a bacterial subunit I. Biochimica et biophysica acta. 2008;1777:904–911. doi: 10.1016/j.bbabio.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Mick DU, Fox TD, Rehling P. Inventory control: cytochrome c oxidase assembly regulates mitochondrial translation. Nat Rev Mol Cell Biol. 2011;12:14–20. doi: 10.1038/nrm3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koch HG, Winterstein C, Saribas AS, Alben JO, Daldal F. Roles of the ccoGHIS gene products in the biogenesis of the cbb(3)-type cytochrome c oxidase. J Mol Biol. 2000;297:49–65. doi: 10.1006/jmbi.2000.3555. [DOI] [PubMed] [Google Scholar]
- 67.Kulajta C, Thumfart JO, Haid S, Daldal F, Koch HG. Multi-step assembly pathway of the cbb3-type cytochrome c oxidase complex. J Mol Biol. 2006;355:989–1004. doi: 10.1016/j.jmb.2005.11.039. [DOI] [PubMed] [Google Scholar]
- 68.Preisig O, Zufferey R, Hennecke H. The Bradyrhizobium japonicum fixGHIS genes are required for the formation of the high-affinity cbb3-type cytochrome oxidase. Arch Microbiol. 1996;165:297–305. doi: 10.1007/s002030050330. [DOI] [PubMed] [Google Scholar]
- 69.Pawlik G, Kulajta C, Sachelaru I, Schroder S, Waidner B, Hellwig P, Daldal F, Koch HG. The putative assembly factor CcoH is stably associated with the cbb3-type cytochrome oxidase. J Bacteriol. 2010;192:6378–6389. doi: 10.1128/JB.00988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schagger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 71.Heuberger EH, Veenhoff LM, Duurkens RH, Friesen RH, Poolman B. Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J Mol Biol. 2002;317:591–600. doi: 10.1006/jmbi.2002.5416. [DOI] [PubMed] [Google Scholar]
- 72.Deshmukh M, Brasseur G, Daldal F. Novel Rhodobacter capsulatus genes required for the biogenesis of various c-type cytochromes. Mol Microbiol. 2000;35:123–138. doi: 10.1046/j.1365-2958.2000.01683.x. [DOI] [PubMed] [Google Scholar]
- 73.Zufferey R, Preisig O, Hennecke H, Thony-Meyer L. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J Biol Chem. 1996;271:9114–9119. doi: 10.1074/jbc.271.15.9114. [DOI] [PubMed] [Google Scholar]
- 74.Koch HG, Moser M, Muller M. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev Physiol Biochem Pharmacol. 2003;146:55–94. doi: 10.1007/s10254-002-0002-9. [DOI] [PubMed] [Google Scholar]
- 75.Koch HG. Folding, Assembly, and Stability of Transmembrane Cytochromes. Current Chemical Biology. 2007;1:59–74. [Google Scholar]
- 76.Khalimonchuk O, Ostermann K, Rodel G. Evidence for the association of yeast mitochondrial ribosomes with Cox11p, a protein required for the Cu(B) site formation of cytochrome c oxidase. Curr Genet. 2005;47:223–233. doi: 10.1007/s00294-005-0569-1. [DOI] [PubMed] [Google Scholar]
- 77.Hannappel A, Bundschuh FA, Ludwig B. Characterization of heme-binding properties of Paracoccus denitrificans Surf1 proteins. FEBS J. 2011;278:1769–1778. doi: 10.1111/j.1742-4658.2011.08101.x. [DOI] [PubMed] [Google Scholar]
- 78.Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP. Assembly of cytochrome-c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J Biol Chem. 2005;280:17652–17656. doi: 10.1074/jbc.C500061200. [DOI] [PubMed] [Google Scholar]
- 79.Bundschuh FA, Hoffmeier K, Ludwig B. Two variants of the assembly factor Surf1 target specific terminal oxidases in Paracoccus denitrificans. Biochimica et biophysica acta. 2008;1777:1336–1343. doi: 10.1016/j.bbabio.2008.05.448. [DOI] [PubMed] [Google Scholar]
- 80.Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M. Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet. 1998;63:1609–1621. doi: 10.1086/302150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Werner C, Richter OM, Ludwig B. A novel heme a insertion factor gene cotranscribes with the Thermus thermophilus cytochrome ba3 oxidase locus. J Bacteriol. 2010;192:4712–4719. doi: 10.1128/JB.00548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Banci L, Bertini I, Cantini F, Ciofi-Baffoni S. Cellular copper distribution: a mechanistic systems biology approach. Cell Mol Life Sci. 2011;67:2563–2589. doi: 10.1007/s00018-010-0330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lutsenko S. Human copper homeostasis: a network of interconnected pathways. Curr Opin Chem Biol. 14:211–217. doi: 10.1016/j.cbpa.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Odermatt A, Suter H, Krapf R, Solioz M. Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 85.Chillappagari S, Miethke M, Trip H, Kuipers OP, Marahiel MA. Copper acquisition is mediated by YcnJ and regulated by YcnK and CsoR in Bacillus subtilis. J Bacteriol. 2009;191:2362–2370. doi: 10.1128/JB.01616-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tottey S, Rich PR, Rondet SA, Robinson NJ. Two Menkes-type atpases supply copper for photosynthesis in Synechocystis PCC 6803. J Biol Chem. 2001;276:19999–20004. doi: 10.1074/jbc.M011243200. [DOI] [PubMed] [Google Scholar]
- 87.Nouet C, Motte P, Hanikenne M. Chloroplastic and mitochondrial metal homeostasis. Trends Plant Sci. 2011;16:395–404. doi: 10.1016/j.tplants.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 88.Beaudoin J, Ioannoni R, Lopez-Maury L, Bahler J, Ait-Mohand S, Guerin B, Dodani SC, Chang CJ, Labbe S. Mfc1 is a novel forespore membrane copper transporter in meiotic and sporulating cells. J Biol Chem. 2011 doi: 10.1074/jbc.M111.280396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat Rev Mol Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- 90.Linz R, Lutsenko S. Copper-transporting ATPases ATP7A and ATP7B: cousins, not twins. J Bioenerg Biomembr. 2007;39:403–407. doi: 10.1007/s10863-007-9101-2. [DOI] [PubMed] [Google Scholar]
- 91.Lutsenko S, Barnes NL, Bartee MY, Dmitriev OY. Function and regulation of human copper-transporting ATPases. Physiol Rev. 2007;87:1011–1046. doi: 10.1152/physrev.00004.2006. [DOI] [PubMed] [Google Scholar]
- 92.Arguello JM, Eren E, Gonzalez-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. Biometals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- 93.Palmgren MG, Axelsen KB. Evolution of P-type ATPases. Biochimica et biophysica acta. 1998;1365:37–45. doi: 10.1016/s0005-2728(98)00041-3. [DOI] [PubMed] [Google Scholar]
- 94.Solioz M, Vulpe C. CPx-type ATPases: a class of P-type ATPases that pump heavy metals. Trends Biochem Sci. 1996;21:237–241. [PubMed] [Google Scholar]
- 95.Gonzalez-Guerrero M, Hong D, Arguello JM. Chaperone-mediated Cu+ delivery to Cu+ transport ATPases: requirement of nucleotide binding. J Biol Chem. 2009;284:20804–20811. doi: 10.1074/jbc.M109.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gonzalez-Guerrero M, Arguello JM. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci U S A. 2008;105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gourdon P, Liu XY, Skjorringe T, Morth JP, Moller LB, Pedersen BP, Nissen P. Crystal structure of a copper-transporting PIB-type ATPase. Nature. 2011;475:59–64. doi: 10.1038/nature10191. [DOI] [PubMed] [Google Scholar]
- 98.Raimunda D, Gonzalez-Guerrero M, Leeber BW, 3rd, Arguello JM. The transport mechanism of bacterial Cu+-ATPases: distinct efflux rates adapted to different function. Biometals. 2011;24:467–475. doi: 10.1007/s10534-010-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gonzalez-Guerrero M, Raimunda D, Cheng X, Arguello JM. Distinct functional roles of homologous Cu+ efflux ATPases in Pseudomonas aeruginosa. Mol Microbiol. 2010;78:1246–1258. doi: 10.1111/j.1365-2958.2010.07402.x. [DOI] [PubMed] [Google Scholar]
- 100.Hassani BK, Astier C, Nitschke W, Ouchane S. CtpA, a copper-translocating P-type ATPase involved in the biogenesis of multiple copper-requiring enzymes. J Biol Chem. 2010;285:19330–19337. doi: 10.1074/jbc.M110.116020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Barker PD, Ferguson SJ. Still a puzzle: why is haem covalently attached in c-type cytochromes? Structure. 1999;7:R281–290. doi: 10.1016/s0969-2126(00)88334-3. [DOI] [PubMed] [Google Scholar]
- 102.Thony-Meyer L. Haem-polypeptide interactions during cytochrome c maturation. Biochim Biophys Acta. 2000;1459:316–324. doi: 10.1016/s0005-2728(00)00167-5. [DOI] [PubMed] [Google Scholar]
- 103.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 2010;18:266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kranz RG, Richard-Fogal C, Taylor JS, Frawley ER. Cytochrome c biogenesis: mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol Mol Biol Rev. 2009;73:510–528. doi: 10.1128/MMBR.00001-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee JH, Harvat EM, Stevens JM, Ferguson SJ, Saier MH., Jr Evolutionary origins of members of a superfamily of integral membrane cytochrome c biogenesis proteins. Biochimica et biophysica acta. 2007;1768:2164–2181. doi: 10.1016/j.bbamem.2007.04.022. [DOI] [PubMed] [Google Scholar]
- 106.Goldman BS, Beck DL, Monika EM, Kranz RG. Transmembrane heme delivery systems. Proc Natl Acad Sci U S A. 1998;95:5003–5008. doi: 10.1073/pnas.95.9.5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Goldman BS, Beckman DL, Bali A, Monika EM, Gabbert KK, Kranz RG. Molecular and immunological analysis of an ABC transporter complex required for cytochrome c biogenesis. J Mol Biol. 1997;268:724–738. doi: 10.1006/jmbi.1997.0992. [DOI] [PubMed] [Google Scholar]
- 108.Thony-Meyer L. A heme chaperone for cytochrome c biosynthesis. Biochemistry. 2003;42:13099–13105. doi: 10.1021/bi035598c. [DOI] [PubMed] [Google Scholar]
- 109.Feissner RE, Richard-Fogal CL, Frawley ER, Kranz RG. ABC transporter-mediated release of a haem chaperone allows cytochrome c biogenesis. Mol Microbiol. 2006;61:219–231. doi: 10.1111/j.1365-2958.2006.05221.x. [DOI] [PubMed] [Google Scholar]
- 110.Christensen O, Harvat EM, Thony-Meyer L, Ferguson SJ, Stevens JM. Loss of ATP hydrolysis activity by CcmAB results in loss of c-type cytochrome synthesis and incomplete processing of CcmE. Febs J. 2007;274:2322–2332. doi: 10.1111/j.1742-4658.2007.05769.x. [DOI] [PubMed] [Google Scholar]
- 111.Schulz H, Fabianek RA, Pellicioli EC, Hennecke H, Thony-Meyer L. Heme transfer to the heme chaperone CcmE during cytochrome c maturation requires the CcmC protein, which may function independently of the ABC-transporter CcmAB. Proc Natl Acad Sci U S A. 1999;96:6462–6467. doi: 10.1073/pnas.96.11.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kadokura H, Katzen F, Beckwith J. Protein disulfide bond formation in prokaryotes. Annu Rev Biochem. 2003;72:111–135. doi: 10.1146/annurev.biochem.72.121801.161459. [DOI] [PubMed] [Google Scholar]
- 113.Nakamoto H, Bardwell JC. Catalysis of disulfide bond formation and isomerization in the Escherichia coli periplasm. Biochim Biophys Acta. 2004;1694:111–119. doi: 10.1016/j.bbamcr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 114.Beckman DL, Kranz RG. Cytochromes c biogenesis in a photosynthetic bacterium requires a periplasmic thioredoxin-like protein. Proc Natl Acad Sci U S A. 1993;90:2179–2183. doi: 10.1073/pnas.90.6.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Monika EM, Goldman BS, Beckman DL, Kranz RG. A thioreduction pathway tethered to the membrane for periplasmic cytochromes c biogenesis; in vitro and in vivo studies. J Mol Biol. 1997;271:679–692. doi: 10.1006/jmbi.1997.1227. [DOI] [PubMed] [Google Scholar]
- 116.Turkarslan S, Sanders C, Ekici S, Daldal F. Compensatory thio-redox interactions between DsbA, CcdA and CcmG unveil the apocytochrome c holdase role of CcmG during cytochrome c maturation. Mol Microbiol. 2008;70:652–666. doi: 10.1111/j.1365-2958.2008.06441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sanders C, Turkarslan S, Lee DW, Onder O, Kranz RG, Daldal F. The cytochrome c maturation components CcmF, CcmH, and CcmI form a membrane-integral multisubunit heme ligation complex. J Biol Chem. 2008;283:29715–29722. doi: 10.1074/jbc.M805413200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ren Q, Ahuja U, Thony-Meyer L. A bacterial cytochrome c heme lyase. CcmF forms a complex with the heme chaperone CcmE and CcmH but not with apocytochrome c. J Biol Chem. 2002;277:7657–7663. doi: 10.1074/jbc.M110979200. [DOI] [PubMed] [Google Scholar]
- 119.Sanders C, Boulay C, Daldal F. Membrane-spanning and periplasmic segments of CcmI have distinct functions during cytochrome c Biogenesis in Rhodobacter capsulatus. J Bacteriol. 2007;189:789–800. doi: 10.1128/JB.01441-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sanders C, Deshmukh M, Astor D, Kranz RG, Daldal F. Overproduction of CcmG and CcmFH(Rc) fully suppresses the c-type cytochrome biogenesis defect of Rhodobacter capsulatus CcmI-null mutants. J Bacteriol. 2005;187:4245–4256. doi: 10.1128/JB.187.12.4245-4256.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lang SE, Jenney FE, Jr, Daldal F. Rhodobacter capsulatus CycH: a bipartite gene product with pleiotropic effects on the biogenesis of structurally different c-type cytochromes. J Bacteriol. 1996;178:5279–5290. doi: 10.1128/jb.178.17.5279-5290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fabianek RA, Hofer T, Thony-Meyer L. Characterization of the Escherichia coli CcmH protein reveals new insights into the redox pathway required for cytochrome c maturation. Arch Microbiol. 1999;171:92–100. doi: 10.1007/s002030050683. [DOI] [PubMed] [Google Scholar]
- 123.Deshmukh M, May M, Zhang Y, Gabbert KK, Karberg KA, Kranz RG, Daldal F. Overexpression of ccl1-2 can bypass the need for the putative apocytochrome chaperone CycH during the biogenesis of c-type cytochromes. Mol Microbiol. 2002;46:1069–1080. doi: 10.1046/j.1365-2958.2002.03212.x. [DOI] [PubMed] [Google Scholar]
- 124.Akiyama S, Takahashi S, Ishimori K, Morishima I. Stepwise formation of alpha-helices during cytochrome c folding. Nat Struct Biol. 2000;7:514–520. doi: 10.1038/75932. [DOI] [PubMed] [Google Scholar]
- 125.Colon W, Elove GA, Wakem LP, Sherman F, Roder H. Side chain packing of the N- and C-terminal helices plays a critical role in the kinetics of cytochrome c folding. Biochemistry. 1996;35:5538–5549. doi: 10.1021/bi960052u. [DOI] [PubMed] [Google Scholar]
- 126.Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 2009;106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Swem DL, Swem LR, Setterdahl A, Bauer CE. Involvement of SenC in assembly of cytochrome c oxidase in Rhodobacter capsulatus. J Bacteriol. 2005;187:8081–8087. doi: 10.1128/JB.187.23.8081-8087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Banci L, Bertini I, Ciofi-Baffoni S, Katsari E, Katsaros N, Kubicek K, Mangani S. A copper(I) protein possibly involved in the assembly of CuA center of bacterial cytochrome c oxidase. Proc Natl Acad Sci U S A. 2005;102:3994–3999. doi: 10.1073/pnas.0406150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abriata LA, Banci L, Bertini I, Ciofi-Baffoni S, Gkazonis P, Spyroulias GA, Vila AJ, Wang S. Mechanism of Cu(A) assembly. Nat Chem Biol. 2008;4:599–601. doi: 10.1038/nchembio.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deshmukh M, Turkarslan S, Astor D, Valkova-Valchanova M, Daldal F. The dithiol:disulfide oxidoreductases DsbA and DsbB of Rhodobacter capsulatus are not directly involved in cytochrome c biogenesis, but their inactivation restores the cytochrome c biogenesis defect of CcdA-null mutants. J Bacteriol. 2003;185:3361–3372. doi: 10.1128/JB.185.11.3361-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Frangipani E, Haas D. Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosa PAO1. FEMS Microbiol Lett. 2009;298:234–240. doi: 10.1111/j.1574-6968.2009.01726.x. [DOI] [PubMed] [Google Scholar]
- 132.Glerum DM, Shtanko A, Tzagoloff A. SCO1 and SCO2 act as high copy suppressors of a mitochondrial copper recruitment defect in Saccharomyces cerevisiae. J Biol Chem. 1996;271:20531–20535. doi: 10.1074/jbc.271.34.20531. [DOI] [PubMed] [Google Scholar]
- 133.Banci L, Bertini I, Ciofi-Baffoni S, Kozyreva T, Mori M, Wang S. Sco proteins are involved in electron transfer processes. J Biol Inorg Chem. 2011;16:391–403. doi: 10.1007/s00775-010-0735-x. [DOI] [PubMed] [Google Scholar]
- 134.Buhler D, Rossmann R, Landolt S, Balsiger S, Fischer HM, Hennecke H. Disparate pathways for the biogenesis of cytochrome oxidases in Bradyrhizobium japonicum. J Biol Chem. 2011;285:15704–15713. doi: 10.1074/jbc.M109.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Masuda S, Matsumoto Y, Nagashima KV, Shimada K, Inoue K, Bauer CE, Matsuura K. Structural and functional analyses of photosynthetic regulatory genes regA and regB from Rhodovulum sulfidophilum, Roseobacter denitrificans, and Rhodobacter capsulatus. J Bacteriol. 1999;181:4205–4215. doi: 10.1128/jb.181.14.4205-4215.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Seib KL, Jennings MP, McEwan AG. A Sco homologue plays a role in defence against oxidative stress in pathogenic Neisseria. FEBS Lett. 2003;546:411–415. doi: 10.1016/s0014-5793(03)00632-x. [DOI] [PubMed] [Google Scholar]
- 137.Borsetti F, Tremaroli V, Michelacci F, Borghese R, Winterstein C, Daldal F, Zannoni D. Tellurite effects on Rhodobacter capsulatus cell viability and superoxide dismutase activity under oxidative stress conditions. Res Microbiol. 2005;156:807–813. doi: 10.1016/j.resmic.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 138.Onder O, Turkarslan S, Sun D, Daldal F. Overproduction or absence of the periplasmic protease DegP severely compromises bacterial growth in the absence of the dithiol: disulfide oxidoreductase DsbA. Mol Cell Proteomics. 2008;7:875–890. doi: 10.1074/mcp.M700433-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Saier MH, Jr, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 140.Puig S, Lee J, Lau M, Thiele DJ. Biochemical and genetic analyses of yeast and human high affinity copper transporters suggest a conserved mechanism for copper uptake. J Biol Chem. 2002;277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 141.Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Dowhan W, Bogdanov M. Lipid-protein interactions as determinants of membrane protein structure and function. Biochem Soc Trans. 2011;39:767–774. doi: 10.1042/BST0390767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tiefenbrunn T, Liu W, Chen Y, Katritch V, Stout CD, Fee JA, Cherezov V. High Resolution Structure of the ba3 Cytochrome c Oxidase from Thermus thermophilus in a Lipidic Environment. PLoS One. 2011;6:e22348. doi: 10.1371/journal.pone.0022348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- 147.Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. Age-dependent decline in the cytochrome c oxidase activity in rat heart mitochondria: role of cardiolipin. FEBS Lett. 1997;406:136–138. doi: 10.1016/s0014-5793(97)00264-0. [DOI] [PubMed] [Google Scholar]
- 148.Aygun-Sunar S, Mandaci S, Koch HG, Murray IV, Goldfine H, Daldal F. Ornithine lipid is required for optimal steady-state amounts of c-type cytochromes in Rhodobacter capsulatus. Mol Microbiol. 2006;61:418–435. doi: 10.1111/j.1365-2958.2006.05253.x. [DOI] [PubMed] [Google Scholar]
- 149.Huang Y, Reimann J, Singh LM, Adelroth P. Substrate binding and the catalytic reactions in cbb3-type oxidases: the lipid membrane modulates ligand binding. Biochimica et biophysica acta. 2010;1797:724–731. doi: 10.1016/j.bbabio.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 150.Aygun-Sunar S, Bilaloglu R, Goldfine H, Daldal F. Rhodobacter capsulatus OlsA is a bifunctional enzyme active in both ornithine lipid and phosphatidic acid biosynthesis. J Bacteriol. 2007;189:8564–8574. doi: 10.1128/JB.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Minnikin DE, Abdolrahimzadeh H. The replacement of phosphatidylethanolamine and acidic phospholipids by an ornithine-amide lipid and a minor phosphorus-free lipid in Pseudomonas fluorescens NCMB 129. FEBS Lett. 1974;43:257–260. doi: 10.1016/0014-5793(74)80655-1. [DOI] [PubMed] [Google Scholar]
- 152.Benning C, Huang ZH, Gage DA. Accumulation of a novel glycolipid and a betaine lipid in cells of Rhodobacter sphaeroides grown under phosphate limitation. Arch Biochem Biophys. 1995;317:103–111. doi: 10.1006/abbi.1995.1141. [DOI] [PubMed] [Google Scholar]
- 153.Geiger O, Rohrs V, Weissenmayer B, Finan TM, Thomas-Oates JE. The regulator gene phoB mediates phosphate stress-controlled synthesis of the membrane lipid diacylglyceryl-N,N,N-trimethylhomoserine in Rhizobium (Sinorhizobium) meliloti. Mol Microbiol. 1999;32:63–73. doi: 10.1046/j.1365-2958.1999.01325.x. [DOI] [PubMed] [Google Scholar]
- 154.Elsen S, Swem LR, Swem DL, Bauer CE. RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol Mol Biol Rev. 2004;68:263–279. doi: 10.1128/MMBR.68.2.263-279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hunter NC, Daldal F, Thurnauer M, Beaty JT. The Purple Photorophic Bacteria. Springer; Dordrecht: 2008. [Google Scholar]