Abstract
Ataxia telangiectasia mutated (ATM) and DNA-PKcs are members of the PI-3-like family of serine/threonine kinases that phosphorylate serines or threonines when positioned adjacent to a glutamine residue (SQ/TQ). Both kinases are activated rapidly by DNA double strand breaks (DSBs) and regulate the function of proteins involved in DNA damage responses (DDR). In developing lymphocytes, DSBs are generated during V(D)J recombination, which is required to assemble the second exon of all antigen receptor genes. This reaction is initiated through a DNA cleavage step by the RAG1 and RAG2 proteins, which together comprise an endonuclease that generates DSBs at the border of two recombining gene segments and their flanking recombination signals. This DNA cleavage step is followed by a joining step, during which pairs of DNA coding and signal ends are ligated to form a coding joint and a signal joint respectively. ATM and DNA-PKcs are integrally involved in the repair of both signal and coding ends, but the targets of these kinases involved in the repair process have not been fully elucidated. In this regard, the RAG1 and RAG2 proteins, which each have several SQ/TQ motifs, have been implicated in the repair of RAG-mediated DSBs. Here we use a previously developed approach for studying chromosomal V(D)J recombination that has been modified to allow for the analysis of RAG1 and RAG2 function. Using this approach we show that phosphorylation of RAG1 or RAG2 by ATM or DNA-PKcs at SQ/TQ consensus sites is dispensable for the joining step of V(D)J recombination.
Introduction
Lymphocyte antigen receptor genes are assembled by the process of V(D)J recombination, whereby different variable (V), diversity (D) and joining (J) gene segments are appended to generate the second exon of all antigen receptor genes (1). The V(D)J recombination reaction can be divided into DNA cleavage and joining steps. The DNA cleavage step is carried out by the RAG1 and RAG2 proteins, which together form the RAG endonuclease that introduces DNA double strand breaks (DSBs) at the borders of two recombining gene segments and their associated RAG recognition sites, termed recombination signals (RSs) (2). Proteins belonging to the non-homologous end-joining (NHEJ) pathway of DNA DSB repair process and join the resulting pair of hairpin-sealed coding ends and blunt phosphorylated signal ends to generate a coding joint and a signal joint respectively (3, 4).
The ATM and DNA-PKcs kinases are members of the PI-3-like family of serine/threonine kinases and are activated early in the DSB response (5–7). Once activated, ATM and DNA-PKcs phosphorylate and regulate a host of downstream proteins that function in DNA damage responses and DSB repair (5–10). ATM and DNA-PKcs specifically phosphorylate serine and threonine residues that are directly followed by glutamine (SQ/TQ motifs). Both kinases are activated by RAG DSBs and are integrally involved in the processing and joining of coding and signal ends (3, 4, 11–19). ATM functions to stabilize coding ends in post-cleavage complexes until they can be joined (16). DNA-PKcs promotes the hairpin-opening activity of the Artemis nuclease (3, 4, 20, 21). ATM and DNA-PKcs also have overlapping activities that are critical for the efficient repair of signal ends (12, 13).
A majority of the known functions attributed to ATM and DNA-PKcs during the process of V(D)J recombination depend on their kinase activities, suggesting that they modulate downstream targets in DSB repair pathways (3, 4, 12, 13, 16). In this regard, many proteins involved in the repair of RAG DSBs can be phosphorylated by ATM or DNA-PKcs either in vitro or in vivo. In addition to ATM and DNA-PKcs themselves, these proteins include Ku70, Ku80, XRCC4, DNA Ligase IV, Artemis, XLF, H2AX and the components of the MRN complex (Mre11, Rad50, and Nbs1) (3, 4, 9, 19, 22–36).
In addition to their central role in DNA cleavage, the RAG proteins also have been implicated in repairing the DSBs generated by their endonuclease function (37–39). After DNA cleavage in vitro, the RAG proteins remain closely associated with signal ends in post-cleavage complexes (40, 41). Subsequent dissociation of the RAG proteins from signal ends is required for the joining of these DNA ends in vitro (42). In contrast, the role of the RAG proteins during coding joint formation remains unclear. RAG2 possesses three SQ/TQ motifs, and RAG1 has ten, any of which could be phosphorylated by ATM and/or DNA-PKcs to modulate RAG function during the repair steps of V(D)J recombination. In this regard, DNA-PKcs has been shown to phosphorylate RAG2 on a conserved SQ motif (serine 365) in vitro (43). However, no significant effects on V(D)J recombination were observed in cells expressing RAG2 containing a serine 365 to alanine mutation (44). In addition, cells expressing a mutant form of RAG1 with two conserved SQ motifs (S479 and S913) mutated to alanine also exhibited no defects in V(D)J recombination (44). Since only a subset of RAG1 and RAG2 SQ/TQ motifs were analyzed, it remains possible that phosphorylation of the RAG proteins by ATM or DNA-PKcs at other SQ/TQ motifs is required for the normal repair of RAG-mediated DSBs. Moreover, in this study, V(D)J recombination was analyzed on extrachromosomal plasmid substrates where the requirements for repair may be different than for RAG DSBs generated within the context of the chromosome. Indeed, although neither ATM nor Mre11 deficiency leads to defects in the repair of RAG DSBs generated on extrachromosomal plasmid substrates, repair of chromosomal RAG DSBs is defective in both mutant backgrounds (14–16, 18, 29, 30, 45, 46).
We have previously developed an experimental approach that allows for the induction of chromosomal V(D)J recombination in abelson transformed pre-B cells, hereafter referred to as abl pre-B cells(16). Treatment of abl pre-B cells with the abl kinase inhibitor STI571 leads to (i) G1 cell cycle arrest, (ii) induction of RAG gene expression and (iii) rearrangement of the endogenous IgLk locus, and (iv) robust recombination at chromosomally integrated retroviral recombination substrates (16, 47). Here we modify this approach using RAG1−/− and RAG2−/− abl pre-B cells to study RAG1 and RAG2 function. When treated with STI571, RAG1−/− and RAG2−/− abl pre-B cells undergo G1 cell cycle arrest, but do not generate RAG DSBs. Constitutive expression of RAG1 or RAG2 by retroviral transduction of RAG1−/− or RAG2−/− abl pre-B cells, respectively, restores STI571-inducible chromosomal V(D)J recombination. By comparing cells that have been reconstituted with either wild type or mutant versions of each RAG, we now analyze the effects of distinct mutations in RAG SQ/TQ motifs on chromosomal V(D)J recombination.
Materials and Methods
Generation of retroviral vectors encoding wild type and mutant RAG proteins
N-terminal FLAG-tagged versions of RAG1 and RAG2 were generated in the pSP72 shuttle vector (Promega, P2191). To this end we first generated a version of pSP72 (pSP72-FLAG) that contained a linker composed of the FLAG-sense and FLAG-antisense oligonucleotides that encode a single FLAG tag (Supplemental Table I). The wild type RAG2 cDNA was modified by PCR to have a 5′ HindIII site that allowed for cloning of the RAG2 cDNA in-frame and downstream of the FLAG sequence in pSP72-FLAG to generate pSP72-FLAG-RAG2WT. To generate pSP72-FLAG-RAG1WT the wild type RAG1 cDNA (RAG1WT) was similarly cloned into pSP72-FLAG except for use of BmtI restrictions sites in RAG1WT and pSP72-FLAG.
To generate the FLAG-RAG2WT-GFP cDNA a previously published RAG2-GFP cDNA was digested with BstXI and BglII to release a fragment that contained the 3′ portion of RAG2 in frame with a complete GFP sequence (48). This fragment was then cloned into pSP72-FLAG-RAG2WT to generate pSP72-FLAG-RAG2WT-GFP. To generate the FLAG-GFP-RAG1WT cDNA the GFP cDNA was modified to have 5′ and 3′ BmtI sites and was subcloned into pSP72-FLAG-RAG1WT to generate pSP72-FLAG-GFP-RAG1WT.
The mutant RAG2 cDNA with the 2 SQ and lone TQ motifs converted to AQ (RAG23AQ) was generated using a PCR based mutagenesis approach. This was done through sequential overlapping PCR using complementary oligonucleotide pairs that contained the three mutations. These sense and antisense pairs included S165A, T264A and S365A olignucleotides (Supplemental Table I). A mutant BstXI/SacI 0.9 kb fragment was generated using these oligonucleotides coupled with the 5′ RAG2-HindIII and the PSP72DS oligonucleotide (Supplemental Table I). pSP72-FLAG-RAG23AQ and pSP72-FLAG-RAG23AQ-GFP were generated by replacing the 0.9 kb BstXI/SacI fragment in pSP72-FLAG-RAG2WT and pSP72-FLAG-RAG2WT-GFP with the mutant PCR fragment.
The mutant RAG1 cDNA with the 6SQ and 4 TQ motifs changed to AQ (RAG110AQ) was also generated using a sequential overlapping PCR mutagenesis approach. The overlapping pairs of oligonucleotides included sense and antisense oligonucleotide pairs T76A, T136A, S191A, T250A, S479A, S635A, S738A, T859A, S913A and S1034A oligonucleotides. These mutations were clustered in three fragments; BmtI/SphI 1.5 kb fragment (T76A, T136A, S191A, T250A and S479A), SphI/MluI 1.5 kb fragment (S635A, S738A, T859A and S913A) and a MluI/BglII 0.2 kb fragment (S1034A). The oligonucleotides used to flank the BmtI/SphI 1.5 kb fragment were RAG1-BmtI and S635A antisense. The oligonucleotides used to flank the SphI/MluI 1.5 kb fragment were S479A sense and PSP72DS. The oligonucleotides used to flank the MluI/SfiI 0.2 kb fragment were S913A sense and PSP72DS. These fragments were shuttle cloned into pSP72-FLAG-RAG1WT and pSP72-FLAG-GFP-RAG1WT to generate pSP72-FLAG-RAG110AQ and pSP72-FLAG-GFP-RAG110AQ. Conditions for all PCR reactions were 95°C for 5 minutes followed by 15 cycles of 92°C for 1 minute, 57°C for 1.5 minutes, and 72°C for 1.5 minutes. The different RAG1 and RAG2 cDNAs were then subcloned into the pCST-iThy1.1 retroviral vector and then introduced into abl pre-B cells by retroviral transduction as previously described(16, 49).
Mice
Animals were housed in a specific pathogen-freeanimal facility at Washington University. Animal protocols were approved by the Washington University Institutional Animal Care and Use Committee.
Generation and culture of abl pre-B cell lines
V-abl-transformed pre-B cells were generated by culturing bone marrow from 3–5 week old mice with the pMSCV v-abl retrovirus as described previously (16). To generate RAG2−/−: INV and RAG1−/−: INV cell lines, RAG2−/− and RAG1−/− abl pre-B cells were infected with pMX-INV, and cells that had integrated the recombination substrate were sorted based on CD4 expression. These cells were then subcloned, and individual subclones were analyzed by Southern blotting to identify cells with single pMX-INV integrants. Cells that underwent robust pMX-INV rearrangement when reconstituted with RAG1 (for RAG1−/−: INV abl pre-B cells) or RAG2 (for RAG1−/−: INV abl pre-B cells) were chosen for analysis. RAG2−/−: DELSJ and RAG1−/−: DELSJ cell lines were made identically except cells were infected with pMX-DELSJ. Retroviral transduction and maintenance of abl pre-B cell cultures was carried out as previously described (16). Cells were treated with 3.0 μM STI571 (Novartis) for the indicated times at 106 cells/ml as previously described (16).
Southern blotting and PCR analysis
Southern blot analyses for pMX-INV and pMX-DELSJ rearrangements were carried out using the C4 and C4b probes as previously described (13, 16). Quantitative PCR analyses for VkJk rearrangements using the VkD and Jk2-3′ oligonucleotides (Supplemental Table I). PCR products were hybridized with the probe Jk2P (Supplemental Table I). Quantitative loading control PCR for the IL-2 gene was performed as described previously (16). Conditions for both PCR reactions were 95°C for 5 minutes followed by 30 cycles of 92°C for 1 minute, 60°C for 1.5 minutes, and 72°C for 1.5 minutes. Densitometric measurements of band intensities in Fig. 2D were determined using ImageJ software (rsbweb.nih.gov/ij/).
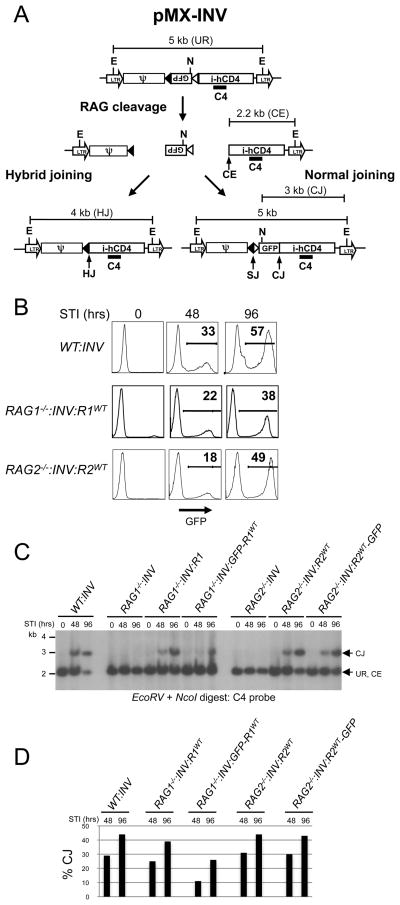
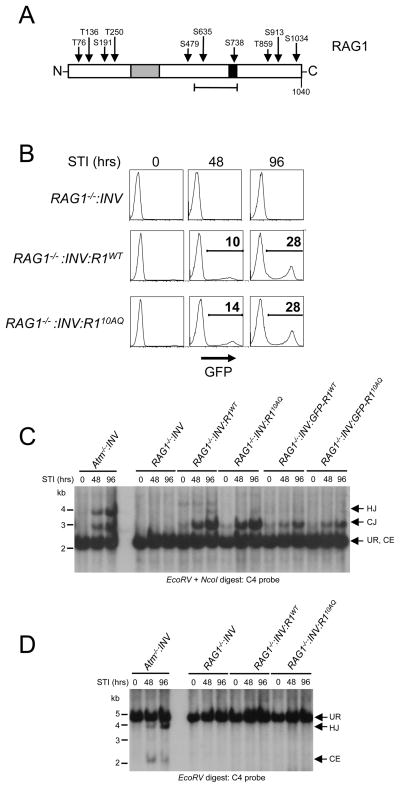
Figure 2. Rescue of chromosomal V(D)J recombination in RAG1−/− and RAG2−/− abl pre-B cells.
A, Schematic of the pMX-INV retroviral recombination substrate. Unrearranged pMX-INV (UR) and pMX-INV with a coding end intermediate (CE), and completed coding joint (CJ), signal joint (SJ) and hybrid joint (HJ) products are shown. The approximate positions of the EcoRV (E) and NcoI (N) sites, and C4 probe are indicated. B, Flow cytometric analysis of GFP expression at various times (hours, hrs) after treatment with STI571 for wild type (WT:INV) abl pre-B cells, (RAG1−/−:INV) pre-B cells transduced with pCST-FLAG-RAG1WT-iThy1.1 (RAG1−/−:INV:R1WT), RAG2−/− abl pre-B cells transduced with pCST-FLAG-RAG2WT-iThy1.1 (RAG2−/−:INV:R2WT) each containing a single pMX-INV genomic integrant. C, EcoRV + NcoI-digested genomic DNA probed with the C4 probe. DNA samples from abl pre-B cells from B and RAG1−/− pre-B cells transduced with pCST-FLAG-GFP-RAG1WT-iThy1.1 (RAG1−/−:INV:GFP-R1WT) and RAG2−/− pre-B cells transduced with pCST-FLAG-RAG2WT-GFP-iThy1.1 (RAG2−/−:INV:R2WT-GFP) and treated with STI571 for the indicated times. The rearrangement products (HJ and CJ) and intermediates (CE) for pMX-INV are shown. Molecular weight markers are shown. D, Relative quantification of coding joints (% CJ) formed in C.
Northern blotting analysis
Northern blot analysis was carried out as previously described using a 0.5 kb EcoRV/SphI RAG1 cDNA fragment or a 0.8 kb PstI RAG2 cDNA fragment (16).
Immunoprecipitation and Western blot analyses
Immunoprecipitation was carried out from 1 × 109 abl pre-B cells using anti-FLAG monoclonal antibody (Sigma, F3040) using a previous protocol except that anti-FLAG antibody (1:500) was pre-bound to Protein A agarose resin beads (50). Immunoprecipitates were separated on a 10% SDS-PAGE gel and transferred onto Immobilon-P PVDF Membrane (Millipore, IPVH00010). Primary antibodies for blotting with were anti-RAG1, 1:500 (Santa Cruz, H-300 sc5599) or anti-RAG2, 1:200 (Santa Cruz, C-19 sc7623). Secondary antibodies were used at the following concentration dilutions: donkey anti-rabbit F(ab′)2 Fragment, 1:5000 (Fisher, 45-000-683) for anti-RAG1 primary; and goat anti-mouse, 1:5000 for anti-RAG2 primary (Invitrogen, 62–6520). Development was performed using the ECL Plus horseradish peroxidase detection kit per the manufacturer’s instructions (Amersham, RPN2132).
Flow cytometry
Flow cytometric analyses were carried out using a BD FACS Caliber and data analyzed using FlowJo 4.6.2 for Macintosh (Tree Star). Flow cytometric cell sorting was carried out using a BD Aria Cell Sorter. Rat anti-Thy1.1-PE antibody, 1:2000 (BD, #551401) was used to detect cells expressing Thy1.1.
Results
Experimental approach to study RAG function in chromosomal DSB repair
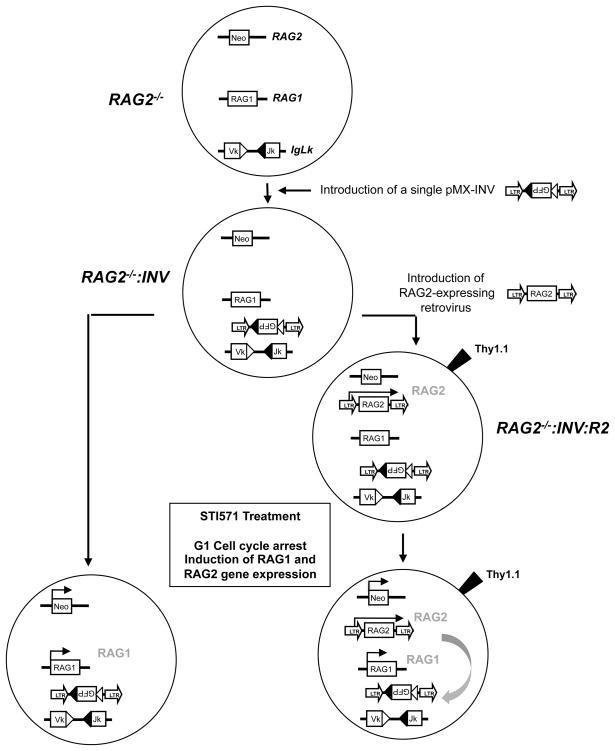
Our strategy to reconstitute inducible chromosomal V(D)J recombination in RAG2−/− abl pre-B cells is shown in Figure 1. A similar strategy was used for reconstituting inducible chromosomal V(D)J recombination in RAG1−/− abl pre-B cells. Initially, several independently derived RAG1−/− and RAG2−/− abl pre-B cell lines were generated with single integrants of the pMX-INV retroviral recombination substrate (RAG1−/−: INV and RAG2−/−: INV abl pre-B cells, respectively) (Fig. 2A) (16). pMX-INV has a single pair of RSs that flank an anti-sense GFP cDNA (Fig. 2A). Recombination of pMX-INV occurs by inversion, placing the GFP cassette in the sense orientation, which permits GFP expression as an indicator of successful rearrangement. Full-length wild type RAG1 and RAG2 cDNAs with single N-terminal FLAG tags were introduced into the pCST-iThy1.1 retroviral vector, generating pCST-FLAG-RAG1WT-iThy1.1 and pCST-FLAG-RAG2WT-iThy1.1 respectively (Supplemental Fig. 1A). pCST-iThy1.1 has an IRES-Thy1.1 cassette, permitting flow cytometric purification of cells with retroviral integrants based on Thy1.1 expression (Fig. 1 and Supplemental Fig. 1A and B).
Figure 1. Experimental approach for studying RAG2 mutations.
RAG2−/− abl pre-B cells are first infected with pMX-INV and cells lines with single pMX-INV integrants (RAG2−/−:INV) were isolated. RAG2−/−:INV abl pre-B cells were then infected with retroviruses encoding wild type RAG2 (or RAG2 mutants) with Thy1.1 as an indicator of retroviral transduction to generate RAG2−/−:INV:R2 abl pre-B cells. These retroviruses permit constitutive expression of RAG2 (shown in gray). Treatment with STI571 leads to G1 cell cycle arrest and induction of transcription at the RAG1 and RAG2 loci. As the RAG2 gene has been replaced by the neomycin resistance gene treatment with STI571 leads only to RAG1 protein expression (shown in gray). This coupled with RAG2 expression from the retrovirus leads to rearrangement of the pMX-INV and the endogenous IgLk locus (gray arrow).
Thy1.1-expressing RAG2−/−: INV abl pre-B cells infected with pCST-FLAG-RAG2WT-iThy1.1 (referred to as RAG2−/−: INV:R2WT abl pre-B cells) constitutively express wild type RAG2 transcripts from the retrovirus (Supplemental Fig. 1C). Treatment of these cells with STI571 leads to G1 cell cycle arrest and induction of RAG1 gene expression, which, when coupled with retroviral RAG2 expression, leads to rearrangement of pMX-INV (Fig. 2B–D and Supplemental Fig. 1). Rearrangement of pMX-INV is evidenced by GFP expression in 18% and 49% of RAG2−/−: INV:R2WT abl pre-B cells treated with STI571 for 48 and 96 hours respectively (Fig. 2B). Moreover, Southern blot analyses revealed a 3kb EcoRV/NcoI C4 probe hybridizing fragment, indicative of pMX-INV coding joint formation in STI571-treated RAG2−/−: INV:R2WT abl pre-B cells, but not in RAG2−/−: INV abl pre-B cells (Figs. 2C and 2D). Rearrangement of pMX-INV in RAG2−/−: INV:R2WT abl pre-B cells approached levels observed in wild type abl pre-B cells (WT:INV) (Figs. 2B–D). Using an identical approach, RAG1−/−: INV abl pre-B cells were reconstituted with wild type RAG1 (RAG1−/−: INV:R1WT abl pre-B cells) using the pCST-FLAG-RAG1WT-iThy1.1 retroviral vector (Fig. 2A). Treatment of RAG1−/−: INV:R1WT abl pre-B cells with STI571 also led to inducible rearrangement of pMX-INV (Figs. 2B–D).
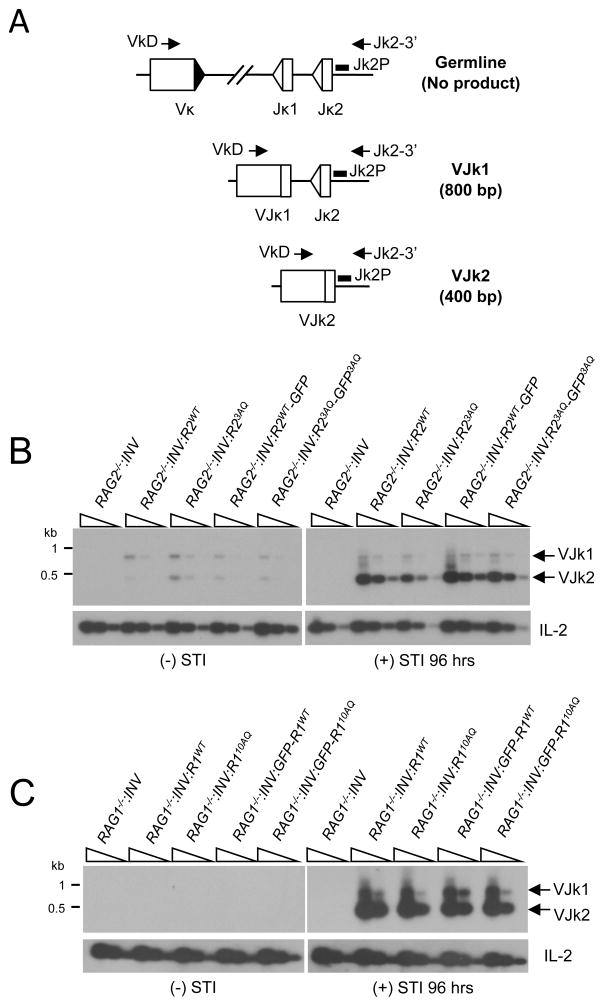
Abl pre-B cells undergo inducible rearrangement of the endogenous IgLk locus upon inhibition of the abl kinase with STI571. The murine IgLk locus contains approximately 250 Vk gene segments and 4 functional Jk gene segments. To determine if IgLk gene rearrangement is also inducible in RAG2−/−: INV:R2WT and RAG1−/−: INV:R1WT abl pre-B cells treated with STI571, genomic DNA from these cells was assayed by PCR using a degenerate Vk oligonucleotide (VkD) and an oligonucleotide downstream of Jk2 (Jk2-3′). The combination of these two oligonulceotides detects Vk rearrangements to Jk1 and Jk2 (Fig. 3A). VkJk rearrangements were readily detected in STI571-treated RAG2−/−: INV:R2WT and RAG1−/−: INV:R1WT abl pre-B cells (Figs. 3B and 3C). Prior to STI571 treatment, these cells exhibit only low levels of VkJk rearrangements (Figs. 3B and 3C). Finally, RAG2−/−: INV and RAG1−/−: INV abl pre-B cells had no detectable IgLk rearrangements regardless of whether or not they were treated with STI571 (Figs. 3B and 3C). Together, these findings demonstrate that chromosomal V(D)J recombination at either pMX-INV or the endogenous IgLk locus is readily inducible in RAG2−/−: INV or RAG1−/−: INV abl pre-B cells reconstituted with wild type RAG2 or RAG1 respectively.
Figure 3. IgLk rearrangement in abl pre-B cells expressing RAG1 and RAG2 mutants.
A, Schematic of PCR strategy for amplifying VkJk1 and VkJk2 coding joints. The RSs (triangles) and relative positions of the VkD and Jk2-3′ oligonucleotides used for PCR amplification and the Jk2P oligonucleotide used as a probe are shown. VkJk1 and VkJk2 coding joint formation results in 800bp and 400bp PCR products respectively. B–C, Southern blot analysis of VkJk1 and VkJk2 coding joint PCR products amplified from genomic DNA (five fold dilutions) obtained before and after 96 hrs in culture with STI571 from (B) RAG2−/−:INV, RAG2−/−:INV:R2WT, RAG2−/−:INV:R23AQ, RAG2−/−:INV:R2WT-GFP and RAG2−/− :INV:R23AQ-GFP abl pre-B cells and (C) RAG1−/−:INV, RAG1−/−:INV:R1WT, RAG1−/− :INV:R110AQ, RAG1−/−:INV:GFP-R1WT, or RAG1−/−:INV:GFP-R110AQ abl pre-B cells. IL-2 gene PCR is also shown as a loading control.
Chromosomal V(D)J recombination catalyzed by RAG1 and RAG2 GFP fusion proteins
To assay the effects of different RAG1 and RAG2 mutations on V(D)J recombination, we required cells with equivalent levels of wild type and mutant RAG proteins. To this end, we generated a version of RAG2 with an N-terminal FLAG tag and a C-terminal GFP fusion and introduced this cDNA into the pCST-iThy1.1 retroviral vector (Supplemental Fig. 1D and E). RAG2WT-GFP expression was readily detected by flow cytometric analyses of RAG2−/−:INV pre-B cells infected with the pCST-RAG2WT-GFP-iThy1.1 retroviral vector (RAG2−/−:INV:R2WT-GFP) (Supplemental Fig. 1D). Moreover, treatment of RAG2−/−:INV:R2WT-GFP abl pre-B cells with STI571 led to inducible rearrangement of both pMX-INV and the IgLk locus (Figs. 2C, 2D and 3B). In contrast, the RAG1WT-GFP C-terminal fusion was not expressed at significant levels and did not rescue rearrangement in RAG1−/−:INV cells (data not shown). Accordingly, we generated and expressed a FLAG-tagged N-terminal GFP-RAG1WT fusion protein in RAG1−/−:INV abl pre-B cells (RAG1−/−:INV:GFP-R1WT) (Supplemental Fig. 1E). The N-terminal GFP-RAG1 fusion variant rescued STI571-inducible rearrangement, albeit at lower levels than those observed in wild type or RAG1−/−:INV:R1WT abl pre-B cells (Figs. 2C, 2D, 3C and Supplemental Fig. 1E). Together, these findings demonstrate that retroviral introduction of the relevant RAG/GFP fusion into RAG-deficient abl pre-B cells rescues STI571-inducible rearrangement of pMX-INV and the IgLk locus. Importantly, the levels of wild type GFP-RAG1WT, RAG2WT-GFP, and their corresponding mutants can be compared by flow cytometry.
Chromosomal V(D)J recombination in cells expressing an SQ/TQ RAG2 mutant
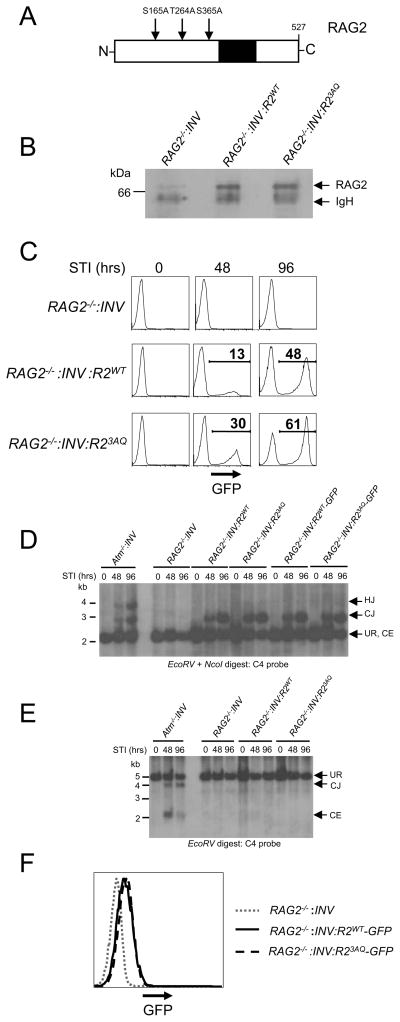
The murine RAG2 protein is 527 amino acids long and contains two SQ (S165 and S365) motifs and one TQ (T264) motif (Fig. 4A). These residues do not overlap with the PHD domain, which functions to tether RAG2 to methylated histone H3 present in chromatin (Fig. 4A) (51)(2). A version of FLAG-tagged RAG2 was generated in which the three SQ/TQ motifs were mutated to AQ (RAG23AQ). This mutant was introduced retrovirally into RAG2−/−:INV abl pre-B cells, yielding a cell line that expresses RAG23AQ (RAG2−/−:INV:R23AQ) at levels equivalent to wild type RAG2 in RAG2−/−:INV:R2WT abl pre-B cells (Fig. 4B). After treatment with STI571, robust rearrangement of pMX-INV was observed in both RAG2−/−:INV:R2WT and RAG2−/−:INV:R23AQ abl pre-B cells as evidenced by both flow cytometric analyses and Southern blotting (Figs. 4C and D). Analyses of another independently generated RAG2−/−:INV abl pre-B cell line that expressed either RAG2 or RAG23AQ yielded similar findings (Supplemental Fig. 2A and B). Moreover, STI571-treated RAG2−/−:INV:R2WT and RAG2−/−:INV:R23AQ abl pre-B cells have equivalent levels of IgLk Vk to Jk rearrangements (Fig. 3B).
Figure 4. SQ/TQ motifs in RAG2 are not required for efficient coding joint formation.
A, Schematic showing positions of the two SQ motifs and the single TQ motif in the RAG2 protein. The PHD domain is also shown (black). B, Western blot analysis of RAG2 protein expression in RAG2−/−:INV, RAG2−/−:INV:R2WT or RAG2−/−:INV:R23AQ pre-B cells. Samples were immunoprecipitated with an anti-FLAG antibody followed by immunoblotting with an anti-RAG2 antibody. C, Flow cytometric analysis of GFP expression after treatment of RAG2−/−:INV, RAG2−/−:INV:R2WT, and RAG2−/−:INV:R23AQ abl pre-B cells with STI571 for 0, 48 or 96 hours. D–E, Southern blot analysis of genomic DNA digested with EcoRV and NcoI (D) or EcoRV (E) and probed with the C4 probe. Atm−/−:INV, RAG2−/−:INV, RAG2−/−:INV:R2WT, RAG2−/−:INV:R23AQ, RAG2−/−:INV:R2WT-GFP, and RAG2−/−:INV:R23AQ-GFP abl pre-B cells were treated with STI571 for 0, 48 or 96 hours prior to harvesting DNA. Bands representing pMX-INV UR, CJ, HJ and CE are indicated. Molecular weight markers are also shown. F, Flow cytometric analysis of GFP expression in RAG2−/−:INV, (dotted line), RAG2−/−:INV:R2WT-GFP (solid line), and RAG2−/−:INV:R23AQ-GFP (dashed line) pre-B cells.
During chromosomal V(D)J recombination, deficiency of ATM leads to a 10–20% loss of coding ends from post-cleavage complexes (16). This dissociation results in diminished coding joint formation, an accumulation of unrepaired coding ends, and the formation of pMX-INV hybrid joints, the latter of which are produced by aberrant ligation of chromosomal pMX-INV coding and signal ends (Figs. 2A, 4D and 4E). To test whether these defects are linked to a loss of ATM-mediated RAG2 phosphorylation, we performed Southern blot analysis of genomic DNA from RAG2−/−:INV:R23AQ abl pre-B cells treated with STI571. We did not detect unrepaired pMX-INV coding ends or pMX-INV hybrid joints, which were both readily apparent in ATM-deficient abl pre-B cells (Figs. 4D and E). Similar results were observed when analyzing another independently derived RAG2−/−:INV abl pre-B cell reconstituted with RAG2WT or RAG23AQ (Supplemental Fig. 2B). Finally, V(D)J recombination was similar in RAG2−/−:INV abl pre-B cells that express RAG23AQ-GFP (RAG2−/−:INV-R23AQ-GFP) or RAG2WT-GFP (RAG2−/−:INV-R2WT-GFP) (Figs. 4D and 4F). Together, these data demonstrate that phosphorylation of RAG2 at SQ/TQ motifs is dispensable for efficient formation of chromosomal coding joints.
Coding joint formation in cells expressing SQ/TQ mutant RAG1
The mouse RAG1 protein is 1040 amino acids long and contains 4 TQ (T76, T136, T250 and T859) and 6 SQ (S191, S479, S635, S738, S913 and S1034) motifs (Fig. 5A). Two serines (S635 and S738) reside within a region that mediates RS heptamer binding and RAG2 interactions with one of these serines (S738) present in a zinc finger region (2). To investigate the potential function of these sites in coding joint formation, we developed mutant versions of FLAG-tagged RAG1 (RAG110AQ) and GFP-RAG1 (GFP-RAG110AQ) where all 10 of these SQ/TQ motifs are mutated to AQ. These proteins were expressed in RAG1−/−:INV abl pre-B cells, and stable clones with equivalent levels of RAG110AQ (RAG1−/−:INV:R110AQ) and RAG1WT (RAG1−/−:INV:R1WT) or GFP-RAG110AQ (RAG1−/−:INV:GFP-R110AQ) and GFP-RAG1WT (RAG1−/−:INV:GFP-R1WT) were selected for analysis (Fig. 5B and F).
Figure 5. SQ/TQ motifs in RAG1 are not required for efficient coding joint formation.
A, Schematic showing positions of the 6 SQ motifs and the 4 TQ motifs in the RAG1 protein. The zinc finger A (gray) and B (black) regions are shown. The heptamer-binding/RAG2 interacting region is also shown (bracket). B, Western blot analysis of RAG1 protein expression in RAG1−/−:INV, RAG1−/− :INV:R1WT or RAG1−/−:INV:R110AQ pre-B cells. C, Flow cytometry of GFP expression at various times in culture with STI571 in RAG1−/−, RAG1−/−:INV:R1WT, and RAG1−/−:INV:R110AQ pre-B cells. D-E, Southern blot analysis of genomic DNA from Atm−/−:INV, RAG1−/−:INV, RAG1−/−:INV:R1WT, RAG1−/−:INV:GFP-R1WT, and RAG1−/−:INV:GFP-R110AQ abl pre-B cells treated with STI571 for 0, 48 or 96 hours. Analysis was carried out as described in Fig. 4D and E. Molecular weight markers are shown. F, Flow cytometric analysis of GFP expression in RAG1−/−:INV, (dotted line), RAG1−/−:INV:GFP-R1WT (solid line), and RAG2−/−:INV:GFP-R110AQ (dashed line) pre-B cells analyzed in Fig. 5D.
After induction of V(D)J recombination, coding joint formation at pMX-INV and the IgLk locus proceeded with similar kinetics in RAG1−/−:INV:R110AQ and RAG1−/−:INV:R1WT abl pre-B cells (Figs. 3C, 5C and 5D). Similar results were obtained upon analysis of a second set of independently derived RAG1−/−:INV:R110AQ and RAG1−/−:INV:R1WT abl pre-B cell lines (Supplemental Fig. 2C and D). Moreover, neither unrepaired pMX-INV coding ends nor pMX-INV hybrid joints were detected in RAG1−/−:INV:R110AQ abl pre-B cells (Figs. 5D and 5E and Supplemental Fig. 2D). Similar results were obtained upon analysis of IgLk locus and pMX-INV rearrangement in RAG1−/−:INV:GFP-R1WT and RAG1−/−:INV:GFP-R110AQ abl pre-B cells (Figs. 3C and 5D and data not shown). Thus, we conclude that phosphorylation of RAG1 at any of the 10 SQ/TQ motifs is not essential for efficient coding joint formation.
Function of RAG1 and RAG2 during signal joint formation
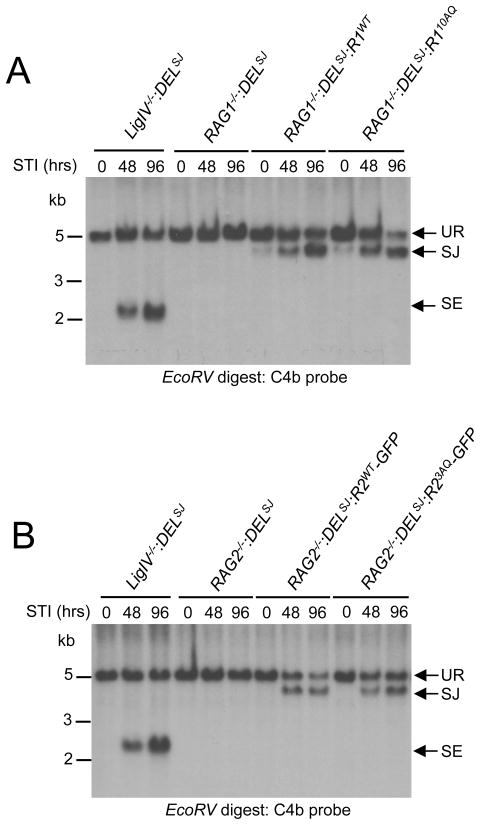
ATM and DNA-PKcs have redundant functions during signal joint formation, suggesting that they phosphorylate common downstream targets important for the repair of signal ends (12, 13). To determine if RAG1 or RAG2 are important targets of ATM and DNA-PKcs during this process, we generated several RAG1−/− and RAG2−/− abl pre-B cell lines with single integrants of the pMX-DELSJ retroviral recombination substrate (RAG1−/−:DELSJ and RAG2−/−:DELSJ). pMX-DELSJ is identical to pMX-INV except that the RSs have been reoriented such that rearrangement results in the formation of a chromosomal signal joint (Supplemental Fig. 3A). Southern blot analyses can be carried out to detect unrepaired pMX-DELSJ chromosomal signal ends. In this regard, inducing rearrangement in DNA Ligase IV-deficient abl pre-B cells containing pMX-DELSJ (LigIV−/−:DELSJ) leads to an accumulation of unrepaired signal ends due to the deficiency in DNA Ligase IV (Fig. 6 and Supplemental Fig. 3).
Figure 6. Signal joint formation in abl pre-B cells expressing RAG1 or RAG2 SQ/TQ mutants.
A-B Southern blot analyses of EcoRV-digested genomic DNA probed with the C4b probe. (A) RAG1−/−:DELSJ abl pre-B cells and RAG1−/−:DELSJ abl pre-B cells transduced with pCST-FLAG-RAG1WT-iThy1.1 (RAG1−/−:DELSJ:R1WT) or a pCST-FLAG-RAG110AQ-iThy1.1 (RAG1−/−:DELSJ:R110AQ) and (B) RAG2−/−:DELSJ abl pre-B cells and RAG2−/−:DELSJ abl pre-B cells transduced with pCST-FLAG-RAG2WT-GFP-iThy1.1 (RAG2−/−:DELSJ:R2WT-GFP) or pCST-FLAG-RAG23AQ-GFP-iThy1.1 (RAG2−/−:DELSJ: R23AQ-GFP) pre-B cells were treated with STI571 for 0, 48 or 96 hours. Bands representing unrearranged (UR) pMX-DELSJ and normal signal joint (SJ) and unrepaired signal end (SE) are indicated. Molecular weight markers are shown.
RAG1−/−:DELSJ abl pre-B cells were transduced with retroviruses encoding RAG1WT and RAG110AQ, and RAG2−/−:DELSJ abl pre-B cells were transduced with retroviruses encoding RAG2WT-GFP and RAG23AQ-GFP Robust pMX-DELSJ signal joint formation was observed after induction of V(D)J recombination in both RAG1−/−:DELSJ:R1WT and RAG2−/−:DELSJ:R2WT-GFP abl pre-B cells treated with STI571 (Fig. 6 and Supplemental Fig. 3). Treatment of RAG1−/−:DELSJ:R110AQ and RAG2−/−:DELSJ:R23AQ-GFP abl pre-B cells also led to efficient pMX-DELSJ signal joint formation with no detectable accumulation of unrepaired signal ends (Fig. 6 and Supplemental Fig. 3). Thus, chromosomal signal joining appears unimpaired in the presence of RAG1 or RAG2 proteins that are crippled for phosphorylation at their SQ/TQ motifs.
Discussion
Here we have shown that constitutive expression of wild type RAG1 in RAG1−/− abl pre-B cells and wild type RAG2 in RAG2−/− abl pre-B cells rescues STI571-inducible V(D)J recombination. Indeed, the level of pMX-INV and IgLk rearrangement in these cells after treatment with STI571 is similar to what is observed in wild type abl pre-B cells treated with STI571. Thus, this approach can be used to assess the activity of mutant forms of RAG1 and RAG2 during chromosomal V(D)J recombination.
We have used this approach to determine whether phosphorylation of any serine or threonine residues in the three SQ/TQ motifs in RAG2 or the ten SQ/TQ motifs in RAG1 may be responsible for any of the defects observed in signal and coding joint formation in cells deficient in ATM or DNA-PKcs. ATM-deficient abl pre-B cells exhibit defects in coding joint formation with an accumulation of unrepaired coding ends and significant levels of hybrid joint formation during inversional rearrangements(16). In contrast, these defects were not observed during V(D)J recombination in RAG2−/−:INV:R23AQ or RAG1−/−:INV:R110AQ abl pre-B cells. Additionally, although efficient repair of signal ends depends on the overlapping activities of ATM and DNA-PKcs, analysis of RAG2−/−:DELSJ:R23AQ or RAG1−/−: DELSJ:R110AQ abl pre-B cells revealed no defects in signal joint formation(12, 13). Together, these findings demonstrate that the observed defects in the repair of RAG DSBs in cells deficient in ATM and/or DNA-PKcs cannot solely reflect a requirement for these kinases to phosphorylate RAG1 or RAG2 at consensus SQ/TQ motifs. ATM or DNA-PKcs could phosphorylate other non-consensus serines or threonines in RAG23AQ or RAG110AQ that exert either a direct or compensatory effect on RAG function during the joining step of the reaction. However, recent analyses of over 700 ATM targets in response to ionizing radiation reveal that essentially all of the target serines and threonines were part of SQ/TQ motifs (9). Our studies are in agreement with previous analyses of mutants of the conserved SQ/TQ motifs in RAG1 and RAG2 and, importantly, extend these findings to show that none of the RAG1 and RAG2 SQ/TQ motifs are required for the normal repair of RAG DSBs (44).
There are several important features of the experimental approach described here that can be used to examine the biologic relevance of RAG1 or RAG2 mutations. Most notably, this method permits evaluation of RAG function during both the DNA cleavage and joining steps of chromosomal V(D)J recombination specifically in G1-phase cells. Previous studies have assayed rearrangements at the IgH and IgLk loci in RAG-deficient abl pre-B cells reconstituted with RAG mutants, but unlike the approach presented here, these abl pre-B cells were continuously cycling (50, 51). Analyses of rearrangements had to be carried out after several weeks of RAG expression likely due to several factors including; (i) RAG1 and RAG2 gene expression is repressed by the abl kinase; (ii) in dividing cells, RAG2 is phosphorylated and degraded upon entry into S-phase; and (iii) cycling cells spend little time in G1 where RAG DSBs are efficiently repaired by NHEJ. Additionally, detecting unrepaired RAG DSBs in proliferating abl pre-B cells is challenging because these genetic lesions induce cell cycle arrest or cell death in addition to being potentially resolved aberrantly when these cells attempt to pass through the cell cycle.
In contrast, the approach described here makes use of the abl kinase inhibitor STI571, which causes G1 cell cycle arrest, induction of endogenous RAG gene expression, and robust inducible V(D)J recombination at the IgLk locus and chormosomally integrated retroviral substrates over a short period of time (2–4 days). Thus, this approach more closely mimics V(D)J recombination in vivo, which occurs in developing lymphocytes arrested in the G1 phase of the cells cycle. Together, our findings establish the utility of this approach for understanding the regulation of RAG1 and RAG2 function in the DNA cleavage and joining steps of chromosomal V(D)J recombination.
Supplementary Material
Acknowledgments
We thank Dr. Eugene Oltz for critical review of the manuscript and Dr. Peter J. McKinnonfor providing the DNA Ligase IV−/− mice.
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair (Amst) 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Rooney S, Chaudhuri J, Alt FW. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol Rev. 2004;200:115–131. doi: 10.1111/j.0105-2896.2004.00165.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 6.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol. 2001;13:225–231. doi: 10.1016/s0955-0674(00)00201-5. [DOI] [PubMed] [Google Scholar]
- 7.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 8.Tomimatsu N, Mukherjee B, Burma S. Distinct roles of ATR and DNA-PKcs in triggering DNA damage responses in ATM-deficient cells. EMBO Rep. 2009;10:629–635. doi: 10.1038/embor.2009.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 10.Callen E, Jankovic M, Wong N, Zha S, Chen HT, Difilippantonio S, Di Virgilio M, Heidkamp G, Alt FW, Nussenzweig A, Nussenzweig M. Essential role for DNA-PKcs in DNA double-strand break repair and apoptosis in ATM-deficient lymphocytes. Mol Cell. 2009;34:285–297. doi: 10.1016/j.molcel.2009.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matei IR, Guidos CJ, Danska JS. ATM-dependent DNA damage surveillance in T-cell development and leukemogenesis: the DSB connection. Immunol Rev. 2006;209:142–158. doi: 10.1111/j.0105-2896.2006.00361.x. [DOI] [PubMed] [Google Scholar]
- 12.Zha S, Jiang W, Fujiwara Y, Patel H, Goff PH, Brush JW, Dubois RL, Alt FW. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc Natl Acad Sci U S A. 2011;108:2028–2033. doi: 10.1073/pnas.1019293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gapud EJ, Dorsett Y, Yin B, Callen E, Bredemeyer A, Mahowald GK, Omi KQ, Walker LM, Bednarski JJ, McKinnon PJ, Bassing CH, Nussenzweig A, Sleckman BP. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc Natl Acad Sci U S A. 2011;108:2022–2027. doi: 10.1073/pnas.1013295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matei IR, Gladdy RA, Nutter LM, Canty A, Guidos CJ, Danska JS. ATM deficiency disrupts TCR{alpha} locus integrity and the maturation of CD4+CD8+ thymocytes. Blood. 2006 doi: 10.1182/blood-2006-05-020917. [DOI] [PubMed] [Google Scholar]
- 15.Vacchio MS, Olaru A, Livak F, Hodes RJ. ATM deficiency impairs thymocyte maturation because of defective resolution of T cell receptor {alpha} locus coding end breaks. Proc Natl Acad Sci U S A. 2007;104:6323–6328. doi: 10.1073/pnas.0611222104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 17.Bredemeyer AL, Huang CY, Walker LM, Bassing CH, Sleckman BP. Aberrant V(D)J recombination in ataxia telangiectasia mutated-deficient lymphocytes is dependent on nonhomologous DNA end joining. J Immunol. 2008;181:2620–2625. doi: 10.4049/jimmunol.181.4.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang CY, Sharma GG, Walker LM, Bassing CH, Pandita TK, Sleckman BP. Defects in coding joint formation in vivo in developing ATM-deficient B and T lymphocytes. J Exp Med. 2007;204:1371–1381. doi: 10.1084/jem.20061460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zha S, Guo C, Boboila C, Oksenych V, Cheng HL, Zhang Y, Wesemann DR, Yuen G, Patel H, Goff PH, Dubois RL, Alt FW. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature. 2011;469:250–254. doi: 10.1038/nature09604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodarzi AA, Yu Y, Riballo E, Douglas P, Walker SA, Ye R, Harer C, Marchetti C, Morrice N, Jeggo PA, Lees-Miller SP. DNA-PK autophosphorylation facilitates Artemis endonuclease activity. Embo J. 2006;25:3880–3889. doi: 10.1038/sj.emboj.7601255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 22.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 23.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen HT, Bhandoola A, Difilippantonio MJ, Zhu J, Brown MJ, Tai X, Rogakou EP, Brotz TM, Bonner WM, Ried T, Nussenzweig A. Response to RAG-mediated VDJ cleavage by NBS1 and gamma-H2AX. Science. 2000;290:1962–1965. doi: 10.1126/science.290.5498.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Ranganathan V, Weisman DS, Heine WF, Ciccone DN, O’Neill TB, Crick KE, Pierce KA, Lane WS, Rathbun G, Livingston DM, Weaver DT. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 26.Lim DS, Kim ST, Xu B, Maser RS, Lin J, Petrini JH, Kastan MB. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 27.Gatei M, Young D, Cerosaletti KM, Desai-Mehta A, Spring K, Kozlov S, Lavin MF, Gatti RA, Concannon P, Khanna K. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 28.Stiff T, O’Driscoll M, Rief N, Iwabuchi K, Lobrich M, Jeggo PA. ATM and DNA-PK function redundantly to phosphorylate H2AX after exposure to ionizing radiation. Cancer Res. 2004;64:2390–2396. doi: 10.1158/0008-5472.can-03-3207. [DOI] [PubMed] [Google Scholar]
- 29.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, Sleckman BP. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J Exp Med. 2009;206:669–679. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, Mahaney BL, Yano K, Ye R, Fang S, Douglas P, Chen DJ, Lees-Miller SP. DNA-PK and ATM phosphorylation sites in XLF/Cernunnos are not required for repair of DNA double strand breaks. DNA Repair (Amst) 2008;7:1680–1692. doi: 10.1016/j.dnarep.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas P, Gupta S, Morrice N, Meek K, Lees-Miller SP. DNA-PK-dependent phosphorylation of Ku70/80 is not required for non- homologous end joining. DNA Repair (Amst) 2005;4:1006–1018. doi: 10.1016/j.dnarep.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y, Wang W, Ding Q, Ye R, Chen D, Merkle D, Schriemer D, Meek K, Lees-Miller SP. DNA-PK phosphorylation sites in XRCC4 are not required for survival after radiation or for V(D)J recombination. DNA Repair (Amst) 2003;2:1239–1252. doi: 10.1016/s1568-7864(03)00143-5. [DOI] [PubMed] [Google Scholar]
- 34.Wang YG, Nnakwe C, Lane WS, Modesti M, Frank KM. Phosphorylation and regulation of DNA ligase IV stability by DNA- dependent protein kinase. J Biol Chem. 2004;279:37282–37290. doi: 10.1074/jbc.M401217200. [DOI] [PubMed] [Google Scholar]
- 35.Weterings E, Chen DJ. DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J Cell Biol. 2007;179:183–186. doi: 10.1083/jcb.200705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 37.Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarnell Schultz H, Landree MA, Qiu JX, Kale SB, Roth DB. Joining-deficient RAG1 mutants block V(D)J recombination in vivo and hairpin opening in vitro. Mol Cell. 2001;7:65–75. doi: 10.1016/s1097-2765(01)00155-1. [DOI] [PubMed] [Google Scholar]
- 39.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 40.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 41.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 42.Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc Natl Acad Sci U S A. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hah YS, Lee JH, Kim DR. DNA-dependent protein kinase mediates V(D)J recombination via RAG2 phosphorylation. J Biochem Mol Biol. 2007;40:432–438. doi: 10.5483/bmbrep.2007.40.3.432. [DOI] [PubMed] [Google Scholar]
- 44.Lin JM, Landree MA, Roth DB. V(D)J recombination catalyzed by mutant RAG proteins lacking consensus DNA-PK phosphorylation sites. Mol Immunol. 1999;36:1263–1269. doi: 10.1016/s0161-5890(99)00099-1. [DOI] [PubMed] [Google Scholar]
- 45.Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh CL, Arlett CF, Lieber MR. V(D)J recombination in ataxia telangiectasia, Bloom’s syndrome, and a DNA ligase I-associated immunodeficiency disorder. J Biol Chem. 1993;268:20105–20109. [PubMed] [Google Scholar]
- 47.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 48.Monroe RJ, Seidl KJ, Gaertner F, Han S, Chen F, Sekiguchi J, Wang J, Ferrini R, Davidson L, Kelsoe G, Alt FW. RAG2:GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 1999;11:201–212. doi: 10.1016/s1074-7613(00)80095-3. [DOI] [PubMed] [Google Scholar]
- 49.Hawley RG, Lieu FH, Fong AZ, Hawley TS. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 50.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 51.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.