Abstract
Aptamers are artificial oligonucleotides (DNA or RNA) selected in vitro that bind a broad range of targets with high affinity and specificity; a sensitive yet simple method to utilize aptamers as recognition elements for the development of biosensors (aptasensors) is to transduce the signal electrochemically. So far, aptasensors have been applied to clinical diagnostics and several technologies are in development. Aptasensors will extend the limits of current clinical diagnostics. Although the potential diagnostic applications are unlimited, the most current applications are foreseen in the areas of biomarker detection, cancer clinical testing, detection of infectious microorganisms and viruses. This review attempts to list examples of the research progresses of aptamers in biosensor platforms that have been published in recent years; in particular, we display cases of aptasensors that are already incorporated in clinical diagnostics or have potential applications in clinical diagnostics.
Keywords: biosensor, aptamer, aptasensors, clinical diagnostics
1. Introduction
High requirements regarding human health have resulted in an increasing number of clinical tests, so there is a rising need to develop more sensitive, reliable, time-efficient and inexpensive methods of analysis. Conventional techniques, such as molecular assays (immunological or nucleic acid technologies) and microbial culture-based tests, are either time consuming or require sophisticated equipment and expensive. Biosensor technology is probably one of the fastest growing areas to solve some of the problems concerning sensitive, rapid and cheap measurement [1].
A biosensor is defined as a compact analytical device incorporating a molecular recognition entity associated with or integrated with a physicochemical transducer [2]. It consists of two main components, a biological component, which reacts with a target substance, and a signal-generating component, which detects the resulting products. More specifically, we can consider a biosensor as a type of biomolecular probe that weighs the presence or concentration of biological molecules or biological structures by translating a biochemical interaction at the probe surface into a quantifiable physical signal. Biosensors are already used for several clinical applications, such as for electrochemical measurement of blood glucose concentrations. The first generation of biosensors were catalytic systems that integrated particularly enzymes with transducers that transferred the biological reaction into an electronic signal. The next generation of biosensors, affinity biosensors, utilized different biological elements, such as antibodies, receptors (natural or synthetic). Furthermore, the reaction between the target and the immobilized biomolecule on the transduction element is controlled by affinity interactions, such as the antigen-antibody, the DNA-DNA.
Recognition molecules are the key element of biosensors since the sensitivity and specificity of the sensing elements will play an important role in the success of the sensor device [3]. A range of recognition molecules have been employed for clinical diagnosis. However, the use of antibodies in biosensor detection methods and in the analysis of very complex samples could partially be hampered by the nature and synthesis of these protein receptors. In order to circumvent some of these drawbacks, other recognition molecules, such as aptamers, are being explored as alternative. This review attempts to list examples of the research progress of aptamers in biosensor platforms that have been published in recent years; in particular, we display cases of aptasensors that are already incorporated in clinical diagnostics or have potential applications in clinical diagnostics.
2. Aptasensors
2.1. Aptamers and Biosensors
Aptamers (derived from the latin aptus, meaning to fit) are artificial nucleic acid (DNA or RNA) ligands which can be selected from combinatorial libraries of synthetic nucleic acids, possessing specific binding characteristics to their targets. Using the systematic evolution of ligands (SELEX) process, aptamers can be isolated from randomly synthesized RNA or DNA pools and produced. Numerous high-affinity and highly specific aptamers have been generated against a wide variety of target molecules including small organics, peptides, proteins, and even complex-target such as whole cell [4,5]. In addtion, aptamers can be selected against non-immunogenic and toxic targets, which make the technique superior to antibodies, enabling the design and synthesis of probes or capture molecules for such targets.
Biosensors that employ aptamers as a recognition element are called aptasensors [6]. DNA and RNA aptamers can be modified chemically to undergo analyte-dependent conformational changes. For instance, Aptamers can be modified for immobilization purposes and incorporated particular reporters, without influencing their affinity, which has aided varieties of design methods [7–12]. Aptamers can also be engineered to withstand repeated cycles of denaturation and renaturation; this opens up a possibility to regenerate the immobilized biocomponent function for reuse [6,13]. In addition, they can be easily labeled for their use in diagnostics [14].
2.2. Signal-Transduction
Development of aptasensors has been carried out with various detection schemes, from label-free methods, such as surface plasmon resonance (SPR) [15], and quartz crystal microbalance (QCM) measurements, to other methods often requiring labels, such as electrochemistry[16], fluorescence [17–19], chemiluminescence [20], field effect transistors [10,16], which have been reported and enhanced the advancement of this field. Alternatively, sensors often utilize labeled target molecules or secondary capture agents (in the case of a sandwich-type assay) which themselves contain some forms of label or reporter molecule. So far, there are mainly two types of aptasensors to apply, including electrochemical and optical aptasensors.
2.2.1. Electrochemical Aptasensors
A typical electrochemical aptasensor makes use of an electrode surface as the platform to immobilize biological sensing aptamer, for which the analyte-binding event is monitored based on electrochemical current variations. Electrochemical transduction offers the advantages of high sensitivity, which can be enhanced by attaching biocatalytic labels to the aptamer-target complexes to amplify the detection signal, readily miniaturized, and low cost of production since they do not require expensive optical instruments [16,21]. Additionally, it is possible to use label-free and reusable detection systems.
2.2.2. Optical Aptasensors
Optical aptasensors include aptamers labeled with fluorescence, luminophore, enzyme, nanoparticles) or aptamer with label-free detection systems (e.g., SPR, optical resonance) [22]. Most methods are mainly based on a labeling approach. Some targets can be optically detected using not only colorimetry, chemiluminescence or the most developed fluorescence mode but also more recent non-conventional optical methods such as surface plasmon-coupled directional emission (SPCDE) [22].
Different label-free techniques have recently been shown to be suitable for developing aptasensors or aptamer-based microarrays, such as surface plasmon resonance (SPR), diffraction grating, evanescent-field-coupled (EFC) waveguide-mode, optical resonance or Brewster angle straddle interferometry (BASI) [22]. SPR biosensors prepared using optical fibers can be used as a cost-effective and relatively simple-to-implement alternative to well established biosensor platforms for monitoring biomolecular interactions in situ or possibly in vivo [23].
3. Application of Aptasensors in Clinical Diagnosis
Beyond all these features, the applications of aptamers as bio-compotents have offered extraordinary prospects in diagnostic assays [24]. Here we briefly describe a few examples of the use of aptasensors in the areas of biomarker detection, cancer clinical testing, and detection of infectious microorganisms.
3.1. Application of Aptasensors for Detection of Biomarkers
The analysis of biomarkers in blood, urine and other body fluids is one of the methods applied in the early detection of diseases. Aptasensors have been used for detection of biomarkers, such as thrombin, Immunoglobulin (Ig) E. A list of examples of the available reports of aptasensors for different biomarkers is provided in Table 1. Furthermore, representative examples of aptasensors for detection of thrombin are discussed in the following section.
Table 1.
Examples of application of aptasensors for detection of biomarkers.
| Target biomarker | Aptamer | Detection type | Signal transduction | Linear range/LOD | Reference |
|---|---|---|---|---|---|
| Thrombin | DNA labeled with the HCoPt-RGs conjugates | Sandwich-type | Electrochemical | 1.0 × 10−12 to 5.0 × 10−8 M/3.4 × 10−13 M | Wang et al. (2011) [25] |
| Thrombin | DNA dual labeled with AuNPs and HRP | Sandwich-type | Electrochemical | 0.1 to 60 pM/30 fM | Zhao et al. (2011) [26] |
| Thrombin | DNA | Label free detection | Fluorescence | 0 to 0.02 μM/0.1 nM | Pu et al. (2009)[27] |
| Thrombin | DNA labeled with a hole injector, naphthalimide, and a flurophore, Alexa532 | DNA charge transport | Fluorescence | 5 pM to 5 nM/1.2 pM | Zhang et al. (2011) [28] |
| Thrombin | DNA SA-ALP and biotinylated labled | Sandwich-type | ECL | 1 × 10−15 to 1 × 10−8 M/0.33 fM | Liao et al. (2011) [29] |
| IgE | DNA streptavidin labled | ELISA-like array | SPR | n.s./n.s. | Wang et al. (2008) [30] |
| IgE | DNA labeled with avidin monolayer | Direct detection | QCM chemiluminescence | 2.5 to 200 μg L−1/2.5 μg L−1 | Yao et al. (2009) [31] |
| IgE | DNA labeled with single PPy nanowire-based microfluidic | One step electrochemical deposition method | Electrochemical | 0.1 to 100 nM/0.01 nM | Huang et al. (2011) [32] |
| IgE | DNA attached to carboxyl (COOH)-modified NCD surface | Direct and label-free detection | EIS | 0.03 to 42.8 mug/mL/0.03 mug/mL | Tran et al. (2011) [33] |
| RBP4 | single-stranded DNA | Label free detection | SPR | 0.2 to 0.5 μg mL−1/75 nM | Lee et al. (2008) [34] |
| CRP | RNA biotinylated | Direct detection | Optical | n.s./0.005 ppm | Bini et al. (2008) [35] |
| CRP | DNA | Sandwich-type | Optical | 10 microg/L to 100 mg/L/n.s. | Pultar et al. (2009) [36] |
| NT-proBNP | DNA with cocaine-binding | Sandwich-type | ECL | 0.01 to 500 ng mL−1/0.77 pg mL−1 | Mao et al. (2011) [37] |
| IFN gamma | DNA thiolated/MB redox tag | Direct detection | Electrochemical | 10 nM/0.06 nM | Liu et al. (2011) [38] |
Abbreviations: LOD, limit of detection; HCoPt-RGs, hollow CoPt alloy nanoparticle onto reduced graphene oxide sheet; AuNP, Au nanoparticles; HRP, horseradish peroxidase; IgE, Immunoglobulin E; QCM, quartz crystal microbalance; SPR, surface plasmon resonance; PPy, polypyrrole ; EIS, Electrochemical impedance spectroscopy; NCD, nanocrystalline; RBP4, retinol binding protein 4; CRP, C-reactive protein; NT-proBNP, N-terminal pro-brain natriuretic peptide; ECL, electrochemiluminescence; MB, methylene blue; IFN, interferon; n.s., not specified.
Thromin is an enzymatic active, circulating biomarker for blood coagulation activity levels. Clinical studies, applying assays and strategies applicable for endogenous plasma thrombin measurements, are needed to evaluate the clinical impact of thrombin. Nowadays, many assays, based on the thrombin-binding aptamer for the detection of thrombin, have been developed in recent years. Mostly, thrombin has been used as a model for detection in the aptasensors assays. The below are three cases.
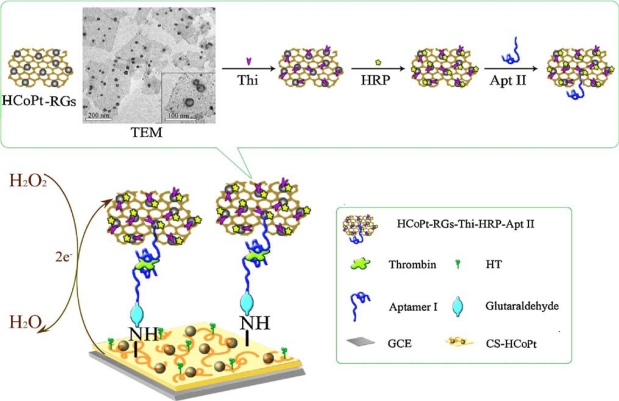
An advanced sandwich-type electrochemical aptasensor assay coupled to use the hollow CoPt alloy nanoparticle (HCoPt)-RGs conjugates as secondary label demonstrated a good sensitivity and selectivity, with a limit of detection (LOD) of 3.4 × 10−13 M. Scheme 1 illustrates the fabrication of electrochemical strategy with the aptamer-linked sandwich assay for the ultrasensitive detection of thrombin. The formed conjugates provided large surface area for loading plentiful redox probe thionine (Thi), horseradish peroxidase (HRP) and secondary aptamer (Apt II) with good stability and friendly biocompatibility [25].
Scheme 1.
Schematic diagrams of preparation of the sandwich-type electrochemical aptasensor and TEM images of the hollow CoPt alloy nanoparticles decorated graphene. Reproduced with permission form Reference [25] (Abbreviations: HT, hexanethiol; GCE, glassy carbon electrode; CS-HCoPt, the chitosan-hollow CoPt alloy nanoparticle).
Also, a sandwich electrochemical aptasensor assay was developed for thrombin based on amplification of aptamer-gold nanoparticles-horseradish peroxidase (aptamer-AuNPs-HRP) conjugates [26]. In this electrochemical protocol, aptamer1 (Apt1) was immobilized on core/shell Fe3O4/Au magnetic nanoparticles (AuMNPs) and served as capture probe. Aptamer2 (Apt2) was dual labeled with AuNPs and HRP and used as detection probe. In the presence of thrombin, the sandwich format of AuMNPs-Apt1/thrombin/Apt2-AuNPs-HRP was fabricated. Remarkable signal amplification was realized by taking the advantage of AuNPs and catalytic reactions of HRP. Other proteins, such as human serum albumin, lysozyme, fibrinogen, and IgG did not show significant interference with the assay for thrombin. Linear response to thrombin concentration in the range of 0.1–60 pM and the LOD of 30 fM (S/N = 3) was obtained with the proposed method. This electrochemical aptasensor was simple, rapid (the whole detection period for a thrombin sample is less than 35 min), sensitive and highly specific, it showed promising potential in protein detection and disease diagnosis [26].
Meanwhile, aptasensor assays can be used for detection with various working principles. Pu et al. [27] have displayed a smart polymeric transducer and aptamer/intercalating dye system that allows the label-free detection of thrombin with high sensitivity and selectivity. Thrombin detection was completed through the simple “mix and detect” mode. In addition, the methodology didn’t need any chemical modification on the probes or the analytes. The amount of the thrombin could be measured by the fluorescence intensity changes. The LOD was estimated to be 0.1 nM. This strategy might offer a new strategy to detect a wide spectrum of analytes and would be highly beneficial in future clinical applications.
3.2. Application of Aptasensors for Cancer Clinical Testing
With the growing number of cancer cases being diagnosed worldwide, cancer is the second most common cause of mortality and morbidity in the World due to late disease detection. Early diagnosis is crucial in all cancers to improve patient survival and disease prognosis, and may lead to cancer prevention- and for this reason sensitive and specific methods are required for early cancer diagnosis.
Detection and identification of tumor cells rely on the identification of some markers that appear only on the tumor cells such as lymphoma (Ramos) cells, leukemia cells. Identification of these cancer markers requires specific probes to bind in order to find the potential risk factor. In blood cells, plasma proteins or free DNA may be tumor markers. A number of recent studies have successfully used aptamers for targeting tumor markers.
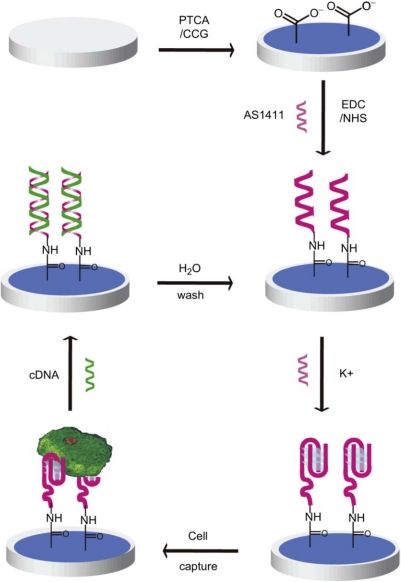
Feng et al. [39] reported an example for label-free cancer cell detection by using an electrochemical sensor based on the first clinical oncology trial II used aptamer AS1411 and graphene-modified electrode. The aptamer-PTCA (perylenetetracarboxylic acid) nanocomposite was utilized as nanoscale anchorage substrates to effectively capture cells on electrode surface through the specific binding between cell surface nucleolin and the aptamer AS1411 (Scheme 2). The electrochemical aptasensor can distinguish cancer cells and normal ones and detect as low as one thousand cells. With DNA hybridization technique, this E-DNA sensor can be regenerated and reusable for cancer cell detection.
Scheme 2.
Schematic representation of the reusable aptamer/graphene-based aptasensor. The sensor is constructed based on graphene-modified electrode and the first clinical trials II used aptamer, AS1411. AS1411 and its complementary DNA are used as a nanoscale anchorage substrate to capture/release cells. Reproduced with permission form Reference [39]. (Abbreviations: CCG, chemical converted grapheme; EDC, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide; NHS, N-hydroxysuccinimide).
Platelet-derived growth factor B chain (PDGF-BB) is a potential cancer marker and is known to be related to tumor growth and transformation. Recently, Chai et al. [40] developed a sensitive and low cost ECL aptasensor for PDGF-BB by assembling N-(aminobutyl)-N-ethylisoluminol functionalized gold nanoparticles (ABEI-AuNPs) with aptamers as nanoprobes. The biotinylated aptamer capture probes were first immobilized on a streptavidin coated gold nanoparticle (AuNPs) modified electrode, then, a sandwich conjugate modified electrode was carried out by successively attached the target PDGF-BB and the ABEI-AuNPs tagged aptamer signal probe. ECL measurement was run with a double-step potential in carbonate buffer solution containing H2O2, showing high sensitivity and specificity. The detection limit was as low as 2.7 × 10−14 M. In addition, the ABEI-AuNPs signal amplification was more simple, stable, practical, and sensitive compared to other reported signal amplification strategies based on aptamer-related polymerase chain reaction or functionalized nanoparticles. Moreover, 7 human serum samples were measured by using the proposed aptasensor. The obtained result were in good agreement those determined by the PDGF-BB ELISA as the reference method, which indicated that it was feasible to apply the proposed ECL aptasensor to detect PDGF-BB in real human serum samples. Table 2 lists examples of aptasensors for cancer clinical testing in recent years.
Table 2.
Examples of application of aptasensors for cancer clinical testing.
| Cancer marker detected | Aptamer | Detection type | Signal transduction | Linear range/LOD | Reference |
|---|---|---|---|---|---|
| HeLa cells, K562 cells, MDA-231 cells | DNA | Label free detection | Electrochemical | n.s./n.s. | Feng et al. (2011) [39] |
| PDGF-BB | DNA labeled with biotin | A sandwich conjugate modified electrode | ECL | 1.0 × 10−13 to 1.0 × 10−11 M/2.7 × 10−14 M | Chai et al. (2011) [40] |
| Ramos cancer cell, CEM cells | DNA | ECL array with a novel cycle-amplifying technique | ECL | n.s./n.s. | Jie et al. (2011) [41] |
| Multi-marker or Ramos cells, CCRF-CEM cells, Toledo Cells | DNA-conjugated FRET NP | Simultaneous multiplexed analysis | fluorescence | n.s./n.s. | Chen et al. (2009) [42] |
| Ramos cancer cell | DNA | Label free detection | ECL | 100 to 1,000 cells mL−1/58 cells mL−1 | Hun et al. (2011) [43] |
| Leukemia cells | DNA conjugated apt-MBs | a magnet-quartz crystal microbalance system | QCM | 1 × 104 to 1.5 × 105 cells mL−1/8 × 103 cells mL−1 | Pan et al. (2010) [44] |
| PSA | DNA labeled with FITC | Aptamer blotting assay | Chemiluminescence | 40 to 100 nM/n.s. | Savory et al. 2010 [45] |
| MUC1 | DNA labeled with single PPy nanowire-based microfluidic | One step electrochemical deposition method | Electrochemical | n.s./2.66 nM | Huang et al. (2011) [33] |
| MUC1 | DNA labeled with QD | Aptamer-based detection with quantum-dot based fluorescence readout | Fluorescence | n.s./250 nM | Cheng et al. (2009) [46] |
| GSH | RNA | SPR analysis and isocratic affinity chromatography | SPR | n.s./n.s. | Bala et al. (2011) [47] |
| VEGF | RNA conjugated CPNTs | FET-type biosensor based on CPNTs-aptamer | Electrochemical | n.s./400 fM | Kwon et al. (2010) [48] |
Abbreviations: PDGF-BB, platelet-derived growth factor B chain; PSA, prostate specific antigen; MUC1, Mucin 1; FRET NP, fluorescence resonance energy transfer nanoparticles; GSH, Glutathione; SPR, surface plasmon resonance; ECL, electrogenerated chemiluminescence; QD NCs, quantum dot nanoclusters; VEGF, Vascular Endothelial Growth Factor; CPNTs, carboxylated polypyrrole nanotubes; FET, field-effect transistor; QCM, quartz crystal microbalance; apt-MBs, conjugated magnetic beads; QD, quantum dot; n.s., not specified.
3.3. Aptasensors for Detection of Microorganisms and Viruses
Detection, identification and quantification of microbial pathogens are crucial for public health protection. Similarly, identification of virally infected cells is essential for the study of the mechanism of viral diagnosis. Virus-infected cells display certain viral proteins, which are detected using probes for the detection of infection. Up to date, the ability to design probes for specific viral targets on the host cell surface is very limited. Several aptasensors have been developed to detect microorganisms and viral proteins. One representative example of HIV-1 Tat aptamers developed for detection is discussed in the following part. In addition, Table 3 lists examples of aptasensors for detection of microorganisms and viruses.
Table 3.
Examples of application of aptasensors for detection of microorganisms and viruses.
| Microbial and viral targets | Aptamer | Detection type | Signal transduction | Linear range/LOD | Reference |
|---|---|---|---|---|---|
| Bacillus thuringiensis | DNA | Aptamer-functionalized QDs | Fluorescence | n.s./1,000 CFU/mL | Ikanovic et al. (2007) [49] |
| E. coli DH5a | DNA | Aptamer-functionalized SWNT-FET arrays | Fluorescence | n.s./n.s. | So et al. (2008) [50] |
| HIV-1 Tat | RNA biotinylated | A QCM –based and an SPR-based biosensor | QCM SPR |
QCM (0–1.2 ppm) SPR (0–2.5 ppm)/QCM (0.25 ppm) |
Tombelli et al. (2005) [51] |
| HCV core antigen | RNA | Chip-based detection | Fluorescence | n.s./n.s. | Lee et al. (2007) [52] |
| Prion | RNA | 3-dimensional analysis | Fluorescence | n.s./n.s. | Mashima et al. (2009) [53] |
Abbreviations: QD, quantum dots; CFU, colony forming units; SWNT-FET, single-walled carbon nanotube field-effect transistor; QCM, quartz crystal microbalance; SPR, surface plasmon resonance; n.s., not specified.
Microgravimetric methods on piezoelectric quartz crystals are based on the change of the oscillation frequency of the crystal upon mass change at its surface owing to receptor-target binding (QCM). Then, this change of oscillation frequency is the signal that is detected. With this method, a label-free detection of the target is possible. Tombelli et al. [51] developed an aptasensor platform to detect HIV-1 Tat protein by immobilizing an RNA aptamer on a piezoelectric quartz crystal. The QCM-based aptasensor has also been compared with the corresponding SPR-based aptasensor. The two aptasensors were constructed using biotin–avidin linking onto the gold surface of the transducers (quartz crystals or chips) for the immobilization chemistry. Both platforms showed similar reproducibility, sensitivity and specificity. The linear range of SPR (1–2.5 ppm) was higher that of QCM (0–1.25 ppm) [51].
4. Conculsions
Aptamer science has now reached maturity, through its growing impact on biology and medicine. The therapeutic use of aptamers is now well established, as in December 2004 the first aptamer compound named Pegaptanib was approved for clinical use [54]. Conversely, the different fields of clinical diagnostics, when dealing with systems based on biomolecular interactions, are still under the supremacy of immunoassays but diagnostics studies are now showing that some of the limits of current clinical diagnostics can be circumvented by taking advantage of the differences between aptamers and antibodies, such as cost-effective synthesis, flexibility for signal transduction and detection [55]. The good news is that there are currently some more aptamers in clinical phase 2/3 trials, such as factor IXa (FIXa), Von Willebrand factor (Vwf). However, the potential values of those aptamers for clinical diagnosis have not been fully investigated. Up to now, there is still no good answers to the question why aptamers have not yet penetrated into the clinical laboratory [56] and only a few aptasensors for clinical diagnostics were described [56,57]. As long as more aptamers will be developed and specified, the more aptasensors promise to play an important role in the future development of diagnostic methods.
References
- 1.Lazcka O., del Campo F.J., Munoz F.X. Pathogen detection: A perspective of traditional methods and biosensors. Biosens. Bioelectron. 2007;22:1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Newman J.D., Tigwell L.J., Turner A.P.F., Warner P.J. Biosensors—A Clearer View. Cranfield University Publication; Cranfield, UK: 2004. [Google Scholar]
- 3.Tothill I.E. Biosensors for cancer markers diagnosis. Semin. Cell Dev. Biol. 2009;20:55–62. doi: 10.1016/j.semcdb.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 5.Shamah S.M., Healy J.M., Cload S.T. Complex target SELEX. Acc. Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 6.O’Sullivan C.K. Aptasensors—The future of biosensing. Anal. Bioanal. Chem. 2002;372:44–48. doi: 10.1007/s00216-001-1189-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z., Wilkop T., Xu D., Dong Y., Ma G., Cheng Q. Surface plasmon resonance imaging for affinity analysis of aptamer-protein interactions with PDMS microfluidic chips. Anal. Bioanal. Chem. 2007;389:819–825. doi: 10.1007/s00216-007-1510-x. [DOI] [PubMed] [Google Scholar]
- 8.So H.M., Won K., Kim Y.H., Kim B.K., Ryu B.H., Na P.S., Kim H., Lee J.O. Single-walled carbon nanotube biosensors using aptamers as molecular recognition elements. J. Am. Chem. Soc. 2005;127:11906–11907. doi: 10.1021/ja053094r. [DOI] [PubMed] [Google Scholar]
- 9.Polsky R., Gill R., Kaganovsky L., Willner I. Nucleic acid-functionalized Pt nanoparticles: Catalytic labels for the amplified electrochemical detection of biomolecules. Anal. Chem. 2006;78:2268–2271. doi: 10.1021/ac0519864. [DOI] [PubMed] [Google Scholar]
- 10.Zayats M., Huang Y., Gill R., Ma C.A., Willner I. Label-free and reagentless aptamer-based sensors for small molecules. J. Am. Chem. Soc. 2006;128:13666–13667. doi: 10.1021/ja0651456. [DOI] [PubMed] [Google Scholar]
- 11.Phillips J.A., Xu Y., Xia Z., Fan Z.H., Tan W. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal. Chem. 2009;81:1033–1039. doi: 10.1021/ac802092j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balamurugan S., Obubuafo A., Soper S.A., Spivak D.A. Surface immobilization methods for aptamer diagnostic applications. Anal. Bioanal. Chem. 2008;390:1009–1021. doi: 10.1007/s00216-007-1587-2. [DOI] [PubMed] [Google Scholar]
- 13.Jayasena S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 14.Ulrich H., Wrenger C. Disease-specific biomarker discovery by aptamers. Cytometry A. 2009;75:727–733. doi: 10.1002/cyto.a.20766. [DOI] [PubMed] [Google Scholar]
- 15.Ostatna V., Vaisocherova H., Homola J., Hianik T. Effect of the immobilisation of DNA aptamers on the detection of thrombin by means of surface plasmon resonance. Anal. Bioanal. Chem. 2008;391:1861–1869. doi: 10.1007/s00216-008-2133-6. [DOI] [PubMed] [Google Scholar]
- 16.Willner I., Zayats M. Electronic aptamer-based sensors. Angew. Chem. Int. Ed. Engl. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 17.Rupcich N., Nutiu R., Li Y., Brennan J.D. Entrapment of fluorescent signaling DNA aptamers in sol-gel-derived silica. Anal. Chem. 2005;77:4300–4307. doi: 10.1021/ac0506480. [DOI] [PubMed] [Google Scholar]
- 18.Rankin C.J., Fuller E.N., Hamor K.H., Gabarra S.A., Shields T.P. A simple fluorescent biosensor for theophylline based on its RNA aptamer. Nucleos. Nucleot. Nucleic Acids. 2006;25:1407–1424. doi: 10.1080/15257770600919084. [DOI] [PubMed] [Google Scholar]
- 19.Rupcich N., Nutiu R., Li Y., Brennan J.D. Solid-phase enzyme activity assay utilizing an entrapped fluorescence-signaling DNA aptamer. Angew. Chem. Int. Ed. Engl. 2006;45:3295–3299. doi: 10.1002/anie.200504576. [DOI] [PubMed] [Google Scholar]
- 20.Wang X., Zhou J., Yun W., Xiao S., Chang Z., He P., Fang Y. Detection of thrombin using electrogenerated chemiluminescence based on Ru(bpy)3(2+)-doped silica nanoparticle aptasensor via target protein-induced strand displacement. Anal. Chim. Acta. 2007;598:242–248. doi: 10.1016/j.aca.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.O., So H.M., Jeon E.K., Chang H., Won K., Kim Y.H. Aptamers as molecular recognition elements for electrochemical nanobiosensors. Anal. Bioanal. Chem. 2008;390:1023–1032. doi: 10.1007/s00216-007-1643-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sassolas A., Blum L.J., Leca-Bouvier B.D. Optical detection systems using immobilized aptamers. Biosens. Bioelectron. 2011;26:3725–3736. doi: 10.1016/j.bios.2011.02.031. [DOI] [PubMed] [Google Scholar]
- 23.Shevchenko Y., Francis T.J., Blair D.A., Walsh R., DeRosa M.C., Albert J. In situ biosensing with a surface plasmon resonance fiber grating aptasensor. Anal. Chem. 2011;83:7027–7034. doi: 10.1021/ac201641n. [DOI] [PubMed] [Google Scholar]
- 24.Tombelli S., Minunni M., Mascini M. Analytical applications of aptamers. Biosens. Bioelectron. 2005;20:2424–2434. doi: 10.1016/j.bios.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Yuan R., Chai Y., Yuan Y., Bai L., Liao Y. A multi-amplification aptasensor for highly sensitive detection of thrombin based on high-quality hollow CoPt nanoparticles decorated graphene. Biosens. Bioelectron. 2011;30:61–66. doi: 10.1016/j.bios.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 26.Zhao J., Zhang Y., Li H., Wen Y., Fan X., Lin F., Tan L., Yao S. Ultrasensitive electrochemical aptasensor for thrombin based on the amplification of aptamer-AuNPs-HRP conjugates. Biosens. Bioelectron. 2011;26:2297–2303. doi: 10.1016/j.bios.2010.09.056. [DOI] [PubMed] [Google Scholar]
- 27.Pu F., Huang Z., Hu D., Ren J., Wang S., Qu X. Sensitive, selective and label-free protein detection using a smart polymeric transducer and aptamer/ligand system. Chem. Commun. (Camb) 2009;47:7357–7359. doi: 10.1039/b918241a. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X., Zhao Z., Mei H., Qiao Y., Liu Q., Luo W., Xia T., Fang X. A fluorescence aptasensor based on DNA charge transport for sensitive protein detection in serum. Analyst. 2011;22:4764–4769. doi: 10.1039/c1an15265c. [DOI] [PubMed] [Google Scholar]
- 29.Liao Y., Yuan R., Chai Y., Zhuo Y., Yuan Y., Bai L., Mao L., Yuan S. In situ produced ascorbic acid as coreactant for an ultrasensitive solid-state tris(2,2′-bipyridyl) ruthenium(II) electrochemiluminescence aptasensor. Biosens. Bioelectron. 2011;26:4815–4818. doi: 10.1016/j.bios.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Lv R., Xu J., Xu D., Chen H. Characterizing the interaction between aptamers and human IgE by use of surface plasmon resonance. Anal. Bioanal. Chem. 2008;390:1059–1065. doi: 10.1007/s00216-007-1697-x. [DOI] [PubMed] [Google Scholar]
- 31.Yao C., Qi Y., Zhao Y., Xiang Y., Chen Q., Fu W. Aptamer-based piezoelectric quartz crystal microbalance biosensor array for the quantification of IgE. Biosens. Bioelectron. 2009;24:2499–2503. doi: 10.1016/j.bios.2008.12.036. [DOI] [PubMed] [Google Scholar]
- 32.Huang J., Luo X., Lee I., Hu Y., Cui X.T., Yun M. Rapid real-time electrochemical detection of proteins using single conducting polymer nanowire-based microfluidic aptasensor. Biosens. Bioelectron. 2011;30:306–309. doi: 10.1016/j.bios.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tran D.T., Vermeeren V., Grieten L., Wenmackers S., Wagner P., Pollet J., Janssen K.P., Michiels L., Lammertyn J. Nanocrystalline diamond impedimetric aptasensor for the label-free detection of human IgE. Biosens. Bioelectron. 2011;26:2987–2993. doi: 10.1016/j.bios.2010.11.053. [DOI] [PubMed] [Google Scholar]
- 34.Lee S.J., Youn B.S., Park J.W., Niazi J.H., Kim Y.S., Gu M.B. ssDNA aptamer-based surface plasmon resonance biosensor for the detection of retinol binding protein 4 for the early diagnosis of type 2 diabetes. Anal. Chem. 2008;80:2867–2873. doi: 10.1021/ac800050a. [DOI] [PubMed] [Google Scholar]
- 35.Bini A., Centi S., Tombelli S., Minunni M., Mascini M. Development of an optical RNA-based aptasensor for C-reactive protein. Anal. Bioanal. Chem. 2008;390:1077–1086. doi: 10.1007/s00216-007-1736-7. [DOI] [PubMed] [Google Scholar]
- 36.Pultar J., Sauer U., Domnanich P., Preininger C. Aptamer-antibody on-chip sandwich immunoassay for detection of CRP in spiked serum. Biosens. Bioelectron. 2009;24:1456–1461. doi: 10.1016/j.bios.2008.08.052. [DOI] [PubMed] [Google Scholar]
- 37.Mao L., Yuan R., Chai Y., Zhuo Y., Xiang Y. Signal-enhancer molecules encapsulated liposome as a valuable sensing and amplification platform combining the aptasensor for ultrasensitive ECL immunoassay. Biosens. Bioelectron. 2011;26:4204–4208. doi: 10.1016/j.bios.2011.02.035. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y., Tuleouva N., Ramanculov E., Revzin A. Aptamer-based electrochemical biosensor for interferon gamma detection. Anal. Chem. 2010;82:8131–8136. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng L., Chen Y., Ren J., Qu X. A graphene functionalized electrochemical aptasensor for selective label-free detection of cancer cells. Biomaterials. 2011;32:2930–2937. doi: 10.1016/j.biomaterials.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 40.Chai Y., Tian D., Gu J., Cui H. A novel electrochemiluminescence aptasensor for protein based on a sensitive N-(aminobutyl)-N-ethylisoluminol-functionalized gold nanoprobe. Analyst. 2011;136:3244–3251. doi: 10.1039/c1an15298j. [DOI] [PubMed] [Google Scholar]
- 41.Jie G., Wang L., Yuan J., Zhang S. Versatile electrochemiluminescence assays for cancer cells based on dendrimer/CdSe-ZnS-quantum dot nanoclusters. Anal. Chem. 2011;83:3873–3880. doi: 10.1021/ac200383z. [DOI] [PubMed] [Google Scholar]
- 42.Chen X., Estevez M.C., Zhu Z., Huang Y.F., Chen Y., Wang L., Tan W. Using aptamer-conjugated fluorescence resonance energy transfer nanoparticles for multiplexed cancer cell monitoring. Anal. Chem. 2009;81:7009–7014. doi: 10.1021/ac9011073. [DOI] [PubMed] [Google Scholar]
- 43.Hun X., Chen H., Wang W. A electrogenerated chemiluminescence biosensor for Ramos cancer cell using DNA encapsulated Ru(bpy)(3)Cl(2) as signal probe. Biosens. Bioelectron. 2011;26:3887–3893. doi: 10.1016/j.bios.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 44.Pan Y., Guo M., Nie Z., Huang Y., Pan C., Zeng K., Zhang Y., Yao S. Selective collection and detection of leukemia cells on a magnet-quartz crystal microbalance system using aptamer-conjugated magnetic beads. Biosens. Bioelectron. 2010;25:1609–1614. doi: 10.1016/j.bios.2009.11.022. [DOI] [PubMed] [Google Scholar]
- 45.Savory N., Abe K., Sode K., Ikebukuro K. Selection of DNA aptamer against prostate specific antigen using a genetic algorithm and application to sensing. Biosens. Bioelectron. 2010;26:1386–1391. doi: 10.1016/j.bios.2010.07.057. [DOI] [PubMed] [Google Scholar]
- 46.Cheng A.K., Su H., Wang Y.A., Yu H.Z. Aptamer-based detection of epithelial tumor marker mucin 1 with quantum dot-based fluorescence readout. Anal. Chem. 2009;81:6130–6139. doi: 10.1021/ac901223q. [DOI] [PubMed] [Google Scholar]
- 47.Bala J., Bhaskar A., Varshney A., Singh A.K., Dey S., Yadava P. In vitro selected RNA aptamer recognizing glutathione induces ROS mediated apoptosis in the human breast cancer cell line MCF 7. RNA Biol. 2011;8:101–111. doi: 10.4161/rna.8.1.14116. [DOI] [PubMed] [Google Scholar]
- 48.Kwon O.S., Park S.J., Jang J. A high-performance VEGF aptamer functionalized polypyrrole nanotube biosensor. Biomaterials. 2010;31:4740–4747. doi: 10.1016/j.biomaterials.2010.02.040. [DOI] [PubMed] [Google Scholar]
- 49.Ikanovic M., Rudzinski W.E., Bruno J.G., Allman A., Carrillo M.P., Dwarakanath S., Bhahdigadi S., Rao P., Kiel J.L., Andrews C.J. Fluorescence assay based on aptamer-quantum dot binding to Bacillus thuringiensis spores. J. Fluoresc. 2007;17:193–199. doi: 10.1007/s10895-007-0158-4. [DOI] [PubMed] [Google Scholar]
- 50.So H.M., Park D.W., Jeon E.K., Kim Y.H., Kim B.S., Lee C.K., Choi S.Y., Kim S.C., Chang H., Lee J.O. Detection and titer estimation of Escherichia coli using aptamer-functionalized single-walled carbon-nanotube field-effect transistors. Small. 2008;4:197–201. doi: 10.1002/smll.200700664. [DOI] [PubMed] [Google Scholar]
- 51.Tombelli S., Minunni M., Luzi E., Mascini M. Aptamer-based biosensors for the detection of HIV-1 Tat protein. Bioelectrochemistry. 2005;67:135–141. doi: 10.1016/j.bioelechem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 52.Lee S., Kim Y.S., Jo M., Jin M., Lee D.K., Kim S. Chip-based detection of hepatitis C virus using RNA aptamers that specifically bind to HCV core antigen. Biochem. Biophys. Res. Commun. 2007;358:47–52. doi: 10.1016/j.bbrc.2007.04.057. [DOI] [PubMed] [Google Scholar]
- 53.Mashima T., Matsugami A., Nishikawa F., Nishikawa S., Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gragoudas E.S., Adamis A.P., Cunningham E.T., Jr., Feinsod M., Guyer D.R. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 55.Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baird G.S. Where are all the aptamers? Am. J. Clin. Pathol. 2010;134:529–531. doi: 10.1309/AJCPFU4CG2WGJJKS. [DOI] [PubMed] [Google Scholar]
- 57.Zhang P., Zhao N., Zeng Z., Chang C.C., Zu Y. Combination of an aptamer probe to CD4 and antibodies for multicolored cell phenotyping. Am. J. Clin. Pathol. 2010;134:586–593. doi: 10.1309/AJCP55KQYWSGZRKC. [DOI] [PubMed] [Google Scholar]