Abstract
Background
The metabolic activation of clopidogrel is a two-step process. It has been suggested that paraoxonase-1 (PON1) is a rate-limiting enzyme in the conversion of 2-oxo- clopidogrel to an active thiol metabolite. Conflicting results have been reported in regard to (1) the association of a common polymorphism of PON1 (Q192R) with reduced rates of coronary stent thrombosis in patients taking clopidogrel and (2) its effects on platelet inhibition in patient populations of European descent.
Methods
Blood samples from 151 subjects of mixed racial background with established coronary artery disease and who received clopidogrel were analyzed. Platelet aggregation was determined with light transmittance aggregometry and VerifyNow® P2Y12 assay. Genotyping for cytochrome P450 2C19 (CYP2C19)*2 and *3 and PON1 (Q192R) polymorphisms was performed.
Results
Carriers of CYP2C19*2 alleles exhibited lower levels of platelet inhibition and higher on-treatment platelet aggregation than noncarriers. There was no significant difference in platelet aggregation among PON1 Q192R genotypes. Homozygous carriers of the wild-type variant of PON1 (QQ192) had similar on-treatment platelet reactivity to carriers of increased-function variant alleles during maintenance clopidogrel dosing, as well as after administration of a clopidogrel 600 mg loading dose.
Conclusion
CYP2C19*2 allele is associated with impaired platelet inhibition by clopidogrel and high on-treatment platelet aggregation. PON1 (Q192R) polymorphism does not appear to be a significant determinant of clopidogrel response.
Keywords: PON1, platelet, aggregation, cytochrome P450 enzymes
Introduction
Interindividual variations of ex vivo platelet aggregation during clopidogrel therapy have been correlated to reduced exposure of active clopidogrel thiol metabolite and differences in P2Y12 receptor occupancy.1 High on-treatment platelet reactivity during therapy with clopidogrel and low platelet inhibition are associated with risk of ischemic events after coronary stenting – in particular, stent thrombosis.1–5 As compared with extensive metabolizers, carriers of cytochrome P450 2C19 (CYP2C19) nonfunctional alleles have been shown to have reduced platelet inhibition by clopidogrel and are at increased risk of adverse events and stent thrombosis after coronary.5–9 Recent advances in the understanding of clopidogrel metabolism suggest that the inactive prodrug undergoes two steps of bioactivation. First, clopidogrel undergoes oxidation to 2-oxo-clopidogrel by hepatic CYP450 enzymes (1A2, 2B6, 2C9, 2C19, and 3A4/5 isoenzymes). Then, in the second step of the biotransformation, thiol metabolite is generated.10 A recent study employing sophisticated metabolomic methods suggested a hydrolytic process involving paraoxonase-1 (PON1) as the rate-limiting enzyme.10 A common polymorphism of PON1 (Q192R) is associated with reduced paraoxonase activity in carriers of the QQ (AA) haplotype. In two cohorts of patients with coronary stenting, and using a case study design, the investigators found the PON1 (QQ192) genotype was associated with increased risk of stent thrombosis.10 A group of patients with a history of prior stent thrombosis but who were not receiving current therapy with clopidogrel were administered a clopidogrel 600 mg loading dose. Within this group, reduced plasma activity of PON1 and PON1 (QQ192) genotype were associated with reduced active thiol metabolite exposure and reduced platelet inhibition by clopidogrel.10
Only one genome-wide association study (GWAS) has been performed examining the association of common genetic polymorphisms on platelet inhibition.11 This GWAS examined platelet aggregation data from 429 generally healthy white subjects of Amish descent after administration of a clopidogrel 300 mg loading dose, followed by maintenance clopidogrel dosing of 75 mg daily for 6 days. Platelet aggregation was assessed before administration of clopidogrel and again after 6 days of maintenance dosing. The results from the GWAS only found single nucleotide polymorphisms (SNPs) on chromosome 10q24 in linkage equilibrium with the CYP2C19*2 loss-of-function variant to be significantly associated with clopidogrel platelet inhibition.11 The GWAS did not find evidence of a significant association of SNPs located on or near the PON1 gene on chromosome 7 with variation in platelet inhibition by clopidogrel.11
The aim of the current study was to assess the impact of PON1 (Q192R) and CYP2C19*2 polymorphisms on clopidogrel platelet inhibition, as measured by light transmittance aggregometry and VerifyNow® (VN) P2Y12 assay, in a North American study population of mixed racial background.
Methods
Patients
The Indiana University School of Medicine’s Institutional Review Board for Research approved the study protocols. Informed consent was obtained from all subjects. Subjects were eligible to be enrolled if they had established coronary disease and were on dual antiplatelet therapy with clopidogrel and aspirin 81–325 mg daily for secondary prevention. Subjects were included in this study either if they had been taking clopidogrel 75 mg for at least 14 days prior to enrollment or if they had received a clopidogrel 600 mg loading dose during a percutaneous coronary intervention (PCI). Subjects were excluded if they had a history of drug or alcohol abuse, bleeding disorder, myelodysplastic or myeloproliferative disorders, chronic liver disease, or current warfarin use. Subjects were also excluded if they were pregnant, if the platelet count was less than 150,000/mm3, and if there was planned glycoprotein IIb/IIIa antagonist use during PCI. See Table 1 for baseline characteristics of the study subjects.
Table 1.
Baseline characteristics according to genotypes
| Variable | PON1 QQ192 (AA) (n = 63) | PON1 QR192 (AG) (n = 67) | PON1 RR192 (GG) (n = 21) | P-value | CYP2C19 *1/*1 (n = 107) |
CYP2C19 *1/*2 (n = 39) |
CYP2C19 *2/*2 (n = 5) |
P-value |
|---|---|---|---|---|---|---|---|---|
| Age (years) (mean ± SD) | 58.1 ± 8.6 | 59.2 ± 10.7 | 57.1 ± 10.7 | 0.67 | 58.5 ± 9.6 | 58.8 ± 8.9 | 54.7 ± 19.8 | 0.68 |
| BMI (kg/m2) (mean ± SD) | 31.3 ± 6.6 | 31.6 ± 7.2 | 34.9 ± 7.3 | 0.10 | 32.5 ± 6.7 | 30.4 ± 7.5 | 33.3 ± 9.5 | 0.25 |
| Men [n (%)] | 43 (72) | 41 (62) | 11 (52) | 0.41 | 65 (61) | 28 (72) | 2 (40) | 0.28 |
| Women [n (%)] | 20 (28) | 25 (38) | 10 (48) | 0.41 | 41 (39) | 11 (28) | 3 (60) | 0.28 |
| White [n (%)] | 57 (90) | 49 (74) | 9 (45) | <0.001 | 77 (73) | 34 (89) | 4 (80) | 0.1 |
| African-American [n (%)] | 6 (10) | 17 (26) | 11 (55) | <0.001 | 29 (27) | 4 (11) | 1 (20) | 0.1 |
| Clopidogrel 75 mg daily maintenance dose [n (%)] | 38 (60) | 42 (64) | 16 (76) | 0.42 | 68 (64) | 24 (62) | 4 (80) | 0.72 |
| Clopidogrel 600 mg loading dose [n (%)] | 25 (40) | 25 (37) | 5 (24) | 0.42 | 39 (36) | 15 (38) | 1 (20) | 0.72 |
| Risk factors [n (%)] | ||||||||
| Diabetes mellitus | 21 (33) | 27 (41) | 13(62) | 0.07 | 46 (43) | 14 (36) | 1 (20) | 0.45 |
| Hypertension | 60 (95) | 64 (97) | 21 (100) | 0.57 | 104 (98) | 36 (92) | 5 (100) | 0.21 |
| Dyslipidemia | 60 (95) | 65 (98) | 20 (95) | 0.55 | 102 (96) | 38 (97) | 5 (100) | 0.86 |
| Previous CABG | 12 (19) | 11 (17) | 3 (14) | 0.87 | 19 (18) | 7 (18) | 0 (0) | 0.58 |
| Peripheral vascular disease | 14 (22) | 9 (14) | 4 (19) | 0.44 | 19 (18) | 7 (18) | 1 (20) | 0.99 |
| Current smoker | 32 (51) | 30 (45) | 5 (24) | 0.1 | 43 (41) | 21 (54) | 3 (60) | 0.28 |
| Medications [n (%)] | ||||||||
| Aspirin dose (mean daily dose, mg) | 245 | 247 | 179 | 0.49 | 253 | 200 | 325 | 0.27 |
| Angiotensin inhibition-ACEI, ARB | 50 (81) | 50 (76) | 17 (81) | 0.76 | 83 (78) | 30 (79) | 4 (80) | 0.99 |
| Beta-blocker | 57 (92) | 65 (98) | 20 (95) | 0.22 | 103 (97) | 34 (89) | 5 (100) | 0.14 |
| Calcium channel antagonist | 11 (18) | 14 (21) | 4 (19) | 0.88 | 22 (21) | 7 (18) | 0 (0) | 0.51 |
| Statin | 53 (84) | 56 (85) | 20 (95) | 0.42 | 90 (85) | 35 (90) | 4 (80) | 0.7 |
| Proton pump inhibitor | 20 (33) | 25 (38) | 7 (33) | 0.78 | 36 (35) | 14 (37) | 2 (40) | 0.95 |
| SSRI antidepressant | 14 (22) | 11 (17) | 7 (35) | 0.22 | 23 (22) | 9 (24) | 0 (0) | 0.47 |
Abbreviations: ACEI, angiotensin converting enzyme inhibitor; ARB, angiotensin receptor blocker; BMI, body mass index; CABG, coronary artery bypass grafting; CYP2C19, cytochrome P450 2C19; PON1, paraoxonase-1; SD, standard deviation; SSRI, selective serotonin reuptake inhibitor.
Blood samples
After 14 days of taking clopidogrel 75 mg daily, peripheral venous blood samples were obtained from subjects prior to the next dose of clopidogrel and aspirin to determine on-treatment platelet aggregation. For patients who received clopidogrel 600 mg at the time of a PCI, a baseline blood sample was obtained from the arterial access sheath prior to administration of the clopidogrel loading dose and administration of heparin or bivalirudin. Peripheral venous blood samples were drawn at 4 and 16–24 hours after administration of the clopidogrel loading dose. For subjects who received a clopidogrel 600 mg loading dose, the 16- to 24-hour sample was used to determine the final on-treatment platelet aggregation used for the primary analysis of platelet aggregation among genotypes. All blood samples were directly transferred into vacutainer tubes containing 3.2% sodium citrate and were analyzed within 2 hours.
Platelet aggregation studies
Ex vivo platelet function was assessed by light transmittance aggregometry (LTA) at 37°C with an Optical Lumi- Aggregometer (Model 700 with AGGRO/LINK 8 software; Chrono-Log Corporation, Havertown, PA). Platelet-rich and platelet-poor plasma were obtained by differential centrifugation, as previously described.12,13 Platelet aggregation in platelet-rich plasma was induced with adenosine diphosphate (ADP) at 5, 10, and 20 μmol/L.
A VN-P2Y12 point-of-care assay (Accumetrics, Inc, San Diego, CA) was used to assess platelet inhibition in whole blood in subjects during maintenance clopidogrel dosing, as previously described.14
Genotyping
Genomic DNA was isolated from whole blood with the aid of the QIAamp® DNA Blood Midi Kit (Qiagen, Germantown, MD). Subjects were genotyped for CYP2C19*2 (681G>A; rs4244285), CYP2C19*3 (636G>A; rs4986893), and PON1 (Q192R; 575 A>G; rs662) SNPs using a Bio-Rad Laboratories real-time iCycler thermal cycler (Bio-Rad Laboratories, Inc, Hercules, CA). Sequence-specific primers were used to amplify the alleles of interest, along with two allele-specific TaqMan® probes (Applied Biosystems, Foster City, CA). Allelic discrimination software (Optical System, v 3.1; Bio-Rad Laboratories) was used to determine individual genotypes.
Statistical analysis
Statistical significance was defined as P < 0.05. All statistical tests were two-sided, and values are represented as the mean plus or minus the standard deviation, unless otherwise specified. Categorical variables were compared using the chi-square test. The Kolmogorov–Smirnov test was used to assess the normal distribution of continuous data. Unpaired two-sided Student’s t-test was used to compare normally distributed continuous data between two groups and for genotype group comparisons using one-way analysis of variance. Non-normally distributed continuous data were compared across two groups with the two-sided unpaired Wilcoxon test and genotype group comparisons with the Kruskal–Wallis test. Genotypes for CYP2C19 and PON1 were identified and included in univariate and multivariate linear regression, along with clinical variables that are known to affect platelet reactivity and response to clopidogrel and variables with P < 0.1 in univariate analysis (Table 2).
Table 2.
Platelet aggregation
| LTA 5 μmol/L ADP (%) |
LTA 10 μmol/L ADP (%) |
LTA 20 μmol/L ADP (%) |
VN-P2Y12 result (PRU) | VN-P2Y12 inhibition (%) | |
|---|---|---|---|---|---|
| PON1 QQ192 (AA) (n = 63) | 31.1 ± 12.2 | 37.8 ± 13.7 | 45.2 ± 10.8 | 192.9 ± 70 | 23.8 ± 20 |
| PON1 QR192 (AG) (n = 67)# | 30.8 ± 15.7 | 37.7 ± 16 | 45.3 ± 13.6 | 205.9 ± 88 | 27.4 ± 27 |
| PON1 RR192 (GG ) (n = 21)# | 32 ± 12.8 | 43.6 ± 10.9 | 48.6 ± 11.7 | 239.1 ± 53 | 12.6 ± 17 |
| P-value (univariate) | 0.7 | 0.74 | 0.96 | 0.167 | 0.913 |
| P-value (multivariate) | 0.81 | 0.89 | 0.87 | 0.081 | 0.638 |
|
| |||||
| 2C19*2 Carriers (n = 44)# | 37.9 ± 12 | 45.0 ± 13.1 | 51.3 ± 11 | 234.6 ± 67 | 12.9 ± 14 |
| 2C19*2 Noncarriers (n = 107)# | 29.9 ± 11.2 | 36.2 ± 14.3 | 43.6 ± 12 | 195.0 ± 78 | 27.7 ± 25 |
| P-value (univariate) | 0.037 | 0.034 | 0.034 | 0.027 | 0.013 |
| P-value (multivariate) | 0.04 | 0.036 | 0.034 | 0.014 | 0.012 |
Note:
Values represent the mean plus or minus the standard deviation.
Abbreviations: ADP, adenosine diphosphate; LTA, light transmittance aggregometry; PON1, paraoxonase-1; PRU, platelet reactivity units; VN, VerifyNow®.
Results
Baseline characteristics of the study subjects are described according to CYP2C19*2 and PON1 Q192R genotypes in Table 1. Overall clinical variables were well balanced among genotype groups. A total of 96 subjects on maintenance clopidogrel therapy for over 14 days and 55 subjects who received a clopidogrel 600 mg loading dose at the time of PCI were enrolled. The prevalence of African-Americans was significantly different across PON1 Q192R genotypes, with a higher prevalence of African-Americans in carriers of PON1 QQ192 (Table 1). These findings are consistent with the increased prevalence of QQ192 genotypes previously described in the African population as compared with populations of European descent.15
Among the 151 patients included in this study, 63 (42%) were carriers of the QQ192 genotype, 67 (44%) were heterozygous allele carriers (QR192), and 21 patients (14%) were homozygous RR192 genotype carriers. For the CYP2C19*2 allele, 107 (71%) were wild-type homozygous for the *2 allelic variant (*1/*1), 39 (26%) were heterozygous *2 allele carriers (*1/*2), and five patients (3%) were homozygous *2 allele carriers (*2/*2). For both genotype distributions, no significant deviations from the Hardy– Weinberg equilibrium were observed (P = 0.64 for PON1 Q192R; P = 0.54 for CYP2C19*2 genotypes).
There were no CYP2C19*3 alleles detected in the study population, which is consistent with the very rare frequency (<1%) of this loss-of-function allele in Caucasians and African- Americans. Platelet aggregation induced by ADP demonstrated wide interindividual variability during therapy with clopidogrel, as previously documented (Table 2, Figures 1–4).1–5
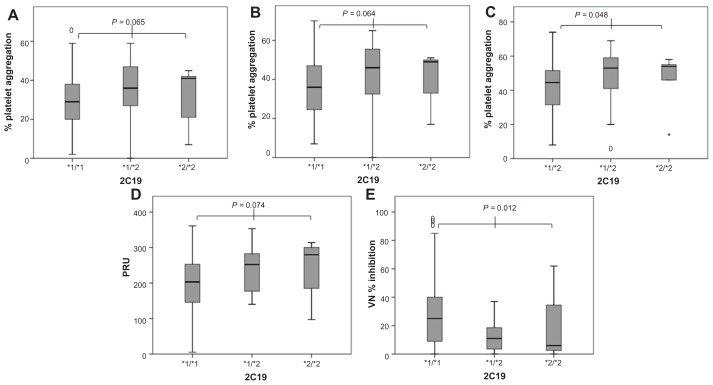
Figure 1.
Platelet aggregation represented as box plots and grouped according to cytochrome P450 2C19*2 genotype (*1/*1 [n = 107] vs *1/*2 [n = 39] vs *2/*2 [n = 5]). (A) Maximal platelet aggregation induced by adenosine diphosphate (ADP) 5 μmol/L (light transmittance aggregometry [LTA]); (B) maximal platelet aggregation induced by ADP 10 μmol/L (LTA); (C) maximal platelet aggregation induced by ADP 20 μmol/L (LTA); (D) VerifyNow® (VN) P2Y12 reactivity; (E) platelet inhibition measured by VN-P2Y12 assay.
Abbreviation: PRU, platelet reactivity units.
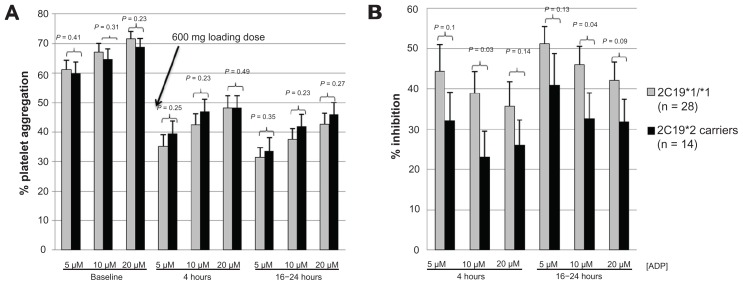
Figure 4.
Panels A and B represent data from patients not previously exposed to clopidogrel (n = 42) who received a clopidogrel 600 mg loading dose prior to coronary intervention and data are grouped according to 2C19*2 genotype (*1/*1 vs *2 carriers). (A) Light transmittance aggregometry before and at 4 and 16–24 hours after a 600 mg loading dose of clopidogrel; (B) platelet inhibition (percent change from baseline measurement) at 4 and 16–24 hours after a clopidogrel 600 mg loading dose.
Note: Values represent the mean plus or minus the standard error of the mean.
Abbreviation: ADP, adenosine diphosphate.
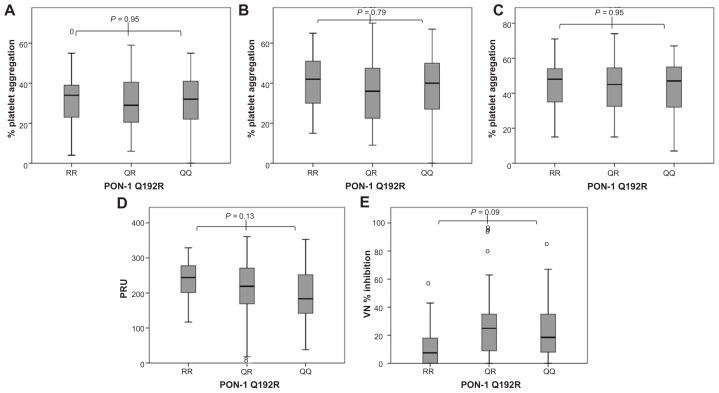
PON1 QQ192 was not associated with increased platelet aggregation induced by ADP at various concentrations during clopidogrel therapy before and after adjustment for 2C19*2, diabetes mellitus, obesity, smoking, and proton pump inhibitor therapy (Table 2). PON1 QQ192 was also not associated with increased VN-P2Y12 reactivity; in fact, there was a trend towards lower platelet reactivity in PON1 QQ192 individuals. No significant difference in the percentage of platelet inhibition measured by VN was documented among individuals of varying PON1 genotype (Table 2, Figure 2). Carriers of 2C19*2 alleles had higher platelet aggregation and higher VN-P2Y12 reactivity with significantly lower VN platelet inhibition than noncarriers (Table 2, Figure 1).
Figure 2.
Platelet aggregation represented as box plots and grouped according to paraoxonase-1 (PON1) Q192R genotype (RR192 [n = 21] vs QR192 [n = 67] vs QQ192 [n = 63]). (A) Maximal platelet aggregation induced by adenosine diphosphate (ADP) 5 μmol/L (light transmittance aggregometry [LTA]); (B) maximal platelet aggregation induced by ADP 10 μmol/L (LTA); (C) maximal platelet aggregation induced by ADP 20 μmol/L (LTA); (D) VerifyNow® (VN) P2Y12 reactivity; (E) platelet inhibition measured by VN-P2Y12 assay.
Abbreviation: PRU, platelet reactivity units.
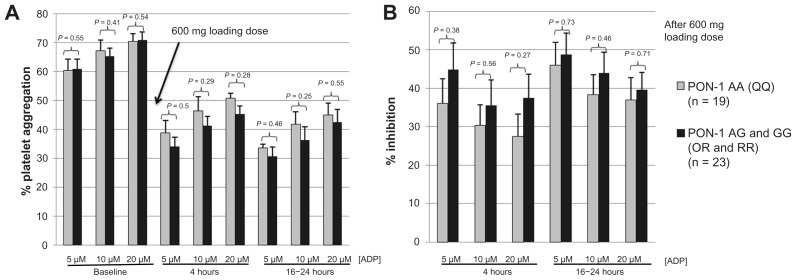
Among the subgroup of patients who received a clopidogrel 600 mg loading dose prior to undergoing PCI, platelet aggregation measured before and after administration of clopidogrel was not significantly different between subjects of PON1 QQ192 and QR192/RR192 genotypes (Figure 3). There was no significant difference in platelet inhibition among homozygous carriers of reduced-function PON1 (QQ192) and QR192 and RR192 genotypes at 4 and 16–24 hours after administration of a clopidogrel 600 mg loading dose (Figure 3). In the same subgroup of patients who had received a clopidogrel 600 mg loading dose and who had a baseline platelet aggregation measurement, 2C19*2 carriers status was associated with significantly lower percentage of platelet inhibition measured by ADP 10 μmol/L, without reaching statistical significance for other concentrations of ADP (Figure 4).
Figure 3.
Panels A and B represent data from patients not previously exposed to clopidogrel (n = 42) who received a clopidogrel 600 mg loading dose prior to coronary intervention and data are grouped according to PON1 genotype (QQ192 vs QR/RR192). (A) Light transmittance aggregometry before and at 4 and 16–24 hours after a 600 mg loading dose of clopidogrel; (B) platelet inhibition (percent change from baseline measurement) at 4 and 16–24 hours after a clopidogrel 600 mg loading dose.
Note: Values represent the mean plus or minus the standard error of the mean.
Abbreviation: ADP, adenosine diphosphate.
In the univariate linear regression analysis, only 2C19*2 carrier status (ADP 10 μmol/L; P = 0.034), current smoking (ADP 10 μmol/L; P = 0.025), and weight (ADP 10 μmol/L; P = 0.018) were significantly associated with ADP-induced platelet aggregation. PON1 Q192R homozygous reduced-function genotype (QQ192) (ADP 10 μmol/L; P = 0.7), diabetes (ADP 10 μmol/L; P = 0.22), and proton pump inhibitor use (ADP 10 μmol/L; P = 0.65) were not significantly associated with ADP-induced platelet aggregation (Table 2). Covariates with P < 0.1 were included in a multivariate regression analysis, as well as variables such as diabetes and proton pump inhibitor use that have previously been established as affecting clopidogrel platelet inhibition.
In a multivariate linear regression analysis that included diabetes, smoking, obesity, and proton pump inhibitor use as variables in addition to PON1 Q192R and CYP2C19*2 genotypes, CYP2C19*2 carrier status (ADP 10 μmol/L; P = 0.036), but not PON1 Q192R (ADP 10 μmol/L; P = 0.89), were associated with significant differences in ADP-induced platelet aggregation (Table 2).
Discussion
Clopidogrel bioactivation involves several enzymatic steps that have been delineated recently. In essence, clopidogrel is transformed into 2-oxo-clopidogrel by CYP450 enzymes and, in a second step, into the active thiol metabolite. 10 A recent metabolomic study suggested that PON1, a serum esterase, may represent the rate-limiting enzyme by hydrolysis of 2-oxo-clopidogrel into thiol clopidogrel. Common SNPs of PON1 have been suggested to affect enzymatic activity of the enzyme. In Bouman et al’s10 study, PON1 activity was significantly reduced in subjects homozygous for the wild-type allele (PON1 QQ192) compared with carriers of the mutant allele. In addition, in a group of patients with stent thrombosis and matched controls without stent thrombosis, PON1 QQ192 was associated with decreased platelet inhibition by clopidogrel and decreased plasma exposure to active thiol metabolite after a clopidogrel 600 mg loading dose. Also, PON1 QQ192 genotype was associated with an odds ratio of 3.3 for the occurrence of stent thrombosis as compared with QR192 or RR192 genotypes.10 The only GWAS on clopidogrel response had previously not found an association with SNPs located on or near the PON1 gene, but the platelet assays in that study were performed after 7 days of therapy with clopidogrel.11 The patients included in the GWAS were generally healthy white subjects, who had received a clopidogrel 300 mg loading dose followed by 75 mg daily for 6 days prior to analysis of platelet aggregation.11
Similar to findings from other investigators since the publication of the study by Bouman et al,10 the current study could not document an association of PON1 (Q192R) genotype with platelet inhibition by clopidogrel in patients of mixed racial background with established coronary artery disease during treatment with clopidogrel.16–21 Neither platelet inhibition measured by VN-P2Y12 assay nor on-treatment platelet aggregation using various doses of ADP showed significant differences among genotypes of PON1 Q192R. Among the group of patients who received a clopidogrel 600 mg loading dose immediately before PCI, platelet studies performed before and after administration of 600 mg of clopidogrel showed no significant differences in platelet aggregation among genotypes of PON1 Q192R (Figure 3).
The results of the current study are consistent with findings of Shuldiner et al,11 who did not demonstrate an association of SNPs in or near the PON1 gene with clopidogrel nonresponse during clopidogrel maintenance dosing, and they confirm the association of 2C19*2 polymorphism with reduced platelet inhibition by clopidogrel.22,23
In the current study, CYP2C19*2 carrier status was associated with reduced platelet inhibition, as measured by VN-P2Y12 assay and LTA 10 μmol/L, and increased on-treatment platelet reactivity, as measured by both LTA and VN-P2Y12 assay (Table 2, Figures 1 and 4).
CYP2C19*2 polymorphism has been demonstrated to be a strong determinant of reduced active clopidogrel metabolite formation, increased on-treatment ADP-induced platelet reactivity, and clinical ischemic events after coronary stenting.5–9,11,22,23 In a meta-analysis investigating the effect of reduced-function 2C19 alleles on recurrent ischemic events in patients receiving clopidogrel after coronary stenting, the presence of one reduced-function allele was associated with a hazard ratio of 2.67 and the presence of two reduced-function alleles was associated with a hazard ratio of 3.97 for the occurrence of stent thrombosis. These findings support the clinical importance of the reduced-function CYP2C19*2 polymorphism and clopidogrel nonresponse after coronary stenting.9
Although not supported by the findings of this study, that PON1 Q192R polymorphisms could affect the generation of active thiol metabolite with administration of 600 mg or greater loading doses (as suggested by the findings of Bouman et al)10 and that it could be associated with small differences in platelet inhibition cannot be excluded.10 However, in contrast to Bouman et al’s10 study, several investigators were unable to confirm the association between PON1 Q192R genotypes and stent thrombosis in other independent large cohorts of patients undergoing PCI who were treated with clopidogrel.16–21
Limitations of the study include a relatively small sample size and a lack of pharmacokinetic analysis to detect differences in active metabolite concentrations. In addition, the authors did not directly measure PON1 activity in plasma. Although considered the gold standard for measuring platelet aggregation and clopidogrel platelet inhibition, LTA is subject to operator variability, and on-treatment platelet aggregation may not be a direct reflection of P2Y12 receptor inhibition by active clopidogrel metabolites.
Conclusion
This study confirms the previously reported impact of reduced-function CYP2C19*2 polymorphism on platelet inhibition by clopidogrel. Polymorphisms of PON1 Q192R appear not to be predictive of clopidogrel response when measured by LTA and VN-P2Y12 assay in patients with coronary artery disease.
Acknowledgments
This study was supported in part by the Indiana Clinical and Translational Sciences Institute; funded in part by grant number RR025761 from the National Institutes of Health, National Center for Research Resources, Clinical and Translational Sciences Award; and through internal funding from the Department of Medicine, Indiana University School of Medicine, Indianapolis, to Rolf Kreutz and Yan Jin.
Footnotes
Disclosure
This study was performed while Yan Jin was employed at the Indiana University School of Medicine. Yan Jin is currently an employee of Eli Lilly and Company, Indianapolis, IN. The other authors report no conflicts of interest in this work.
References
- 1.Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol. 2007;49(14):1505–1516. doi: 10.1016/j.jacc.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 2.Gurbel PA, Bliden KP, Guyer K, et al. Platelet reactivity in patients and recurrent events post-stenting: results of the PREPARE POST-STENTING study. J Am Coll Cardiol. 2005;46(10):1820–1826. doi: 10.1016/j.jacc.2005.07.041. [DOI] [PubMed] [Google Scholar]
- 3.Buonamici P, Marcucci R, Migliorini A, et al. Impact of platelet reactivity after clopidogrel administration on drug-eluting stent thrombosis. J Am Coll Cardiol. 2007;49(24):2312–2317. doi: 10.1016/j.jacc.2007.01.094. [DOI] [PubMed] [Google Scholar]
- 4.Breet NJ, van Werkum JW, Bouman HJ, et al. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA. 2010;303(8):754–762. doi: 10.1001/jama.2010.181. [DOI] [PubMed] [Google Scholar]
- 5.Trenk D, Hochholzer W, Fromm MF, et al. Cytochrome P450 2C19 681G>A polymorphism and high on-clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug-eluting or bare-metal stents. J Am Coll Cardiol. 2008;51(20):1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 6.Mega JL, Close SL, Wiviott SD, et al. Cytochrome P-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 7.Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet. 2009;373(9660):309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 8.Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360(4):363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 9.Mega JL, Simon T, Collet JP, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304(16):1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bouman HJ, Schömig E, van Werkum JW, et al. Paraoxonase-1 is a major determinant of clopidogrel efficacy. Nat Med. 2011;17(1):110–116. doi: 10.1038/nm.2281. [DOI] [PubMed] [Google Scholar]
- 11.Shuldiner AR, O’Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreutz RP, Tantry US, Bliden KP, Gurbel PA. Inflammatory changes during the ‘common cold’ are associated with platelet activation and increased reactivity of platelets to agonists. Blood Coagul Fibrinolysis. 2007;18(8):713–718. doi: 10.1097/MBC.0b013e328201c77e. [DOI] [PubMed] [Google Scholar]
- 13.Kreutz RP, Alloosh M, Mansour K, et al. Morbid obesity and metabolic syndrome in Ossabaw miniature swine are associated with increased platelet reactivity. Diabetes Metab Syndr Obes. 2011;4:99–105. doi: 10.2147/DMSO.S17105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakubowski JA, Payne CD, Li YG, et al. The use of the VerifyNow P2Y12 point-of-care device to monitor platelet function across a range of P2Y12 inhibition levels following prasugrel and clopidogrel administration. Thromb Haemost. 2008;99(2):409–415. doi: 10.1160/TH07-09-0575. [DOI] [PubMed] [Google Scholar]
- 15.International HapMap Project [database] [Accessed September 26, 2011]. Available from: http://www.hapmap.org.
- 16.Sibbing D, Koch W, Massberg S, et al. No association of paraoxonase- 1 Q192R genotypes with platelet response to clopidogrel and risk of stent thrombosis after coronary stenting. Eur Heart J. 2011;32(13):1605–1613. doi: 10.1093/eurheartj/ehr155. [DOI] [PubMed] [Google Scholar]
- 17.Lewis JP, Fisch AS, Ryan K, et al. Paraoxonase 1 (PON1) gene variants are not associated with clopidogrel response. Clin Pharmacol Ther. 2011;90(4):568–574. doi: 10.1038/clpt.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rideg O, Komócsi A, Magyarlaki T, et al. Impact of genetic variants on post-clopidogrel platelet reactivity in patients after elective percutaneous coronary intervention. Pharmacogenomics. 2011;12(9):1269–1280. doi: 10.2217/pgs.11.73. [DOI] [PubMed] [Google Scholar]
- 19.Fontana P, James R, Barazer I, et al. Relationship between paraoxonase- 1 activity, its Q192R genetic variant and clopidogrel responsiveness in the ADRIE study. J Thromb Haemost. 2011;9(8):1664–1666. doi: 10.1111/j.1538-7836.2011.04409.x. [DOI] [PubMed] [Google Scholar]
- 20.Trenk D, Hochholzer W, Fromm MF, et al. Paraoxonase-1 Q192R polymorphism and antiplatelet effects of clopidogrel in patients undergoing elective coronary stent placement. Circ Cardiovasc Genet. 2011;4(4):429–436. doi: 10.1161/CIRCGENETICS.111.960112. [DOI] [PubMed] [Google Scholar]
- 21.Simon T, Steg PG, Becquemont L, et al. Effect of paraoxonase-1 polymorphism on clinical outcomes in patients treated with clopidogrel after an acute myocardial infarction. Clin Pharmacol Ther. 2011;90(4):561–567. doi: 10.1038/clpt.2011.193. [DOI] [PubMed] [Google Scholar]
- 22.Geisler T, Schaeffeler E, Dippon J, et al. CYP2C19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics. 2008;9(9):1251–1259. doi: 10.2217/14622416.9.9.1251. [DOI] [PubMed] [Google Scholar]
- 23.Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost. 2008;6(8):1439–1441. doi: 10.1111/j.1538-7836.2008.03050.x. [DOI] [PubMed] [Google Scholar]