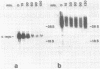
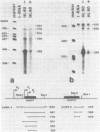
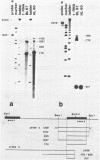
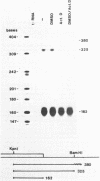
Abstract
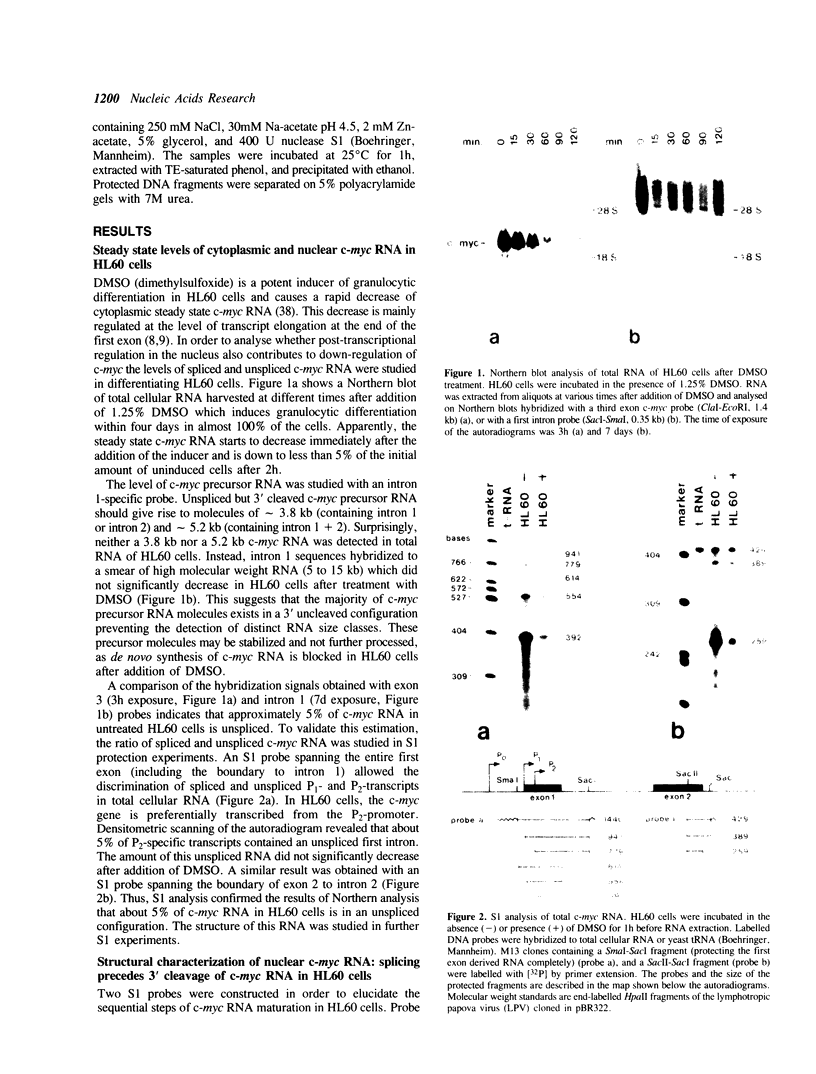
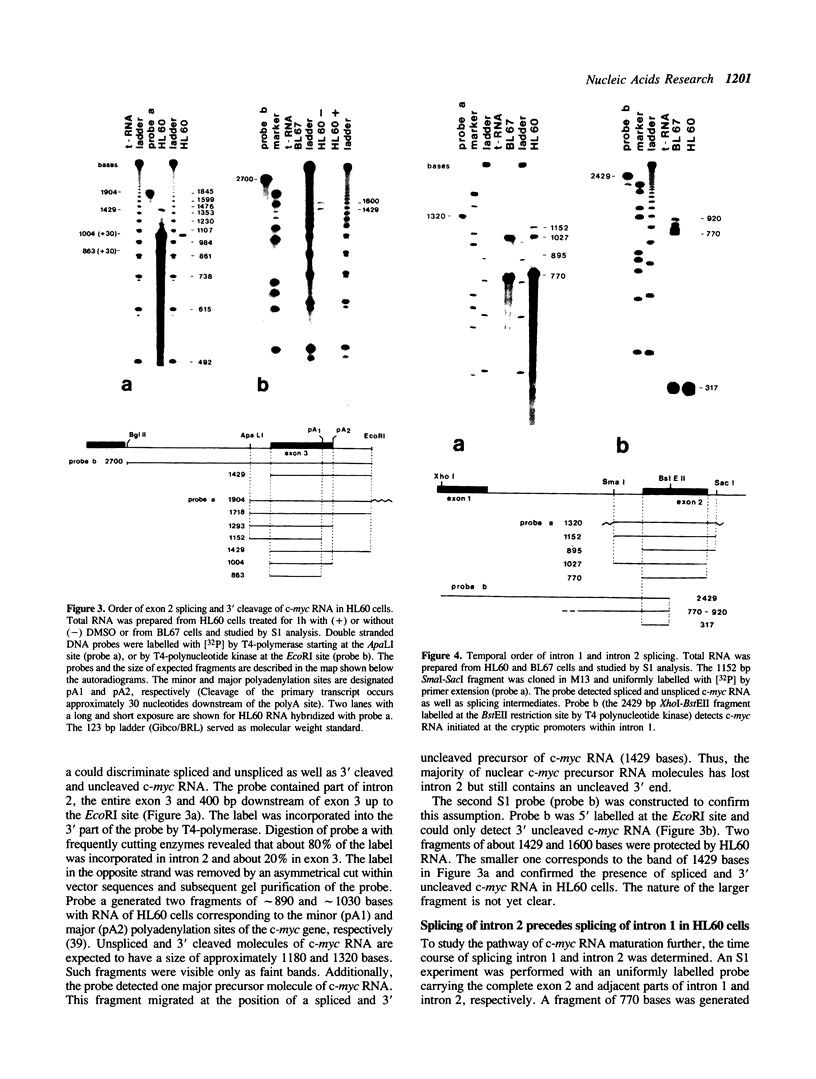
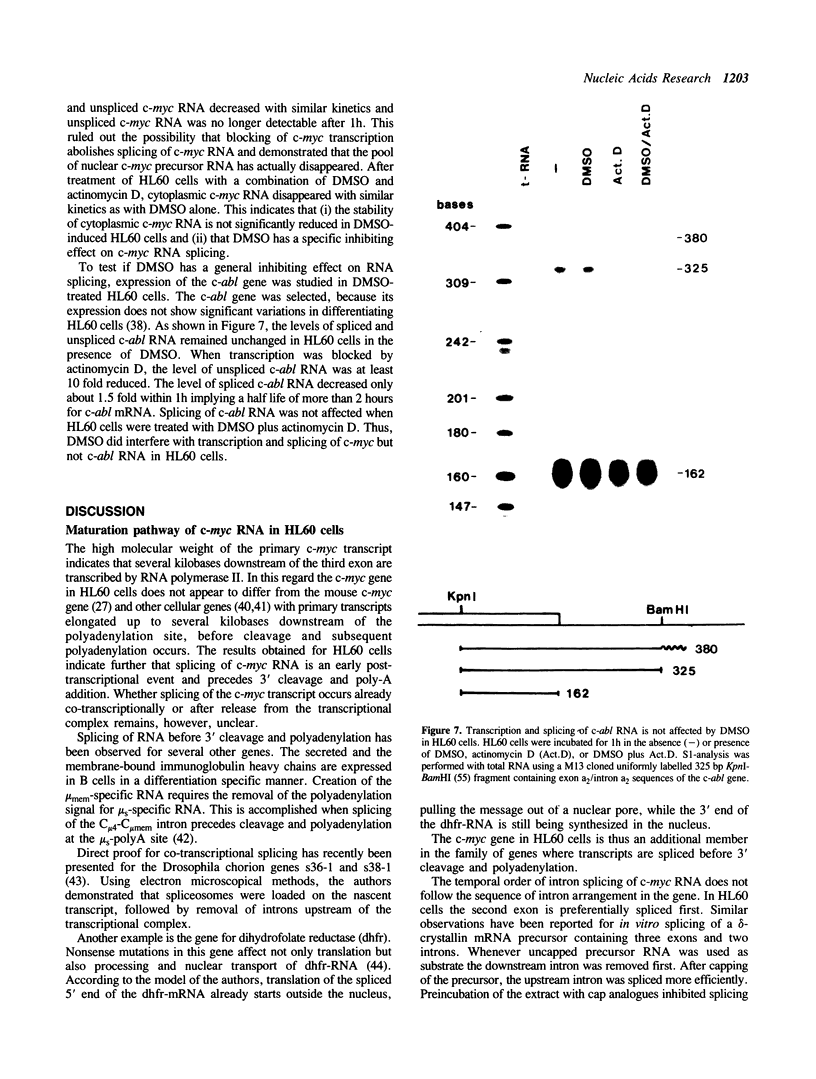
Maturation of c-myc mRNA proceeds in a given order in HL60 cells. It starts with splicing of intron 2, continues with splicing of intron 1 and ends with 3' cleavage and polyadenylation of the primary transcript. This process is inhibited, when HL60 cells were induced to terminal differentiation by dimethylsulfoxide (DMSO). DMSO interferes specifically with maturation of c-myc but not c-abl RNA in HL60 cells. Simultaneously, DMSO induces a block to RNA elongation at the boundary of c-myc exon 1 and intron 1 in HL60 cells. Participation of the same factor(s) in the regulation of c-myc RNA elongation and splicing is supposed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Auffray C., Rougeon F. Purification of mouse immunoglobulin heavy-chain messenger RNAs from total myeloma tumor RNA. Eur J Biochem. 1980 Jun;107(2):303–314. doi: 10.1111/j.1432-1033.1980.tb06030.x. [DOI] [PubMed] [Google Scholar]
- Battey J., Moulding C., Taub R., Murphy W., Stewart T., Potter H., Lenoir G., Leder P. The human c-myc oncogene: structural consequences of translocation into the IgH locus in Burkitt lymphoma. Cell. 1983 Oct;34(3):779–787. doi: 10.1016/0092-8674(83)90534-2. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature. 1986 Jun 12;321(6071):702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Groudine M. Sequence requirements for premature termination of transcription in the human c-myc gene. Cell. 1988 Apr 22;53(2):245–256. doi: 10.1016/0092-8674(88)90386-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Beyer A. L., Osheim Y. N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988 Jun;2(6):754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Bishop J. M. The molecular genetics of cancer. Science. 1987 Jan 16;235(4786):305–311. doi: 10.1126/science.3541204. [DOI] [PubMed] [Google Scholar]
- Blanchard J. M., Piechaczyk M., Dani C., Chambard J. C., Franchi A., Pouyssegur J., Jeanteur P. c-myc gene is transcribed at high rate in G0-arrested fibroblasts and is post-transcriptionally regulated in response to growth factors. Nature. 1985 Oct 3;317(6036):443–445. doi: 10.1038/317443a0. [DOI] [PubMed] [Google Scholar]
- Brinster R. L., Allen J. M., Behringer R. R., Gelinas R. E., Palmiter R. D. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988 Feb;85(3):836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman A. R., Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988 Oct;8(10):4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Sinn E., Reed R. R., Leder P. Trans-acting elements modulate expression of the human c-myc gene in Burkitt lymphoma cells. Proc Natl Acad Sci U S A. 1986 Oct;83(20):7918–7922. doi: 10.1073/pnas.83.20.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Sussman D. J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987 Dec 24;51(6):1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. J., Ruscetti F. W., Gallagher R. E., Gallo R. C. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc Natl Acad Sci U S A. 1978 May;75(5):2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. [DOI] [PubMed] [Google Scholar]
- Dani C., Blanchard J. M., Piechaczyk M., El Sabouty S., Marty L., Jeanteur P. Extreme instability of myc mRNA in normal and transformed human cells. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7046–7050. doi: 10.1073/pnas.81.22.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dani C., Mechti N., Piechaczyk M., Lebleu B., Jeanteur P., Blanchard J. M. Increased rate of degradation of c-myc mRNA in interferon-treated Daudi cells. Proc Natl Acad Sci U S A. 1985 Aug;82(15):4896–4899. doi: 10.1073/pnas.82.15.4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dony C., Kessel M., Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985 Oct 17;317(6038):636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- Eick D., Berger R., Polack A., Bornkamm G. W. Transcription of c-myc in human mononuclear cells is regulated by an elongation block. Oncogene. 1987;2(1):61–65. [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Expression of normal and translocated c-myc alleles in Burkitt's lymphoma cells: evidence for different regulation. EMBO J. 1989 Jul;8(7):1965–1972. doi: 10.1002/j.1460-2075.1989.tb03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Bornkamm G. W. Transcriptional arrest within the first exon is a fast control mechanism in c-myc gene expression. Nucleic Acids Res. 1986 Nov 11;14(21):8331–8346. doi: 10.1093/nar/14.21.8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick D., Piechaczyk M., Henglein B., Blanchard J. M., Traub B., Kofler E., Wiest S., Lenoir G. M., Bornkamm G. W. Aberrant c-myc RNAs of Burkitt's lymphoma cells have longer half-lives. EMBO J. 1985 Dec 30;4(13B):3717–3725. doi: 10.1002/j.1460-2075.1985.tb04140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Grosveld G., Verwoerd T., van Agthoven T., de Klein A., Ramachandran K. L., Heisterkamp N., Stam K., Groffen J. The chronic myelocytic cell line K562 contains a breakpoint in bcr and produces a chimeric bcr/c-abl transcript. Mol Cell Biol. 1986 Feb;6(2):607–616. doi: 10.1128/mcb.6.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss P., Khoury G. Rescue of a splicing defective mutant by insertion of an heterologous intron. Nature. 1980 Aug 7;286(5773):634–637. doi: 10.1038/286634a0. [DOI] [PubMed] [Google Scholar]
- Gruss P., Lai C. J., Dhar R., Khoury G. Splicing as a requirement for biogenesis of functional 16S mRNA of simian virus 40. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbüchle O., Wellauer P. K., Cribbs D. L., Schibler U. Termination of transcription in the mouse alpha-amylase gene Amy-2a occurs at multiple sites downstream of the polyadenylation site. Cell. 1984 Oct;38(3):737–744. doi: 10.1016/0092-8674(84)90269-1. [DOI] [PubMed] [Google Scholar]
- Hamer D. H., Leder P. Splicing and the formation of stable RNA. Cell. 1979 Dec;18(4):1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- Hay N., Bishop J. M., Levens D. Regulatory elements that modulate expression of human c-myc. Genes Dev. 1987 Sep;1(7):659–671. doi: 10.1101/gad.1.7.659. [DOI] [PubMed] [Google Scholar]
- Hiebert S. W., Lipp M., Nevins J. R. E1A-dependent trans-activation of the human MYC promoter is mediated by the E2F factor. Proc Natl Acad Sci U S A. 1989 May;86(10):3594–3598. doi: 10.1073/pnas.86.10.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer E., Darnell J. E., Jr The primary transcription unit of the mouse beta-major globin gene. Cell. 1981 Feb;23(2):585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- Kakkis E., Riggs K. J., Gillespie W., Calame K. A transcriptional repressor of c-myc. Nature. 1989 Jun 29;339(6227):718–721. doi: 10.1038/339718a0. [DOI] [PubMed] [Google Scholar]
- Kelly K., Siebenlist U. The regulation and expression of c-myc in normal and malignant cells. Annu Rev Immunol. 1986;4:317–338. doi: 10.1146/annurev.iy.04.040186.001533. [DOI] [PubMed] [Google Scholar]
- Kerppola T. K., Kane C. M. Intrinsic sites of transcription termination and pausing in the c-myc gene. Mol Cell Biol. 1988 Oct;8(10):4389–4394. doi: 10.1128/mcb.8.10.4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G., Klein E. Evolution of tumours and the impact of molecular oncology. Nature. 1985 May 16;315(6016):190–195. doi: 10.1038/315190a0. [DOI] [PubMed] [Google Scholar]
- Lindsten T., June C. H., Thompson C. B. Multiple mechanisms regulate c-myc gene expression during normal T cell activation. EMBO J. 1988 Sep;7(9):2787–2794. doi: 10.1002/j.1460-2075.1988.tb03133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipp M., Schilling R., Bernhardt G. Trans-activation of human MYC: the second promoter is target for the stimulation by adenovirus E1a proteins. Oncogene. 1989 May;4(5):535–541. [PubMed] [Google Scholar]
- Lipp M., Schilling R., Wiest S., Laux G., Bornkamm G. W. Target sequences for cis-acting regulation within the dual promoter of the human c-myc gene. Mol Cell Biol. 1987 Apr;7(4):1393–1400. doi: 10.1128/mcb.7.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechti N., Piechaczyk M., Blanchard J. M., Marty L., Bonnieu A., Jeanteur P., Lebleu B. Transcriptional and post-transcriptional regulation of c-myc expression during the differentiation of murine erythroleukemia Friend cells. Nucleic Acids Res. 1986 Dec 22;14(24):9653–9666. doi: 10.1093/nar/14.24.9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nepveu A., Levine R. A., Campisi J., Greenberg M. E., Ziff E. B., Marcu K. B. Alternative modes of c-myc regulation in growth factor-stimulated and differentiating cells. Oncogene. 1987;1(3):243–250. [PubMed] [Google Scholar]
- Nepveu A., Marcu K. B., Skoultchi A. I., Lachman H. M. Contributions of transcriptional and post-transcriptional mechanisms to the regulation of c-myc expression in mouse erythroleukemia cells. Genes Dev. 1987 Nov;1(9):938–945. doi: 10.1101/gad.1.9.938. [DOI] [PubMed] [Google Scholar]
- Ohno M., Sakamoto H., Shimura Y. Preferential excision of the 5' proximal intron from mRNA precursors with two introns as mediated by the cap structure. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5187–5191. doi: 10.1073/pnas.84.15.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson M. L., Perry R. P. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989 Feb;9(2):726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmers E. F., Yang J. Q., Marcu K. B. A negative transcriptional control element located upstream of the murine c-myc gene. EMBO J. 1986 May;5(5):899–904. doi: 10.1002/j.1460-2075.1986.tb04301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Schaulies J., Schimpl A., Wecker E. Kinetics of cellular oncogene expression in mouse lymphocytes. II. Regulation of c-fos and c-myc gene expression. Eur J Immunol. 1987 May;17(5):713–718. doi: 10.1002/eji.1830170521. [DOI] [PubMed] [Google Scholar]
- Showe L. C., Croce C. M. The role of chromosomal translocations in B- and T-cell neoplasia. Annu Rev Immunol. 1987;5:253–277. doi: 10.1146/annurev.iy.05.040187.001345. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Bressler P., Kelly K. Two distinct mechanisms of transcriptional control operate on c-myc during differentiation of HL60 cells. Mol Cell Biol. 1988 Feb;8(2):867–874. doi: 10.1128/mcb.8.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Mitchell P. J., Ciudad C. J., Chasin L. A. Nonsense mutations in the dihydrofolate reductase gene affect RNA processing. Mol Cell Biol. 1989 Jul;9(7):2868–2880. doi: 10.1128/mcb.9.7.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisinger G., Remmers E. F., Hearing P., Marcu K. B. Multiple negative elements upstream of the murine c-myc gene share nuclear factor binding sites with SV40 and polyoma enhancers. Oncogene. 1988 Dec;3(6):635–646. [PubMed] [Google Scholar]
- Westin E. H., Wong-Staal F., Gelmann E. P., Dalla-Favera R., Papas T. S., Lautenberger J. A., Eva A., Reddy E. P., Tronick S. R., Aaronson S. A. Expression of cellular homologues of retroviral onc genes in human hematopoietic cells. Proc Natl Acad Sci U S A. 1982 Apr;79(8):2490–2494. doi: 10.1073/pnas.79.8.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. Q., Remmers E. F., Marcu K. B. The first exon of the c-myc proto-oncogene contains a novel positive control element. EMBO J. 1986 Dec 20;5(13):3553–3562. doi: 10.1002/j.1460-2075.1986.tb04682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]