Abstract
Background and the purpose of the study
Artemisinin is one of the most effective medicine against malaria, which is produced naturally by Artemisia annua in low yield. It is produced in a metabolic pathway, in which several genes and gene products are involved. One of the key genes in this pathway is am1, which encodes amorpha-4, 11-diene synthase (ADS), a key enzyme in artemisinin biosynthesis pathway. The aim of this study was to determine the presence of this gene in ten Artemisia species in order to increase the yield of production of Artemisinin.
Methods
The experiments were carried out using PCR. Specific primers were designed based on the published am1 gene sequence obtained from A. annua (NCBI, accession number AF327527).
Results
The amplification of this gene by the specific primers was considered as a positive sign for the potentiality of artemisinin production. Since the entire am1 gene was not amplified in any of the 10 species used, four parts of the gene, essential in ADS enzyme function, corresponding to a) pair site of Arg10-Pro12 in the first 100 amino acids, b) aspartate rich motif (DDXXD), c) active site final lid and d) active site including farnesyl diphosphate (FDP) ionization sites and catalytic site in the ADS enzyme, were investigated.
Major conclusion
The sequence corresponding to ADS active site was amplified only in A. annua, A. aucheri and A. chamaemelifolia. The negative results obtained with other species could be due to some sequence alteration, such as point mutations or INDELs. We propose A. aucheri and A. chamaemelifolia as two potential candidate species for further characterization, breeding and transferring am1 gene for artemisinin overproduction.
Keywords: amorpha-4,11-diene synthase; Artemisia (sweet wormwood); artemisinin; Asteraceae; PCR
INTRODUCTION
Human malaria is an important cause of mortality in tropical regions and have been reported to affect 300–500 million people worldwide annually. Infection with Plasmodium falciparum accounts for 200–250 million cases, of which around 2 million are fatal (1). Artemisia annua L. is the main source for Artemisinin extraction. At the present, the World Health Organization (WHO) has recognised this sesquiterpene 3-oxane and its derivates (arteether, artesunate, artemether) obtained from A. annua as one of the most effective medicines for curing malaria. Artemisinin is a novel drug in the treatment of malaria, especially in those areas where resistance to quinine derivatives is common (2, 3). Artemisinin is the last effective drug against deadly resistant strains of P. falciparum (4). The first step in artemisinin biosynthesis is the cyclisation of farnesyl diphosphate (FDP) by amorpha-4,11- diene synthase (ADS) (5). At the present time the extraction of artemisinin from A. annua plants seems to be the only source of this medicine (6).
The chemical synthesis of artemisinin is not cost effective and the toxicity of by-products is also another problem (7, 8). Employment of cell and callus culture has not been promising, since a certain level of differentiation is required for artemisinin production (9). The yield of Artemisinin production by the plant A. annua is relatively low ranging from 0.01% to 0.5% on the basis of dry weight (6). An alternative for the production of artemisinin in higher amounts have been proposed as the selection and breeding of high artemisinin yielding cultivars (10). In this investigation presence of ADS encoding gene (am1) in ten different species of Artemisia growing in Iran was examined to discover potential candidate species for further characterization, and possibly am1 gene transfer to increase artemisinin.
MATERIALS AND METHODS
Plant materials
The seeds of all Artemisia species except A. draconculus, were gifted by Dr. Razban (Research Institute for Forests and Rangelands; RIFR). The seeds had been collected during summer 2008, from different parts of Iran (Table 1). Seeds were sterilized in 5% (v/v) bleach for 10 min, followed by 3 times washings in double sterilised distilled water. For breaking seed dormancy, they were stored at 4°C for 72 hrs, then germinated in a 0.5 X MS medium (11), supplemented with 3% (w/v) sucrose of pH 5.9 and 0.8% (w/v) plant agar. After germination, seeds were subcultured in 1 X MS medium containing 1.5% (w/v) sucrose, 1.5% (w/v) glucose and incubated at 25°C ± 1 and 16/8 h light/dark photoperiod with 55.5 µmol m-2 s-1 illumination, according to the report of Weathers et al. (12).
Table 1.
Artemisia species used in this study.
| Species | Collection place | Specimen* No. | Species | Collection place | Specimen No. |
|---|---|---|---|---|---|
| A. annua | Gorgan | 1595 | A. fragrance | Jolfa | 19969 |
| A. aucheri | Yazd | 4500 | A. cina | Karaj | 923 |
| A. siberi | Isfahan | 10132 | A. austerica | Sarab | 2953 |
| A. vulgaris | Urmiah | 3709 | A. draconculus** | --------- | ---------- |
| A. scoparia | Zanjan | 1298 | A. chamaemelifolia | Tabriz | 15385 |
The specimen numbers are available in the Research Institute of Forests and Rangelands of Iran.
This species was purchased from local shops.
DNA extraction
Genomic DNA extraction from fresh medicinal plants such as Artemisia is a difficult task due to the presence of high amounts of secondary metabolites. A method (13) was chosen and carried out with some modifications to reduce the secondary metabolites and polysaccharides contents.
About 0.1 g of the leaf tissue was grounded to a fine powder by liquid nitrogen in a pestle and mortar. The powder was transferred into a microtube containing 350 µl of the extraction buffer [2% (w/v) CTAB, 100 mM Tris-HCl, 20 mM EDTA, 1.4 M NaCl, 2% (w/v) PVP, 2% (v/v) 2-mercaptoethanol] and 350 µl 8 M lithium chloride, both pre-warmed at 65 °C and incubated for 45 min, with occasional inversions at 10 min intervals. An equal volume of chloroform: isoamyl alcohol 24: 1 was added and tubes were shaken at 50 rpm for 20 min. Microtubes were centrifuged at 13000 rpm (Labnet, UK) for 5 min at room temperature and supernatant was transferred into a new tube. To each tube were added, 0.15 M sodium acetate (5 M) and 0.6 vol ice cold isopropanol and then inverted gently, untill DNA bundles appeared. Tubes were transferred to a -20 °C freezer and left for one hour and then centrifuged at 13000 rpm (Labnet, UK) for 10 min and supernatant was poured out. An aliquot of 300 µl of 70% ethanol (v/v) was added to the pellets and centrifuged at 13000 rpm (Labnet, UK) for 5 min. Supernatant was poured out, pellet were air dried and 10-50 µl sterilized double distilled water was added to each tube. DNA samples were stored at -70 °C for future uses.
Primers
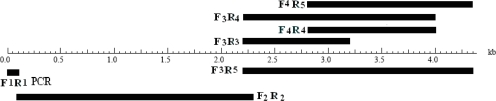
Primers were designed purely based on the DNA sequences (NCBI accession number AF327527) on the am1 gene (with the length of 4392 bp) corresponding to amino acid residues, important for ADS function (14). Also conserved sequences in mono and sesquiterpene synthases (3, 4), and sesquiterpene cyclases in dicotiledons (2) were considered in primer design (Table 2). Position of the primers on the am1 gene is shown in figure 1.
Table 2.
Primers designed according to am1 gene sequence.
| Primer | Sequence 5’ to 3’ | Amplification region | Tm °C |
|---|---|---|---|
| F1 | CCTCCTTCAACCGTTACCCCG | Arg 10 –Pro12 site | 75 |
| R1 | GCGAGAAGGATACCAAGGCAG | Conserved sequence | 73.2 |
| F2 | CTTCTCGCCAGTGGTAGGGTCA | Conserved sequence | 75 |
| R2 | GAAGATACTCCCATCGACCCCT | Conserved sequence | 73.2 |
| F3 | GCTAACGAACTTGCGAGGTAGA | FDP ionization site | 71.3 |
| R3 | CGTTTCCTCCCTTCTTGTCTAG | Conserved sequence | 71.3 |
| F4 | CGGACTTGGATCAGGGGTTTTC | Active site | 73.2 |
| R4 | ATGGTTAGGAAGCACGTATCGG | Catalytic site | 71.3 |
| F5 | GCTTAAAGGGAAACGGCAAC | Start point | 68.9 |
| R5 | CATGATGTGTATAGCGTGC | Catalytic site | 65.4 |
Figure 1.
Primers positions in am1 gene sequence and their expected products.
Following 4 parts were investigated in am1 gene:
Pair site of Arg10-Pro12 in the first 100 amino acids using primers F1-R1. The expected amplification product was a 412 bp fragment.
Aspartate rich motif (DDXXD), using primers F2-R2. This motif is the binding site of substrate. Primers F2 and R2 lie on exons 2 and 3, respectively. The expected amplification fragment was 1870 bp long and includes the intronic part of the gene.
Active site, using primers F3-R3, F3-R4, F4-R4 and F3-R5. This site is formed by three amino acids; Asp 444, Tyr 520 and Asp 524 and includes FDP ionization and catalytic sites. Primers F3-R3, F3- R4 and F4-R4 amplify the FDP ionization site and primer F3-R5 amplify the catalytic site; the expected amplification bands are 1196 bp, 1750 bp, 997 bp and 2144 bp respectively.
Active site final lid, using F4 and R5 primers. This site is formed by three amino acids; Trp 271, Tyr 520 and Asp 524. Primer F4 sits around Trp 271 and primer R5 sits around Asp 524. The expected amplification band is 1544 bp long.
PCR conditions
PCR reactions were carried out in a total volume of 20 µl reaction mixture containing: 2 µl DNA polymerase buffer (Sib enzyme, Russia), 0.2 mM dNTPs (Sib enzyme, Russia), 20 pmol of each primer, 2 units of Taq DNA polymerase (Sib enzyme, Russia) and 50 ng of genomic DNA. Thermal cycler apparatus (Techne, Model TC512, UK) was programmed at 94°C for 4 min, 25 cycles of 94°C for 40 s, 56°C for 60 s, 72°C for 2 min and the final extension at 72°C for 8 min. Different annealing temperatures were used for each pair of primers according to Gene Runner software recommendations (Table 1). PCR products were run on a 1% (w/v) agarose gel along with a 1kb DNA size marker (Cinnagen), stained by ethidium bromide (0.5 µg/ml) and visualized in a gel documentation system (UVP, USA)
Digestion of PCR products
PCR products were digested by HindII and BglII enzymes. The cutting sites and the expected number of fragments were obtained using Gene Runner software. Reactions were carried out in a 20 µl final volume. Each reaction contained 1 µg DNA, 2 µl of buffer (10 X), 0.2 µg BSA, 1 unit of each of restriction enzymes (Sibenzyme, Russia) and distilled sterile water.
RESULTS
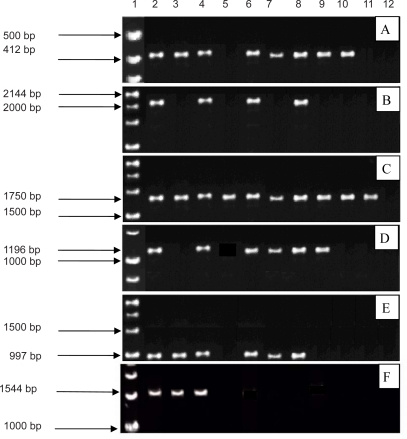
Amplification of entire am1 gene in the 10 mentioned Artemisia species, was the main aim of this study. Nevertheless, despite applying different annealing temperatures, extension times and PCR components concentrations, this amplification did not happen. In the amplification of aspartate rich motif (DDXXD), all species gave a 412 bp amplification band except A. scoparia and A. draconculus (Fig 2a). Aspartate rich motif is the binding site of substrate to ADS enzyme (14). The expected band (1870 bp) was not observed in any of the species. Several unspecific bands were amplified in A. annua, A. austrica and A. chamaemelifolia (data not shown).
Figure 2.
PCR products obtained from the designed primers based on the am1 gene sequence with the following primer pairs: A) F1-R1 B) F3-R5 C) F3-R4 D) F3-R3 E) F4-R4 F) F4-R5 in 10 wild artemisia species. The lanes show in order (1) 1kb DNA size marker, (2) A. annua, (3) A. austerica, (4) A. aucheri, (5) A. scoparia, (6) A. chamaemelifolia, (7) A. vulgaris, (8) A. siberi, (9) A. cina, (10) A. fragrance, (11) A. draconculus and (12) negative control.
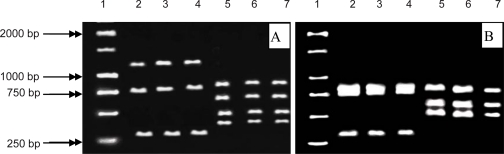
The F3 primer position was designed around Arg 262 and R5 primer position around Tyr 520 and Asp 524 residues to find catalytic site. The expected amplification band was observed in A. annua, A. aucheri, A. chamaemelifolia and A. siberi (Fig. 2b). To investigate FDP ionization site, primers F3 and F4 were used. Primer F3 contains the Arg 262 position and Primer R4 corresponds to the positions of Arg 440 and Asp 444 on the ADS enzyme. R3 primer sits on a conserved sequence in terpene synthase genes (14). Primers F3-R4 amplified the 1750 bp fragment in A. annua, A. siberi, A. austrica, A. vulgaris, A. scoparia, A. draconculus and A. cina (Fig. 2c). F3- R3 primers could amplify the 1196 bp fragment in A. annua, A. aucheri, A. chamaemelifolia, A. vulgaris, A. siberi and A. cina (Fig. 2d). Primers F4- R4 amplified the 997 bp fragment in A. annua, A. aucheri, A. chamaemelifolia, A. siberi, A. vulgaris and A. austrica (Fig. 2e). By using primers F4 and R5, the expected 1524 bp fragment was observed only in A. annua, A. aucheri and A. chamaemelifolia (Fig. 1f). To ensure that the PCR fragments contained the correct sequences, the products of F3-R4 (FDP ionization site) and F3-R5 (catalytic site) were amplified in A. annua, A. chammelifoli and A. aucheri were digested with BglII and HindII. The restriction fragments expected from F3-R4 product digestion with BglII were 802, 728 and 264 bp and the expected restriction fragments obtained from HindII digestion were 761, 575 and 458 bp long. The restriction fragments expected from F3- R5 digestion with BglII and HindII which also obtained were 1151, 728, 264 bp, and 575, 480,327 bp long respectively. Indicating that correct sequences had been amplified (Figs. 3A, B). Results are summarized in (Table 3).
Figure 3.
Restriction digestion of F3-R3 (A) and F3-R5 (B) amplification products. Lane (1), 1 kb DNA size marker, lanes 2-4, PCR products from A. annua, A. aucheri and A. chamaemelifolia, digested with BglII and lanes 5-7 PCR products from A. annua, A. aucheri and A. chamaemelifolia digested with HindII, respectively.
Table 3.
The summary of results obtained with different primer pairs on the am1 gene.
| Artemisia species | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| primer | annua | austrica | aucheri | scoparia | chamaemelifolia | vulgaris | siberi | cina | fragrance | draconculus | |
| F1-R1 | + | + | + | − | − | + | + | + | + | + | |
| F3-R3 | + | − | + | − | + | + | + | + | − | − | |
| F3-R4 | + | + | + | + | + | + | + | + | + | + | |
| F3-R5 | + | - | + | − | + | - | + | − | − | − | |
| F4-R4 | + | + | + | − | + | + | + | − | − | − | |
| F4-R5 | + | − | + | − | + | − | − | − | − | − | |
DISCUSSION
There are different medicines for treatment of malaria. A novel medicine on this respect is artemisinin, obtained from A. annua. This study was conducted to discover Artemisia species containing the am1 gene, an important gene in artemisinin synthesis pathway. Since most of the studied species grow wildly in different areas, it was not possible to investigate artemisinin production in those species, especially at different growth stages. In addition, some of the species needed a stage of adaptation to the glasshouse or field conditions to grow. To solve each one of these problems, further studies were required. Therefore an initial study was conducted to investigate the presence of am1 gene. PCR was employed and primers were designed to amplify the entire am1 gene as well as 4 important sequences, essential for the correct ADS enzyme function. Positive results were obtained from several pairs of primers, amplifying 5’, 3’ and the middle part of the am1 gene in some of the Artemisia species. Nevertheless, despite applying a wide range of PCR conditions, the whole gene (4392 bp) was not amplified. However, the primers designed to amplify both 5’ and 3’ ends of the am1 gene, showed positive results. This means that forward and reverse primers were matching the corresponding sequences at the two ends of the gene, but probably changes in some sequences in the gene would not allow whole gene amplification. This may be as a result of INDELs somewhere in the am1 gene sequence that prevented the whole gene amplification.
CONCLUSION
Result of this investigation showed that two Artemisia species contain the DNA sequences to encode the important parts of ADS enzyme for correct folding and function. In conclusion two Artemisia species, A. aucheri and A. chamaemelifolia are recommended for further investigation and possibly gene transfer with the aim of increasing artemisinin production.
ACKNOWLEDGMENTS
We thank Dr Razban for the donation of Artemisia seeds, technical support and invaluable suggestions.
REFERENCES
- 1.Francell WA, Cinza MB, Alessandra V. Isoprenoid biosynthesis in Artemisia annua cloning heterologous expression of germacrene synthase from glandular trichomes cDNA library. Arch. Biochem Biophys. 2006;448:3–12. doi: 10.1016/j.abb.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 2.Gelder EV, Pauw ID, Montagu MV, Eekhout EV. Cloning and molecular analysis of two new sesquiterpene cyclases from Artemisia annua. Plant Sci. 2000;158:163–171. doi: 10.1016/s0168-9452(00)00322-8. [DOI] [PubMed] [Google Scholar]
- 3.Mereke P, Benetsson M, Boumeester HJ, Bordelious P. Molecular cloning and charactetrization of Amorpha-4,11-diene synthase in artemisinin biosynthesis in Artemisia annua. Arch Biochem Biophys. 2000;381:173–180. doi: 10.1006/abbi.2000.1962. [DOI] [PubMed] [Google Scholar]
- 4.Chang YJ, Song SH, Kim SV. Amorpha diene synthase of Artemisia annua: cDNA isolation and bacterial expression of a terpene synthase involved in Artemisinin biosynthesis. Arch Biochem Biophys. 2000;383:178–184. doi: 10.1006/abbi.2000.2061. [DOI] [PubMed] [Google Scholar]
- 5.Brodelius PE, Picaud S, Bordelius M. Expression, purification, and characterization of recombinant amorpha-4, 11-diene synthase from Artemisia annua. Arch Biochem Biophys. 2005;436:215–226. doi: 10.1016/j.abb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Delabays N, Simonet X, Gaudin M. The genetics of artemisinin content in Artemisia annua L. and the breeding of high yielding cultivars. Curr Med Chem. 2001;18:1795–1801. doi: 10.2174/0929867013371635. [DOI] [PubMed] [Google Scholar]
- 7.Duke MV, Paul RN, Elsophy NH, Duke SO. Localisation of artemisinin and artemisinic acid in foliar tissues of gland and glandless biotypes of Artemisia annua. J Plant Sci. 1999;155:365–372. [Google Scholar]
- 8.Sabitha G, Yadau JS, Satheesh RB. Total synthesis of artemisinin. Arch Organ Chem. 2003;12:125–139. [Google Scholar]
- 9.Ferreira JFS, Janick J. Roots as an enhancing factor for the production of artemisinin in shoot cultures of Artemisia annua. Plant Cell. Tiss Org Cult. 1996;44:211–217. [Google Scholar]
- 10.Wallart TE, Boumeester HJ, Hill J, Maijres CA. Amorpha diene synthase cloning and functional expression of a key enzyme in the biosynthetic pathway of a novel antimalarial drug Artemisinin. Planta. 2001;212:460–465. doi: 10.1007/s004250000428. [DOI] [PubMed] [Google Scholar]
- 11.Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plantarum. 1962;15:473–497. [Google Scholar]
- 12.Weathers PJ, Jesus LD, Gonzales J, Kim YJ. Alternation of biomass and artemisinin production in Artemisia annua hairy roots by media sterilization methods and sugars. Plant Cell Rep. 2004;23:414–418. doi: 10.1007/s00299-004-0837-4. [DOI] [PubMed] [Google Scholar]
- 13.Kamaranen T, Pirttila MA, Hirskoppi M, Kamaranen T. DNA isolation methods for medicinal and aromatic plants. Plant Mol Biol Rep. 2001;19:273. a-f. [Google Scholar]
- 14.Chen DH, Hes HC, Li GF. Expression of chimeric FPPS gene in Artemisia annua via Agrobacterium- mediated transformation. Plant Sci. 2000;155:179–185. doi: 10.1016/s0168-9452(00)00217-x. [DOI] [PubMed] [Google Scholar]