Abstract
Background:
The Hedgehog (Hh) signalling pathway functions as an organiser in embryonic development. Recent studies have shown constitutive activation of this pathway in various malignancies, but its role in bladder cancer remains poorly studied.
Methods:
Expression levels of 31 genes and 9 microRNAs (miRNAs) involved in the Hh pathway were determined by quantitative real-time RT–PCR in 71 bladder tumour samples (21 muscle-invasive (MIBC) and 50 non-muscle-invasive (NMIBC) bladder cancers), as well as in 6 bladder cancer cell lines.
Results:
The SHH ligand gene and Gli-inducible target genes (FOXM1, IGF2, OSF2, H19, and SPP1) were overexpressed in tumour samples as compared with normal bladder tissue. SHH overexpression was found in 96% of NMIBC and 52% of MIBC samples, as well as in two bladder cancer cell lines. Altered expression of miRNAs supported their oncogene or tumour-suppressor gene status. In univariate analysis, high expression levels of PTCH2, miRNA-92A, miRNA-19A, and miRNA-20A were associated with poorer overall survival in MIBC (P=0.02, P=0.012, P=0.047, and P=0.036, respectively).
Conclusion:
We observed constitutive activation of the Hh pathway in most NMIBC and about 50% of MIBC. We also found that some protein-coding genes and miRNAs involved in the Hh pathway may have prognostic value at the individual level.
Keywords: bladder cancer, Hedgehog signalling pathway, molecular marker, prognostic factor, RT–PCR
In western countries, bladder cancer is the fourth and ninth most common malignancy in men and women, respectively. About 90% of malignancies arising in the urothelium are of epithelial origin (transitional cell carcinoma, TCC). About two-thirds of newly diagnosed cases of TCC are superficial papillary tumours, which are frequently recurrent. The TCC is muscle-invasive at diagnosis in about one-third of cases and metastatic in about 7% of cases. Patients with a given tumour stage and grade may have different outcomes, which cannot be predicted by current prognostic factors, namely TNM stage and pathological grade. New prognostic molecular markers, which might also serve as therapeutic targets, are therefore needed.
The Hedgehog (Hh) family of proteins regulates a wide variety of developmental processes, and Hh pathway defects have been implicated in many developmental disorders (Ingham and McMahon, 2001). However, continuous Hh pathway activity has a role in the growth of various malignancies, that together account for approximately one-quarter of all cancer deaths (Berman et al, 2003; Pasca di Magliano and Hebrok, 2003; Scott, 2003; Beachy et al, 2004; Fan et al, 2004; Kayed et al, 2004; Liu et al, 2007).
Three Hh genes have been described in mammals: Sonic (SHH), Indian (IHH), and Desert (DHH; McMahon, 2000; Nybakken and Perrimon, 2002). The Hh proteins are ligands for the patched receptor (Ptch), which negatively regulates smoothened protein (Smo). The Ptch binds to the Hh proteins, resulting in Ptch internalisation in endosomes and lifting Ptch-mediated repression. This allows Smo to move from an intracellular compartment to the cell surface, resulting in Smo activation and signal transmission. Two homologous Ptch receptors, Ptch1 and Ptch2, have been described, both of which are able to interact with the Hh ligands and Smo protein. Downstream of Smo, the Hh signal activates target genes through the Gli family of zinc-finger transcription factors (including Gli1, Gli2, Gli3, and Gli4 in vertebrates). The Ptch1 is also a target of this pathway, forming a negative feedback mechanism and thus maintaining pathway activity at an appropriate level in a given cell.
Mutational activation of the Hh pathway, whether sporadic or constitutional as in Gorlin's syndrome, is associated with tumorigenesis in a variety of tissues, but predominantly in skin, the cerebellum and skeletal muscle. The Hh pathway activation, whether triggered by Hh binding (ligand overexpression) or by Ptch mutational inactivation (Ptch is unable to restrain Smo-mediated activation of transcriptional targets through the Gli family even when not bound by the Hh protein), requires Smo regulation (Johnson et al, 1996). Smo, which is inactivated by the pathway antagonist cyclopamine, is also a candidate therapeutic target (Incardona et al, 1998; Rudin et al, 2009; Von Hoff et al, 2009).
The role of the Hh pathway in TCC remains poorly studied (Fei et al, 2010; Mechlin et al, 2010; He et al, 2011). The PTCH1 gene, located in chromosome region 9q22, is a candidate tumour-suppressor gene, as loss of heterozygosity on chromosome arm 9q occurs in more than 50% of TCC (Linnenbach et al, 1993; Habuchi et al, 1995; Hirao et al, 2005), and PTCH1 mRNA expression is low, compared with normal urothelium, in early-stage tumours exhibiting LOH in the 9q22 region (Aboulkassim et al, 2003; Hirao et al, 2005). However, few mutations have been detected in the retained PTCH1 allele (McGarvey et al, 1998). Other mechanisms, such as PTCH1 haploinsufficiency or alterations in other genes regulating the Hh signalling, could be involved in urothelial development. Furthermore, gene amplification of part of chromosome region 12q13-q15, which encompasses GLI1, has been found in a subset of bladder cancers (Simon et al, 2002). Altered expression of Gli proteins seems to be associated with a more invasive phenotype of bladder tumours in vitro (Fei et al, 2010; Mechlin et al, 2010).
More recently, another mechanism leading to abnormal Hh pathway activation – an autocrine or paracrine loop initiated by Shh overexpression – was detected in gastrointestinal (Berman et al, 2003), pancreatic (Thayer et al, 2003), and small-cell lung cancer (Watkins et al, 2003).
Finally, microRNAs (miRNAs) were recently described as a class of small non-coding cellular RNAs that bind to cis-regulatory elements mainly present in the 3′-untranslated regions (3′-UTRs) of their target-protein-coding mRNAs, resulting in translational regulation (Stefani and Slack, 2008). MicroRNAs are crucial post-transcriptional regulators of gene expression, controlling cell differentiation and proliferation, and being implicated in tumour formation (Calin and Croce, 2006). However, little is known of how miRNAs target specific developmental pathways, including the Hh pathway (Ferretti et al, 2008; Uziel et al, 2009).
In an attempt to identify new molecular markers in TCC, we analysed a large panel of genes (n=31) and miRNAs (n=9) involved in the Hh pathway, in a series of 71 urothelial bladder tumours. Using real-time quantitative RT–PCR, we determined expression levels of the selected gene mRNAs and miRNAs in each bladder sample. In this pilot study, the prognostic value of these molecular markers for patient survival was examined retrospectively.
Patients and methods
Patients and samples
Normal bladder samples (n=5, group I) were obtained during prostatic adenomectomy from patients with no history of bladder cancer.
Bladder cancer samples were obtained from patients who underwent transurethral bladder resection (TURB) or radical cystectomy at Cochin Hospital, Paris, France, between January 2001 and December 2002. All patients signed an informed consent. During TURB, tumour fragments comprising both visible urothelium and underlying muscle were selected for RNA extraction and immediately stored in liquid nitrogen at −80 °C. Remaining fragments were fixed in formaldehyde for pathological analysis. The similar nature of frozen and formaldehyde-fixed samples was confirmed by examining frozen sections of each cryopreserved sample.
Cystectomy specimens were immediately reviewed by the pathologist, who visually selected the tumour zone to be frozen in liquid nitrogen. The rest of the cystectomy specimen was fixed in formaldehyde and subjected to standard pathological analysis after step-sectioning. If insufficient material was available for both nitrogen and formaldehyde storage, the latter took priority.
Each tumour was reviewed by the same pathologist. All the tumours were of urothelial origin. Tumour stage was determined with the 2002 UICC TNM classification of bladder cancer, and tumour grade using the OMS 2004 grading scheme (IUAC, 2002; Molinie, 2006).
There were 11 women and 60 men, with a median age of 68 years (range 42–88 years). Pathological staging showed non-muscle-invasive bladder cancer (NMIBC) in 50 patients (21 low-grade pTa, 10 high-grade pTa, and 19 high-grade pT1) and high-grade muscle-invasive bladder cancer (MIBC, ⩾pT2) in 21 patients.
Outcomes were obtained from the patients’ medical records. After a median follow-up of 71.5 months (range, 1–104), 24 NMIBC patients had recurrences and 4 patients progressed to muscle-invasive disease; statistical analysis was not possible in this group, because of the small number of events. Five NMIBC patients were lost to follow-up and were therefore excluded from the prognostic analysis. Eleven MIBC patients had local or metastatic relapses after a median follow-up of 36.2 months (range, 4–97 months), and all died.
Clinical and histological parameters and outcomes in the NMIBC and MIBC populations are shown in Tables 1a and 1b. These population characteristics are consistent with bladder cancer presentation and evolution.
Table 1a. Clinical and pathological characteristics of the NMIBC subpopulation (n=45a).
| No recurrence (n=17) |
Recurrence (n=24)
|
Muscle-invasive progression (n=4) | ||
|---|---|---|---|---|
| Number (%) | Number (%) | P-values b | Number (%) | |
| Median age, years (range) | 66 (44–86) | 68 (42–83) | 0.47c | 78 (77–87) |
| Sex | 0.36 | |||
| Male | 13 (76.5) | 22 (91.7) | 2 (50.0) | |
| Female | 4 (23.5) | 2 (8.3) | 2 (50.0) | |
| Smoking status | 0.56 | |||
| Non-smoker | 12 (70.6) | 20 (83.3) | 3 (75.0) | |
| Smoker | 5 (29.4) | 4 (16.7) | 1 (25.0) | |
| History of NMIBC | 0.001 | |||
| No | 15 (88.2) | 8 (33.3) | 3 (75.0) | |
| Yes | 2 (11.8) | 16 (66.7) | 1 (25.0) | |
| Associated pTis | 0.63 | |||
| No | 16 (94.1) | 23 (95.8) | 4 (100) | |
| Yes | 1 (5.9) | 1 (4.2) | 0 (0) | |
| Multifocality | 0.85 | |||
| No | 11 (64.7) | 15 (62.5) | 3 (75.0) | |
| Yes | 6 (35.3) | 9 (37.5) | 1 (25.0) | |
| Grade | 0.77 | |||
| Low grade | 8 (47.1) | 9 (37.5) | 1 (25.0) | |
| High grade | 9 (52.9) | 15 (62.5) | 3 (75.0) | |
| Tumour stage | 0.93 | |||
| Ta | 10 (58.8) | 15 (62.5) | 2 (50.0) | |
| T1 | 7 (41.2) | 9 (37.5) | 2 (50.0) | |
Abbreviations: NMIBC=non-muscle-invasive bladder cancer; pTis=carcinoma in situ.
Five patients lost to follow-up and excluded from the analysis.
χ2-test.
Student's t-test.
Bold value indicates significant P-value (<0.05).
Table 1b. Clinical and pathological characteristics of the MIBC subpopulation (n=21).
|
Disease-free survival
|
|||
|---|---|---|---|
| Number of patients (%) | Number of events (%) a | P-values b | |
| Age | 0.63 | ||
| <50 years | 1 (4.8) | 0 (0) | |
| 50–70 years | 9 (42.8) | 5 (55.6) | |
| >70 years | 11 (52.4) | 6 (54.5) | |
| Sex | 0.15 | ||
| Male | 19 (90.5) | 11 (57.9) | |
| Female | 2 (9.5) | 0 (0) | |
| Smoking status | 0.23 | ||
| Non-smoker | 6 (28.6) | 4 (66.7) | |
| Smoker | 15 (71.4) | 7 (46.7) | |
| History of NMIBC | 0.27 | ||
| No | 16 (76.2) | 7 (43.7) | |
| Yes | 5 (23.8) | 4 (80.0) | |
| Associated pTis | 0.30 | ||
| No | 17 (80.9) | 9 (52.9) | |
| Yes | 4 (19.1) | 2 (50.0) | |
| Multifocality | 0.68 | ||
| No | 18 (85.7) | 10 (55.6) | |
| Yes | 3 (14.3) | 1 (33.3) | |
| Tumour stage | 0.0020 | ||
| T2 | 13 (61.9) | 5 (38.5) | |
| >T2 | 8 (38.1) | 6 (75) | |
| Lymph node status | 0.0015 | ||
| N− | 15 (71.4) | 5 (33.3) | |
| N+ | 6 (28.6) | 6 (100) | |
Abbreviations: MIBC=muscle-invasive bladder cancer; NMIBC=non-muscle-invasive bladder cancer; pTis=carcinoma in situ.
First recurrence (local or metastatic).
Log rank test.
Bold values indicate significant P-values (<0.05).
Bladder cancer cell lines
We also analysed six bladder cancer cell lines of different origins (CRL1472, CRL1749, CRL2169, HTB2, HTB4, and HTB9) obtained from the American Tissue Type Culture Collection.
Gene and micro-RNA selection
After examining the literature on the Hh pathway and bladder carcinogenesis, we selected 31 protein-coding genes, including the three Hh pathway ligands (SHH, IHH, DHH), the two homologous Ptch receptors (PTCH1 and PTCH2), transduction and transcription factors, target genes, and 9 miRNAs (Supplementary Data 1 and 2).
Real-time quantitative RT–PCR analysis of protein-coding genes
The theoretical basis and practical aspects of real-time quantitative RT–PCR (primers and PCR consumables; RNA extraction, cDNA synthesis, and PCR reaction conditions) are described in detail elsewhere (Pignot et al, 2009). Quantitative values are obtained from the cycle threshold (Ct), at which the increase in the signal associated with the exponential growth of PCR products begins to be detected. Two endogenous RNA control genes involved in different metabolic pathways were chosen, namely TBP (Gen-Bank accession Number NM_003194), which encodes the TATA-box-binding protein, and RPLP0 (Gen-Bank accession Number NM_001002), which encodes human acidic ribosomal phosphoprotein P0. Each sample was normalised on the basis of its TBP (or RPLP0) content. Results, expressed as N-fold differences in target gene expression relative to the TBP (or RPLP0) gene, and termed ‘Ntarget’, were determined as Ntarget=2ΔCtsample, where the ΔCt value of the sample was determined by subtracting the average Ct value of the target gene from the average Ct value of the TBP (or RPLP0) gene. The Ntarget values of the samples were subsequently normalised such that the median of the five normal-bladder Ntarget values was 1. For each investigated gene, mRNA values of 3 or more were considered to represent marked overexpression and mRNA values of 0.3 or less were considered to represent marked under-expression. We have previously used the same cutoff points for altered tumour gene expression (Pignot et al, 2009). Primers were chosen with the Oligo 6.0 computer programme (National Biosciences, Plymouth, MN, USA). For each primer pair, we performed no-template control and no-RT control (RT negative) assays, which produced negligible signals (Ct values usually >40), suggesting that primer-dimer formation and genomic DNA contamination effects were negligible (Supplementary Data 1). Experiments were performed with duplicates for each data point.
Real-time quantitative RT–PCR assay of mature miRNAs
MicroRNAs were isolated with the extraction procedure used for the protein-coding genes (total RNA extraction). Reverse transcription was performed with the QIAGEN miScript Reverse Transcription kit, according to the manufacturer's protocol (QIAGEN, GmbH, Hilden, Germany). Specific miRNAs were quantified by real-time PCR with the QIAGEN miScript SYBR Green PCR kit (QIAGEN). The small nucleolar RNA U44 was used as an internal control. The Δ-Δ Ct method was used to determine miRNA expression, as for protein coding gene expression.
Statistical analysis
Clinical and pathological features of NMIBC and MIBC were tested for their association with tumour recurrence and patient survival, using Student's t-test for continuous variables and the χ2-test for qualitative variables. The distribution of mRNA (or miRNA) levels was analysed using the median and range. Relationships between mRNA (or miRNA) levels and clinical and histological parameters were identified with the Kruskal–Wallis non-parametric H-test (link between one qualitative parameter and one quantitative parameter). Overall survival (OS) was calculated from the date of surgery to death from bladder cancer or last follow-up. Survival curves were derived from the Kaplan–Meier estimates. The log-rank test was used to compare survival distributions between subgroups. Differences between two populations were judged significant at confidence levels greater than 95% (P<0.05) and all tests were two-sided.
Results
mRNA expression in normal bladder tissue
To determine the cutpoint for altered gene expression in tumour samples, the Ntarget value of the 31 genes, calculated as described in Patients and Methods, were first determined in 5 normal bladder samples. All the genes had quantifiable mRNA levels by real-time quantitative RT–PCR (Ct value <38), suggesting basal expression of this pathway in normal bladder (Table 2).
Table 2. mRNA values of Hedgehog signalling pathway authors.
| NMIBC | ||||||||
|---|---|---|---|---|---|---|---|---|
| Normal Group 1 | pTaG1–G2 Group 2 | pTaG3 Group 3 | pT1G3 Group 4 | MIBC ⩾pT2 Group 5 | All NMIBC 2+3+4 | All tumours 2+3+4+5 | ||
| n=5 | n=21 | n=10 | n=19 | n=21 | P-values a | n=50 | n=71 | |
| Ligands | ||||||||
| SHH mRNA values: median (range) | 1 (0.37–1.81) | 121.9 (14.88–566.1) | 85.2 (1.16–189.4) | 89.9 (9.29–721.6) | 3.92 (0.22–293.1) | <10−5 | 93.9 (1.16–721.6) | 78.0 (0.22–721.6) |
| Overexpression, n (%) | — | 21 (100) | 8 (80) | 19 (100) | 11 (52.4) | — | 48 (96.0) | 59 (83.1) |
| IHH mRNA values: median (range) | 1 (0.39–1.53) | 0.12 (0.01–2.73) | 1.22 (0.08–7.29) | 0.32 (0.00–16.3) | 0.07 (0.01–1.12) | 0.01 | 0.30 (0.00–16.3) | 0.23 (0.00–16.3) |
| Overexpression, n (%) | — | 0 (0) | 2 (20) | 5 (26.3) | 0 (0) | — | 7 (14.0) | 7 (13.5) |
| DHH mRNA values: median (range) | 1 (0.17–1.92) | 1.52 (0.48–3.56) | 1.22 (0.40–8.05) | 1.83 (0.52–17.9) | 1.55 (0.13–5.42) | 0.61 | 1.62 (0.40–17.9) | 1.61 (0.13–17.9) |
| Overexpression, n (%) | — | 2 (9.5) | 2 (20) | 3 (15.8) | 5 (23.8) | — | 7 (14.0) | 12 (16.9) |
| Receptors | ||||||||
| PTCH1 mRNA values: median (range) | 1 (0.82–1.54) | 0.61 (0.15–1.59) | 0.31 (0.15–1.11) | 0.42 (0.07–1.54) | 0.39 (0.05–2.19) | 0.34 | 0.47 (0.07–1.59) | 0.45 (0.05–2.19) |
| Under-expression, n (%) | — | 4 (19) | 4 (40) | 6 (31.6) | 8 (38.1) | — | 14 (28.0) | 22 (31.0) |
| PTCH2 mRNA values: median (range) | 1 (0.50–1.83) | 0.25 (0.00–2.03) | 0.39 (0.10–2.76) | 0.27 (0.01–1.23) | 0.60 (0.06–1.37) | 0.003 | 0.26 (0.00–2.76) | 0.31 (0.00–2.76) |
| Under-expression, n (%) | — | 13 (61.9) | 4 (40) | 11 (57.9) | 3 (14.3) | — | 28 (56.0) | 31 (43.7) |
| Transduction factors | ||||||||
| SMOH mRNA values: median (range) | 1 (0.94–1.07) | 0.14 (0.02–0.33) | 0.13 (0.02–0.42) | 0.20 (0.03–0.60) | 0.23 (0.02–0.61) | 0.001 | 0.15 (0.02–0.60) | 0.17 (0.02–0.61) |
| Under-expression, n (%) | 19 (90.5) | 7 (70) | 15 (78.9) | 12 (57.1) | 41 (82.0) | 53 (74.6) | ||
| HHIP mRNA values: median (range) | 1 (0.68–1.25) | 0.26 (0.01–1.48) | 0.10 (0.00–0.46) | 0.17 (0.01–0.59) | 0.03 (0.00–1.22) | 0.0002 | 0.18 (0.00–1.48) | 0.12 (0.00–1.48) |
| Under-expression, n (%) | — | 12 (57.1) | 9 (90) | 14 (73.7) | 20 (95.2) | — | 35 (70.0) | 55 (77.5) |
| SUFU mRNA values: median (range) | 1 (0.18–1.48) | 0.30 (0.13–1.45) | 0.88 (0.39–4.26) | 0.48 (0.14–3.24) | 0.29 (0.08–1.39) | 0.01 | 0.42 (0.13–4.26) | 0.36 (0.08–4.26) |
| Under-expression, n (%) | — | 10 (47.6) | 0 (0) | 6 (31.6) | 11 (52.4) | — | 16 (32.0) | 27 (38.0) |
| DISP1 mRNA values: median (range) | 1 (0.89–1.33) | 0.99 (0.40–2.54) | 0.96 (0.46–1.45) | 0.86 (0.28–2.95) | 0.48 (0.19–1.81) | 0.67 | 0.89 (0.28–2.95) | 0.76 (0.19–2.95) |
| Under-expression, n (%) | 0 (0) | 0 (0) | 1 (5.3) | 3 (14.3) | 1 (2.0) | 4 (5.6) | ||
| DISP2 mRNA values: median (range) | 1 (0.84–2.57) | 3.19 (0.07–23.5) | 9.35 (1.17–46.1) | 3.40 (0.18–22.0) | 2.54 (0.00–25.7) | 0.04 | 3.70 (0.07–46.1) | 3.42 (0.00–46.1) |
| Overexpression, n (%) | — | 11 (52.4) | 8 (80) | 12 (63.2) | 9 (42.9) | — | 31 (62.0) | 40 (56.3) |
| GAS1 mRNA values: median (range) | 1 (0.47–1.24) | 0.07 (0.03–1.19) | 0.12 (0.04–2.35) | 0.09 (0.02–0.85) | 0.50 (0.03–2.18) | 2 × 10−5 | 0.09 (0.02–2.35) | 0.10 (0.02–2.35) |
| Under-expression, n (%) | — | 19 (90.5) | 8 (80) | 16 (84.2) | 8 (38.1) | — | 43 (86.0) | 51 (71.8) |
| STK36 mRNA values: median (range) | 1 (0.81–1.59) | 1.11 (0.46–2.13) | 1.18 (0.54–2.08) | 0.75 (0.33–2.51) | 0.89 (0.41–3.23) | 0.62 | 0.97 (0.33–2.51) | 0.96 (0.33–3.23) |
| Overexpression, n (%) | — | 0 (0) | 0 (0) | 0 (0) | 1 (4.8) | — | 0 (0) | 1 (1.4) |
| KIF7 mRNA values: median (range) | 1 (0.87–1.48) | 0.95 (0.09–2.12) | 0.61 (0.15–2.09) | 0.67 (0.19–2.86) | 0.51 (0.18–1.34) | 0.95 | 0.70 (0.09–2.86) | 0.68 (0.09–2.86) |
| Under-expression, n (%) | — | 1 (4.8) | 1 (10) | 1 (5.3) | 1 (4.8) | — | 3 (6.0) | 4 (5.6) |
| KIF27 mRNA values: median (range) | 1 (0.76–1.26) | 0.64 (0.21–2.36) | 0.63 (0.25–1.83) | 0.63 (0.17–3.34) | 0.55 (0.11–3.35) | 0.52 | 0.64 (0.17–3.34) | 0.63 (0.11–3.35) |
| Under-expression, n (%) | — | 4 (19) | 2 (20) | 1 (5.3) | 2 (9.5) | — | 7 (14.0) | 9 (12.7) |
| Overexpression, n (%) | 0 (0) | 0 (0) | 1 (5.3) | 1 (4.8) | 1 (2.0) | 2 (2.8) | ||
| RAB23 mRNA values: median (range) | 1 (0.84–1.24) | 0.16 (0.05–0.95) | 0.17 (0.07–0.61) | 0.11 (0.05–0.75) | 0.21 (0.08–1.27) | 2 × 10−5 | 0.13 (0.05–0.95) | 0.16 (0.05–1.27) |
| Under-expression, n (%) | — | 20 (95.2) | 6 (60) | 18 (94.7) | 15 (71.4) | — | 44 (88.0) | 59 (83.1) |
| BTRC mRNA values: median (range) | 1 (0.93–1.21) | 1.10 (0.40–1.93) | 0.91 (0.60–1.77) | 0.72 (0.36–1.21) | 0.49 (0.25–1.28) | 0.62 | 0.87 (0.36–1.93) | 0.74 (0.25–1.93) |
| Under-expression, n (%) | — | 0 (0) | 0 (0) | 0 (0) | 1 (4.8) | — | 0 (0) | 1 (1.4) |
| Metabolic enzymes | ||||||||
| HHAT mRNA values: median (range) | 1 (0.90–1.55) | 1.05 (0.47–2.14) | 0.99 (0.32–1.61) | 0.64 (0.11–2.04) | 0.25 (0.04–0.95) | <10−5 | 0.95 (0.11–2.14) | 0.78 (0.04–2.14) |
| Under-expression, n (%) | — | 0 (0) | 0 (0) | 1 (5.3) | 11 (52.4) | — | 1 (2.0) | 12 (16.9) |
| Transcription factors | ||||||||
| GLI mRNA values: median (range) | 1 (0.82–1.12) | 0.22 (0.03–0.72) | 0.18 (0.04–3.58) | 0.27 (0.00–1.01) | 0.27 (0.01–1.38) | 0.048 | 0.23 (0.00–3.58) | 0.24 (0.00–3.58) |
| Under-expression, n (%) | — | 15 (71.4) | 7 (70) | 12 (63.2) | 11 (52.4) | — | 34 (68.0) | 45 (63.4) |
| GLI2 mRNA values: median (range) | 1 (0.47–1.43) | 0.08 (0.01–0.61) | 0.07 (0.01–1.51) | 0.07 (0.02–0.62) | 0.21 (0.01–0.68) | 3 × 10−5 | 0.07 (0.01–1.51) | 0.10 (0.01–1.51) |
| Under-expression, n (%) | — | 20 (95.2) | 7 (70) | 18 (94.7) | 13 (61.9) | — | 46 (92.0) | 59 (83.1) |
| GLI3 mRNA values: median (range) | 1 (0.50–1.31) | 0.44 (0.09–1.23) | 0.67 (0.18–3.67) | 0.28 (0.03–1.99) | 0.40 (0.03–1.77) | 0.07 | 0.45 (0.03–3.67) | 0.42 (0.03–3.67) |
| Under-expression, n (%) | — | 6 (28.6) | 2 (20) | 11 (57.9) | 7 (33.3) | — | 19 (38.0) | 26 (36.6) |
| GLI4 mRNA values: median (range) | 1 (0.72–1.48) | 1.52 (0.64–4.21) | 1.82 (0.62–4.24) | 2.00 (0.52–5.15) | 1.39 (0.49–4.13) | 0.14 | 1.63 (0.52–5.15) | 1.59 (0.49–5.15) |
| Overexpression, n (%) | — | 1 (4.8) | 3 (30) | 5 (26.3) | 2 (9.5) | — | 9 (18.0) | 11 (15.5) |
| GLIS1 mRNA values: median (range) | 1 (0.62–2.09) | 0.01 (0.00–0.15) | 0.01 (0.00–0.20) | 0.02 (0.00–0.07) | 0.08 (0.00–0.27) | <10−5 | 0.01 (0.00–0.20) | 0.02 (0.00–0.27) |
| Under-expression, n (%) | — | 21 (100) | 10 (100) | 19 (100) | 21 (100) | — | 50 (100) | 71 (100) |
| GLIS2 mRNA values: median (range) | 1 (0.83–1.10) | 0.17 (0.04–0.59) | 0.26 (0.07–0.87) | 0.28 (0.09–0.74) | 0.68 (0.04–2.02) | 0.0002 | 0.22 (0.04–0.87) | 0.27 (0.04–2.02) |
| Under-expression, n (%) | — | 18 (85.7) | 5 (50) | 11 (57.9) | 5 (23.8) | — | 34 (68.0) | 39 (54.9) |
| Target genes | ||||||||
| FOXM1 mRNA values: median (range) | 1 (0.62–1.14) | 3.42 (0.74–26.8) | 12.2 (6.94–23.0) | 19.9 (6.21–82.4) | 37.4 (11.84–98.7) | <10−5 | 13.4 (0.74–82.4) | 17.8 (0.74–98.7) |
| Overexpression, n (%) | — | 13 (61.9) | 10 (100) | 19 (100) | 21 (100) | — | 42 (84.0) | 63 (88.7) |
| SPP1 mRNA values: median (range) | 1 (0.85–2.51) | 1.16 (0.06–14.0) | 1.62 (0.60–13.1) | 2.87 (0.21–214.5) | 5.54 (0.44–43.0) | 0.02 | 1.65 (0.06–214.5) | 2.58 (0.06–214.5) |
| Overexpression, n (%) | — | 5 (23.8) | 3 (30) | 10 (52.6) | 13 (61.9) | — | 18 (36.0) | 31 (43.7) |
| IGF2 mRNA values: median (range) | 1 (0.70–2.37) | 44.0 (0.07–209.4) | 54.5 (7.09–217.5) | 1.02 (0.05–232.3) | 0.26 (0.04–13.8) | <10−5 | 11.5 (0.05–232.3) | 2.93 (0.04–232.3) |
| Under-expression, n (%) | — | 3 (14.3) | 0 (0) | 5 (26.3) | 11 (52.4) | — | 8 (16.0) | 19 (26.8) |
| Overexpression, n (%) | — | 16 (76.2) | 10 (100) | 7 (36.8) | 2 (9.5) | — | 33 (66.0) | 35 (49.3) |
| OSF2 mRNA values: median (range) | 1 (0.64–1.10) | 0.45 (0.08–5.19) | 0.45 (0.01–19.0) | 0.55 (0.01–7.31) | 3.43 (0.13–39.3) | 0.0005 | 0.52 (0.01–19.0) | 0.77 (0.01–39.3) |
| Under-expression, n (%) | — | 9 (42.9) | 4 (40) | 6 (31.6) | 3 (14.3) | — | 19 (38.0) | 22 (31.0) |
| Overexpression, n (%) | — | 1 (4.8) | 1 (10) | 2 (10.5) | 12 (57.1) | — | 4 (8.0) | 16 (22.5) |
| EPHA7 mRNA values: median (range) | 1 (0.49–1.46) | 0.12 (0.00–0.77) | 0.08 (0.05–1.25) | 0.11 (0.00–0.59) | 0.07 (0.00–0.79) | 0.0008 | 0.10 (0.00–1.25) | 0.09 (0.00–1.25) |
| Under-expression, n (%) | — | 19 (90.5) | 7 (70) | 16 (84.2) | 15 (71.4) | — | 42 (84.0) | 57 (80.3) |
| PTHR1 mRNA values: median (range) | 1 (0.98–1.54) | 0.14 (0.03–0.76) | 0.13 (0.04–0.50) | 0.10 (0.00–0.37) | 0.08 (0.01–0.55) | 4 × 10−5 | 0.12 (0.00–0.76) | 0.11 (0.00–0.76) |
| Under-expression, n (%) | — | 19 (90.5) | 8 (80) | 18 (94.7) | 17 (81.0) | — | 45 (90.0) | 62 (87.3) |
| H19 mRNA values: median (range) | 1 (0.85–2.12) | 16.9 (0.03–196.7) | 10.9 (0.89–186.1) | 0.76 (0.02–105.8) | 2.38 (0.31–101.8) | 0.02 | 6.74 (0.02–196.7) | 3.35 (0.02–196.7) |
| Overexpression, n (%) | — | 14 (66.7) | 7 (70) | 7 (36.8) | 8 (38.1) | — | 28 (56.0) | 36 (50.7) |
| MTSS1 mRNA values: median (range) | 1 (0.82–1.62) | 2.01 (0.64–5.15) | 3.39 (1.28–7.49) | 1.83 (0.38–6.28) | 1.65 (0.47–5.28) | 0.21 | 2.00 (0.38–7.49) | 1.91 (0.38–7.49) |
| Overexpression, n (%) | — | 8 (38.1) | 5 (50) | 6 (31.6) | 4 (19.0) | — | 19 (38.0) | 23 (32.4) |
Abbreviations: DHH=Desert Hedgehog; IHH=Indian Hedgehog; MIBC=muscle-invasive bladder cancer; NMIBC=non-muscle-invasive bladder cancer; SHH=Sonic Hedgehog.
χ2-test (comparison of under- or overexpression in group 1 vs group 2 vs group 3 vs group 4 vs group 5).
Bold values indicate significant P-values (<0.05).
The mRNA values were between 0.3 and 3 in normal bladder samples, which helped define values of overexpression and under-expression in tumour samples (mRNA values of 3 or more were considered to represent marked overexpression, and mRNA values of 0.3 or less were considered to represent marked under-expression). We have previously used the same cutoff points for altered tumour gene expression (Pignot et al, 2009).
mRNA expression in bladder tumours according to pathological stage
Table 2 shows mRNA expression levels of the 31 genes relative to the TBP endogenous control, according to pathological stage. Similar results were obtained when the endogenous control was RPLP0.
Ligands
SHH showed marked overexpression in bladder cancer compared with normal tissue (P<10−5), with a median expression level of 78.0 (range 0.22–721.6). The SHH overexpression was observed in 96.0% of NMIBC and 52.4% of MIBC, and the median SHH mRNA level decreased gradually from pTa low-grade tumours (121.9) to ⩾pT2 tumours (3.92).
IHH and DHH
expression was less variable, with overexpression in, respectively, 13.5 and 16.9% of tumour samples.
Ptch receptors
PTCH1 and PTCH2 were under-expressed in, respectively, 31.0 and 43.7% of tumour samples. The PTCH2 was under-expressed in 56.0% of NMIBC and only 14.3% of MIBC.
Transduction factors and regulators
SMOH was under-expressed in 74.6% of tumour samples (P=0.001).
We observed other markedly under-expressed genes (P<0.05) included tumour-suppressor candidates that negatively regulate the Hh pathway (SUFU, GAS1, RAB23) and genes encoding inhibitory proteins (HHIP, HHAT). The HHAT under-expression was far more frequent in MIBC (52.4%) than in NMIBC (2.0%).
Only DISP2, a protein-encoding gene involved in the regulation of ligand secretion, was overexpressed (P=0.04) both in MIBC (42.9%) and in NMIBC (62.0%).
DISP1, STK36, KIF7, KIF27, and BTRC expression levels did not differ significantly between tumour samples and normal bladder tissue.
Transcription factors (GLI family)
GLI1 and GLI2 showed marked under-expression, whereas GLI3 only tended to be under-expressed. GLI4, often described as an antagonistic factor (Kas et al, 1996), was overexpressed in 15.5% of tumour samples, but not significantly.
Target genes
Five target genes were significantly overexpressed in the tumour samples compared with normal bladder tissue: FOXM1, IGF2, OSF2, and H19, which promote cell proliferation and growth, and SPP1, which is involved in extracellular matrix interactions. Two of these five genes were either overexpressed or under-expressed, depending on the tumour stage. IGF2 was overexpressed in NMIBC, and particularly in pTa tumours, whereas it was under-expressed in more than half the MIBC samples. In contrast, OSF2 was under-expressed in 38% of NMIBC and overexpressed in 57% of MIBC. Two target genes, PTHR1 and EPHA7, were under-expressed, whatever the stage.
miRNA expression in bladder tumours
Table 3 shows expression levels of the nine selected miRNAs relative to the endogenous control U44, according to pathological stage. Similar results were obtained when two other endogenous miRNA controls (U6B and U48) were used (data not shown).
Table 3. Values of the nine miRNAs involved in the Hedgehog signalling pathway.
| NMIBC | MIBC | |||||||
|---|---|---|---|---|---|---|---|---|
| Normal | pTaG1-G2 | pTaG3 | pT1G3 | ⩾pT2 | All NMIBC | All tumours | ||
| Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | 2+3+4 | 2+3+4+5 | ||
| n=5 | n=21 | n=10 | n=19 | n=21 | P-values a | n=50 | n=71 | |
| 125B miRNA values: median (range) | 1 (0.71–2.33) | 0.01 (0.00–0.29) | 0.00 (0.00–0.22) | 0.02 (0.00–0.37) | 0.09 (0.01–1.52) | <10−5 | 0.01 (0.00–0.37) | 0.03 (0.00–1.52) |
| Under-expression, n (%) | — | 21 (100) | 10 (100) | 17 (89.5) | 17 (81.0) | — | 48 (96.0) | 65 (91.5) |
| 326 miRNA values: median (range) | 1 (0.59–1.49) | 0.02 (0.01–0.23) | 0.02 (0.01–0.12) | 0.04 (0.01–0.33) | 0.08 (0.03–3.13) | <10−5 | 0.03 (0.01–0.33) | 0.04 (0.01–3.13) |
| Under-expression, n (%) | — | 21 (100) | 10 (100) | 17 (89.5) | 19 (90.5) | — | 48 (96.0) | 67 (94.4) |
| 324 miRNA values: median (range) | 1 (0.59–2.51) | 0.15 (0.03–0.98) | 0.15 (0.05–0.46) | 0.26 (0.04–1.69) | 0.32 (0.05–1.38) | 0.001 | 0.22 (0.03–1.69) | 0.23 (0.03–1.69) |
| Under-expression, n (%) | — | 18 (85.7) | 8 (80) | 13 (68.4) | 9 (42.9) | — | 39 (78.0) | 48 (67.6) |
| 100 miRNA values: median (range) | 1 (0.36–2.09) | 0.01 (0.00–0.20) | 0.00 (0.00–0.06) | 0.01 (0.00–0.30) | 0.03 (0.00–0.81) | <10−5 | 0.01 (0.00–0.30) | 0.01 (0.00–0.81) |
| Under-expression, n (%) | — | 21 (100) | 10 (100) | 19 (100) | 19 (90.5) | — | 50 (100) | 69 (97.2) |
| 361 miRNA values: median (range) | 1 (0.60–2.38) | 0.19 (0.08–0.98) | 0.16 (0.13–0.30) | 0.32 (0.07–1.59) | 0.36 (0.06–1.13) | 0.0001 | 0.24 (0.07–1.59) | 0.29 (0.06–1.59) |
| Under-expression, n (%) | — | 15 (71.4) | 10 (100) | 8 (42.1) | 6 (28.6) | — | 33 (66.0) | 39 (54.9) |
| 136 miRNA value: median (range) | 1 (0.36–2.23) | 0.03 (0.02–0.34) | 0.02 (0.02–0.18) | 0.06 (0.01–0.50) | 0.12 (0.04–4.68) | 1 × 10−5 | 0.05 (0.01–0.50) | 0.06 (0.01–4.68) |
| Under-expression, n (%) | — | 19 (90.5) | 10 (100) | 17 (89.5) | 17 (81.0) | — | 46 (92.0) | 63 (88.7) |
| 92A miRNA values: median (range) | 1 (0.72–2.22) | 0.29 (0.10–1.94) | 0.33 (0.14–1.02) | 0.63 (0.12–5.66) | 1.44 (0.33–5.67) | 0.003 | 0.41 (0.10–5.66) | 0.54 (0.10–5.67) |
| Under-expression, n (%) | — | 11 (52.4) | 2 (20) | 3 (15.8) | 0 (0) | — | 16 (32.0) | 16 (22.5) |
| Overexpression, n (%) | — | 0 (0) | 0 (0) | 1 (5.3) | 4 (19.0) | — | 1 (2.0) | 5 (7.0) |
| 19A miRNA values: median (range) | 1 (0.68–1.46) | 0.22 (0.03–0.65) | 0.31 (0.12–0.82) | 0.52 (0.13–2.12) | 0.74 (0.11–9.21) | <10−5 | 0.32 (0.03–2.12) | 0.48 (0.03–9.21) |
| Under-expression, n (%) | — | 17 (81.0) | 4 (40) | 4 (21.1) | 2 (9.5) | — | 25 (50.0) | 27 (38.0) |
| Overexpression, n (%) | — | 0 (0) | 0 (0) | 0 (0) | 5 (23.8) | — | 0 (0) | 5 (7.0) |
| 20A miRNA values: median (range) | 1 ([0.64–1.67) | 0.13 (0.05–1.03) | 0.17 (0.11–0.46) | 0.37 (0.09–2.34) | 0.82 (0.14–2.98) | <10−5 | 0.26 (0.05–2.34) | 0.30 (0.05–2.98) |
| Under-expression, n (%) | — | 18 (85.7) | 8 (80) | 8 (42.1) | 3 (14.3) | — | 34 (68.0) | 37 (52.1) |
Abbreviations: MIBC=muscle-invasive bladder cancer; NMIBC=non-muscle-invasive bladder cancer.
χ2-test (comparison of under- or overexpression in group 1 vs group 2 vs group 3 vs group 4 vs group 5).
Bold values indicate significant P-values (<0.05).
Six of the nine miRNAs (miRNA-125B, miRNA-326, miRNA-324, miRNA-100, miRNA-361, and miRNA-136) were significantly under-expressed in both NMIBC and MIBC. All these miRNAs have been described as potential inhibitors of the Hh pathway (Tsuda et al, 2006; Ferretti et al, 2008). Three miRNAs (miRNA-92A, miRNA-19A, and miRNA-20A) were under-expressed in NMIBC, but normally expressed or overexpressed in MIBC, with a gradual increase in expression from pTa low-grade samples to invasive samples. Interestingly, these three miRNA are encoded by the miR-17-92 cluster, a group of miRNA described as oncogenes in several tumours (Northcott et al, 2009; Uziel et al, 2009).
SHH expression and miRNA expression in bladder cancer cell lines
We also measured the SHH mRNA levels in six bladder cancer cell lines: HTB2, HTB4, HTB9, CRL1472, CRL1749, and CRL2169 (Supplementary Data 3). Interestingly, we observed high expression level of SHH in cell line HTB2, which is often used as a model system for non-muscle-invasive bladder tumours.
The expression levels of the nine selected miRNA genes were also measured in the six bladder cancer cell lines. High expression levels of miRNA-92A, miRNA-19A, and miRNA-20A and low expression levels of miRNA-125B, miRNA326, miRNA-324, miRNA-100, miRNA-361, and miRNA136 were observed in several cell lines. These results were in keeping with those obtained with the tumour samples. Interestingly, the three miRNAs encoded by the miR-17-92 cluster (miRNA-92A, miRNA-19A, and miRNA-20A) were markedly overexpressed in cell line CRL1472, which is representative of high-grade urothelial bladder carcinoma.
Correlation between protein-coding mRNA/miRNA expression levels and patient survival
To analyse the expression level as a qualitative variable, the patients were subdivided into equal groups around the median.
MIBC
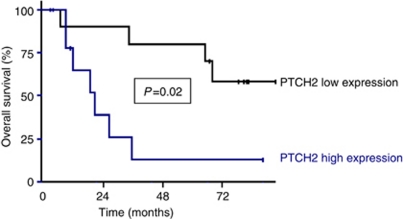
Among the 20 significantly altered mRNAs, PTCH2 was the only one with prognostic value in MIBC; high PTCH2 expression was significantly associated with worse outcome in univariate analysis (P=0.02; Figure 1). The 5-year OS rate was 13.0% (s.e.=12.1%) among patients with high PTCH2 expression and 80.0% (s.e.=12.6%) among patients with low PTCH2 expression.
Figure 1.
Overall survival curves in MIBC according to expression level of PTCH2.
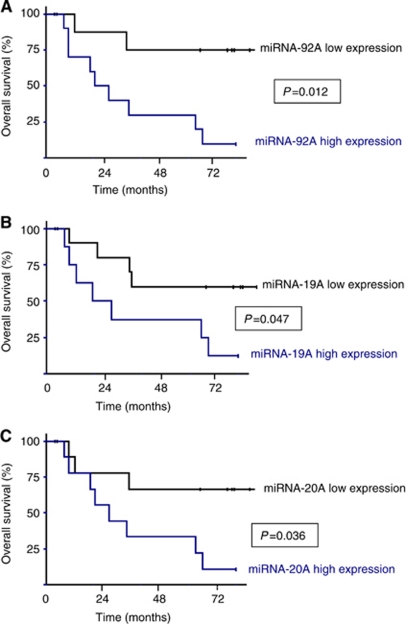
The expression levels of five of the nine selected miRNA genes were associated with OS among the MIBC patients in univariate analysis. Low miRNA-100 and miRNA-361 expression was significantly associated with better outcome (P=0.032 and P=0.044, respectively) (Supplementary Data 4). In contrast, high miRNA-92A, miRNA-19A, and miRNA-20A expression was significantly associated with worse outcome (P=0.012, P=0.047, and P=0.036, respectively; Figure 2A–C). The 5-year OS rates were, respectively, 30.0 (s.e.=14.5%), 37.5 (s.e.=17.1%), and 33.3% (s.e.=15.7%) among patients with high miRNA-92A, miRNA-19A, and miRNA-20A expression, vs 75.0 (s.e.=15.3%), 60.0 (s.e.=15.5%), and 66.7% (s.e.=15.7%) among patients with low expression.
Figure 2.
Overall survival curves in MIBC according to expression level of the three miRNA encoded by the miR-17-92 cluster family. (A) miRNA-92A, (B) miRNA-19A, and (C) miRNA-20A.
Multivariate analysis could not be achieved properly due to the pilot nature of this study, which was performed on a small number of patients (only 21 patients with MIBC).
NMIBC
Neither mRNA nor miRNA levels were associated with recurrence or progression of NMIBC in univariate analysis.
Discussion
Several studies suggest that the Hh signalling may contribute to the development of bladder cancer (McGarvey et al, 1998; Aboulkassim et al, 2003). Here we show that the Hh pathway is activated in TCC, and particularly in NMIBC. SHH (encoding the Sonic Hh ligand) and most of the target genes of the Hh pathway were markedly overexpressed, even in pTa low-grade tumours.
It is interesting to note that the expression levels of some Hh target genes differ depending on tumour stage (NMIBC or MIBC). For instance, IGF2 is overexpressed in 66% of NMIBC, whereas it is under-expressed in 52.4% of MIBC. At the opposite, OSF2 is under-expressed in 38% of NMIBC and overexpressed in 57.1% of MIBC. This supports the theory that there are two distinct molecular pathways in bladder carcinogenesis, that of hyperplasia and low-grade tumours and/or non-invasive, and that of dysplasia and high-grade tumours and/or infiltrating, with two different gene-expression profile as suggested by Wu (2005).
Constitutive activation of the Hh pathway has been found in several tumour types. In a small subset of the brain, skin and muscle tumours, mutations in PTCH1 or SMOH trigger ligand-independent activation of the Hh pathway (Johnson et al, 1996; Cowan et al, 1997). Ligand-dependent activation of the Hh pathway has been shown in small-cell lung carcinoma and digestive tract tumours, such as oesophageal carcinoma, gastric carcinoma (Berman et al, 2003), and pancreatic carcinoma (Thayer et al, 2003; Liu et al, 2007). Ligand-dependent oncogenic Hh signalling is associated with high-level expression of the Hh ligand by tumour cells, as observed here with the Sonic Hh ligand. In bladder cancer, Hh pathway activation thus seems to be initiated by overexpression of the Hh ligands, and especially SHH, which was markedly overexpressed (>80-fold higher than in normal bladder tissue) both in the bladder tumour samples and in two of the six bladder tumour cell lines tested here. These results are consistent with recent published data, which confirm overexpression of SHH at a protein level (He et al, 2011).
As expected (Aboulkassim et al, 2003), the expression level of both PTCH1 and PTCH2, which code for the receptors Ptch1 and Ptch2, was lower in tumour samples than in normal tissue, possibly participating in the Hh pathway activation. However, PTCH2 was re-expressed in MIBC, and this re-expression was associated with poorer OS.
The observed under-expression of SMOH and GLI members, associated with significant overexpression of SHH ligand and of the majority of the Hh target genes (i.e., FOXM1, SPP1, IGF2, OSF2, H19, and MTSS1), could be related to the fact that Smo and Gli activity are essentially regulated by post-transcriptional critical events, such as changes in protein conformation, subcellular localisation, phosphorylation, and dimerisation (Denef et al, 2000; Taipale et al, 2000; Hooper, 2003; Zhu et al, 2003) without marked changes at the mRNA level. Alternatively, marked activation of the Hh pathway could lead to a decrease in SMOH and GLI mRNA levels by a negative feedback.
An additional hypothesis is that tumour cells can produce SHH ligand, stimulating neighbouring stromal cells in paracrine manner, as observed in pancreatic tumours for example (Yauch et al, 2008; Bailey et al, 2009; Scales and de Sauvage, 2009; Tian et al, 2009). Indeed, it may be possible that SHH ligand is overexpressed in urothelial tumour cells and that Hh response occurs in supportive stroma. This paracrine signalling could control bladder tumour growth.
Another mechanism potentially regulating the Hh signalling might involve miRNA-mediated post-transcriptional control, a phenomenon recently described in several studies (Tsuda et al, 2006; Ferretti et al, 2008; Northcott et al, 2009; Uziel et al, 2009). We obtained evidence that overexpression of a miRNA cluster (miR-17-92) might induce specific activation of the Hh pathway. Overexpression of this cluster has been described in several tumour types and was recently identified as a possible regulator of the Hh pathway. For example, Uziel et al (2009) showed that the Hh pathway can be targeted at multiple levels by the same miRNAs in the medulloblastoma. Our findings confirm the existence of a novel regulatory mechanism of Hh signalling in bladder cancer and suggest that misregulation of specific miRNAs may sustain cancer development. Moreover, we found that high expression of 17-92 cluster miRNAs was associated with a poorer vital outcome of MIBC. These results suggest that the Hh pathway activation through overexpression of certain oncogenic miRNAs has prognostic implications, although this needs to be confirmed in a large, independent, and homogenous series of bladder tumours. Indeed, this is a pilot study involving a small number of patients with MIBC, and multivariate analysis (Cox model) could not be performed to confirm the results obtained in univariate analysis.
Moreover, immunochemistry studies are required to confirm these results at a protein level and to precise if the Hh pathway activation effects are epithelial tumour cell specific.
Our finding that the Hh pathway is constitutively activated in bladder tumours raises the possibility of novel therapeutic targets. Indeed, cyclopamine, a plant-derived teratogenic steroidal alkaloid, inhibits the Hh-ligand-dependent and -independent Hh pathway activation by directly interacting with Smo (Incardona et al, 1998; Berman et al, 2002; Kubo et al, 2004). More recently, other drugs, effective in vitro and less toxic than cyclopamine (and thus usable in humans), have been described. Two recent clinical trials tested a new drug, GDC-0449, that inhibits the Hh signalling pathway by targeting Smo, in advanced basal-cell carcinoma and medulloblastoma (Rudin et al, 2009; Von Hoff et al, 2009). Ligand-dependent Hh pathway activation might be blocked by antibodies directly targeting the Sonic Hh ligand or competing for its receptor, as suggested by preclinical studies (Scales and de Sauvage, 2009). However, even if such strategies interfere effectively with the Hh ligand, combination therapy will be needed to deal with other activated oncogenic pathways in the same tumour (Shafaee et al, 2006; Olive et al, 2009).
The miRNA regulation is another potential mechanism of the Hh pathway inhibition. In a recent study, Tsuda et al (2006) showed that a synthetic designer miRNA targeting the 3′-UTRs of Gli-1mRNA effectively inhibited tumour cell proliferation by delaying cell division and activating late apoptosis in pancreatic cell lines.
These recent studies bring compelling evidence that therapies directed against the Hh pathway is a promising new approach for the treatment of several tumours. These new drugs could be useful in the management of non-muscle-invasive bladder tumours, most of which show Hh pathway activation through Shh overexpression, compared with about 50% of muscle-invasive forms. In MIBC, it will be necessary to test tumours for overexpression of the Hh pathway genes, such as the Sonic Hh ligand gene, to select patients who are likely to benefit from these drugs.
Conclusion
We observed constitutive ligand-dependent activation of the Hh pathway in bladder cancer, due to genetic (protein-coding mRNA) and epigenetic (miRNA) dysregulation. The expression levels of PTCH2 and of miRNAs encoded by the miR-17-92 cluster are attractive candidate prognostic factors in MIBC. Finally, rationalised use of targeted therapies against the Hh pathway could be a new therapeutic hope for selected patients.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Aboulkassim TO, LaRue H, Lemieux P, Rousseau F, Fradet Y (2003) Alteration of the PATCHED locus in superficial bladder cancer. Oncogene 22(19): 2967–2971 [DOI] [PubMed] [Google Scholar]
- Bailey JM, Mohr AM, Hollingsworth MA (2009) Sonic hedgehog paracrine signalling regulates metastasis and lymphangiogenesis in pancreatic cancer. Oncogene 28(40): 3513–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachy PA, Karhadkar SS, Berman DM (2004) Tissue repair and stem cell renewal in carcinogenesis. Nature 432(7015): 324–331 [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, Beachy PA (2002) Medulloblastoma growth inhibition by hedgehog pathway blockade. Science 297(5586): 1559–1561 [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman JR, Watkins DN, Beachy PA (2003) Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature 425(6960): 846–851 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM (2006) MicroRNA signatures in human cancers. Nat Rev Cancer 6(11): 857–866 [DOI] [PubMed] [Google Scholar]
- Cowan R, Hoban P, Kelsey A, Birch JM, Gattamaneni R, Evans DG (1997) The gene for the naevoid basal cell carcinoma syndrome acts as a tumour-suppressor gene in medulloblastoma. Br J Cancer 76(2): 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM (2000) Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102(4): 521–531 [DOI] [PubMed] [Google Scholar]
- Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, Gipp J, Shaw A, Lamm ML, Munoz A, Lipinski R, Thrasher JB, Bushman W (2004) Hedgehog signalling promotes prostate xenograft tumor growth. Endocrinology 145(8): 3961–3970 [DOI] [PubMed] [Google Scholar]
- Fei DL, Li H, Kozul CD, Black KE, Singh S, Gosse JA, DiRenzo J, Martin KA, Wang B, Hamilton JW, Karagas MR, Robbins DJ (2010) Activation of Hedgehog signaling by the environmental toxicant arsenic may contribute to the etiology of arsenic-induced tumors. Cancer Res 70(5): 1981–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Miele E, Laneve P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E, Screpanti I, Bozzoni I, Gulino A (2008) Concerted microRNA control of Hedgehog signalling in cerebellar neuronal progenitor and tumour cells. EMBO J 27(19): 2616–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habuchi T, Devlin J, Elder PA, Knowles MA (1995) Detailed deletion mapping of chromosome 9q in bladder cancer: evidence for two tumour suppressor loci. Oncogene 11(8): 1671–1674 [PubMed] [Google Scholar]
- He HC, Chen JH, Chen XB, Qin GQ, Cai C, Liang YX, Han ZD, Dai QS, Chen YR, Zeng GH, Zhu JG, Jiang FN, Zhong WD (2011) Expression of Hedgehog pathway components is associated with bladder cancer progression and clinical outcome. Pathol Oncol Res; e-pub ahead of print 24 August 2011; doi:10.1007/s12253-011-9451-2 [DOI] [PubMed]
- Hirao S, Hirao T, Marsit CJ, Hirao Y, Schned A, Devi-Ashok T, Nelson HH, Andrew A, Karagas MR, Kelsey KT (2005) Loss of heterozygosity on chromosome 9q and p53 alterations in human bladder cancer. Cancer 104(9): 1918–1923 [DOI] [PubMed] [Google Scholar]
- Hooper JE (2003) Smoothened translates Hedgehog levels into distinct responses. Development 130(17): 3951–3963 [DOI] [PubMed] [Google Scholar]
- Incardona JP, Gaffield W, Kapur RP, Roelink H (1998) The teratogenic Veratrum alkaloid cyclopamine inhibits sonic hedgehog signal transduction. Development 125(18): 3553–3562 [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP (2001) Hedgehog signalling in animal development: paradigms and principles. Genes Dev 15(23): 3059–3087 [DOI] [PubMed] [Google Scholar]
- IUAC (2002) TNM Classification of Malignant Tumors. J.W. Sons: New York [Google Scholar]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein Jr EH, Scott MP (1996) Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272(5268): 1668–1671 [DOI] [PubMed] [Google Scholar]
- Kas K, Wlodarska I, Meyen E, Van den Berghe H, Van de Ven WJ (1996) Assignment of the gene encoding human Kruppel-related zinc finger protein 4 (GLI4) to 8q24.3 by fluorescent in situ hybridization. Cytogenet Cell Genet 72(4): 297–298 [DOI] [PubMed] [Google Scholar]
- Kayed H, Kleeff J, Keleg S, Guo J, Ketterer K, Berberat PO, Giese N, Esposito I, Giese T, Buchler MW, Friess H (2004) Indian hedgehog signalling pathway: expression and regulation in pancreatic cancer. Int J Cancer 110(5): 668–676 [DOI] [PubMed] [Google Scholar]
- Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, Kuroki S, Katano M (2004) Hedgehog signalling pathway is a new therapeutic target for patients with breast cancer. Cancer Res 64(17): 6071–6074 [DOI] [PubMed] [Google Scholar]
- Linnenbach AJ, Pressler LB, Seng BA, Kimmel BS, Tomaszewski JE, Malkowicz SB (1993) Characterization of chromosome 9 deletions in transitional cell carcinoma by microsatellite assay. Hum Mol Genet 2(9): 1407–1411 [DOI] [PubMed] [Google Scholar]
- Liu MS, Yang PY, Yeh TS (2007) Sonic hedgehog signalling pathway in pancreatic cystic neoplasms and ductal adenocarcinoma. Pancreas 34(3): 340–346 [DOI] [PubMed] [Google Scholar]
- McGarvey TW, Maruta Y, Tomaszewski JE, Linnenbach AJ, Malkowicz SB (1998) PTCH gene mutations in invasive transitional cell carcinoma of the bladder. Oncogene 17(9): 1167–1172 [DOI] [PubMed] [Google Scholar]
- McMahon AP (2000) More surprises in the Hedgehog signalling pathway. Cell 100(2): 185–188 [DOI] [PubMed] [Google Scholar]
- Mechlin CW, Tanner MJ, Chen M, Buttyan R, Levin RM, Mian BM (2010) Gli2 expression and human bladder transitional carcinoma cell invasiveness. J Urol 184(1): 344–351 [DOI] [PubMed] [Google Scholar]
- Molinie V (2006) [Bladder tumors classification]. Prog Urol FMC 16: 7–10 [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD (2009) The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res 69(8): 3249–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybakken K, Perrimon N (2002) Hedgehog signal transduction: recent findings. Curr Opin Genet Dev 12(5): 503–511 [DOI] [PubMed] [Google Scholar]
- Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, Madhu B, Goldgraben MA, Caldwell ME, Allard D, Frese KK, Denicola G, Feig C, Combs C, Winter SP, Ireland-Zecchini H, Reichelt S, Howat WJ, Chang A, Dhara M, Wang L, Rückert F, Grützmann R, Pilarsky C, Izeradjene K, Hingorani SR, Huang P, Davies SE, Plunkett W, Egorin M, Hruban RH, Whitebread N, Mc Govern K, Adams J, Iacobuzio-Donahue C, Griffiths J, Tuveson DA (2009) Inhibition of Hedgehog signalling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 324(5933): 1457–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca di Magliano M, Hebrok M (2003) Hedgehog signalling in cancer formation and maintenance. Nat Rev Cancer 3(12): 903–911 [DOI] [PubMed] [Google Scholar]
- Pignot G, Bieche I, Vacher S, Guet C, Vieillefond A, Debre B, Lidereau R, Amsellem-Ouazana D (2009) Large-scale real-time reverse transcription-PCR approach of angiogenic pathways in human transitional cell carcinoma of the bladder: identification of VEGFA as a major independent prognostic marker. Eur Urol 56(4): 678–688 [DOI] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, de Sauvage FJ, Low JA (2009) Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med 361(12): 1173–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales SJ, de Sauvage FJ (2009) Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci 30(6): 303–312 [DOI] [PubMed] [Google Scholar]
- Scott MP (2003) Cancer: a twist in a hedgehog's tale. Nature 425(6960): 780–782 [DOI] [PubMed] [Google Scholar]
- Shafaee Z, Schmidt H, Du W, Posner M, Weichselbaum R (2006) Cyclopamine increases the cytotoxic effects of paclitaxel and radiation but not cisplatin and gemcitabine in Hedgehog expressing pancreatic cancer cells. Cancer Chemother Pharmacol 58(6): 765–770 [DOI] [PubMed] [Google Scholar]
- Simon R, Struckmann K, Schraml P, Wagner U, Forster T, Moch H, Fijan A, Bruderer J, Wilber K, Mihatsch MJ, Gasser T, Sauter G (2002) Amplification pattern of 12q13-q15 genes (MDM2, CDK4, GLI) in urinary bladder cancer. Oncogene 21(16): 2476–2483 [DOI] [PubMed] [Google Scholar]
- Stefani G, Slack FJ (2008) Small non-coding RNAs in animal development. Nat Rev Mol Cell Biol 9(3): 219–230 [DOI] [PubMed] [Google Scholar]
- Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, Scott MP, Beachy PA (2000) Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406(6799): 1005–1009 [DOI] [PubMed] [Google Scholar]
- Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernandez del Castillo C, Yajnik V, Antoniu B, McMahon M, Warshaw AL, Hebrok M (2003) Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 425(6960): 851–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Callahan CA, DuPree KJ, Darbonne WC, Ahn CP, Scales SJ, de Sauvage FJ (2009) Hedgehog signalling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci USA 106(11): 4254–4259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda N, Ishiyama S, Li Y, Ioannides CG, Abbruzzese JL, Chang DZ (2006) Synthetic microRNA designed to target glioma-associated antigen 1 transcription factor inhibits division and induces late apoptosis in pancreatic tumor cells. Clin Cancer Res 12(21): 6557–6564 [DOI] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, He L, Ellison D, Gilbertson RJ, Hannon G, Roussel MF (2009) The miR-17∼92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA 106(8): 2812–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, Weiss GJ, Borad MJ, Hann CL, Brahmer JR, Mackey HM, Lum BL, Darbonne WC, Marsters Jr JC, de Sauvage FJ, Low JA (2009) Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med 361(12): 1164–1172 [DOI] [PubMed] [Google Scholar]
- Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB (2003) Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature 422(6929): 313–317 [DOI] [PubMed] [Google Scholar]
- Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, Marshall D, Fu L, Januario T, Kallop D, Nannini-Pepe M, Kotkow K, Marsters JC, Rubin LL, de Sauvage FJ (2008) A paracrine requirement for hedgehog signalling in cancer. Nature 455(7211): 406–410 [DOI] [PubMed] [Google Scholar]
- Wu XR (2005) Urothelial tumorigenesis: a tale of divergent pathways. Nat Rev Cancer 5(9): 713–725 [DOI] [PubMed] [Google Scholar]
- Zhu AJ, Zheng L, Suyama K, Scott MP (2003) Altered localization of Drosophila Smoothened protein activates Hedgehog signal transduction. Genes Dev 17(10): 1240–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.