Abstract
The RNA ligase RtcB is conserved in all domains of life, and is essential for tRNA maturation in archaea and metazoa. Here we show that bacterial and archaeal RtcB catalyze the GTP-dependent ligation of RNA with 3′-phosphate and 5′-hydroxyl termini. Reactions with analogues of RNA and GTP suggest a mechanism in which RtcB heals the 3′-phosphate terminus by forming a 2′,3′-cyclic phosphate before joining it to the 5′-hydroxyl group of a second RNA strand. Thus, RtcB can ligate RNA cleaved by RNA endonucleases, which generate 2′,3′-cyclic phosphate and then 3′-phosphate termini on one strand, and a 5′-hydroxyl terminus on another strand.
RNA ligases catalyze the formation of a phosphodiester bond between RNA strands.1, 2 RtcB has been identified as the enzyme responsible for catalyzing the direct ligation of 2′,3′-cyclic phosphate and 5′-OH RNA termini.3-5 This enzyme is conserved in all domains of life, and its activity is essential in archaea and metazoa for the ligation of transfer RNA (tRNA) molecules after intron removal by the tRNA splicing endonuclease.6 This endonuclease generates RNA fragments analogous to those of the archetypal acid-base catalyst: bovine pancreatic ribonuclease (RNase A).7, 8 These reactions proceed in two separate steps: cleavage of RNA to generate fragments with 2′,3′-cyclic phosphate and 5′-OH termini, and hydrolysis of the 2′,3′-cyclic phosphate to form a 3′-phosphate (3′-P).9, 10
The fate of 3′-P RNA termini in tRNA splicing intermediates is not well understood, but they are presumed to be recyclized by the enigmatic RNA phosphate cyclase (RtcA).11 RtcA is highly conserved in all three domains of life and is also postulated to function in the cyclization of the 3′-P of spliceosomal U6 snRNA.12 Synthesis of 2′,3′-cyclic phosphate termini by the ATP-dependent RtcA occurs in three nucleotidyl transfer steps: (1) reaction of ATP with an active-site histidine residue to form a covalent enzyme–AMP intermediate and release PPi, (2) transfer of the AMP moiety to the terminal 3′-P to form an RNA–adenylylate intermediate, and (3) attack by the terminal 2′-OH on the adenylylated 3′-P to form the 2′,3′-cyclic phosphate product and release AMP.13 This cyclization of 3′-P RNA termini is thought to be a prerequisite for ligation to 5′-OH RNA termini.
We found that Escherichia coli RtcB ligase can accept 3′-P RNA termini as its substrate, consistent with a recent report.14 To assay for ligation, we initially used two single-stranded RNA (ssRNA) oligos that were each 10 nucleotides in length. The 5′ RNA fragment was labeled with 6-carboxyfluorescein (FAM) at its 5′ end and phosphorylated at its 3′ end. The 3′ RNA fragment had hydroxyl groups at each end. We intended to convert the 3′-P into a 2′,3′-cyclic phosphate by incubation with RtcA and ATP, so as to produce a suitable substrate for RtcB. Surprisingly, we found that prior cyclization of the 3′-P was not necessary for ligation to a 5′-OH RNA termini, as long as GTP was included in reaction mixtures (Figures S1 and S2 of the Supporting Information). This result was found even with RtcB produced in an rtcA deletion strain of E. coli, eliminating the possibility of inadvertent contamination.
To explore this reactivity further, we used a substrate that mimics the broken anticodon stem-loop of yeast tRNAGlu(UUC) (Figure 1A), which was ligated more efficiently by RtcB than was ssRNA because of the proximity of its 3′-P and 5′-OH termini. We found that ATP, CTP, UTP, or NAD+ were unable to substitute for GTP (Figure 1B). Ligation was dependent on Mn2+, and a C78A substitution within a predicted metal-binding site abolished ligase activity, as reported previously.14
Figure 1.
E. coli RtcB catalyzed GTP-dependent ligation of 3′-P and 5′-OH RNA termini. (A) RNA ligase substrate that mimics a broken tRNA anti-codon loop. FAM label allows visualization in a urea-polyacrylamide gel. Ligation of 3′-P and 5′-OH termini generates a 35-nt RNA, whereas unligated substrate appears as a 17-nt band. (B) Ligation assays for cofactor- and metal-dependence. Reactions contain Mn2+, except in the “−Mn2+” lane. The lane labeled “C78A” is a ligation reaction performed with RtcB that has a C78A substitution within the predicted metal-binding site. (C) Ligation assays using GTP analogues as the cofactor. (D) Ligation assays for GTP concentration-dependence, enabling estimation of a GTP KM that is <16 μM. (E) Ligation assays for RNA-termini specificity. Modifications to the liga-table ends of the tRNA-mimic substrate (panel A) were made as indicated and tested in ligation reactions.
Then, we tested five GTP analogues as cofactors in ligation reactions, and discovered that bond cleavage between the α and β phosphoryl groups of GTP occurs during catalysis by RtcB. These reactions were performed with high concentrations of GTP analogues to ensure saturation of RtcB. Four of the analogues, β,γ-methylene (GppCp), β,γ-imido (GppNHp), γ-thio (GTPγS), and α-thio (GTPαS), enabled ligation, albeit at a lower efficiency than did GTP (Figure 1C). In contrast, α,β-methylene (GpCpp) did not enable the ligation to proceed, suggesting that cleavage of the phosphoanhydride bond between the α and β phosphoryl groups of GTP is critical for the ligation of 3′-P and 5′-OH RNA termini. GDP and GMP allowed for low levels of ligation, which could arise from low amounts of contaminating GTP. Titrations with GTP revealed that RtcB has a KM of <16 μM for this cofactor (Figure 1D). This value is similar to the ATP KM of 20 μM for E. coli RtcA15 and 6 μM for human RtcA.16
The GTP-dependent ligation activity of RtcB requires a 3′-P/2′-OH on the 5′ side of the ligation junction and a 5′-OH on the 3′ side. We investigated the RNA-termini specificity for ligation by modifying the ends of our tRNA-mimic substrate. First, all four combinations of either a phosphate or hydroxyl group at each termini were sampled while maintaining a 2′-OH on the 5′ side of the ligation junction. RtcB was found to be highly specific for RNA substrates that have 3′-P and 5′-OH termini (Figure 1E). Next, the importance of the terminal ribose 2′-OH for ligation was probed by replacing this hydroxyl group with either hydrogen (2′-H) or fluorine (2′-F). The RNA substrate with a 3′-P/2′-H termini produced no observable ligation product, whereas the 3′-P/2′-F substrate yielded only trace product. The 2′-deoxy-2′-fluororibose will adopt a ring pucker similar to that of the 3′-P/2′-OH substrate,17 but will not allow for phosphate cyclization. These data suggest that the primary mechanism involves cyclization of the 3′-P as prerequisite for ligation to a 5′-OH RNA terminus, though the appearance of a trace of ligation product with the 2′-F substrate (Figure 1E) does not allow us to rule out the possibility of direct ligation to the GMP-activated 3′-P terminus. In that mechanism, the 2′-OH could assist catalysis in a manner similar to that during ribosome-catalyzed peptide-bond formation.18
RtcB from the archaeal domain of life likewise catalyzes the GTP-dependent ligation of 3′-P and 5′-OH RNA termini. The crystal structure of RtcB from a hyperthermophilic archaeon, Pyrococcus horikoshii, was determined as part of a structural genomics project.19 This structure reveals a hydrophilic pocket with a predicted metal-binding site consisting of residues Cys98, His203, His234, and His404. The P. horikoshii homo-logue has 29% amino-acid sequence identity to E. coli RtcB, and 49% identity to the human ligase (HSPC117).
To investigate if RtcB from archaea catalyzes the GTP-dependent ligation of 3′-P and 5′-OH RNA termini, we synthesized a P. horikoshii rtcB gene and produced soluble enzyme in E. coli. RtcB from P. horikoshii catalyzed the same reaction as its E. coli homologue and displayed identical cofactor and metal-ion requirements (Figure 2). The P. horikoshii ligase also required a 2′-OH on the terminal nucleotide of the 5′ RNA fragment. Although RtcB from the archaeon Pyrobaculum aerophilum has been reported to depend on Zn2+ for activity,4 we found that Zn2+ is unable to replace Mn2+ in active P. horikoshii RtcB. We did find that a C98A substitution (Cys98→Ala98) within the predicted metal-binding site abolishes ligase activity, as reported for P. aerophilum RtcB.
Figure 2.
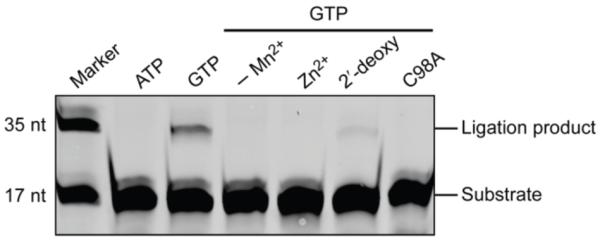
P. horikoshii RtcB catalyzed ligation of 3′-P and 5′-OH RNA termini. P. horikoshii RtcB requires Mn2+ and GTP for ligation. Omission of Mn2+ or replacement with Zn2+ did not allow ligation to proceed. The “2′-deoxy” lane is a ligation reaction using the tRNA-mimic substrate but with a 3′-P/2′-H termini on its 5′ fragment. The “C98A” lane is a ligation reaction with RtcB with a C98A substitution in the predicted metal-binding site.
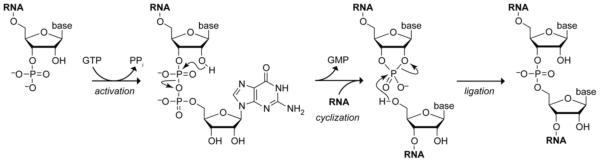
We propose a three-step mechanism for catalysis of the ligation of 3′-P and 5′-OH RNA termini by RtcB (Scheme 1). In the activation step, cleavage of the phosphoanhydride bond between the α and β phosphoryl groups of GTP occurs during transfer of GMP to the 3′-P terminus of an RNA strand to form a guanylylate-activated intermediate. ATP-dependent DNA and RNA ligases that catalyze a similar activation of 5′-P RNA termini first form a covalent enzyme–AMP intermediate with an active-site lysine, prior to transfer of AMP to the 5′-P terminus.1 Likewise, RtcB is known to be radiolabeled with [α-32P]GTP but not [γ-32P]GTP.14 Our data with GTP analogues show without ambiguity that cleavage occurs between the α and β phosphoryl groups of GTP during catalysis (Figure 1C). In the cyclization step of our mechanism, the 2′-OH on the terminal ribose attacks the GMP-activated 3′-P to generate a 2′,3′-cyclic phosphate and release GMP. In the ligation step, the 5′-OH terminus of a second RNA strand attacks the cyclic phosphate to form a 3′,5′-phosphodiester bond. We note that our mechanism accommodates the known competency of an RNA strand with a 2′,3′-cyclic phosphate to be a substrate for RtcB in the absence of GTP.3-5
Scheme 1.
Three sets of RNA termini have been identified as competent substrates for an RNA ligase. Bacteriophage T4 RNA ligase 1 (Rnl1) and RNA ligase 2 (Rnl2) catalyze the ATP-dependent ligation of 3′-OH and 5′-P RNA termini in three nucleotidyl transfer steps similar to those of DNA ligases.1, 20 All three domains of life encode ligases that accept 2′,3′-cyclic phosphate and 5′-OH RNA termini4 and are known to be essential for tRNA maturation and the unconventional splicing of HAC1 mRNA.2 The yeast tRNA ligase is a class I 5′-P RNA ligase and is homologous to T4 Rnl1.2 The multifunctional yeast ligase is unable to join tRNA exon termini directly, but instead has cyclic phosphodiesterase and polynucleotide kinase activities that yield 3′-OH/2′-P and 5′-P termini. The yeast ligase then seals these ends in an ATP-dependent reaction.21 In plants, a similar mechanism catalyzed by the class II 5′-P ligase forms mature tRNAs.22 In contrast, archaea and metazoa can form mature tRNAs via the direct ligation of 2′,3′-cyclic phosphate and 5′-OH termini in a reaction catalyzed by RtcB.3, 4 The relief of strain that accompanies the cleavage of a cyclic phosphate drives this reaction.23, 24 This reaction is unique in that it entails nucleophilic attack by a 5′-OH, which has a much higher pKa value than does a 3′-OH.25
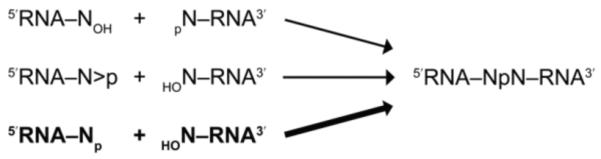
The presence of RtcB in bacteria is mysterious because group 1 introns in bacterial pre-tRNAs self-splice to form mature tRNAs.26 It has been suggested that E. coli RtcB might serve to protect against damage from stress-induced tRNA endonucleases,5 which cleave tRNA using a mechanism similar to that of RNase A.27 We also note that the hairpin and hammerhead ribozymes catalyze the direct ligation of 2′,3′-cyclic phosphate and 5′-OH termini.28, 29 The discovery that RtcB is a multifunctional ligase that can prepare a 3′-P termini for ligation to a 5′-OH, establishes a third pair of RNA termini that are suitable substrates for an RNA ligase (eq 1).
 |
(1) |
Recently, the human RtcB homologue was purified from HeLa cell extracts.3 A double-stranded RNA (dsRNA) substrate containing 3′-P and 5′-OH termini was used to follow human ligase activity after the dsRNA became linked, apparently after cyclization of the 3′-P by RtcA. The covalent linking of 3′-P and 5′-OH dsRNA, upon incubation with human cell extract, has also been observed in other studies.11, 30 Our data are consistent with RtcB being the sole enzyme responsible for these previous observations. In addition, our findings question the importance of RtcA in maintaining the 2′,3′-cyclic phosphate termini of tRNA splicing intermediates.
A search of the NCBI database reveals that RtcB has no homology to other known RNA or DNA ligases and does not have a predictable GTP-binding site. Furthermore, RtcB does not contain the conserved sequence motif KXXG that defines a superfamily of covalent nucleotidyltransferases, which includes DNA and RNA ligases and mRNA-capping enzymes.1 Finally, we note that RtcB is able to ligate RNA strands after cleavage by common ribonucleases or at high pH, which generates 2′,3′-cyclic phosphate and then 3′-P termini on one strand, and 5′-OH termini on the other strand.9, 10 The ability of this ancient enzyme4 to repair damaged RNA and to shuffle transcripts could have implications for organismal survival and evolution. Moreover, the ease of synthesizing RNA strands with 3′-P and 5′-OH termini suggests that RtcB could have practical applications in RNA tagging and cloning.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. G. Miller, J. D. Vasta, R. G. Presler, Jr., T. Hoang, and K. A. Andersen for helpful discussions.
Funding Source
This work was supported by grant R01 CA073808 (NIH).
Footnotes
ASSOCIATED CONTENT
Supporting Information. Figures S1 and S2, and experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).Shuman S, Lima CD. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- (2).Schwer B, Sawaya R, Ho CK, Shuman S. Proc. Natl. Acad. Sci. USA. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lührmann R, Söll D, Martinez J. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- (4).Englert M, Sheppard K, Aslanian A, Yates JR, Söll D. Proc. Natl. Acad. Sci. USA. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Tanaka N, Shuman S. J. Biol. Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Abelson J, Trotta CR, Li H. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- (7).Calvin K, Li H. Cell. Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Cuchillo CM, Nogués MV, Raines RT. Biochemistry. 2011;50:7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Markham R, Smith JD. Biochem. J. 1952;52:552–557. doi: 10.1042/bj0520552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Thompson JE, Venegas FD, Raines RT. Biochemistry. 1994;33:7408–7414. doi: 10.1021/bi00189a047. [DOI] [PubMed] [Google Scholar]
- (11).Filipowicz W, Konarska M, Gross HJ, Shatkin AJ. Nucleic Acids Res. 1983;11:1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Lund E, Dahlberg JE. Science. 1992;255:327–330. doi: 10.1126/science.1549778. [DOI] [PubMed] [Google Scholar]
- (13).Genschik P, Billy E, Swianiewicz M, Filipowicz W. EMBO J. 1997;16:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Tanaka N, Chakravarty AK, Maughan B, Shuman S. J. Biol. Chem. 2011;286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Genschik P, Drabikowski K, Filipowicz W. J. Biol. Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- (16).Vicente O, Filipowicz W. Eur. J. Biochem. 1988;176:431–439. doi: 10.1111/j.1432-1033.1988.tb14300.x. [DOI] [PubMed] [Google Scholar]
- (17).Hakoshima T, Omori H, Tomita K.-i., Miki H, Ikehara M. Nucleic Acids Res. 1981;9:711–729. doi: 10.1093/nar/9.3.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. Nat. Struct. Biol. 2004;11:1101–1106. doi: 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- (19).Okada C, Maegawa Y, Yao M, Tanaka I. Proteins. 2006;63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- (20).Silber R, Malathi VG, Hurwitz J. Proc. Natl. Acad. Sci. USA. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Greer CL, Peebles CL, Gegenheimer P, Abelson J. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- (22).Englert M, Beier H. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Kumamoto J, Cox JR, Jr., Westheimer FH. J. Am. Chem. Soc. 1956;78:4858–4860. [Google Scholar]
- (24).Haake PC, Westheimer FH. J. Am. Chem. Soc. 1961;83:1102–1109. [Google Scholar]
- (25).Izatt RM, Hansen LD, Rytting JH, Christensen JJ. J. Am. Chem. Soc. 1965;87:2760–2761. doi: 10.1021/ja01090a044. [DOI] [PubMed] [Google Scholar]
- (26).Reinhold-Hurek B, Shub DA. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- (27).Tomita K, Ogawa T, Uozumi T, Watanabe K, Masaki H. Proc. Natl. Acad. Sci. 2000;97:8278–8283. doi: 10.1073/pnas.140213797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Hertel KJ, Herschlag D, Uhlenbeck OC. Biochemistry. 1994;33:3374–3385. doi: 10.1021/bi00177a031. [DOI] [PubMed] [Google Scholar]
- (29).Hegg LA, Fedor MJ. Biochemistry. 1995;34:15813–15828. doi: 10.1021/bi00048a027. [DOI] [PubMed] [Google Scholar]
- (30).Martinez J, Patkaniowska A, Urlaub H, Lührmann R, Tuschl T. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.