Abstract
By combining astigmatism imaging with a dual-objective scheme, we improved the image resolution of stochastic optical reconstruction microscopy (STORM) and obtained <10 nm lateral resolution and <20 nm axial resolution when imaging biological specimens. Using this approach, we resolved individual actin filaments in cells and revealed three-dimensional ultrastructure of the actin cytoskeleton. We observed two vertically separated layers of actin networks with distinct structural organizations in sheet-like cell protrusions.
Recent advances in super-resolution fluorescence microscopy have substantially improved the spatial resolution of optical imaging1,2. When applied to biological samples, resolutions down to ~20 nm have been achieved in lateral directions with axial resolutions in the range of 10 to 100 nm1,2. The enhanced resolution has enabled the visualization of various cellular ultrastructures previously inaccessible to optical methods. Nonetheless, the current state-of-the-art resolution still cannot resolve many cellular structures, and further improvement of the resolution is desirable.
For example, among the three major types of cytoskeletal structures, individual microtubules and intermediate filaments can be observed with optical microscopy; by contrast, individual actin filaments have not been resolved in cells by optical means, including super-resolution methods3–6, due to the small diameter and high packing density of actin filaments. On the other hand, actin is of vital importance to many cellular processes7. In particular, the assembly and disassembly of actin filaments in the thin sheet-like cell protrusions drive cell locomotion8. The knowledge of how actin filaments are spatially organized is critical for understanding these processes. While electron microscopy and cryo-electron tomography can resolve individual actin filaments in cells, three-dimensional (3D) reconstructions are still challenging due in part to the structural perturbations induced by the dehydration and embedding treatment required for conventional electron microscopy and the difficulty in reconstructing large volumes by cryo-tomography9,10. Therefore, except for the several hundred nanometers near the cell edge, a full 3D reconstruction of actin has not been achieved for the sheet-like cell protrusion10, and how actin is vertically organized in this region is still unclear (see more details in Supplementary Discussion).
We set out to increase the image resolution of super-resolution fluorescence microscopy by combining 3D stochastic optical reconstruction microscopy (STORM)11,12 with a dual-objective detection scheme13,14 (Fig. 1a). In the original 3D STORM approach12, an optically resolvable subset of fluorescent probes are activated at any given instant and imaged using a single objective. Astigmatism is introduced in the detection path using a cylindrical lens such that the images obtained for individual molecules are elongated in x and y directions for molecules on the proximal and distal sides of the focal plane, respectively. The lateral and axial coordinates of the molecules are determined from the centroid positions and ellipticities of these single-molecule images, respectively. Iteration of the activation and imaging cycles allows the positions of numerous molecules to be determined and a super-resolution image to be reconstructed from these molecular coordinates. It could be reasoned that the total collected fluorescence signal would double by sandwiching the sample between two opposing objectives and detecting the signal from both objectives13,14. Since the localization uncertainty of each molecule scales with the inverse square root of the number of photons detected, doubling the photon count is expected to yield a 1.4-fold improvement in the image resolution.
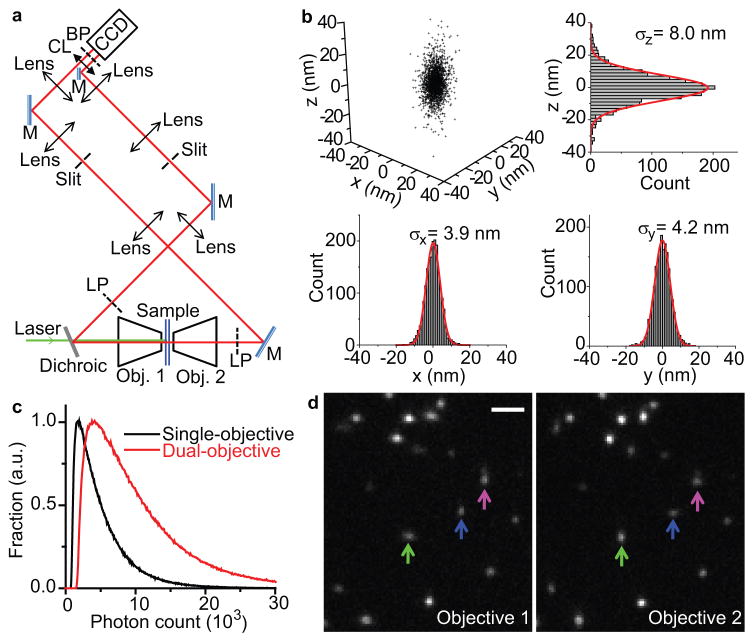
Figure 1. Experimental setup and spatial resolution of dual-objective 3D STORM.
(a) Schematic of the setup. Two microscope objectives are placed opposing each other and focused on the same spot of the sample. Astigmatism is introduced into the images collected by both objectives using a cylindrical lens. M: mirror; Obj.: objective; LP: long-pass filter; CL: cylindrical lens; BP: band-pass filter. (b) Localization precision of Alexa 647 molecules in fixed cells measured with dual-objective STORM. Each molecule gives a cluster of localizations due to repetitive activation of the same molecule. Localizations from 108 clusters (each containing >10 localizations) are aligned by their center of mass to generate the 3D presentation of the localization distribution. Histograms of the distribution in x, y, and z are fit to Gaussian functions, and the resultant standard deviations (σx, σy, and σz) are given in the plots. (c) Distribution of the number of photons detected for individual Alexa 647 molecules through both objectives (red; average: 10,600) and from a single objective (black; average: 5,200). (d) Images of activated Alexa 647 molecules obtained from the two objectives in a single frame. A molecule that appears elongated in x through one objective should appear elongated in y through the opposing objective (green and blue arrows). On the other hand, if two nearby molecules were mistaken as a single molecule, the images obtained through both objectives would appear elongated in the same direction along the line that connects the two molecules (example marked by the magenta arrow). Scale bar: 2 μm.
To characterize the localization precision of the dual-objective 3D STORM setup, we imaged individual Alexa 647 molecules scattered in fixed cells within ~150 nm of the focal plane, a range comparable to the thickness of the sheet-like cell protrusions. In this work, we use the term “cell protrusion” to refer to the thin region of the cell near its edge generally without distinguishing protruding or retracting regions. While we limited our imaging z-range to be comparable to the thickness of cell protrusions here, the imaging depth could be readily increased by stepping the sample in the z-direction. As each Alexa 647 molecule can be switched on and off multiple times, the standard deviation (SD) of repetitive localizations of the same molecule allowed an experimental determination of the localization precision. Surprisingly, the measured localization precisions, ~4 nm and ~8 nm in x-y and z directions, respectively (Fig. 1b), represent a more than two-fold improvement over previously reported values with the same fluorophore using single-objective STORM12. This localization precision corresponds to an image resolution of ~9 nm in the lateral directions and ~19 nm in the axial direction, measured in full width at half maximum (FWHM).
Further examination of the single-molecule images showed that beyond the expected doubling of photon-collection efficiency (Fig. 1c), the combination of astigmatism imaging and dual-objective detection provided a unique noise-cancelling mechanism that further improved the image precision. Molecules that were axially closer to one of the objectives were necessarily farther from the opposing one, resulting in anti-correlated changes in the ellipticity detected by the two objectives (Fig. 1d, green and blue arrows). In contrast, noises (such as those caused by sample drift) and abnormalities (such as two nearby molecules with overlapping images that are misidentified as a single molecule) led to correlated changes (Fig. 1d, magenta arrow). This effect allowed noises to be cancelled by averaging the z measurements from the two channels, and abnormalities to be identified and rejected by examining the difference in the z-positions obtained from the two objectives (see Online Methods). In addition, we found the mechanical stability of the dual-objective setup (with the optical axis of the objectives parallel to the optical table and the sample vertical oriented) to be higher than that of the single-objective setup built on a standard inverted microscope. The synergy of these effects resulted in a substantial improvement of the spatial resolution.
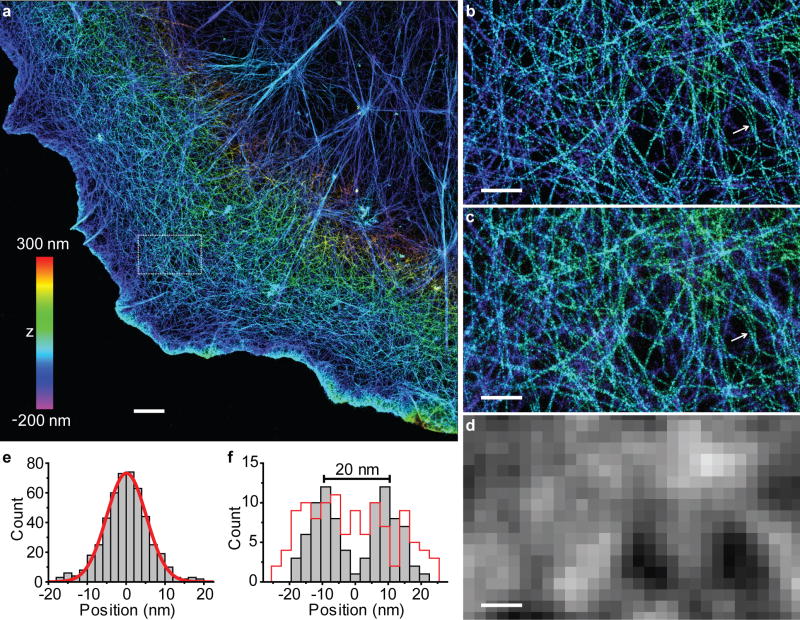
Next, we tested these imaging capabilities on the actin cytoskeleton (Fig. 2). To optimally benefit from the high image resolution, we labeled the target structure using small organic molecules, by staining actin filaments with Alexa 647 dye labeled phalloidin, which binds actin filaments with high specificity. STORM imaging was performed through the direct activation of Alexa 647 (refs 5,15). Individual actin filaments were clearly resolved in dual-objective STORM images of fixed cells (Fig. 2a,b). The cross-sectional profile of individual filaments exhibited a 12 nm FWHM (Fig. 2e). After accounting for the 9-nm lateral image resolution, we derived the width of phalloidin-labeled actin filaments to be ~8 nm (= (122 – 92)1/2), which agrees with the known diameter of actin (5–9 nm). In addition, nearby filaments with a center-to-center distance of ~20 nm were well resolved from each other (Fig. 2f). In comparison, lower resolution was achieved if one only relies on the information collected by one of the two objectives (Fig. 2c) and actin filaments were completely unresolvable in conventional fluorescent images (Fig. 2d).
Figure 2. Dual-objective 3D STORM resolves individual actin filaments in cells.
(a) Dual-objective STORM image of actin (labeled with Alexa 647-phalloidin) in a COS-7 cell. The z-positions are color-coded according to the color bar, with violet indicating positions closest to the substratum and red indicating farthest. (b) Close-up of the boxed region in a. (c) STORM image of the same area obtained by using only the information collected by Objective 1 of the dual-objective setup. (d) Conventional fluorescence image of the same area. (e) The cross-sectional profile of eight filaments overlaid by the center of each filament. The red line is a Gaussian fit with FWHM of 12 nm. (f) The cross-sectional profiles for two nearby filaments in (b) and (c) (white arrows). The grey bars correspond to the dual-objective images in (b) and the red line corresponds to the single-objective image in (c). Scale bars: 2 μm for a, 500 nm for b–d.
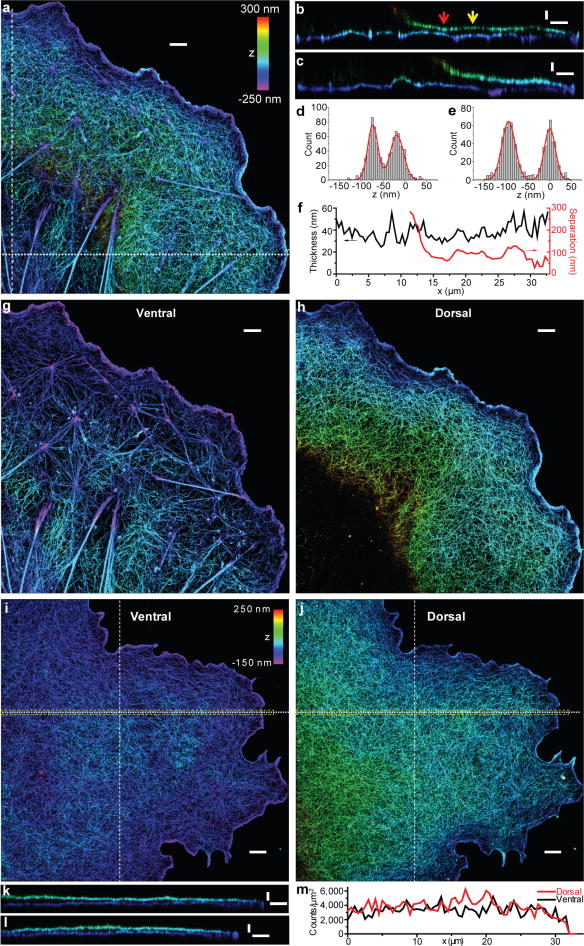
The volumetric imaging capability with the 9-nm lateral and 19-nm axial resolutions further allowed us to obtain a holistic, 3D view of the actin networks with dual-objective STORM. Two vertically separated actin layers were observed in the sheet-like cell protrusion despite its small thickness (Fig. 3a–c). Each of these layers was apparently ~30 – 40 nm thick (Fig. 3d–f). The separation between the two layers was generally ~100 nm, but can be as small as ~ 50 nm (Fig. 3d–f). The separation increased to much larger than 200 nm in the interior region far from the cell edge, suggesting that the two layers evolve into the cortical actin layers in the cell body. We also validated the two layer organization in living cells (Supplementary Fig. 1). Similar to the xy case, only using information obtained from one of the objectives substantially reduced the quality of the xz cross-section images (Supplementary Fig. 2).
Figure 3. Sheet-like cell protrusion is comprised of two layers of actin networks with distinct structures.
(a) Dual-objective STORM image of actin in a BSC-1 cell. The z-positions are color-coded according to the color bar. (b,c) Vertical cross sections (each 500-nm wide in x or y) of the cell in (a) along the dot and dash lines, respectively. Note when far from the cell edge, the z-position of the dorsal layer increases quickly and falls out of the imaging range. (d,e) The z-profiles for two points along the vertical section, corresponding to the red and yellow arrows in (b), respectively. Each histogram is fit to two Gaussians (red curves), yielding the apparent thickness of the ventral and dorsal layers and the peak separation between the two layers. (f) Quantification of the apparent thickness averaged over the two layers and the dorsal-ventral separation obtained from the xz cross-section profile in (b). (g,h) The ventral and dorsal actin layers of the cell in (a). (i,j) The ventral and dorsal actin layers of a COS-7 cell that was treated with blebbistatin. (k,l) Vertical cross sections (each 500-nm wide in x or y) of the cell along the dot and dash lines, respectively. (m) Actin density of the ventral and dorsal layers along the yellow box in (i,j), measured by the localization density. Scale bars: 2 μm for a, g, h, i, and j; 100 nm for z and 2 μm for x/y for b, c, k and l.
Although filaments in the two layers formed well separated networks, thick filament bundles occasionally connected the two layers (Supplementary Fig. 3). Such bundles typically originated from adhesion plaques, ran through the ventral layer, and gradually rose towards and ultimately reached the dorsal layer (Supplementary Fig. 3b–d), as expected for the dorsal stress fibers16. Thick bundles connecting adhesion plaques on the ventral surface, as expected for ventral stress fibers16, were also observed. In addition to these mature focal adhesions at the end of the thick actin bundles17, smaller and more isotropic adhesion complexes were also observed (Supplementary Fig. 3e–h), likely representing nascent adhesions complexes17. Actin filaments attached to these structures diverged in different directions.
Remarkably, the two layers of actin networks often exhibited highly distinct spatial organizations of actin filaments (Fig. 3g,h). While the dorsal layer appeared as a consistently dense and homogeneous meshwork, the ventral layer typically formed a web-like structure with a lower filament density and highly variable organization. The two-layer arrangement was observed in BSC-1 epithelial cells (Fig. 3a–h and Supplementary Fig. 4), as well as in COS-7 fibroblast cells (Supplementary Figs. 5 and 6). The actin density in the dorsal layer can be several times higher than that in the ventral layer (Supplementary Fig. 5f,i,j). Additional analysis suggests that the two layer arrangement spans the lamellum and possibly extends into the lamellipodium (see Supplementary Results).
To explore the molecular mechanisms underlying the structural differences observed for the two actin networks, we investigated how these networks responded to different actomyosin-perturbing drugs. Cytochalasin D, a drug that inhibits actin polymerization, reduced the filament density as expected (Supplementary Fig. 7a)18. Latrunculin A, a drug that sequesters monomeric actin, also reduced the filament density as expected18, but with the dorsal network substantially more disrupted than the ventral layer (Supplementary Fig. 7b), suggesting that the dorsal layer is potentially more dynamic and thus more readily disrupted by actin monomer depletion. Interestingly, blebbistatin, an inhibitor for myosin II, removed the structural differences between the ventral and dorsal networks: both networks became uniform actin meshworks of similar density, reminiscent of the dense, uniform dorsal network observed in untreated cells (Fig. 3i–m). This result suggests that myosin II plays a key role in the structural organization of the ventral actin layer and in maintaining the structural differences between the dorsal and ventral actin networks. This function of myosin II is potentially related to its activities previously found in regulating actin disassembly, actin bundle formation, and focal adhesion maturation (see more detailed discussions in the Supplementary Discussion).
By combining astigmatism imaging, dual-objective detection and small-molecule labeling in STORM, we have achieved an image resolution of < 10 nm in the lateral directions and < 20 nm in the axial direction. This lateral resolution is 2-fold or more higher than previously achieved for biological samples using super-resolution methods. Although higher axial resolutions have been reported using interferometry approaches13,14, the astigmatism-based system reported here is substantially simpler to implement.
The ability to resolve individual actin filaments in cells for the first time using fluorescence microscopy opens a new window for studying numerous actin-related processes in cells. The two layers of continuous actin networks with distinct structures we observed in sheet-like cell protrusions support and extend previous understanding of the 3D ultrastructure of the actin cytoskeleton19 (see Supplementary Discussion). The high image resolution obtained with dual-objective STORM should also find use in many other biological systems.
METHODS
Methods and any associated references are available in the online version of the paper.
Supplementary Material
Supplementary Protocol 1. Setting up the dual-objective 3D STORM system.
Supplementary Protocol 2. Cell fixation and staining for visualization of the actin cytoskeleton.
Supplementary Software. Analysis software
Supplementary Figure 1. Dual-objective STORM images of the actin cytoskeleton in a live BSC-1 cell.
Supplementary Figure 2. Comparison between the vertical cross sections of dual- objective and single-objective STORM images obtained by the dual-objective setup.
Supplementary Figure 3. Relations between the two actin layers, filament bundles, and adhesion plaques.
Supplementary Figure 4. Two other example images of the two-layer actin structure in the sheet-like protrusions of BSC-1 epithelial cells.
Supplementary Figure 5. The two-layer organization of actin networks in a COS-7 fibroblast cell.
Supplementary Figure 6. Two other example images of the two-layer actin structure in the sheet-like protrusions of COS-7 fibroblast cells.
Supplementary Figure 7. Effects of cytochalasin D and latrunculin A on actin organization in the dorsal and ventral layers.
Supplementary Results. The two-layer arrangement of actin spans the lamellum and possibly extends into the lamellipodium.
Supplementary Discussion. Previous understandings of the actin filament organization in the sheet-like cell protrusions and the implications of our results.
Acknowledgments
We thank G. Danuser for helpful discussion. This work is supported in part by the National Institutes of Health, Collaborative Innovation Award (#43667) from Howard Hughes Medical Institute and Gatsby Charitable Foundation (to X.Z.). X.Z. is a Howard Hughes Medical Institute Investigator.
Footnotes
Author contributions
K.X., H.P.B. and X.Z. designed research. K.X. performed experiments and data analysis. H.P.B. assisted with the optical setup. K.X. and X.Z. prepared the manuscript. X.Z. supervised the project.
Competing interests statement.
The authors declare that they have no competing financial interests.
Supplementary Software is available on the Nature Methods website.
References
- 1.Hell SW. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 2.Huang B, Babcock H, Zhuang XW. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Betzig E, et al. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 4.Gould TJ, et al. Nat Methods. 2008;5:1027–1030. doi: 10.1038/nmeth.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heilemann M, et al. Angew Chem-Int Edit. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- 6.Vogelsang J, Cordes T, Forthmann C, Steinhauer C, Tinnefeld P. Proc Natl Acad Sci U S A. 2009;106:8107–8112. doi: 10.1073/pnas.0811875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chhabra ES, Higgs HN. Nat Cell Biol. 2007;9:1110–1121. doi: 10.1038/ncb1007-1110. [DOI] [PubMed] [Google Scholar]
- 8.Pollard TD, Borisy GG. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 9.Svitkina T. Methods Cell Biol. 2007;79:295–319. doi: 10.1016/S0091-679X(06)79012-4. [DOI] [PubMed] [Google Scholar]
- 10.Urban E, Jacob S, Nemethova M, Resch GP, Small JV. Nat Cell Biol. 2010;12:429–435. doi: 10.1038/ncb2044. [DOI] [PubMed] [Google Scholar]
- 11.Rust MJ, Bates M, Zhuang XW. Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang B, Wang WQ, Bates M, Zhuang XW. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shtengel G, et al. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aquino D, et al. Nat Methods. 2011;8:353–359. doi: 10.1038/nmeth.1583. [DOI] [PubMed] [Google Scholar]
- 15.Zhuang XW. Nat Photonics. 2009;3:365–367. doi: 10.1038/nphoton.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrin S, Mellor H. J Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 17.Geiger B, Spatz JP, Bershadsky AD. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 18.Svitkina TM, Borisy GG. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannone G, et al. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Protocol 1. Setting up the dual-objective 3D STORM system.
Supplementary Protocol 2. Cell fixation and staining for visualization of the actin cytoskeleton.
Supplementary Software. Analysis software
Supplementary Figure 1. Dual-objective STORM images of the actin cytoskeleton in a live BSC-1 cell.
Supplementary Figure 2. Comparison between the vertical cross sections of dual- objective and single-objective STORM images obtained by the dual-objective setup.
Supplementary Figure 3. Relations between the two actin layers, filament bundles, and adhesion plaques.
Supplementary Figure 4. Two other example images of the two-layer actin structure in the sheet-like protrusions of BSC-1 epithelial cells.
Supplementary Figure 5. The two-layer organization of actin networks in a COS-7 fibroblast cell.
Supplementary Figure 6. Two other example images of the two-layer actin structure in the sheet-like protrusions of COS-7 fibroblast cells.
Supplementary Figure 7. Effects of cytochalasin D and latrunculin A on actin organization in the dorsal and ventral layers.
Supplementary Results. The two-layer arrangement of actin spans the lamellum and possibly extends into the lamellipodium.
Supplementary Discussion. Previous understandings of the actin filament organization in the sheet-like cell protrusions and the implications of our results.