Abstract
Bone strength is influenced by many properties intrinsic to bone, including its mass, geometry, and mineralization. To further advance our understanding of the genetic basis of bone strength-related traits, we utilized a large (N=815), moderately (G4) advanced intercross line (AIL) of mice derived from a high-runner selection line (HR) and the C57BL/6J inbred strain. In total, 16 quantitative trait loci (QTL) were identified that affected areal bone mineral density (aBMD) and femoral length and width. Four significant (P<0.05) and one suggestive (P<0.10) QTL were identified for three aBMD measurements: total body, vertebral and femoral. A QTL on Chromosome (Chr.) 3 influenced all three aBMD measures, while the other four QTL were unique to a single measure. A total of 10 significant and one suggestive QTL were identified for femoral length (FL) and two measures of femoral width, anterior-posterior (AP) and medial-lateral (ML). FL QTL were distinct from loci affecting AP and ML width, and of the seven AP QTL, only three affected ML. A QTL on Chr. 8 that explained 7.1% and 4.0% of the variance in AP and ML, respectively, was mapped to a six megabase (Mb) region harboring 12 protein-coding genes. The pattern of haplotype diversity across the QTL region and expression profiles of QTL genes, suggested that of the 12, cadherin 11 (Cdh11) was most likely the causal gene. These findings, when combined with existing data from gene knockouts, identify Cdh11 as a strong candidate gene within which genetic variation may affect bone morphology.
Keywords: Bone mineral density, bone morphology, bone geometry, osteoporosis, mouse genetics, advanced intercross line
Introduction
Osteoporosis is a common and complex disorder characterized by bone fragility. Bone fragility is influenced by a myriad of factors, and of those intrinsic to bone, bone mineral density (BMD) and bone geometry are two of the most important (1–3). Although these traits are influenced by both environmental and genetic factors, most (~60–80%) of their variance is genetically based (4). Thus, bone fragility is primarily a genetic disorder, and studies aimed at elucidating its genetic basis are critical for the development of a comprehensive understanding of osteoporosis.
Over the last decade the mouse has been used extensively to investigate the genetic basis of bone traits. Because of its clinical relevance, most studies have focused on BMD (5,6), although other skeletal traits, such as bone geometry, have also been subjected to genetic analysis (7). With regard to BMD, much of this work was recently summarized in the reanalysis of genetic data from 11 F2 crosses which placed over 150 BMD QTL on a standardized mouse genetic map (5). To gauge the usefulness of the mouse for the discovery of BMD genes the authors evaluated the genomic overlap between human BMD genome-wide associations (GWAs) and newly positioned mouse QTLs. Of the 28 human GWAs identified at the time, 26 overlapped with mouse BMD QTL. These data suggest that there may be a significant overlap in genes harboring natural variation that perturbs skeletal development and maintenance in humans and mice. In addition, human GWA studies that include upwards of 20,000 subjects have only been able to explain ~3% of the genetic variance for BMD (8) and there are a number of difficulties inherent in assessing all traits that contribute to bone fragility in human populations. Therefore, it is likely that mouse genetics has much to contribute with regards to the discovery of bone fragility genes.
In the current study, we used a G4 AIL to identify QTL modulating skeletal architecture. This G4 population originated from a reciprocal cross between mice with genetic propensity for increased voluntary exercise [high-runner (HR) line] and the C57BL/6J (B6) inbred strain (10,14). Our analysis revealed a complex genetic architecture for both areal BMD and femoral morphology. Furthermore, as a step towards gene discovery we investigated a QTL affecting femur width on Chr. 8 in more detail. This locus was the most statistically significant and possessed the smallest 1-LOD confidence interval, a region harboring only 12 genes. The combination of gene expression data and an analysis of identity-by-descent (IBD) suggested that of the 12, cadherin 11 (Cdh11) was the most likely candidate. These results increase our understanding of the genetic influences on skeletal architecture and suggest that Cdh11 is involved in the regulation of femur morphology.
Materials and Methods
Methods relevant to the creation, phenotyping, and genotyping of the mapping population utilized here have been previously described (10,14). Additionally, a complete list of the final set of SNPs (N=530) used for the QTL analyses can be found elsewhere (10,14). SNP locations are from Mouse Build 36 of the Mouse Diversity Genotyping Array (http://cgd.jax.org/tools/diversityarray.shtml). Only methods relevant to the current phenotypes and statistical analyses will be presented here.
Phenotypes
A moderately (G4) advanced intercross line (AIL) was generated from a reciprocal cross between mice selectively bred for high voluntary wheel running (HR) and the inbred strain C57BL/6J (B6). The HR line (one out of 4 replicates) is the result of a long-term artificial selection experiment for high voluntary wheel running on days 5 and 6 of 6-day wheel exposure (15). The original progenitors of the selection experiment were outbred, genetically variable house mice (N=224, Mus domesticus, Hsd:ICR, Harlan Sprague Dawley, Indianapolis, IN). At the outset of the selection experiment mice were mated for 2 generations and randomly assigned to 8 closed lines (4 HR lines and 4 control lines). These 8 lines remained closed in each successive generation. HR mice in the current experiment originated from the 44th generation of selection. HR mice have been the focus of numerous investigations including those examining plasticity of hind limb bones (16), bone morphology and mechanics (17), within-bone stiffness (18), and diaphyseal structure (19). To generate the AIL employed here, the F1 generation was intercrossed to produce an F2 and subsequently an F3 generation. Following the F3 generation, a large G4 population (N=815) was generated for broad phenotype and genotype collection. In all generations, both sexes and each reciprocal cross-line population (HR♀ X B6♂ and B6♀ X HR♂) were roughly equally represented. At approximately 9 weeks of age, G4 mice were weighed (±0.1 g), sacrificed, and carcasses stored at −30°C.
Areal bone mineral density (aBMD) measures were determined using a Lunar PIXImus II Densitometer (DXA; GE Healthcare, Piscataway, NJ, USA). Carcasses were thawed overnight at 4°C and placed for DXA imagining in a prostrate fashion, with the head always in the same orientation, on an imaging positioning tray. Subsequently, whole-body scans were analyzed using the manufacturer’s software (v2.0, Lunar Corp, Madison, WI). From each DXA scan we evaluated total aBMD (excluding the skull) (TBMD), vertebral aBMD (L1 – L5) (VBMD), and femoral aBMD [(right + left)/2] (FBMD). VBMD and FBMD were obtained, utilizing the imaging software, by placing a region of interest (ROI) over the lumbar vertebrae and around each of the left and right femora. Previous investigations utilizing PIXImus Densitometers have documented variation in aBMD associated with positioning of the animal during imaging (20). Accordingly, we noted the x and y coordinates for each scan and region of interest to evaluate potential positioning effects in the current investigation.
Following DXA measurements, right femora were removed, partially de-fleshed, wrapped in cheesecloth saturated with phosphate-buffered saline, and stored at −80°C. At a later date, femora were partially thawed and the three morphometric traits were measured to the nearest 0.01 mm with digital calipers. The traits were: femoral length (FL), the proximal tip of the femoral head to the distal most end of the medial condyle; anterior-posterior femoral width (AP) at the mid-diaphysis just below the gluteal tuberosity; and medial-lateral femoral width (ML), also at the mid-diaphysis. Partial correlations were performed in SAS (version 9.1; SAS Institute, Cary, NC) controlling for parent-of-origin type, sex, and body mass. Presented P values were adjusted for multiple comparisons utilizing the false discovery rate procedure (21) in SAS.
QTL analysis
The G4 AIL was produced through intercrossing over multiple generations; as a result, the assumption of independence of individuals is formally incorrect and conventional mapping methods that assume so may lead to potential false positive signals (22,23). Therefore, we employed the Genome Reshuffling for Advanced Intercross Permutation (GRAIP) procedure (22) to generate genome wide significance thresholds that appropriately account for the family structure in the current AIL population. The specific details of the implementation of this procedure, for this population, can be found elsewhere (10,24).
We evaluated the six (TBMD, VBMD, FBMD, FL, AP and ML) skeletal architecture traits for QTL using R/qtl (25) for the R environment (v 2.8.1) (26). Within R/qtl we used the multiple imputation method (27). For the aBMD traits, the additive statistical model included parent-of-origin type [whether a G4 individual was descended from a progenitor (F0) cross of HR♀ X B6♂ or B6♀ X HR♂, coded as 1 or 0 respectively], sex, body mass (at time of sacrifice), and the x and y coordinates for each specific aBMD measure. For the length and width measures, the additive statistical model included parent-of-origin type, sex, body mass, and technician (coded as 0, 1). All factors included in both additive models have known effects on the traits of interest.
Locus-specific P values and genome-wide GRAIP-adjusted significance thresholds were calculated using R/qtl output from the original population and the 50,000 GRAIP-permuted populations as described previously (10,28). As we employed 50,000 permutations to generate genome-wide adjusted significance thresholds, a minimum possible P value for the GRAIP output is 0.00002 (1/50,000) with a corresponding maximum log P of 4.7. Significant and suggestive loci were defined as those that met or exceeded the 95th (P ≥ 0.05) and 90th (P ≥ 0.1) percentiles. Confidence intervals (90 and 95%) were approximated by 1-LOD-drop support intervals (in Mb) relative to the GRAIP-permuted LOD score. In cases where the maximum GRAIP LOD score (4.7) spanned nearly the entire chromosome, confidence intervals were estimated using the naive LOD scores. The percent of phenotypic variation accounted for by each QTL, as well as additive and dominance effects, were estimated in R/qtl.
In addition to the analyses presented above, we also investigated potential QTL X sex interactions. Significant interactions were identified as LODFull – LODAdditive = LOD ≥ 3, where the LODFull model contains the interaction term (27).
Expression Survey
In an effort to prioritize genes within candidate regions identified below (see Results), we examined existing microarray data and evaluated relative expression levels in primary osteoblasts and osteoclasts. For this analysis, we used previously generated microarray data which profiled a large number of mouse tissues and cell-types (29). We downloaded the raw Affymetrix MOE430 microarray data for all osteoblast and osteoclast samples from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo; accession #GSE10246). The data consisted of two replicates for each of four samples: primary osteoblasts at day 5, 14 and 21 of differentiation and primary osteoclasts. The raw data were imported and processed using the affy R package (30). The Robust Multiarray Algorithim (RMA) was used to normalize and generate probe level expression data (31). A t-test was used to determine the significance of differences between time points among the osteoblast samples.
Exclusion mapping analysis
For a subset of the results presented below we further refined candidate gene regions by excluding intervals in which the haplotypes of the parental strains (i.e, HR and B6) are IBD utilizing SNPs from the Mouse Diversity array (32). We genotyped a subset of representative individuals from the F0 parental strains (N=12, HR; N=1, B6) and used SNPs to determine: i) whether the interval is homozygous or heterozygous in each sample and ii) whether samples are IBD in each homozygous interval. Briefly, we determined the frequency of heterozygous calls in windows of 200 consecutive SNPs in each one of the 12 HR individuals independently. Regions with >2% of SNPs with heterozygous calls were considered heterozygous based on the analysis of 101 fully inbred strains from the Jackson Laboratory. We then tested whether regions of homozygosity in all 12 HR founders contain different haplotypes. Finally, we identified intervals that are IBD among all 12 HR founders and B6 using the same 200 SNP window and threshold (>97% genotype identity) approach used to identify the segregating regions.
Results
Descriptive statistics and partial correlations among skeletal traits are presented in Table 1. The pairwise partial correlations between aBMD and femoral geometry traits were positive and statistically significant (P<0.05). As would be expected, TBMD was highly correlated with VBMD (r=0.708) and FBMD (r=0.699). In addition, FBMD and VBMD were also positively correlated (r=0.551). Since aBMD is a measure of bone mineral content per unit area we would not expect that measures of femur size and FBMD would be correlated, however, the correlation between FBMD and FL was significant (r=0.120; P<0.05). Additionally, AP and ML femur widths were even more highly correlated with FBMD (r=0.315 and r=0.294, respectively, P<0.0001), indicating that independent of sex, body mass, and femur length and width, areal FBMD was higher in mice with larger bones. We observed a high correlation between AP and ML (r=0.547), with lower correlations between the width traits and FL (r=0.343 and r=0.253, respectively), suggesting that many of the genetic determinants of AP also affect ML, but these are largely independent of factors affecting FL.
Table 1.
Descriptive statistics and partial correlations for skeletal architecture traits in the G4 population
| Trait | n | Mean | SD | Range | Femoral BMD |
Vertebral BMD |
Total BMD |
Femoral length |
A-P femoral width |
M-L femoral width |
|---|---|---|---|---|---|---|---|---|---|---|
| Femoral BMD | 798 | 0.07 | 0.008 | 0.05–0.10 | 0.551** | 0.699** | 0.120* | 0.315** | 0.294** | |
| Vertebral BMD | 797 | 0.06 | 0.006 | 0.04–0.08 | 0.708** | 0.230** | 0.319** | 0.309** | ||
| Total BMD | 799 | 0.05 | 0.004 | 0.04–0.06 | 0.223** | 0.399** | 0.372** | |||
| Femoral length | 763 | 15.76 | 0.609 | 14.10–17.25 | 0.343** | 0.253** | ||||
| A-P femoral width | 790 | 1.39 | 0.097 | 1.16–1.68 | 0.547** | |||||
| M-L femoral width | 790 | 1.88 | 0.150 | 1.45–2.32 |
Pearson partial correlations (controlling for parent-of-origin, sex, and body mass).
P < 0.05,
P <0.0001 following correction for multiple comparisons utilizing the false discovery rate procedure (Curran-Everett, 2000).
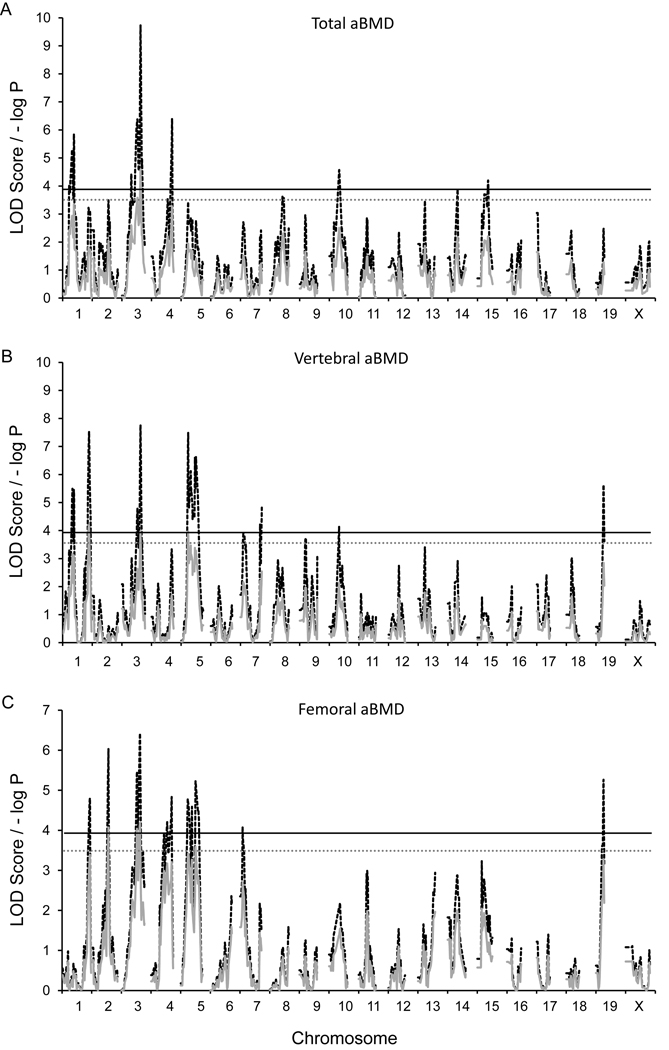
GRAIP-adjusted significant and suggestive loci for all skeletal architecture traits are presented in Table 2. Linkage analysis revealed one GRAIP-adjusted significant QTL on Chr. 3 for TBMD, colocalizing with loci observed for FBMD and VBMD (Figure 1A). This locus explained 3.0% of the total variance in TBMD. For VBMD, statistically significant loci were observed on chromosomes (Chr.) 1, 3, and 5; collectively explaining 14.1% of the total phenotypic variation (Figure 1B). The locus identified on Chr. 3 colocalized with that identified for FBMD. Significant additive effects were noted for the loci identified on Chr. 1 and 5, with opposing allelic effects. In contrast, the contributions from additive and dominance effects were more equally balanced for the Chr. 3 QTL. For FBMD, two significant (P≤0.05, LOD ≥ 3.9) and one suggestive (P≤0.1, LOD ≥ 3.5) QTL were observed (Figure 1C). These QTL collectively explained 5.6% of the total phenotypic variance with additive and dominance effects varying by loci. No significant QTL X sex interactions were observed for TBMD, VBMD or FBMD.
Table 2.
QTL detected and respective statistics for skeletal architecture traits
| Trait | Nearest Marker |
rs | MMU | Peak Position (Mb) |
Naive LOD |
GRAIP LOD |
CI (Mb) | % Variance |
Additive ±SE |
Dominance ±SE |
|---|---|---|---|---|---|---|---|---|---|---|
| Femoral BMD | JAX00098814 | 27504412 | 2 | 113 | 6.0 | 4.1* | 107–115 | 2.2 | −0.0018±0.0004 | −0.001±0.001 |
| JAX00189293 | 50963474 | 3 | 125 | 6.4 | 4.2* | 115–130 | 1.8 | 0.001±0.001 | 0.001±0.001 | |
| JAX00133397 | 29583351 | 5 | 100 | 5.2 | 3.6 | 97–123 | 1.6 | −0.0015±0.0004 | 0.0003±0.0006 | |
| Vertebral BMD | JAX00277411 | 48732938 | 1 | 179 | 7.5 | 4.2* | 172–184 | 7.3 | 0.0022±0.0003† | 0.0003±0.0004 |
| JAX00189293 | 50963474 | 3 | 126 | 7.8 | 4.0* | 121–130 | 3.9 | 0.0014±0.0003 | 0.0013±0.0004 | |
| JAX00581045 | 49523785 | 5 | 47 | 7.5 | 4.0* | 43–52 | 2.9 | −0.0014±0.0003† | 0.0004±0.0004 | |
| Total BMD | JAX00189293 | 50963474 | 3 | 126 | 9.7 | 4.6* | 122–130 | 3.0 | 0.0007±0.0002 | 0.0008±0.0003 |
| Femoral length | JAX00002741 | 31930716 | 1 | 40 | 9.0 | 4.7* | 35–63 | 0.3 | −0.04±0.03 | −0.05±0.04 |
| JAX00582506 | 48305016 | 5 | 53 | 9.9 | 4.7* | 47–60 | 2.1 | 0.11±0.03 | 0.09±0.04 | |
| A-P femoral width | JAX00005495 | 31575493 | 1 | 76 | 16.3 | 4.7* | 72–80‡ | 0.6 | −0.01±0.01 | 0.01±0.01 |
| JAX00567938 | 27582053 | 4 | 135 | 13.7 | 4.6* | 124–139 | 3.5 | 0.02±0.01 | 0.02±0.01 | |
| JAX00153077 | 31789816 | 7 | 76 | 10.0 | 4.4* | 71–80 | 4.7 | 0.03±0.01† | −0.01±0.01 | |
| JAX00165438 | 32321713 | 8 | 103 | 15.7 | 4.7* | 101–107‡ | 7.1 | −0.034±0.004† | −0.0001±0.0068 | |
| JAX00021324 | 13480722 | 10 | 97 | 9.2 | 4.4* | 87–103 | 1.4 | −0.017±0.005 | 0.003±.007 | |
| JAX00041702 | 29736244 | 13 | 10 | 8.1 | 3.9* | −19 | 0.7 | 0.0002±0.0049 | 0.02±0.01 | |
| JAX00395686 | 33859224 | 15 | 24 | 7.8 | 3.9* | −30 | 4.0 | 0.017±0.004 | −0.03±0.01† | |
| M-L femoral width | JAX00100848 | 27189926 | 2 | 140 | 7.1 | 3.6 | 130–159 | 3.1 | −0.03±0.01† | 0.02±0.01 |
| JAX00107680 | 30641053 | 3 | 56 | 8.0 | 4.4* | 46–68 | 1.2 | 0.02±0.01 | −0.003±0.011 | |
| JAX00165438 | 32321713 | 8 | 103 | 12.1 | 4.7* | 100–107‡ | 4.0 | −0.04±0.01† | 0.02±0.01 | |
| JAX00021324 | 13480722 | 10 | 97 | 9.2 | 4.7* | 90–107 | 1.4 | −0.02±0.01 | −0.01±0.01 | |
| JAX00041702 | 29736244 | 13 | 10 | 11.6 | 4.7* | −14‡ | 0.1 | −0.01±0.01 | 0.01±0.01 |
LOD exceeding the 95% (P = 0.05, LOD ≥ 3.9) permutation threshold are denoted by *; other QTL exceeded the 90% (P = 0.1, LOD ≥ 3.5) threshold. Confidence intervals (CIs) for QTL positions were obtained using a 1.0 LOD drop in Mb. Marker positions are based on Mouse Build 36 of the Mouse Diversity Genotyping Array.
Confidence intervals (CI) are relative to the GRAIP permuted LOD score with the exception of those denoted by ‡, which are relative to the naive LOD score. % variance is the percentage of phenotypic variance accounted for by the QTL effect. For additive and dominance effects: positive values indicate increasing effect of the HR allele or increasing effect of the heterozygote, respectively.
†Indicates additive and dominance effects were statistically significant at P < 0.05.
Figure 1.
G4 QTL maps of (A) total aBMD, (B) vertebral aBMD, and (C) femoral aBMD. Red traces are the simple mapping output, and black traces are GRAIP permutation output. For the GRAIP output, a minimum possible P value with 50,000 permutations is 0.00002 (1/50,000), so the maximum −log P = 4.7. The black and gray lines represent the permuted 95% (LOD≥3.9, P≤0.05) and 90% (LOD≥3.5, P≤0.1) LOD thresholds, respectively.
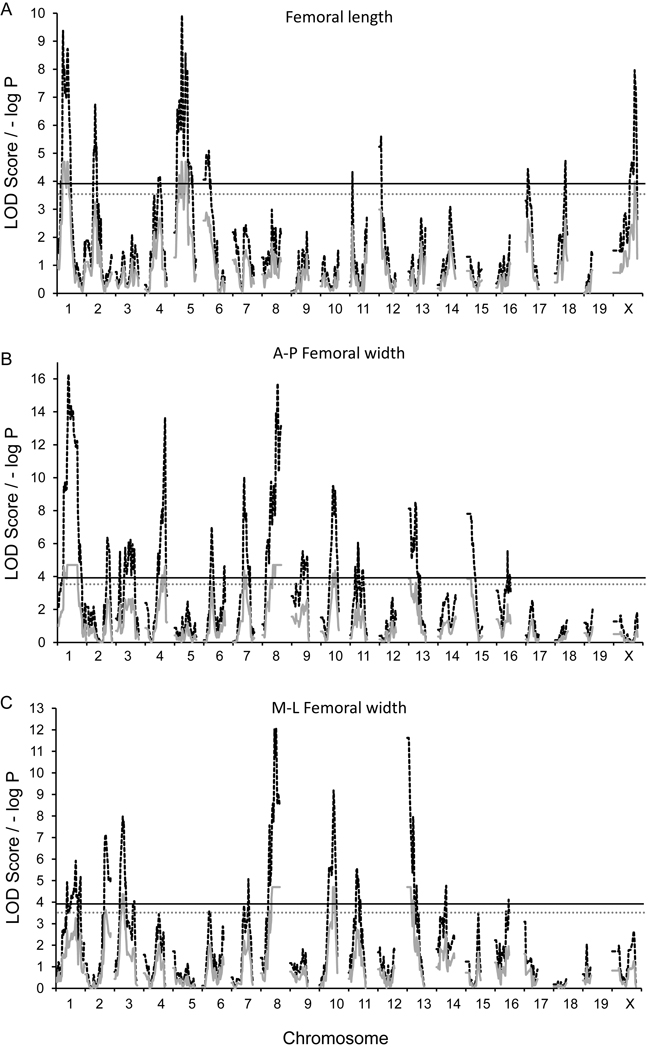
Regarding FL, significant loci were observed on Chr. 1 and 5, collectively explaining 2.4% of the total phenotypic variation with relatively large average additive and dominance effects (Figure 2A). Additionally, analyses of AP and ML femoral width resulted in 8 significant and one suggestive QTL on Chrs. 1, 2, 3, 4, 7, 8, 10, 13, and 15; with naive LOD scores ranging from 7.1 to 16.3 (Figure 2B and C). Collectively, AP and ML width loci explained 22% and 9.8% of the total phenotypic variation. For AP width, allelic effects varied by locus and additive and dominance effects were generally large and balanced. However, large average dominance effects, with little additively, were noted on Chrs. 13 and 15. While the significant dominance effect on Chr. 15 represented a decreasing effect in the heterozygote, a reversal was seen on Chr. 13, with an increasing effect of the heterozygote. For ML width, two (Chrs. 2, and 3) of the five loci were trait-specific QTL, while signals observed on Chrs. 8, 10, and 13 were congruent with those observed for AP width. No significant QTL X sex interactions were observed for AP or ML femoral width.
Figure 2.
G4 QTL maps of (A) femoral length (B) anterior-posterior femoral width, and (C) medial-lateral femoral width. Red traces are the simple mapping output, and black traces are GRAIP permutation output. For the GRAIP output, a minimum possible P value with 50,000 permutations is 0.00002 (1/50,000), so the maximum −log P = 4.7. The black and gray lines represent the permuted 95% (LOD≥3.9, P≤0.05) and 90% (LOD≥3.5, P≤0.1) LOD thresholds, respectively.
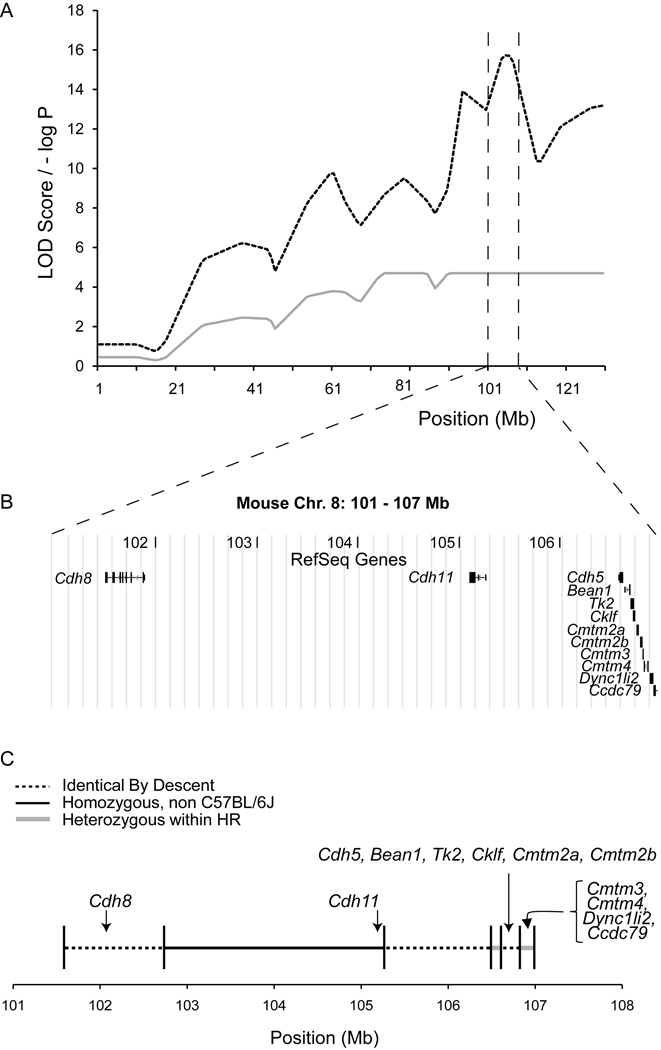
The 1-LOD drop CIs for all QTL ranged from 7 to 30 Mb. To identify putative candidate genes we examined QTL with narrow (<10 Mbp) CIs. Based on this analysis we focused on the AP/ML QTL on Chr. 8 (Table 2), since it i) was the most statistically significant (naive LOD=16.3), ii) had the most narrow CI (101–107 Mb) and iii) was gene sparse. Based on the NCBI37.1 genome build, this region harbored a total of 12 RefSeq protein-coding genes (Cdh8, Cdh11, Cdh5, Bean1, Tk2, Cklf, Cmtm2a, Cmtm2b, Cmtm3, Cmtm4, Dync1li2 and Ccdc79) (Figure 3B). The QTL peak was located just upstream of Cdh11 at ~104 Mb (Figure 3B).
Figure 3.
Haplotype diversity analysis narrows QTL to three non-IBD regions. The panels present (A) the naive and GRAIP adjusted AP LOD score profiles across Chr. 8, (B) the RefSeq gene content within and (C) the IBD assignments across the 6 Mb 1-LOD QTL CI.
Next, we characterized the pattern of haplotype diversity in the founders for this region to prioritize candidate genes. Approximately half of the region (49.5%) is IBD among B6 and all 12 HR founders. Given that these regions are IBD, they are not likely to harbor QTL; therefore our analysis confined the QTL to three regions that account for the other half of the original candidate region (Figure 3C). Of the 12 candidate genes, the coding regions of seven (Cdh8, Cdh5, Bean1, Tk2, Cklf, Cmtm2a, Cmtm2b) were located within IBD regions and can be excluded, while five (Cdh11, Cmtm3, Cmtm4, Dync1li2 and Ccdc79) were located in segregating regions and are, therefore, now of a higher priority (Figure 3C).
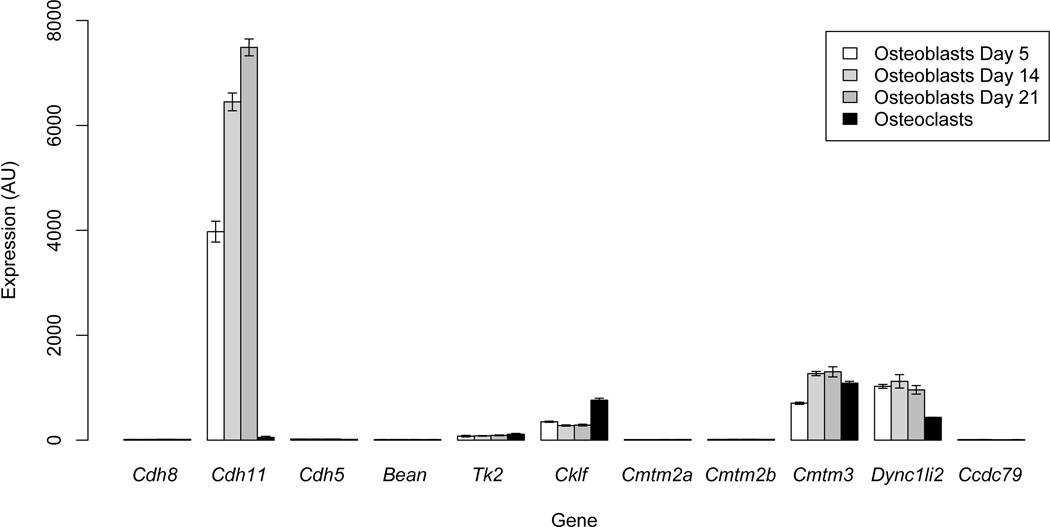
It is likely that the gene responsible for the effects on femoral width on Chr. 8 is expressed in osteoblasts and/or osteoclasts, the cells responsible for the bone modeling and remodeling that determines femoral morphology in adult mice (33). Therefore, we surveyed publically available microarray expression data on primary osteoblasts (at 5, 14 and 21 days of differentiation) and osteoclasts (http://biogps.gnf.org (29,34)) to determine which, if any, of the 12 candidates were expressed in these cells. Of the 12, microarray data were available for all except Cmtm4. As shown in Figure 4, five (Cdh11, Tk2, Cklf, Cmtm3, and Dync1li2) were expressed in osteoblasts and/or osteoclasts. Cdh11 demonstrated the highest expression in any of the two cell-types and its expression was significantly (P<0.05) increased as a function of osteoblast differentiation (expression at day 14 and 21 was increased relative to day 5). The expression of the other four did not differ statistically (P>0.05) across the osteoblast differentiation time course. Together, the IBD data and gene expression profiles of the genes in the QTL CI are consistent with variation within Cdh11 being the basis of the Chr. 8 AP and ML QTL.
Figure 4.
Gene expression analysis for QTL genes in primary calvarial osteoblasts at 5, 14 and 21 days of differentiation and osteoclasts. The mean±sem of two microarray replicates per sample are plotted. Of the five genes that show detectable expression (Cdh11, Tk2, Cklf, Cmtm3 and Dync1li2) in osteoblasts, Cdh11 is the only gene that is statistically different (P<0.05) across the time course.
Discussion
Traits that contribute to bone strength are under strong genetic regulation (4). Therefore, identification of novel genes that regulate these phenotypes promises to highlight the key biological processes that contribute to bone fragility. In the current study, we utilized a novel mouse AIL to identify 16 QTL affecting aBMD and femoral geometry phenotypes. One of the key observations from this study was that an AP/ML femoral width QTL on Chr. 8 was most likely the result of variation in Cdh11.
One of the limitations of genetic analysis in the mouse is that the most commonly used experimental crosses (e.g., an F2 cross) suffer from a lack of resolution (9). Recently, a number of groups have begun to explore the use of higher-resolution approaches that identify QTL with more narrow confidence intervals using populations such as advanced intercross lines (AILs) (as examples (10–12)). AILs are created by randomly mating mice derived from two parental strains for multiple generations (13). The accumulation of recombination events during each round of meiosis results in mice whose genomes are fined-grained mosaics of the two founder strains. These extra recombinations increase the precision of QTL localization, resulting in the confinement of QTL to smaller genomic intervals, which aids in the elucidation of the underlying genes [13].
As discussed above, the G4 AIL has the significant advantage of improved mapping resolution over more traditional approaches. Due to the accumulation of additional recombination events, the genetic map in the G4 was expanded by approximately a factor of three, relative to an equivalent F2 (10). To put this into context, we calculated the size of the 95% CIs for the 150 BMD QTL indentified in F2 crosses recently reported by Ackert-Bicknell et al. (35). The average F2 CI was 49.8±2.3 Mb compared to 12.4±6.5 Mb for the aBMD QTL identified in our G4 AIL. Although this comparison is not entirely appropriate, due to differences in statistical power and the way the CIs were calculated, it still demonstrates the improvement in resolution. To our knowledge, only one other group has used an AIL to study the genetics of bone traits. Recently, Norgard et al. used an F2–F3 population (N=~2000) derived from the LG/J and SM/J inbred strains of mice to identify QTL affecting the lengths of the humerus, ulna, femur, and tibia (7). The authors identified seventy QTL affecting bone length traits. None of the femoral length QTL identified by Norgard et al. overlapped with FL loci identified in this study. It is, however, not uncommon for distinct sets of QTL to be identified in different mouse crosses and this is likely due to the segregation of unique genetic variation in each cross, different ages at measurement, different environmental conditions, and/or differences in statistical power in the two studies. In a subsequent study, the bone length QTL identified by Norgard et al. were fine-mapped using F9–F10 AIL mice from the same population (36). The use of F9–F10 mice resulted in a significant increase in mapping resolution. Their average 95% CI was ~1.7 F2 cM or ~3.4 Mb and encompassed on average 35 genes as compared to 275 per CI in the F2–F3 population (36). This later study demonstrates the significant improvement in mapping resolution that can be obtained using later-generation AIL mice.
Differences in the genetic regulation of aBMD at various skeletal sites have been observed in previous mouse mapping studies (37–40). We observed similar results for TBMD, VBMD, and FBMD in this study. Only the locus on Chr. 3 regulated all three traits. In some cases this may be due to differences in effect sizes across traits. For example, on the distal end of Chr. 1 the LOD score for FBMD is just below the significance threshold in the location of a significant VBMD QTL (Figure 1). We did, however, observe examples of site specific QTL, such as the Chr. 2 FBMD QTL. The naive LOD score for FBMD on Chr. 2 at 113 Mbp was 6.0, whereas, the LOD score in the same location was <1.0 for VBMD (Figure 1). These results confirm that aBMD at different skeletal sites are under both common and unique genetic regulation.
A number of QTL mapping studies for aBMD have been performed in the mouse, resulting in the identification of hundreds of loci (5,6). Based on data from a recent review (6) and meta-analysis (5), we determined that all five peak aBMD QTL markers identified herein overlapped with the 95% CI for a previously identified BMD QTL. In particular, the Chr. 1 VBMD QTL overlapped with QTL identified in at least seven different studies (6). This region appears to be a hotspot for BMD QTL, and congenic strain analyses have demonstrated that the distal region of Chr.1 harbors multiple linked BMD loci (41,42). One such locus is the bone mineral density 1 (Bmd1) QTL discovered by Beamer et al. in an F2 cross between the CAST/EiJ and B6 inbred strains. Recently, the Duffy Antigen Receptor for Chemokines (Darc) gene was identified to be at least partially responsible for the effects of Bmd1 (43). In addition, the same group has used congenic strains to fine map a second separate QTL, just distal of Darc, down to a 0.152 Mbp region containing two candidates, absent in melanoma 2 (Aim2) and AC084073.22 (41).
Although fewer studies have investigated the genetic basis of femur geometry, we did identify overlap between loci identified in this study and previously detected QTL affecting the same or similar traits. To identify previously known QTL, we searched the Mouse Genome Informatics (MGI; http://www.informatics.jax.org) QTL database. This search did not identify any overlap with previously identified QTL for FL. However, of the nine unique femur width QTL, the peak markers for six were found to lie within the CIs of previously identified QTL affecting femur geometry and/or femoral biomechanical traits. Of the six, overlaps with the QTL on Chr. 4 and 8 were of particular interest. In a number of studies, the distal end of Chr. 4 has been found to harbor large-effect QTL influencing many bone strength phenotypes (37,42,44–48). Most QTLs in this region have peaks between 130 and 140 Mb. In our study, we identified a significant AP QTL at 135 Mb (Table 2). In addition, QTL with significant naive LOD scores that failed to reach significant after the GRAIP adjustment were identified for FBMD and TBMD at 139 Mb (data not shown). Although this region has been identified as a bone QTL hotspot, the underlying gene(s) have not been identified. Mapping efforts are complicated in this region by an extremely high gene density. The 10 Mb interval from 130 to 140 Mb on Chr. 4 contains 213 RefSeq genes, a density (21.3 genes/Mb) that is roughly three times the genome average (~7 genes/Mb). Given that the G4 segregates for variation affecting femoral bone traits in this region, the analysis of mice from the G10 or later may provide the added resolution needed to identify the responsible gene(s). This is especially important given that the syntenic human region has been implicated in the regulation of BMD in recent GWA studies (8,49).
Although previously identified QTL specifically for AP and ML have not observed for the Chr. 8 AP/ML locus, this region of Chr. 8 appears to be a hotspot for similar bone structure and strength trait QTL. Among the overlaps, Klein et al. identified the femoral cross-sectional area 2 (Fcsa2) QTL at 89 Mb affecting femoral cross-sectional area (FCSA) in an F2 cross between B6 and DBA/2J mice (50). In that study as well as in the present study, the B6 allele increased trait means. A second FCSA locus was identified at 90 Mb by Volkman et al. in the progeny of a cross between (BALB/cJ X C57BL/6J) F1 females and (C3H/HeJ X DBA/2J) F1 males (51). Additionally, Koller et al. identified the femoral bone trait QTL 3 (Fbtq3) located at 119 Mb in a cross between B6 and C3H/HeJ (46). This locus affected femoral polar moment of interia (Ip), stiffness (S) and load to failure (Fu). B6 alleles were associated with larger Ip, S and Fu. This region has also been found to harbor QTL affecting changes in tibial BMD, periosteal circumference, and cortical thickness, and the transcript levels of the bone marker genes bone sialoprotein (Bsp) and alkaline phosphatase (Akp2) in response to mechanical loading (52,53). These data suggest that this region of Chr. 8 contains variants that affect many aspects of bone density, structure, strength, and mechanical responsiveness. As will be discussed below, our genetic data suggest that Cdh11 is at least partially responsible for these effects.
To identify candidate genes, we performed a detailed investigation of the genes within the Chr. 8 AP/ML QTL CI. This locus was chosen because it was the most statistically significant and had the smallest CI. It also turned out that this region was gene sparse, increasing our odds of being able to prioritize among candidates. A total of 12 genes were located in the CI, and at ~2 genes/Mb it had a lower gene density than the genome average (~7 genes/Mb). Three lines of evidence suggest that of the 12, Cdh11 is likely the causal gene for the bone QTL discovered in these (and possibly other) studies: i) it was located within a non-B6 haplotype in the HR founders, ii) it was highly expressed in primary osteoblasts and was the only gene whose expression differed as a function of osteoblast differentiation, and iii) it was the only gene in the region previously implicated in bone development. It is important to note that Cdh11 has also been suggested as a candidate for the mouse tibial mechanical responsiveness QTL described above (52,53) and for a QTL affecting alterations in the contour of the navicular bones in horses (54).
Cdh11 encodes cadherin-11 (also known as osteoblast-cadherin) and is thought to play an important role in mediating cell-cell adhesion in the skeleton (55). Cadherin-11 along with N-cadherin (encoded by Cdh2) are the major cadherin family members expressed in bone forming osteoblasts (56,57). A role for Cdh11 in osteoblast function was recently demonstrated by the observation that osteoblast differentiation was defective in primary calvarial osteoblasts isolated from Cdh11 deficient (Cdh11−/−) mice (58,59). However, in adult Cdh11−/− mice the only observed skeletal defect was modest osteopenia (58,59). It has been hypothesized that the lack of a more severe in vivo skeletal phenotype may be due to functional compensation by N-cadherin in osteoblasts (59). In support of this hypothesis, double Cdh2/Cdh11 knockout mice have significantly reduced BMD and femoral cross-sectional area, and the latter is a phenotype that would be captured by the AP and/or ML measurements obtained in the present study (59). The double knockout data indicate that in the “correct” genetic background, perturbation of Cdh11 alters femoral width. Therefore, it is possible that the genetic background in the G4 (due to mutations in Cdh2 or other unknown “enabling” loci) provides an environment in which functional variation within Cdh11 leads to alterations in femur width. Although these data strongly suggest that Cdh11 is the causal gene, other possibilities do exist. For instance, one of the four other genes in the non-IBD regions could be causal, or regulatory variation in the non-IBD regions may be altering the expression of other genes in the region. It is also possible that multiple causal linked genes exist. Additional work is needed to confirm our hypothesis that Cdh11 is the responsible gene.
In conclusion, we have used a G4 AIL to identify regions of the mouse genome that regulate aBMD and femur morphology. The utilization of an AIL resulted in significantly reduced confidence intervals as compared to an F2 cross. In addition, by combining QTL mapping data, microarray data, and an IBD analysis, Cdh11 was identified as a likely regulator of femoral morphology. These results lay the groundwork for the ultimate discovery of causal genes, and their identification promises to significantly increase our understanding of genes and pathways that regulate bone strength.
Acknowledgements
We thank Z. Yun for assistance with animal care and data collection and K.M. Middleton for insightful comments on the manuscript. This work was supported by NIH grant DK076050 to DP. SAK was supported through a NIMH funded (5T32MH075854-04) Interdisciplinary Obesity Training (IDOT) program. Phenotypes were collected in part using the Animal Metabolism Phenotyping core facility within UNC’s Clinical Nutrition Research Center (funded by NIDDK grant DK056350).
Footnotes
Disclosures
The authors declare no conflict of interest.
References
- 1.Gilsanz V, Loro ML, Roe TF, Sayre J, Gilsanz R, Schulz EE. Vertebral size in elderly women with osteoporosis. Mechanical implications and relationship to fractures. J Clin Invest. 1995;95(5):2332–2337. doi: 10.1172/JCI117925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ng AH, Wang SX, Turner CH, Beamer WG, Grynpas MD. Bone quality and bone strength in BXH recombinant inbred mice. Calcif Tissue Int. 2007;81(3):215–223. doi: 10.1007/s00223-007-9047-8. [DOI] [PubMed] [Google Scholar]
- 3.Johnell O, Kanis JA, Oden A, Johansson H, De Laet C, Delmas P, Eisman JA, Fujiwara S, Kroger H, Mellstrom D, Meunier PJ, Melton LJ, 3rd, O'Neill T, Pols H, Reeve J, Silman A, Tenenhouse A. Predictive value of BMD for hip and other fractures. J Bone Miner Res. 2005;20(7):1185–1194. doi: 10.1359/JBMR.050304. [DOI] [PubMed] [Google Scholar]
- 4.Peacock M, Turner CH, Econs MJ, Foroud T. Genetics of osteoporosis. Endocr Rev. 2002;23(3):303–326. doi: 10.1210/edrv.23.3.0464. [DOI] [PubMed] [Google Scholar]
- 5.Ackert-Bicknell CL, Karasik D, Li Q, Smith RV, Hsu Y-H, Churchill GA, Paigen BJ, Tsaih S-W. Mouse BMD quantitative trait loci show improved concordance with human genome wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010 doi: 10.1002/jbmr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong Q, Jiao Y, Hasty KA, Canale ST, Stuart JM, Beamer WG, Deng H-W, Baylink D, Gu W. Quantitative trait loci, genes, and polymorphisms that regulate bone mineral density in mouse. Genomics. 2009;93(5):401–414. doi: 10.1016/j.ygeno.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norgard EA, Roseman CC, Fawcett GL, Pavlicev M, Morgan CD, Pletscher LS, Wang B, Cheverud JM. Identification of quantitative trait loci affecting murine long bone length in a two-generation intercross of LG/J and SM/J Mice. J Bone Miner Res. 2008;23(6):887–895. doi: 10.1359/JBMR.080210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HA, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FM, Wilson SG, Zhou Y, Ralston SH, van Duijn CM, Spector T, Kiel DP, Stefansson K, Ioannidis JP, Uitterlinden AG. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat Rev Genet. 2005;6(4):271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 10.Kelly SA, Nehrenberg DL, Peirce JL, Hua K, Steffy BM, Wiltshire T, Pardo-Manuel de Villena F, Garland T, Jr, Pomp D. Genetic architecture of voluntary exercise in an advanced intercross line of mice. Physiol Genomics. 2010;42(2):190–200. doi: 10.1152/physiolgenomics.00028.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fawcett GL, Jarvis JP, Roseman CC, Wang B, Wolf JB, Cheverud JM. Fine-mapping of obesity-related quantitative trait loci in an F9/10 advanced intercross line. Obesity (Silver Spring) 2010;18(7):1383–1392. doi: 10.1038/oby.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu X, Bauer K, Wernhoff P, Ibrahim SM. Using an advanced intercross line to identify quantitative trait loci controlling immune response during collagen-induced arthritis. Genes Immun. 2007;8(4):296–301. doi: 10.1038/sj.gene.6364385. [DOI] [PubMed] [Google Scholar]
- 13.Darvasi A, Soller M. Advanced intercross lines, an experimental population for fine genetic mapping. Genetics. 1995;141(3):1199–1207. doi: 10.1093/genetics/141.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelly SA, Nehrenberg DL, Hua K, Gordon RR, Garland T, Jr, Pomp D. Parent-of-origin effects on voluntary exercise levels and body composition in mice. Physiol Genomics. 2010;40(2):111–120. doi: 10.1152/physiolgenomics.00139.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swallow JG, Carter PA, Garland T., Jr Artificial selection for increased wheel-running behavior in house mice. Behav Genet. 1998;28(3):227–237. doi: 10.1023/a:1021479331779. [DOI] [PubMed] [Google Scholar]
- 16.Kelly SA, Czech PP, Wight JT, Blank KM, Garland T., Jr Experimental evolution and phenotypic plasticity of hindlimb bones in high-activity house mice. J Morphol. 2006;267(3):360–374. doi: 10.1002/jmor.10407. [DOI] [PubMed] [Google Scholar]
- 17.Middleton KM, Shubin CE, Moore DC, Carter PA, Garland T, Jr, Swartz SM. The relative importance of genetics and phenotypic plasticity in dictating bone morphology and mechanics in aged mice: evidence from an artificial selection experiment. Zoology (Jena) 2008;111(2):135–147. doi: 10.1016/j.zool.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Middleton KM, Goldstein BD, Guduru PR, Waters JF, Kelly SA, Swartz SM, Garland T., Jr Variation in within-bone stiffness measured by nanoindentation in mice bred for high levels of voluntary wheel running. J Anat. 2010;216(1):121–131. doi: 10.1111/j.1469-7580.2009.01175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wallace IJ, Middleton KM, Lublinsky S, Kelly SA, Judex S, Garland T, Jr, Demes B. Functional significance of genetic variation underlying limb bone diaphyseal structure. Am J Phys Anthropol. 2010;143(1):21–30. doi: 10.1002/ajpa.21286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez Franco GE, O'Neil TK, Litscher SJ, Urban-Piette M, Blank RD. Accuracy and precision of PIXImus densitometry for ex vivo mouse long bones: comparison of technique and software version. J Clin Densitom. 2004;7(3):326–333. doi: 10.1385/jcd:7:3:326. [DOI] [PubMed] [Google Scholar]
- 21.Curran-Everett D. Multiple comparisons: philosophies and illustrations. Am J Physiol Regul Integr Comp Physiol. 2000;279(1):R1–R8. doi: 10.1152/ajpregu.2000.279.1.R1. [DOI] [PubMed] [Google Scholar]
- 22.Peirce JL, Broman KW, Lu L, Chesler EJ, Zhou G, Airey DC, Birmingham AE, Williams RW. Genome Reshuffling for Advanced Intercross Permutation (GRAIP): simulation and permutation for advanced intercross population analysis. PLoS One. 2008;3(4):e1977. doi: 10.1371/journal.pone.0001977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valdar W, Holmes CC, Mott R, Flint J. Mapping in structured populations by resample model averaging. Genetics. 2009;182(4):1263–1277. doi: 10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107(44):18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19(7):889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- 26.Team RDC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 27.Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159(1):371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peirce JL, Broman KW, Lu L, Williams RW. A simple method for combining genetic mapping data from multiple crosses and experimental designs. PLoS One. 2007;2(10):e1036. doi: 10.1371/journal.pone.0001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lattin JE, Schroder K, Su AI, Walker JR, Zhang J, Wiltshire T, Saijo K, Glass CK, Hume DA, Kellie S, Sweet MJ. Expression analysis of G Protein-Coupled Receptors in mouse macrophages. Immunome Res. 2008;4:5. doi: 10.1186/1745-7580-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 31.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 32.Yang H, Ding Y, Hutchins LN, Szatkiewicz J, Bell TA, Paigen BJ, Graber JH, de Villena FP, Churchill GA. A customized and versatile high-density genotyping array for the mouse. Nat Methods. 2009;6(9):663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeman E, Delmas PD. Bone quality--the material and structural basis of bone strength and fragility. N Engl J Med. 2006;354(21):2250–2261. doi: 10.1056/NEJMra053077. [DOI] [PubMed] [Google Scholar]
- 34.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackert-Bicknell CL, Karasik D, Li Q, Smith RV, Hsu YH, Churchill GA, Paigen BJ, Tsaih SW. Mouse BMD quantitative trait loci show improved concordance with human genome-wide association loci when recalculated on a new, common mouse genetic map. J Bone Miner Res. 2010;25(8):1808–1820. doi: 10.1002/jbmr.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norgard EA, Jarvis JP, Roseman CC, Maxwell TJ, Kenney-Hunt JP, Samocha KE, Pletscher LS, Wang B, Fawcett GL, Leatherwood CJ, Wolf JB, Cheverud JM. Replication of long-bone length QTL in the F9–F10 LG,SM advanced intercross. Mamm Genome. 2009;20(4):224–235. doi: 10.1007/s00335-009-9174-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16(7):1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 38.Ishimori N, Li R, Walsh KA, Korstanje R, Rollins JA, Petkov P, Pletcher MT, Wiltshire T, Donahue LR, Rosen CJ, Beamer WG, Churchill GA, Paigen B. Quantitative trait loci that determine BMD in C57BL/6J and 129S1/SvImJ inbred mice. J Bone Miner Res. 2006;21(1):105–112. doi: 10.1359/JBMR.050902. [DOI] [PubMed] [Google Scholar]
- 39.Ishimori N, Stylianou IM, Korstanje R, Marion MA, Li R, Donahue LR, Rosen CJ, Beamer WG, Paigen B, Churchill GA. Quantitative trait loci for BMD in an SM/J by NZB/BlNJ intercross population and identification of Trps1 as a probable candidate gene. J Bone Miner Res. 2008;23(9):1529–1537. doi: 10.1359/JBMR.080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masinde GL, Li X, Gu W, Wergedal J, Mohan S, Baylink DJ. Quantitative trait loci for bone density in mice: the genes determining total skeletal density and femur density show little overlap in F2 mice. Calcif Tissue Int. 2002;71(5):421–428. doi: 10.1007/s00223-001-1113-z. [DOI] [PubMed] [Google Scholar]
- 41.Beamer WG, Shultz KL, Coombs HF, 3rd, DeMambro VE, Reinholdt LG, Ackert-Bicknell CL, Canalis E, Rosen CJ, Donahue LR. BMD regulation on mouse distal chromosome 1, candidate genes, and response to ovariectomy or dietary fat. J Bone Miner Res. 2011;26(1):88–99. doi: 10.1002/jbmr.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shultz KL, Donahue LR, Bouxsein ML, Baylink DJ, Rosen CJ, Beamer WG. Congenic strains of mice for verification and genetic decomposition of quantitative trait loci for femoral bone mineral density. J Bone Miner Res. 2003;18(2):175–185. doi: 10.1359/jbmr.2003.18.2.175. [DOI] [PubMed] [Google Scholar]
- 43.Edderkaoui B, Baylink DJ, Beamer WG, Wergedal JE, Porte R, Chaudhuri A, Mohan S. Identification of mouse Duffy antigen receptor for chemokines (Darc) as a BMD QTL gene. Genome Res. 2007;17(5):577–585. doi: 10.1101/gr.6009507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiao F, Chiu H, Jiao Y, de Rijk WG, Li X, Eckstein EC, Beamer WG, Gu W. Quantitative trait loci for tibial bone strength in C57BL/6J and C3H/HeJ inbred strains of mice. J Genet. 2010;89(1):21–27. doi: 10.1007/s12041-010-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saless N, Lopez Franco GE, Litscher S, Kattappuram RS, Houlihan MJ, Vanderby R, Demant P, Blank RD. Linkage mapping of femoral material properties in a reciprocal intercross of HcB-8 and HcB-23 recombinant mouse strains. Bone. 2010;46(5):1251–1259. doi: 10.1016/j.bone.2010.01.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koller DL, Schriefer J, Sun Q, Shultz KL, Donahue LR, Rosen CJ, Foroud T, Beamer WG, Turner CH. Genetic effects for femoral biomechanics, structure, and density in C57BL/6J and C3H/HeJ inbred mouse strains. J Bone Miner Res. 2003;18(10):1758–1765. doi: 10.1359/jbmr.2003.18.10.1758. [DOI] [PubMed] [Google Scholar]
- 47.Bouxsein ML, Uchiyama T, Rosen CJ, Shultz KL, Donahue LR, Turner CH, Sen S, Churchill GA, Muller R, Beamer WG. Mapping quantitative trait loci for vertebral trabecular bone volume fraction and microarchitecture in mice. J Bone Miner Res. 2004;19(4):587–599. doi: 10.1359/JBMR.0301255. [DOI] [PubMed] [Google Scholar]
- 48.Farber CR, van Nas A, Ghazalpour A, Aten JE, Doss S, Sos B, Schadt EE, Ingram-Drake L, Davis RC, Horvath S, Smith DJ, Drake TA, Lusis AJ. An integrative genetics approach to identify candidate genes regulating BMD: combining linkage, gene expression, and association. J Bone Miner Res. 2009;24(1):105–116. doi: 10.1359/JBMR.080908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters GB, Ingvarsson T, Jonsdottir T, Saemundsdottir J, Center JR, Nguyen TV, Bagger Y, Gulcher JR, Eisman JA, Christiansen C, Sigurdsson G, Kong A, Thorsteinsdottir U, Stefansson K. Multiple genetic loci for bone mineral density and fractures. N Engl J Med. 2008;358(22):2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 50.Klein RF, Turner RJ, Skinner LD, Vartanian KA, Serang M, Carlos AS, Shea M, Belknap JK, Orwoll ES. Mapping quantitative trait loci that influence femoral cross-sectional area in mice. J Bone Miner Res. 2002;17(10):1752–1760. doi: 10.1359/jbmr.2002.17.10.1752. [DOI] [PubMed] [Google Scholar]
- 51.Volkman SK, Galecki AT, Burke DT, Miller RA, Goldstein SA. Quantitative trait loci that modulate femoral mechanical properties in a genetically heterogeneous mouse population. J Bone Miner Res. 2004;19(9):1497–1505. doi: 10.1359/JBMR.040506. [DOI] [PubMed] [Google Scholar]
- 52.Kesavan C, Baylink DJ, Kapoor S, Mohan S. Novel loci regulating bone anabolic response to loading: expression QTL analysis in C57BL/6JXC3H/HeJ mice cross. Bone. 2007;41(2):223–230. doi: 10.1016/j.bone.2007.04.185. [DOI] [PubMed] [Google Scholar]
- 53.Kesavan C, Mohan S, Srivastava AK, Kapoor S, Wergedal JE, Yu H, Baylink DJ. Identification of genetic loci that regulate bone adaptive response to mechanical loading in C57BL/6J and C3H/HeJ mice intercross. Bone. 2006;39(3):634–643. doi: 10.1016/j.bone.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Diesterbeck US, Hertsch B, Distl O. Genome-wide search for microsatellite markers associated with radiologic alterations in the navicular bone of Hanoverian warmblood horses. Mamm Genome. 2007;18(5):373–381. doi: 10.1007/s00335-007-9021-9. [DOI] [PubMed] [Google Scholar]
- 55.Mbalaviele G, Shin CS, Civitelli R. Cell-cell adhesion and signaling through cadherins: connecting bone cells in their microenvironment. J Bone Miner Res. 2006;21(12):1821–1827. doi: 10.1359/jbmr.060811. [DOI] [PubMed] [Google Scholar]
- 56.Cheng SL, Lecanda F, Davidson MK, Warlow PM, Zhang SF, Zhang L, Suzuki S, St John T, Civitelli R. Human osteoblasts express a repertoire of cadherins, which are critical for BMP-2-induced osteogenic differentiation. J Bone Miner Res. 1998;13(4):633–644. doi: 10.1359/jbmr.1998.13.4.633. [DOI] [PubMed] [Google Scholar]
- 57.Okazaki M, Takeshita S, Kawai S, Kikuno R, Tsujimura A, Kudo A, Amann E. Molecular cloning and characterization of OB-cadherin, a new member of cadherin family expressed in osteoblasts. J Biol Chem. 1994;269(16):12092–12098. [PubMed] [Google Scholar]
- 58.Kawaguchi J, Azuma Y, Hoshi K, Kii I, Takeshita S, Ohta T, Ozawa H, Takeichi M, Chisaka O, Kudo A. Targeted disruption of cadherin-11 leads to a reduction in bone density in calvaria and long bone metaphyses. J Bone Miner Res. 2001;16(7):1265–1271. doi: 10.1359/jbmr.2001.16.7.1265. [DOI] [PubMed] [Google Scholar]
- 59.Di Benedetto A, Watkins M, Grimston S, Salazar V, Donsante C, Mbalaviele G, Radice GL, Civitelli R. N-cadherin and cadherin 11 modulate postnatal bone growth and osteoblast differentiation by distinct mechanisms. J Cell Sci. 2010;123(Pt 15):2640–2648. doi: 10.1242/jcs.067777. [DOI] [PMC free article] [PubMed] [Google Scholar]