Abstract
Mammalian pancreatic-type ribonucleases (ptRNases) comprise an enzyme family that is remarkably well suited for therapeutic exploitation. ptRNases are robust and prodigious catalysts of RNA cleavage that can naturally access the cytosol. Instilling cytotoxic activity requires endowing them with the ability to evade a cytosolic inhibitor protein while retaining other key attributes. These efforts have informed our understanding of ptRNase-based cytotoxins, as well as the action of protein-based drugs with cytosolic targets. Here, we address the most pressing problems encountered in the design of cytotoxic ptRNases, along with potential solutions. In addition, we describe assays that can be used to evaluate a successful design in vitro, in cellulo, and in vivo. The emerging information validates the continuing development of ptRNases as chemotherapeutic agents.
1. Introduction
Once, the utility of recombinant DNA technology in generating drugs was limited to producing wild-type human proteins in heterologous hosts. Now, protein engineering is being used to tailor proteins for specific clinical applications. The resulting biologic drugs can provide a level of target-specificity not achievable with small molecules (Szlachcic et al., 2011).
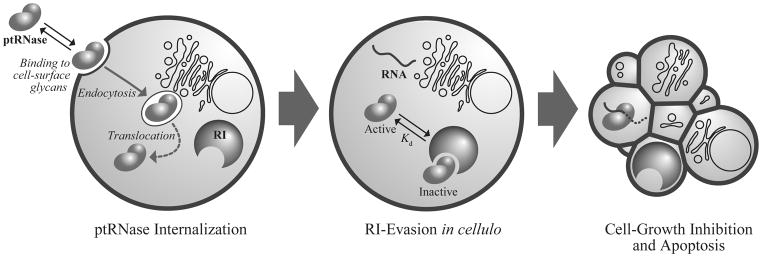
The mammalian pancreatic-type ribonucleases (ptRNases) are especially well suited for exploitation as chemotherapeutic agents. These enzymes circumvent the pitfalls that plague many other protein-based drugs, such as high molecular mass, instability, and immunogenicity. ptRNases comprise a highly conserved family of small (~13 kDa), secreted proteins that catalyze the degradation of RNA with extremely high efficiency (D’Alessio and Riordan, 1997; Raines, 1998). Incredibly, mammalian cells internalize these enzymes readily (Fig. 1); thus, ptRNases are not restricted to the extracellular or cell-surface targets of most other protein-based agents (Carter, 2011). Moreover, whereas many promising proteins never achieve success in the clinic because they are hampered by problems with production, storage, and administration (Szlachcic et al., 2011), ptRNases are produced readily in microbial hosts, have unusually high conformational stability, and maintain their integrity in extracellular fluids and tissues.
Figure 1.
The internalization pathway of ptRNases involves association with glycans on the cell membrane, absorptive endocytosis, and interaction with cytosolic RI. Upon evasion of RI, cytotoxic ptRNases can degrade cellular RNA and induce apoptosis.
Recently, ptRNases have garnered much attention because several, including the dimeric bovine seminal ribonuclease (BS-RNase) and an amphibian ortholog, Onconase® (ONC), have proven to be natural cytotoxins for human cancer cells. ONC is currently in a Phase IIIb confirmatory clinical trial as a second-line chemotherapeutic agent for malignant mesothelioma and has been granted both orphan-drug and fast-track status by the U.S. Food and Drug Administration (Lee and Raines, 2008; Mikulski et al., 2002). Despite their inherent cytotoxicity, both BS-RNase and ONC are of limited clinical utility. For example, ONC is compromised by dose-limiting renal toxicity and high immunogenicity relative to mammalian ribonucleases (Matoušek et al., 2003; Pavlakis and Vogelzang, 2006). Moreover, cytotoxic variants of mammalian ptRNases have greater specificity than ONC for cancer cells (Rutkoski et al., 2005). Efforts to exploit naturally occurring microbial and fungal ribotoxins have been plagued by similar immunogenicity and low specificity. (For reviews, see: Ardelt et al. (2009); Fang and Ng (2011).) Hence, the future of ptRNases as chemotherapeutic agents appears to rely on the strategic development of the mammalian homologues (Leland and Raines, 2001).
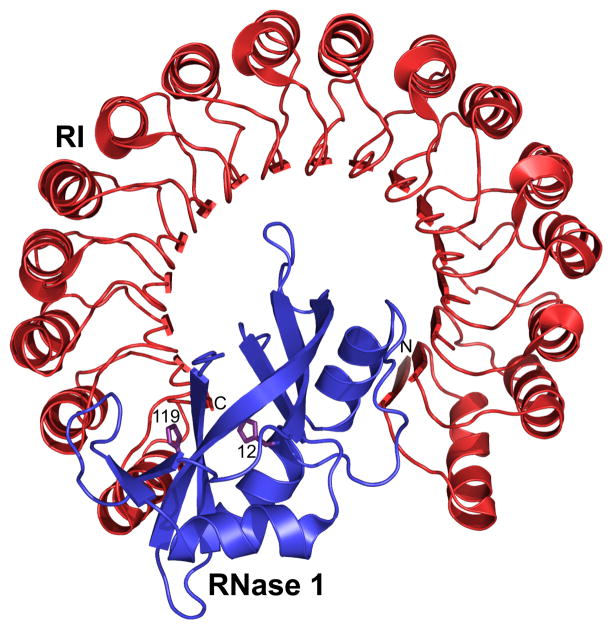
Although mammalian ptRNases possess the necessary stability, catalytic activity, and non-immunogenicity to warrant consideration as potential chemotherapeutic agents, their success is limited by two substantial barriers: internalization into the cytosol of target cells and inhibition by the cytosolic ribonuclease inhibitor protein (RI), which binds with femtomolar affinity to most ptRNases but not BS-RNase or ONC (Fig. 2) (Dickson et al., 2005). To design ptRNase-based agents, we have sought to understand the underlying biophysical and biochemical basis for their mechanism of action, and then to translate that knowledge into optimized proteins.
Figure 2.
Three-dimensional structure of the human RI·RNase 1 complex (Protein Data Bank entry 1z7x). The active-site histidine residues 12 and 119 of RNase 1 are depicted explicitly, and the N and C termini of RI are labeled. The complex has Kd = 2.9 × 10−16 M (Johnson et al., 2007b).
A useful ptRNase-based cytotoxin must catalyze the degradation of RNA within target cells. To do so, it must gain entry to the cytosol, evade RI there, and retain its catalytic activity throughout the process. Here, we report on our current understanding of these requirements and our strategies for engineering ptRNases that achieve maximal therapeutic efficacy. We also provide details on the assays that we use to evaluate relevant attributes of putative ptRNase cytotoxins. Although we focus on the well-known enzymes from cow (RNase A) and human (RNase 1), the methodologies are applicable to other ptRNases as well.
2. Attributes of Cytotoxic ptRNases
2.1 Catalytic activity and proteolytic stability
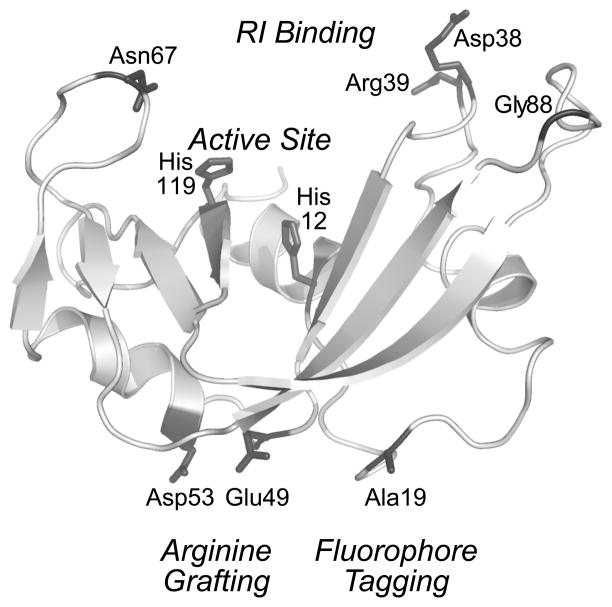
Two important attributes of a cytotoxic ptRNase are its ability to catalyze RNA cleavage and to resist proteolysis. Mammalian ptRNases can catalyze the cleavage of the P–O5′ bond of RNA on the 3′ side of pyrimidine nucleosides with a second-order rate constant (kcat/KM = 3.3 × 109 M−1s−1 (Park and Raines, 2001)) that is among the highest known for an enzyme-catalyzed reaction. This activity leads to cellular apoptosis (Bretscher et al., 2000), and is essential for ptRNase-mediated cytotoxicity (Kim et al., 1995). Proteins with high thermostability tend to have low susceptibility to proteolytic degradation (Parsell and Sauer, 1989). This premise holds true for ptRNases, as installing an additional disulfide bond in RNase A significantly increases its thermostability, resistance to proteolysis, and cytotoxicity (Klink and Raines, 2000). Accordingly, when choosing residues to alter within ptRNases, care must be taken to avoid interference with active-site residues, cysteine residues that participate in disulfide bonds, and other residues critical to structure and function (Fig. 3) (Smith and Raines, 2006). Although no consensus exists regarding the minimal requirements for either parameter, decreasing catalytic activity or thermostability leads to decreases in cytotoxicity (Klink et al., 2000; Leland et al., 2000). Catalytic activity can be measured by using a fluorogenic substrate, such as 6-carboxyfluorescein–dArUdAdA–6-carboxytetramethylrhodamine (Kelemen et al., 1999; Park et al., 2001); thermostability can be monitored by ultraviolet or circular dichroism spectroscopy.
Figure 3.
Three-dimensional structure of RNase A showing residues important in the design and evaluation of an RNase A-based cytotoxin.
2.2. Cellular internalization
ptRNases are especially well suited as biologic drugs due to their endogenous ability to enter cells without requiring any additional delivery strategy. Still, ptRNase internalization remains an inefficient process, and could limit cytotoxicity. To design a ptRNase-based drug that is equipped for more efficient internalization, it is important to understand the pathway by which ptRNases enter cells (Fig. 1).
Mammalian ptRNases undergo endocytosis through an adsorptive process that is non-saturable, non-receptor-mediated, and dynamin-independent (Haigis and Raines, 2003). Certain anionic cell-surface molecules facilitate Coulombic interactions with cationic ptRNases, which are highly positively charged (Table 1). Analyses in vitro and in cellulo reveal that RNase A interacts tightly with abundant anionic cell-surface glycosaminoglycans such as heparan sulfate and chondroitin sulfate, as well as sialic acid-containing glycoproteins. The uptake of RNase A correlates with cell-surface anionicity and could endow mammalian ptRNases with selective cytotoxicity for cancerous cells (Chao et al., 2010). Following endocytosis, a very small fraction of the endosomal ptRNase is able to translocate into the cytosol and catalyze RNA degradation.
Table 1.
Salient Modifications of ptRNases that Enhance Cytotoxicity
| Ribonuclease | Tm (°C)a | Ribonucleolytic Activity (%)b | Ki or Kd (nM)c | IC50 (μM)d | Ze | Reference |
|---|---|---|---|---|---|---|
| Wild-type RNase A | 64 | 100 | 44 × 10−6 | >25 | +4 | Rutkoski et al. (2005) |
| G88R RNase A | 60 | 142 | 2.8 | 6.2 | +5 | Leland et al. (1998) |
| A4C/G88R/V188C RNase A | 69 | 94 | 0.65 | 3 | +5 | Klink and Raines (2000) |
| D38R/R39D/N67R/G88R RNase A | 56 | 75 | 1.4 × 103 | 0.19 | +6 | Rutkoski et al. (2005) |
| E49R/D53R/G88R RNase A | 54 | 5 | 2.6 | 1.9 | +9 | Fuchs et al. (2007) |
| E49R/D53R/G88R RNase A–R9 | 49 | 7 | 3.0 | 0.58 | +18 | Fuchs et al. (2007) |
| (RNase A)2 [SGRSGRSG linker] | 61 | 1.2 | ND | 12.9 | +10 | Leich et al. (2006) |
| (D38R/R39D/N67R/G88C RNase A)3 | ND | 17 | ND | 1.0 | +16 | Rutkoski et al. (2010) |
| Wild-type RNase 1 | 57 | 100 | 29 × 10−8 | >25 | +6 | Johnson et al. (2007b) |
| R39D/N67R/N88R/G89D/R91D RNase 1 | 53 | 81 | 28 | 5.69 | +3 | Johnson et al. (2007b) |
| PE5 (RNase 1–NLS) | 46 | ND | ND | 4.6 | +6 | Tubert et al. (2011) |
| Onconase® | 90 | 100 | ≥103 | 0.27 | +5 | Rutkoski et al. (2005) |
Values of Tm are the temperature at the midpoint of thermal denaturation, which can be monitored by ultraviolet or circular dichroism spectroscopy.
Values of ribonucleolytic activity are relative to the wild-type enzyme.
Values of the equilibrium dissociation constant (or inhibition constant) are for the complex with human RI.
Values of IC50 are for the incorporation of [methyl-3H]thymidine into the DNA of K-562 human leukemia cells.
Values of Z refer to the net molecular charge: Arg + Lys − Asp − Glu − Pyr (where “Pyr” refers to a pyroglutamate residue, which is found at the N-terminus of Onconase®).
Specific modifications to a ptRNase can exploit the Coulombic interactions that likely facilitate RNase internalization. (For a review, see: Rutkoski and Raines (2008).) Recent work has demonstrated that the amount of positive charge, as well as the distribution of that charge, can affect ptRNase adsorption (Turcotte et al., 2009). For example, replacing two anionic surface residues, Glu49 and Asp53 (Fig. 3), with arginines (“arginine grafting”) results in enhanced internalization and cytotoxicity of an RNase A variant (Fuchs et al., 2007). Similarly, chemical cationization of ptRNases by amidation of carboxyl groups with either ethylenediamine or polyethylenimine leads to enhanced internalization and cytotoxicity (Futami and Yamada, 2008). ptRNases can also be fused to cationic cell-penetrating peptides (CPPs) such as nonaarginine to increase internalization (Fuchs and Raines, 2005; Fuchs et al., 2007). These cationic moieties need not be appended to ptRNases, as the addition of a cationic poly(aminoamine) dendrimer in trans increases the internalization and cytotoxicity of a ptRNase (Ellis et al., 2011). We note, however, that increasing the positive charge of a ptRNase can have the adverse effect of increasing its affinity for RI, which is highly anionic (Johnson et al., 2007a).
2.3. Evading the ribonuclease inhibitor protein
Ribonuclease inhibitor (RI) is a ~50-kDa protein found exclusively in the cytosol of mammalian cells. Multiple biological roles for RI have been proposed, including protecting cells from internalized secretory ptRNases and maintaining cellular redox homeostasis (Dickson et al., 2005; Monti et al., 2007). Despite the uncertainty surrounding its precise physiological role(s), RI binds to members of the mammalian ptRNase superfamily with a 1:1 stoichiometry, completely inhibiting their catalytic activity by steric occlusion of the enzymic active site (Fig. 2). Because ribonucleolytic activity is necessary to induce cellular apoptosis, a cytotoxic ptRNase must evade RI. Nevertheless, as the noncovalent complexes formed between RI and its ligands are among the tightest known in biology, instilling RI-evasion is a difficult task.
The goal of RI-evasion strategies is to modify the ptRNase so as to perturb only its interaction with RI. Many strategies are possible. (For a review, see: Rutkoski and Raines (2008).) Recent analyses of various crystallized RI·ptRNase complexes indicate that although the interaction of RI with various ptRNases is similar, evasion strategies should be optimized to recognize the subtle differences that exist in the binding interfaces. Computational analyses can be used to identify which residues of a ptRNase make the most contacts with RI (Rutkoski et al., 2005). These residues can then be targeted for substitution through site-directed mutagenesis. We have found that introducing electrostatic and steric incompatibilities in these regions destabilize the RI·RNase complex, and that disruption is often best achieved by replacing small neutral or anionic residues in a ptRNase with arginine (Fig. 3). Arginine, as the most polar and second largest amino acid, can generate electrostatic repulsion and steric strain while increasing positive molecular charge, thereby enhancing internalization.
Our initial engineering efforts yielded a prototype ptRNase cytotoxin, G88R RNase A. The modification of a single residue imbued native RNase A with 10 thousand-fold lower affinity for RI and cytotoxicity (Leland et al., 1998). Guided by computational algorithms, we have designed second-generation variants of RNase A and RNase 1 that evade RI more efficiently. For example, D38R/R39D/N67R/G88R RNase A demonstrates 20 million-fold lower affinity for RI than does native RNase A with little change to catalytic activity or thermostability (Table 1). This variant is more toxic than ONC to human cancer cells (Rutkoski et al., 2005). In addition to altering the RI-binding interface of RNase A, we find that appending a folate moiety at residue 88 engenders RI-evasion through steric repulsion (Smith et al., 2011). The pendant folate also enhances targeting to and uptake by cells that overexpress the cell-surface folate receptor, as do many types of cancer cells. Benefit is also accrued by attaching a pendant poly(ethylene glycol) (PEG) moiety to RNase A at the same position. Beyond RI-evasion, PEG conjugation elicits markedly decreased renal clearance and increased tumor-growth inhibition in mice (Rutkoski et al., 2011).
Human RNase 1 proved to be a greater challenge, as it binds to RI with 102-fold higher affinity than does RNase A. Although R39D/N67D/N88A/G89D/R91D RNase 1 has 6 billion-fold lower affinity for RI (Johnson et al., 2007b) than does wild-type RNase 1, this variant is not as cytotoxic as D38R/R39D/N67R/G88R RNase A. An alternative strategy to engender RI-evasion is to bypass RI contact altogether. The human ptRNase variant PE5 carries a non-contiguous nuclear localization signal and has been shown to possess potent cytotoxicity that is dependent upon its nuclear uptake (Tubert et al., 2011).
BS-RNase is a naturally dimeric homologue of RNase A that evades RI. But upon entry into the cytosol, the dimer dissociates and the resulting monomers are inhibited by RI (Lee and Raines, 2005). Recently, the endogenous properties of BS-RNase have been recapitulated by creating genetically encoded or chemically conjugated multimers of ptRNases that are unable to dissociate in cellulo (Leich et al., 2006; Leich et al., 2007; Rutkoski et al., 2010). Still, aspects of the mechanism of action of these multimers remain unclear. Multimeric ptRNases can be more cytotoxic than BS-RNase despite being less RI-evasive. Hence, the enhanced cytotoxicity demonstrated could be due, in part, to improved interaction of the multimers with the negatively charged cell membrane, thereby favoring endocytosis (Arnold et al., 2011; Leich et al., 2006; Rutkoski et al., 2010). High cytosolic localization of tandem RNase A dimers supports this hypothesis (Leich et al., 2007).
3. Assays to Evaluate the Cytotoxicity of ptRNases
Just as effort has gone into the rational design of cytotoxic ptRNases, so too has substantial work been done to develop assays to measure and characterize the cytotoxicity of ptRNase-based chemotherapeutic agents. Cytotoxic ptRNases should be characterized thoroughly in vitro before being tested in vivo. Below, we describe the state-of-the-art in quantifying important parameters of ptRNase-mediated cytotoxicity, including cellular internalization, evasion of RI, inhibition of tumor cell proliferation in vitro, and inhibition of tumor growth in vivo. Several of these assays involve the use of fluorophores that can be tethered to ptRNases in a site-specific, non-perturbative manner.
3.1 Utility of small-molecule fluorophores
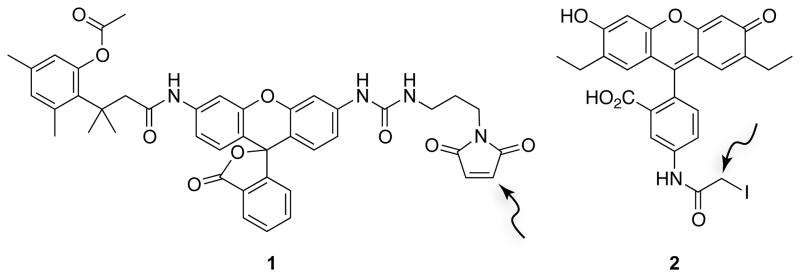
The constitutive fluorescence of traditional fluorophores (e.g., fluorescein) can lead to high background that obscures valuable information. To overcome this limitation, we designed fluorogenic label 1 (Fig. 4), which consists of a rhodamine 110 core enshrouded by an esterase-inducible “trimethyl lock” and a maleimido group for conjugation. Fluorescence is unmasked only in the presence of intracellular esterases. Hence, the endocytic uptake of ptRNase–1 conjugates can be monitored by either fluorescence microscopy or flow cytometry (Lavis et al., 2006).
Figure 4.
Structures of fluorogenic label 1 for monitoring endocytosis (Lavis et al., 2006) and fluorescent probe 2 for monitoring protein–ligand interactions (Lavis et al., 2007). The arrows indicate electrophilic carbons that can form thioether linkages with cysteine residues.
We designed a second fluorescent label that is likewise amenable to thiol-reactive, site-specific conjugation. We had observed that the fluorescence of fluorescein-labeled RNase A decreases upon binding to RI because the protonated, non-fluorescent form of fluorescein is stabilized by the anionic RI (Abel et al., 2002). The discrepancy between the phenolic pKa = 6.30 of fluorescein and physiological pH diminishes the sensitivity of this assay. To address this problem, we synthesized 2′,7′-diethylfluorescein, which has two electron-donating ethyl groups and a phenolic pKa of 6.61. Fluorescent probe 2 (2′,7′-diethylfluorescein-5-iodoacetamide; Fig. 4), which contains an iodoacetamido group for conjugation, enables a highly sensitive assay for the interaction of RI and ptRNases (Lavis et al., 2007).
3.1.1 Site-specific conjugation of ptRNases to fluorophores
Fluorescently labeled ptRNases have proven to be remarkably adaptable tools for a variety of assays. Nonetheless, as mammalian ptRNases contain multiple amino groups—including an essential one for catalysis in an active-site lysine residue (Messmore et al., 1995)—using amine-reactive reagents to cross-link or conjugate ptRNases can result in heterogeneity and inactivation. To overcome this problem, we install cysteine residues at inconsequential positions in ptRNases, allowing for site-specific conjugation to fluorophores.
Both RNase A and RNase 1 contain eight cysteine residues that form four disulfide bonds in the native enzyme. To enable attachment of a thiol-reactive fluorophore, we introduce a cysteine residue at position 19 using site-directed mutagenesis. Position 19 is an optimal location because attachment of fluorophore groups there does not interfere with catalytic activity, RI binding, or cell-surface interactions (Fig. 3). Further, this residue is in a solvated loop that is inconsequential for protein stability (Abel et al., 2002). Free-cysteine variants are produced and purified by methods described previously for other ptRNase variants (Leland et al., 1998), with the following modifications.
To ensure that the free cysteine residue does not suffer irreversible oxidation to a sulfinic or sulfonic acid, O2(g) must be removed from the buffers used in the purification process. Following initial purification, the free thiol group at position 19 is protected as a mixed disulfide by reaction with a 4-fold molar excess of 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB). Immediately following purification by gel-filtration chromatography, pooled protein fractions are adjusted to become 8% (w/v) in 1.0 M Tris–HCl buffer, pH 8.0, containing EDTA (10 mM). DTNB is dissolved in a small quantity of ethanol, then brought to 5 mM with 20 mM HEPES–NaOH buffer, pH 8.0. Upon addition of DTNB to the protein solution, a yellow color is observed due to the production of 2-nitro-5-thiobenzoic acid (NTB). The resulting mixture is dialyzed against several liters of 20 mM sodium acetate buffer, pH 5.0, overnight at 4 °C to remove unreacted NTB. NTB-protected ribonucleases are then purified further with cation-exchange chromatography and stored at 4 °C until needed for conjugation.
Immediately prior to fluorophore attachment, NTB-protected ptRNases are deprotected with a 4-fold molar excess of dithiothreitol and desalted by chromatography. Deprotected ptRNases are incubated for 6 h at 25 °C with a 10-fold molar excess of fluorophore in phosphate-buffered saline (PBS). N,N-Dimethylformamide (≤10% v/v) can be added to the reaction mixture to increase fluorophore solubility. Following the conjugation reaction, the solution is dialyzed against 20 mM sodium acetate buffer, pH 5.0, and then purified again with reverse-phase HPLC to separate labeled and unlabeled protein. The molecular mass of each conjugate is verified by mass spectrometry prior to its use in assays.
3.1.2 Assessing cellular internalization with fluorescence spectroscopy
The internalization of a ptRNase–1 conjugate into live cells can be visualized with fluorescence microscopy. The rate of ptRNase internalization can be quantified with flow cytometry. These analyses can be used with both adherent and nonadherent cell lines.
To quantify internalization, mammalian cells from nearly confluent flasks are collected by centrifugation and resuspended at a density of 106 cells/ml in fresh medium that is appropriate to the cell type. A ptRNase–1 conjugate is added (to 10 μM) to 250 μl of medium containing 106 cells/ml of cells. An unlabeled ptRNase can serve as a negative control. The cells are incubated at 37 °C for known times, typically ≤2 h. During this timeframe, we have observed that only a small fraction of labeled ptRNases are taken up by cells; therefore, we assume that the rate of internalization is not limited by substrate concentration. Further, we do not observe any significant exocytosis of labeled ptRNases by confocal microscopy (Lavis et al., 2006). To quench internalization, cells are collected by centrifugation at 1000 rpm for 5 min at 4 °C, washed once with ice-cold PBS, and resuspended in 250 μl of PBS. Samples are kept on ice until their analysis. The fluorescence of unmasked 1 can be detected by flow cytometry using a 530/30-nm band-pass filter. Total cell viability can be determined by staining with propidium iodide, which can be detected through a 660-nm long-pass filter. The mean channel fluorescence intensity of 20,000 viable cells are determined for each sample and used for subsequent analyses. To determine the steady-state rate constant (kI) for ptRNase internalization, fluorescence intensity data is fitted to Eq. (1), where Fmax is the fluorescence intensity upon reaching the steady state and kI is the first-order rate constant for ptRNase internalization into cells.
| (1) |
3.1.3 Evaluating RI evasion with fluorescence spectroscopy
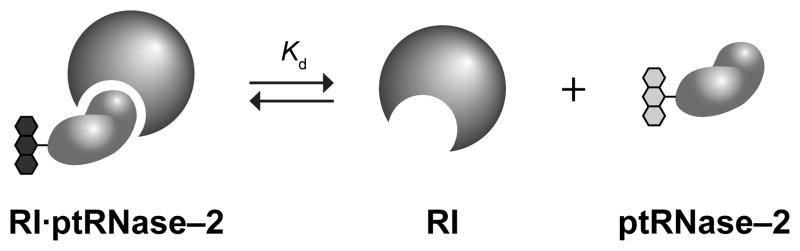
Traditionally, the stability of a RI·RNase complex has been determined by measuring the inhibition of catalytic activity. The Ki values obtained by this method are lower than the concentration of wild-type RNase 1 used in the experiment itself. Accordingly, these values can only be an upper limit for the true Kd value. To more accurately measure the binding of ptRNases to RI, the dissociation rate of an RI·ptRNase complex is determined by monitoring the release of a ptRNase–2 conjugate over time (Fig. 5). To calculate the value of Kd of a RI·ptRNase complex, the value of ka is assumed to be similar to that for the association of hRI with angiogenin or RNase A. These ka values are within twofold of each other and are close to the diffusion limit (Lee et al., 1989). Thus, the ka value of homologous ptRNases is assumed to be equivalent to RNase A.
Figure 5.
Assay for evaluating Kd, which is the equilibrium dissociation constant of an RI·ptRNase complex (Lavis et al., 2007). Dissociation of the complex leads to an increase in fluorescence. The assay can also be used to evaluate the affinity of an unlabeled competitor ptRNase with a Kd value that is higher than the labeled ptRNase.
The dissociation rate of the complex between RI and a ptRNase–2 conjugate can be determined by following the increase in fluorescence upon complex dissociation (Fig. 5). A ptRNase–2 conjugate (100 nM) in PBS containing tris(2-carboxyethyl)phosphine (100 μM) and bovine serum albumin (0.10 mg/ml; Sigma Chemical) is added to a 96-well microtiter plate, and the initial fluorescence is measured with a plate reader. RI is then added at equimolar concentrations and incubated with labeled ptRNase at 25 °C for 5 min. A 50-fold molar excess of human angiogenin or RNase 1 (5 μM) is added to scavenge dissociated complex, and the change in fluorescence is measured at various time points. To insure that the proteins maintain their folded conformation for the duration of the experiment, additional data points should be monitored under the same conditions, only without the addition of the 50-fold molar excess of angiogenin. To account for indeterminate error, data are the mean (± SE) from six solutions normalized for the fluorescence of four solutions of labeled ptRNase (100 nM) in the absence of RI. Fluorescence data are fitted to Eq. (2) to determine the dissociation rate constant (kd), wherein F0 is the fluorescence before the addition of angiogenin, and F∞ is the fluorescence before RI addition. The equilibrium dissociation constant (Kd) for the RI·ptRNase complex can be determined with Eq. (3) and the known value for the association rate constant of RNase A (ka = 3.4 × 108 M−1s−1 (Lee et al., 1989)).
| (2) |
| (3) |
3.2 Measuring inhibition of tumor-cell proliferation in vitro
Multiple assays exist for measuring the effects of ribonucleases on cultured cells in vitro. We choose to use the following cell-proliferation assay because of its very high reproducibility. Cytotoxicity is evaluated by measuring the incorporation of [methyl-3H]thymidine into newly synthesized DNA of either adherent or non-adherent cells. Although this assay measures the synthesis of DNA that is necessary for cell proliferation, the data also report on cell death. As revealed by trypan blue dye exclusion, the number of nonviable cells increases upon treatment with a toxic ptRNase (Leland et al., 2001). A significant portion of these treated cells also display enhanced labeling with the apopotic marker Annexin V (T.-Y. Chao and R.T. Raines, unpublished results). Microscopic analyses of treated cells indicate an apoptotic morphology, including the formation of apoptotic bodies, nuclear condensation, and fragmentation. The extent of cell viability and apoptotic morphology correlates well with [methyl-3H]thymidine incorporation (Haigis et al., 2003). Thus, ptRNases render apoptosis upon target cells, and our assay of the inhibition of DNA synthesis reports on cytotoxicity.
Cells, such as human erythroleukemia line K-562 (ATCC; Manassas, VA), are grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Mediatech, Herndon, VA) supplemented with fetal bovine serum (FBS; 10% v/v), penicillin (100 units/ml), and streptomycin (100 μg/ml), and maintained at 37 °C in a humidified atmosphere containing CO2(g) (5% v/v). Aliquots (95 μl) of cultured cells (105 cells/ml) are placed in a microtiter plate, and sterile solutions (5 μl) of a ptRNase in PBS are added to the aliquots. Cells are incubated in the presence or absence of ribonucleases for 44 h, followed by a 4-h pulse with [methyl-3H]thymidine (0.2 μCi per well). Cells are then harvested onto glass fiber filters by using, for example, a PHD cell harvester (Cambridge Technology, Watertown, MA) and lysed by the passage of several milliliters of water through the filters. The filter retains DNA and other cellular macromolecules, while small molecules, including unincorporated radiolabel, pass through. After washing extensively with water, the filters are dried with methanol and counted by using a liquid scintillation counter. Results from the cytotoxicity assay are reported as the percentage of [methyl-3H]thymidine incorporation into the DNA of PBS-treated control cells. All assays should be repeated at least three times to lessen indeterminate error. Values of IC50, which is the concentration of ribonucleases that decreases cell proliferation by 50%, are calculated by fitting the data using nonlinear regression analysis to a sigmoidal dose–response curve (Eq. 4), in which y is the DNA synthesis following the [methyl-3H]thymidine pulse and h is the slope of the curve.
| (4) |
3.3 Measuring inhibition of tumor-cell proliferation in vivo
The most compelling method to measure the therapeutic effects of a cytotoxic ptRNase comes from in vivo analysis of tumor growth inhibition. This assay utilizes human tumor xenografts grown in the flanks of nude mice. Here, we describe our method for analyzing the effect on the growth of these tumors. We use the human tumor cell lines DU145 (prostate cancer) and A549 (non-small cell lung cancer) for their ability to proliferate in mice, their low rate of spontaneous regression, and their known sensitivity to RI-evasive variants of RNase A (Rutkoski et al., 2005). Moreover, each line represents a clinically relevant target that is used often in the testing of new chemotherapeutic agents.
DU145 cells are grown in Dulbecco’s modified Eagle’s medium (ATCC) containing FBS (10% v/v); A549 cells are grown in F12K medium (ATCC) containing FBS (10% v/v). Cells (~106) are implanted into a rear flank of 5–6 week old male homozygous (nu/nu) nude mice. Tumors are allowed to grow to a volume of ≥75 mm3 before the initiation of treatment. All test compounds are diluted in sterile PBS. All treatments are administered either by intraperitoneal injection (i.p.) or orally with a gavage needle (p.o.) for comparator chemotherapeutics, with the volume of administered solution based upon the body weight of the animal (10 μl/g). It is important to establish a dose and administration schedule that is effective with a specific cytotoxic ptRNase and tumor cell type. We have found that for mice bearing DU145 prostate carcinoma tumors, a dose of 15 mg/kg (i.p., qd × 5) of several monomeric ptRNase variants is optimal for eliciting maximum inhibition of tumor growth while minimizing off-target effects, as monitored by change in body weight. Similarly, we have found that frequent administration (qd × 5) achieves maximal tumor growth inhibition as compared to a single large dose (1 × wk). On the other hand, trimeric conjugates of cytotoxic RNase A variants can be administered less frequently and at a lower dose with comparable effect (Rutkoski et al., 2010). We speculate that the increased hydrodynamic radii of these trimeric conjugates results in enhanced persistence in circulation, as is observed with ptRNase-PEG conjugates (Rutkoski et al., 2011). Animal body weight should be monitored continually throughout the experiment as an indicator of drug tolerance.
Treatment with all agents should be ongoing throughout the entire experiment, with a control set of animals treated with vehicle alone. Comparators can include approved chemotherapeutic agents, such as docetaxel (15 mg/kg; i.p., 1 × wk), cisplatin (6 mg/kg; i.p., 1 × wk), and erlotinib (100 mg/kg; p.o., 2 × wk). Tumor size should be measured twice-weekly using calipers, and tumor volume (mm3) can be estimated by using the formula for a spheroid (Eq. 5). The percent tumor growth inhibition (%TGI) is then calculated with Eq. (6).
| (5) |
| (6) |
4. Prospectus
We have engineered mammalian ptRNases into useful cytotoxins. Through the use of novel, sensitive assays, we have been able to reveal the contribution of various parameters toward cytotoxicity. Still, mechanistic issues remain unclear, involving ptRNase translocation from endosomes to the cytosol and the specific RNA targets of ptRNases. Novel assays to illuminate these issues—and exploit them therapeutically—are being developed in our laboratory. Further work is directed at enhancing the circulating half-life of ptRNases in vivo using pegylation or glycosylation.
ptRNases have shown exceptional applicability as model proteins for multi-faceted drug design. The potential therapeutic value of ptRNases has been extended beyond cancer with the creation of zymogens that can be engineered to be disease-specific. To date, protease-activatable ptRNase zymogens have been developed to combat malaria, hepatitis C, and HIV (Johnson et al., 2006; Plainkum et al., 2003; Turcotte and Raines, 2008). Another member of the ptRNase family, angiogenin, has been designed as a hyperactive variant capable of enhanced neovascularization (Dickson et al., 2009). Continued efforts to engineer this remarkable family of proteins will no doubt add even more therapeutic value.
Acknowledgments
J.E.L. is supported by an NSF Graduate Research Fellowship. Research on ribonucleases as cancer chemotherapeutic agents in the authors’ laboratory is supported by Grant R01 CA073808 (NIH).
Contributor Information
Jo E. Lomax, Email: jelomax@wisc.edu.
Chelcie H. Eller, Email: eller@wisc.edu.
Ronald T. Raines, Email: rtraines@wisc.edu.
References
- Abel RL, Haigis MC, Park C, Raines RT. Fluorescence assay for the binding of ribonuclease A to the ribonuclease inhibitor protein. Anal Biochem. 2002;306:100–107. doi: 10.1006/abio.2002.5678. [DOI] [PubMed] [Google Scholar]
- Ardelt W, Ardelt B, Darzynkiewicz Z. Ribonucleases as potential modalities in anticancer therapy. Eur J Pharmacol. 2009;625:181–189. doi: 10.1016/j.ejphar.2009.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold U, Leich F, Neumann P, Lilie H, Ulbrich-Hofmann R. Crystal structure of RNase A tandem enzymes and their interaction with the cytosolic ribonuclease inhibitor. FEBS J. 2011;278:331–340. doi: 10.1111/j.1742-4658.2010.07957.x. [DOI] [PubMed] [Google Scholar]
- Bretscher LE, Abel RL, Raines RT. A ribonuclease A variant with low catalytic activity but high cytotoxicity. J Biol Chem. 2000;275:9893–9896. doi: 10.1074/jbc.275.14.9893. [DOI] [PubMed] [Google Scholar]
- Carter PJ. Introduction to current and future protein therapeutics: A protein engineering perspective. Exp Cell Res. 2011;317:1261–1269. doi: 10.1016/j.yexcr.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Chao TY, Lavis LD, Raines RT. Cellular uptake of ribonuclease A relies on anionic glycans. Biochemistry. 2010;49:10666–10673. doi: 10.1021/bi1013485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio G, Riordan JF. Ribonucleases: Structures and Functions. Academic Press; New York: 1997. [Google Scholar]
- Dickson KA, Haigis MC, Raines RT. Ribonuclease inhibitor: Structure and function. Prog Nucleic Acid Res Mol Biol. 2005;80:349–374. doi: 10.1016/S0079-6603(05)80009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson KA, Kang DK, Kwon YS, Kim JC, Leland PA, Kim BM, Chang SI, Raines RT. Ribonuclease inhibitor regulates neovascularization by human angiogenin. Biochemistry. 2009;48:3804–3806. doi: 10.1021/bi9005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis GA, Hornung ML, Raines RT. Potentiation of ribonuclease cytotoxicity by a poly(amidoamine) dendrimer. Bioorg Med Chem Lett. 2011;21:2756–2758. doi: 10.1016/j.bmcl.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang EF, Ng TB. Ribonucleases of different origins with a wide spectrum of medicinal applications. Biochim Biophys Acta. 2011;1815:65–74. doi: 10.1016/j.bbcan.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Fuchs SM, Raines RT. Polyarginine as a multifunctional fusion tag. Protein Sci. 2005;14:1538–1544. doi: 10.1110/ps.051393805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Rutkoski TJ, Kung VM, Groeschl RT, Raines RT. Increasing the potency of a cytotoxin with an arginine graft. Protein Eng Des Select. 2007;20:505–509. doi: 10.1093/protein/gzm051. [DOI] [PubMed] [Google Scholar]
- Futami J, Yamada H. Design of cytotoxic ribonucleases by cationization to enhance intracellular protein delivery. Curr Pharm Biotechnol. 2008;9:180–184. doi: 10.2174/138920108784567326. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Kurten EL, Raines RT. Ribonuclease inhibitor as an intracellular sentry. Nucleic Acids Res. 2003;31:1024–1032. doi: 10.1093/nar/gkg163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Raines RT. Secretory ribonucleases are internalized by a dynamin-independent endocytic pathway. J Cell Sci. 2003;116:313–324. doi: 10.1242/jcs.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RJ, Lin SR, Raines RT. A ribonuclease zymogen activated by the NS3 protease of the hepatitis C virus. FEBS J. 2006;273:5457–5465. doi: 10.1111/j.1742-4658.2006.05536.x. [DOI] [PubMed] [Google Scholar]
- Johnson RJ, Chao TY, Lavis LD, Raines RT. Cytotoxic ribonucleases: The dichotomy of Coulombic forces. Biochemistry. 2007a;46:10308–10316. doi: 10.1021/bi700857u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RJ, McCoy JG, Bingman CA, Phillips GN, Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007b;367:434–449. doi: 10.1016/j.jmb.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen BR, Klink TA, Behlke MA, Eubanks SR, Leland PA, Raines RT. Hypersensitive substrate for ribonucleases. Nucleic Acids Res. 1999;27:3696–3701. doi: 10.1093/nar/27.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Souček J, Matoušek J, Raines RT. Catalytic activity of bovine seminal ribonuclease is essential for its immunosuppressive and other biological activities. Biochem J. 1995;308:547–550. doi: 10.1042/bj3080547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink TA, Raines RT. Conformational stability is a determinant of ribonuclese A cytotoxicity. J Biol Chem. 2000;275:17463–17467. doi: 10.1074/jbc.M001132200. [DOI] [PubMed] [Google Scholar]
- Klink TA, Woycechowsky KJ, Taylor KM, Raines RT. Contribution of disulfide bonds to the conformational stability and catalytic activity of ribonuclease A. Eur J Biochem. 2000;267:566–572. doi: 10.1046/j.1432-1327.2000.01037.x. [DOI] [PubMed] [Google Scholar]
- Lavis LD, Chao TY, Raines RT. Fluorogenic label for biomolecular imaging. ACS Chem Biol. 2006;1:252–260. doi: 10.1021/cb600132m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis LD, Rutkoski TJ, Raines RT. Tuning the pKa of fluorescein to optimize binding assays. Anal Chem. 2007;79:6775–6782. doi: 10.1021/ac070907g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee FS, Auld DS, Vallee BL. Tryptophan fluorescence as a probe of placental ribonuclease inhibitor binding to angiogenin. Biochemistry. 1989;28:219–224. doi: 10.1021/bi00427a030. [DOI] [PubMed] [Google Scholar]
- Lee JE, Raines RT. Cytotoxicity of bovine seminal ribonuclease: Monomer versus dimer. Biochemistry. 2005;44:15760–15767. doi: 10.1021/bi051668z. [DOI] [PubMed] [Google Scholar]
- Lee JE, Raines RT. Ribonucleases as novel chemotherapeutics: The ranpirnase example. BioDrugs. 2008;22:53–58. doi: 10.2165/00063030-200822010-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leich F, Köditz J, Ulbrich-Hofman R, Arnold U. Tandemization endows bovine pancreatic ribonuclease with cytotoxic activity. J Mol Biol. 2006;358:1305–1313. doi: 10.1016/j.jmb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Leich F, Stohr N, Rietz A, Ulbrich-Hofmann R, Arnold U. Endocytotic internalization as a crucial factor for the cytotoxicity of ribonucleases. J Biol Chem. 2007;282:27640–27646. doi: 10.1074/jbc.M702240200. [DOI] [PubMed] [Google Scholar]
- Leland PA, Schultz LW, Kim BM, Raines RT. Ribonuclease A variants with potent cytotoxic activity. Proc Natl Acad Sci USA. 1998;95:10407–10412. doi: 10.1073/pnas.95.18.10407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland PA, Staniszewski KE, Kim BM, Raines RT. A synapomorphic disulfide bond is critical for the conformational stability and cytotoxicity of an amphibian ribonuclease. FEBS Lett. 2000;477:203–207. doi: 10.1016/s0014-5793(00)01804-4. [DOI] [PubMed] [Google Scholar]
- Leland PA, Raines RT. Cancer chemotherapy—ribonucleases to the rescue. Chem Biol. 2001;8:405–413. doi: 10.1016/s1074-5521(01)00030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leland PA, Staniszewski KE, Kim BM, Raines RT. Endowing human pancreatic ribonuclease with toxicity for cancer cells. J Biol Chem. 2001;276:43095–43102. doi: 10.1074/jbc.M106636200. [DOI] [PubMed] [Google Scholar]
- Matoušek J, Souček J, Slavík T, Tománek M, Lee JE, Raines RT. Comprehensive comparison of the cytotoxic activities of onconase and bovine seminal ribonuclease. Comp Biochem Physiol. 2003;136C:343–356. doi: 10.1016/j.cca.2003.10.005. [DOI] [PubMed] [Google Scholar]
- Messmore JM, Fuchs DN, Raines RT. Ribonuclease A: Revealing structure – function relationships with semisynthesis. J Am Chem Soc. 1995;117:8057–8060. doi: 10.1021/ja00136a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulski SM, Costanzi JJ, Vogelzang NJ, McCachren S, Taub RN, Chun H, Mittelman A, Panella T, Puccio C, Fine R, Shogen K. Phase II trial of a single weekly intravenous dose of ranpirnase in patients with unresectable malignant mesothelioma. J Clin Oncol. 2002;20:274–281. doi: 10.1200/JCO.2002.20.1.274. [DOI] [PubMed] [Google Scholar]
- Monti DM, Montesano Gesualdi N, Matoušek J, Esposito F, D’Alessio G. The cytosolic ribonuclease inhibitor contributes to intracellular redox homeostasis. FEBS Lett. 2007;581:930–934. doi: 10.1016/j.febslet.2007.01.072. [DOI] [PubMed] [Google Scholar]
- Park C, Kelemen BR, Klink TA, Sweeney RY, Behlke MA, Eubanks SR, Raines RT. Fast, facile, hypersensitive assays for ribonucleolytic activity. Methods Enzymol. 2001;341:81–94. doi: 10.1016/s0076-6879(01)41146-3. [DOI] [PubMed] [Google Scholar]
- Park C, Raines RT. Quantitative analysis of the effect of salt concentration on enzymatic catalysis. J Am Chem Soc. 2001;123:11472–11479. doi: 10.1021/ja0164834. [DOI] [PubMed] [Google Scholar]
- Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibilityin Escherichia coli. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- Pavlakis N, Vogelzang NJ. Ranpirnase—an antitumour ribonuclease: Its potential role in malignant mesothelioma. Expert Opin Biol Ther. 2006;6:391–399. doi: 10.1517/14712598.6.4.391. [DOI] [PubMed] [Google Scholar]
- Plainkum P, Fuchs SM, Wiyakrutta S, Raines RT. Creation of a zymogen. Nat Struct Biol. 2003;10:115–119. doi: 10.1038/nsb884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Rutkoski TJ, Kurten EL, Mitchell JC, Raines RT. Disruption of shape-complementarity markers to create cytotoxic variants of ribonuclease A. J Mol Biol. 2005;354:41–54. doi: 10.1016/j.jmb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Rutkoski TJ, Raines RT. Evasion of ribonuclease inhibitor as a determinant of ribonuclease cytotoxicity. Curr Pharm Biotechnol. 2008;9:185–189. doi: 10.2174/138920108784567344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkoski TJ, Kink JA, Strong LE, Schilling CI, Raines RT. Antitumor activity of ribonuclease multimers created by site-specific covalent tethering. Bioconjugate Chem. 2010;21:1691–1702. doi: 10.1021/bc100292x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkoski TJ, Kink JA, Strong LE, Raines RT. Site-specific PEGylation endows a mammalian ribonuclease with antitumor activity. Cancer Biol Ther. 2011;11:xxx–xxx. doi: 10.4161/cbt.12.3.15959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BD, Raines RT. Genetic selection for critical residues in ribonucleases. J Mol Biol. 2006;362:459–478. doi: 10.1016/j.jmb.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Smith BD, Higgin JJ, Raines RT. Site-specific folate conjugation to a cytotoxic protein. Bioorg Med Chem Lett. 2011;21:xxx–xxx. doi: 10.1016/j.bmcl.2011.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlachcic A, Zakrzewska M, Otlewski J. Longer action means better drug: Tuning up protein therapeutics. Biotechnol Adv. 2011;29:436–441. doi: 10.1016/j.biotechadv.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Tubert P, Rodríguez M, Ribó M, Benito A, Vilanova M. The nuclear transport capacity of a human-pancreatic ribonuclease variant is critical for its cytotoxicity. Invest New Drugs. 2011;29:xxx–xxx. doi: 10.1007/s10637-010-9426-2. [DOI] [PubMed] [Google Scholar]
- Turcotte RF, Raines RT. Design and characterization of an HIV-specific ribonuclease zymogen. AIDS Res Hum Retrovir. 2008;24:1357–1363. doi: 10.1089/aid.2008.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte RF, Lavis LD, Raines RT. Onconase cytotoxicity relies on the distribution of its positive charge. FEBS J. 2009;276:3846–3857. doi: 10.1111/j.1742-4658.2009.07098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]