Abstract
Objective
UBE2L3 is associated with susceptibility to systemic lupus erythematosus (SLE) and rheumatoid arthritis in European ancestry populations, and this locus has not been investigated fully in non-European populations. We studied the UBE2L3 risk allele for association with SLE, interferon-α (IFN-α), and autoantibodies in a predominantly African American SLE cohort.
Methods
We studied 395 patients with SLE and 344 controls. The UBE2L3 rs5754217 polymorphism was genotyped using Taqman primer-probe sets, and IFN-α was measured using a reporter cell assay.
Results
The UBE2L3 rs5754217 T allele was strongly enriched in African American patients with anti-La antibodies as compared to controls, and a recessive model was the best fit for this association (OR 2.55, p = 0.0061). Serum IFN-α also demonstrated a recessive association with the rs5754217 genotype in African American patients, and the TT/anti-La-positive patients formed a significantly high IFN-α subgroup (p = 0.0040). Similar nonstatistically significant patterns of association were observed in the European American patients with SLE. Case-control analysis did not show large allele frequency differences, supporting the idea that this allele is most strongly associated with anti-La-positive patients.
Conclusion
This pattern of recessive influence within a subgroup of patients may explain why this allele does not produce a strong signal in standard case-control studies, and subphenotypes should be included in future studies of UBE2L3. The interaction we observed between UBE2L3 genotype and autoantibodies upon serum IFN-α suggests a biological role for this locus in patients with SLE in vivo.
Keywords: SYSTEMIC LUPUS ERYTHEMATOSUS, GENETICS INTERFERON-α, AUTOANTIBODIES, UBE2L3 GENOTYPE
Recent studies have begun to unravel the genetic architecture of systemic lupus erythematosus (SLE)1. Despite recent successes, much of the heritability of the disorder remains to be described, and most large-scale studies directed at gene discovery to date have been performed exclusively in populations of European ancestry. SLE in African Americans is both more common and more severe than in European Americans2,3, and thus genetic study of SLE in African American populations is a high priority. While some of the genetic risk loci reported in European populations have been replicated in African American populations, there is already clear precedent that some European ancestry genetic risk factors will not be relevant in African Americans4. Additionally, emerging evidence supports the existence of genetic risk factors for SLE that are particular to those of African ancestry5,6,7,8.
Genetic variation in the UBE2L3 gene has been associated with risk of rheumatoid arthritis9, Crohn's disease10, and SLE1,11. These studies have all been performed in people of European ancestry. Interestingly, the autoimmune disease risk allele of UBE2L3 reported in Europeans is more common in African populations in the HapMap dataset (rs5754217 T allele frequency is 0.168 in the European-derived CEU population and 0.496 in the African YRI population). This presents the possibility that this genetic risk factor could be more important in African Americans, and that studies of this allele will be characterized by greater statistical power in this ancestral background. Ubiquitin-conjugating enzyme E2L3 (UBE2L3, also known as UbcH7) attaches ubiquitin molecules to other proteins, targeting them for destruction12. UBE2L3 has been shown to attach ubiquitin molecules to nuclear factor-κB, p53, and Fos13. Additionally, UBE2L3 interacts with Triad3A (RNF216), which can regulate the degradation of Toll-like receptors (TLR)14. In SLE, signaling through the endosomal TLR is thought to be an important pathway for the generation of interferon-α (IFN-α)15.
Much evidence supports the idea that increased IFN-α pathway signaling is causal in human SLE. Serum IFN-α is high in many patients with SLE16, and a number of patients treated with recombinant human IFN-α for malignancy and chronic viral hepatitis have developed de novo SLE, which typically resolves after the IFN-α is discontinued17. Many of the confirmed SLE risk genes function within the IFN-α pathway1,18,19. We have shown previously that some established IFN-α pathway SLE risk genes are associated with increased IFN-α signaling in patients with SLE20,21,22,23. Elevated serum IFN-α is clustered within SLE families in a pattern consistent with a complex trait16, further supporting the idea that a multifactorial heritable tendency toward high IFN-α is a primary pathogenic mediator in SLE24. SLE-associated autoanti-bodies are strongly associated with increased IFN-α in patients with SLE25, and these autoantibodies may also be primary pathogenic factors in SLE26.
We investigated the UBE2L3 rs5754217 autoimmune disease risk allele in our local predominantly African American SLE cohort. We hypothesized that there may be an association between this allele and serum IFN-α or autoantibody traits in SLE, and that by studying genetic associations with these intermediate phenotypes we may be able to better understand the effect of this locus on human disease.
MATERIALS AND METHODS
Patients and samples
We studied serum and genomic DNA samples from 395 patients with SLE from the University of Chicago Translational Research in the Department of Medicine (TRIDOM) registry and Rush University Medical Center. The SLE cohort consisted of 252 African American and 143 European American patients with SLE. All patients met the revised 1982 American College of Rheumatology criteria for the diagnosis of SLE26. Sex-matched controls were also obtained from the TRIDOM registry, including 239 African American and 105 European American subjects. The control subjects were all screened for absence of autoimmune disease by medical record review. The subjects in this study were not related to each other. Informed consent was obtained from all subjects at each site, and the study was approved by the institutional review board at each institution.
Single-nucleotide polymorphism (SNP) genotyping of UBE2L3 rs5754217
Patients with SLE were genotyped at UBE2L3 rs5754217 using ABI Taqman Assays-by-Design primers (Applied Biosystems, Foster City, CA, USA) and probes on an ABI 7900HT polymerase chain reaction (PCR) machine with > 98% genotyping success. Scatterplots were all reviewed individually for quality, and genotype frequencies did not deviate significantly from the expected Hardy-Weinberg proportions (p > 0.01 in each ancestral background).
Reporter cell assay for IFN-α
The reporter cell assay for IFN-α has been described in detail16. Reporter cells were used to measure the ability of patient sera to cause IFN-induced gene expression. The reporter cells (WISH cells, American Type Culture Collection CCL-25) were cultured with 50% patient sera for 6 hours, and then lysed. Messenger RNA (mRNA) was purified from cell lysates, and cDNA was made from total cellular mRNA. Then cDNA was quantified using real-time PCR with the SYBR Green fluorophore system. Forward and reverse primers for the genes MX1, PKR, and IFIT1, which are known to be highly and specifically induced by IFN-α, were used in the reaction16. GAPDH was amplified in the same samples to control for background gene expression.
The amount of PCR product of the IFN-α-induced gene was normalized to the amount of product for the housekeeping gene GAPDH in the same sample. The relative expression of each of the 3 tested IFN-induced genes was calculated as a fold increase compared to its expression in WISH cells cultured with media alone. Results from the IFN-α assay were standardized to a healthy multiancestral reference population as described, and a serum IFN-α activity score was calculated based upon the mean and SD of the reference population16. This assay has been highly informative when applied to SLE as well as other autoimmune disease populations27,28,29.
Measurement of autoantibodies
Antibodies to anti-Ro, anti-La, anti-Sm, and anti-RNP were measured in all samples by ELISA methods using kits from Inova Diagnostics (San Diego, CA, USA), and anti-dsDNA antibodies were measured using Crithidia luciliae immunofluorescence, with detectable fluorescence considered positive. All samples were assayed in a University of Chicago clinical laboratory. Standard cutoff points designated by the manufacturer for a positive test were used to categorize samples as positive or negative.
Statistical analysis
To account for potential differences related to proportional ancestry in admixed populations, we performed a principal component analysis on data from 12 SNP that confer information about genetic ancestry and were genotyped in all cases and controls as described in Kariuki, et al6, similar to the approach outlined in Parra, et al30. The first principal component in this analysis provided strong separation of self-reported European versus African American ancestry, and this component was included in logistic regression analyses as a covariate to control for any differences in the degree of admixture between cases and controls that could otherwise potentially confound genetic association analyses.
Logistic regression models were used to detect associations between genotype at rs5754217 and cases versus control status, and the presence or absence of each of the 5 tested autoantibodies in the SLE cases (case-case analysis). The IFN-α data was non-normally distributed, and the nonpara-metric Mann-Whitney U test was used to compare quantitative IFN-α data in patients with SLE between genotype subgroups. P values shown here are uncorrected for multiple comparisons. To account for multiple comparisons, we used a threshold p value of 0.01 or smaller to control the family-wise type I error rate at 0.05 using a Bonferroni correction when testing differences in allele frequencies between different patient groups defined by autoantibodies. For the serum IFN-α studies, p values < 0.017 would withstand a Bonferroni correction for the number of comparisons possible between the different genotype groups presented on the graphs in Figures 1 and 2.
Figure 1.
Serum interferon-α in patients with SLE stratified by UBE2L3 rs5754217 genotype. A. African American patients with SLE. B. European American patients with SLE. Serum IFN-α activity is shown on the Y-axis. Lines indicate the median and error bars show the interquartile range. P values were calculated using the Mann-Whitney U test, comparing TT genotype to the GG and GG genotypes (recessive model).
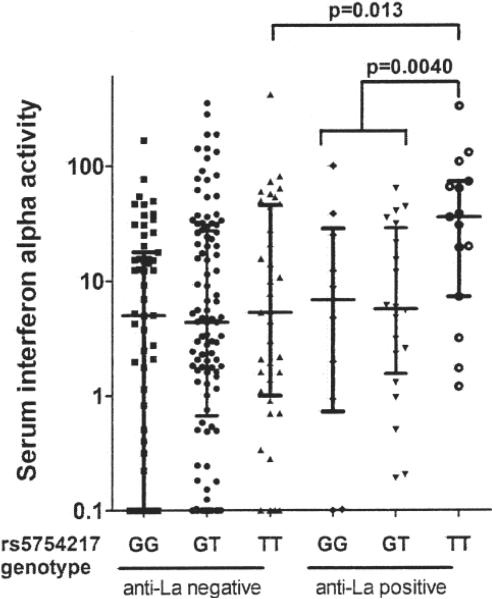
Figure 2.
Serum interferon-α (IFN-α) in African American patients with SLE stratified by UBE2L3 rs5754217 genotype and anti-La autoantibodies. Serum IFN-α activity is shown on the Y-axis. Lines indicate the median and error bars show the interquartile range. P values are calculated using the Mann-Whitney U test.
RESULTS
UBE2L3 rs5754217 T association with SLE in African American patients with anti-La autoantibodies
In logistic regression models, we detected an association between rs5754217 T and anti-La autoantibodies in African Americans, and the recessive model produced the strongest association. Table 1 shows the case-case and case-control analyses examining allele frequencies in anti-La-positive patients versus anti-La-negative patients and anti-La-positive patients versus controls (OR 2.55, p = 0.0061). The other SLE-associated autoantibodies did not show evidence for association with rs5754217 genotype. As shown in Table 1, the majority (63%) of excess T alleles in African American cases were contributed by the relatively small anti-La-positive group (18% of patients). Thus, the anti-La-positive subgroup can largely account for the allele frequency difference between African American SLE cases and controls. A similar nonsignificant tendency was observed in the European American patients with SLE, but the lower prevalence of the allele in European ancestry and the low frequency of anti-La antibodies prevents any definitive comment about whether the anti-La association extends to European Americans. In overall case-control analysis in African Americans, we observed a modest effect size (OR 1.16, p = 0.24) that is similar to the effect observed in European ancestry cohorts to date (OR of 1.20–1.22)1,11. Thus, in African Americans the recessive effect within the anti-La-positive group was clearly a stronger genetic model for UBE2L3 rs5754217 than the overall additive case-control model (OR 2.55, p = 0.0061, vs OR 1.16, p = 0.24).
Table 1.
UBE2L3 rs5754217 genotype and allele frequencies in controls and cases stratified by anti-La autoantibody status.
| Background | Clinical Category | N | GG | GT | TT | MAF (T) | La+ Cases vs LA− Cases, Additive | La+ Cases vs LA− Cases, Recessive | La+ Cases vs Controls, Additive | La+ Cases vs Controls, Recessive |
|---|---|---|---|---|---|---|---|---|---|---|
| African American | Controls | 239 | 74 | 127 | 38 | 0.425 | OR 1.57, p = 0.050 | OR 2.21, p = 0.027 | OR 1.69, p = 0.022 | OR 2.55, p = 0.0061 |
| Cases | 252 | 71 | 129 | 52 | 0.462 | |||||
| La+ Cases | 46 | 10 | 21 | 15 | 0.554 | |||||
| La− Cases | 206 | 61 | 108 | 37 | 0.442 | |||||
| European American | Controls | 105 | 64 | 36 | 5 | 0.219 | OR 1.32, p = 0.47 | OR 3.52, p = 0.27 | OR 1.31, p = 0.46 | OR 1.66, p = 0.38 |
| Cases | 143 | 88 | 51 | 4 | 0.206 | |||||
| La+ Cases | 13 | 7 | 5 | 1 | 0.269 | |||||
| La− Cases | 130 | 81 | 46 | 3 | 0.200 |
La+ indicates patients with a positive test for anti-La antibodies. La− indicates patients who lack anti-La antibodies. MAF: minor allele frequency.
Serum IFN-α is increased in patients with SLE who have the rs5754217 TT genotype
We next examined serum IFN-α in patients with SLE in the context of the UBE2L3 rs5754217 genotype. As shown in Figure 1A, elevated serum IFN-α is observed only in African American subjects with the TT geno-type, and there is no evidence for increased IFN-α in the GT genotype category. This supports a recessive effect of the rs5754217 SNP upon serum IFN-α in African Americans, similar to the effect observed upon anti-La antibodies. European American patients with SLE demonstrated a similar nonsignificant tendency toward a recessive effect upon serum IFN-α (Figure 1B).
IFN-α is higher in anti-La positive individuals with the rs5754217 TT genotype
When serum IFN-α was examined in African American subjects stratified by anti-La autoantibodies, the difference in serum IFN-α related to rs5754217 geno-type was observed only in anti-La-positive subjects (Figure 2). The data again fit a recessive model, and the recessive effect upon IFN-α observed in African American patients illustrated in Figure 1 is wholly restricted to the anti-La-positive subjects. In European Americans, only 1 subject had both anti-La antibodies and TT genotype, so these data are not shown and it is not clear whether this pattern extends beyond African American patients with SLE.
DISCUSSION
These data demonstrate a recessive influence of the rs5754217 T allele upon important pathogenic subphenotypes in SLE in African American patients. While our European American dataset was more limited and the risk allele is less common in people with this ancestral background, we observed similar nonsignificant patterns there as well. The UBE2L3 rs5754217 T allele is characterized by a modest overall association with SLE in European populations, with an OR of about 1.2 in additive model case-control studies1,11 and 1.30 in a family-based study31. A similar effect size was demonstrated in our African American population in overall case-control analysis, while a much greater effect was demonstrated using a recessive model in the anti-La-positive subgroup (OR > 2.5). This recessive effect upon a subgroup of patients would not be strongly detected in case-control study designs. Our findings support the idea that heterogeneity in the pathogenesis of SLE underlies some of the modest effect size and missing heritability observed in case-control genetic studies of SLE to date.
We have shown that a number of genetic polymorphisms are associated with increased serum IFN-α in SLE20,21,22,32. This supports the idea that a number of genetic factors converge upon IFN-α as a common pathogenic mediator. We have also been able to use unbiased genome-wide techniques to identify a number of novel polymorphisms related to both autoantibodies and serum IFN-α in SLE in multiple different ancestral backgrounds7. These studies support the relevance of these subphenotypes in genetic studies of SLE, and also suggest that both of these phenotypes are primary pathogenic factors in SLE. We find a strong recessive influence of UBE2L3 rs5754217 T upon the serum IFN-α phenotype, and interestingly this is also dependent upon anti-La autoantibodies. We have observed this pattern with the SLE risk-associated polymorphisms of interferon regulatory factors (IRF)-5 and IRF-720,22, and both of these IRF proteins function in the IFN-α pathway downstream of endosomal TLR signaling. SLE-associated autoantibodies can activate the endosomal TLR pathway of IFN-α generation33, and this chronic endogenous stimulation of TLR may emphasize the genetic effects of these polymorphisms upon IFN-α generation in patients with SLE in vivo15. It is possible that the autoantibody-dependent influence of UBE2L3 genotype upon serum IFN-α also indicates a role for the UBE2L3 risk variant downstream of endosomal TLR signals in human SLE.
The exact molecular function of the rs5754217 polymorphism and the reason for a recessive association are not clear. It is possible that this SNP is in linkage disequilibrium with a functional element and is not the true causal allele, and future fine-mapping studies will assist in this determination. Regarding the recessive effect, UBE2L3 functions normally in targeting proteins involved in inflammatory signaling for destruction, and the risk allele may result in decreased function of this protein. In that case, it is possible that 1 nonrisk allele could compensate for the risk allele, and a decrease in UBE2L3 function may only occur when 2 risk alleles coincide. This possibility would have to be addressed in future mechanistic studies.
We do not have data regarding disease activity for the patients in our study, and cannot comment upon whether the rs5754217 allele is associated with SLE disease activity. While a cross-sectional association between disease activity and serum IFN-α has been observed34, longitudinal studies of IFN-α in SLE to date have not supported a strong relationship between changes in serum IFN-α and fluctuations in disease activity35,36. In previous work, we have shown that serum IFN-α is heritable within families in which SLE is present, in both affected and unaffected members16, and that serum levels of IFN-α are correlated with fixed genetic factors24,37. These data support the concept of a relatively stable threshold for circulating IFN-α levels in the individual. The potential effect of the UBE2L3 rs5754217 polymorphism upon disease activity, as either a primary or secondary factor, would be an area of interest for future work.
Given our findings, we would predict that larger-scale genetic studies of this locus will confirm an association between rs5754217 T and SLE in African Americans. Additionally, our study will inform future genetic studies of this locus, as we find strong recessive association in the anti-La-positive subgroup that may not be evident in analyses that are not stratified by autoantibodies. The gene-antibody interaction we describe affects pathogenic cytokine levels in SLE, providing some insight into the biological function of this locus in patients with SLE in vivo.
Acknowledgments
Supported by research grants from the Lupus Clinical Trials Consortium to Dr. Utset, and research grants from the National Institutes of Health [K08 AI083790, P30 DK42086, LRP AI071651, and CTSA Pilot Grant from UL1 RR024999], the Lupus Research Institute, the Alliance for Lupus Research, and the Arthritis National Research Foundation Eng Tan Scholar Award to Dr. Niewold.
REFERENCES
- 1.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harley JB, Kelly JA, Kaufman KM. Unraveling the genetics of systemic lupus erythematosus. Springer Semin Immunopathol. 2006;28:119–30. doi: 10.1007/s00281-006-0040-5. [DOI] [PubMed] [Google Scholar]
- 3.Cooper GS, Parks CG, Treadwell EL, Clair EW, Gilkeson GS, Cohen PL, et al. Differences by race, sex and age in the clinical and immunologic features of recently diagnosed systemic lupus erythematosus patients in the southeastern United States. Lupus. 2002;11:161–7. doi: 10.1191/0961203302lu161oa. [DOI] [PubMed] [Google Scholar]
- 4.Chapman SJ, Khor CC, Vannberg FO, Maskell NA, Davies CW, Hedley EL, et al. PTPN22 and invasive bacterial disease. Nat Genet. 2006;38:499–500. doi: 10.1038/ng0506-499. [DOI] [PubMed] [Google Scholar]
- 5.Lodolce JP, Kolodziej LE, Rhee L, Kariuki SN, Franek BS, McGreal NM, et al. African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. J Immunol. 2010;184:7001–9. doi: 10.4049/jimmunol.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kariuki SN, Franek BS, Mikolaitis RA, Utset TO, Jolly M, Skol AD, et al. Promoter variant of PIK3C3 is associated with autoimmunity against Ro and Sm epitopes in African-American lupus patients. J Biomed Biotechnol. 2010;2010:826434. doi: 10.1155/2010/826434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kariuki SN, Franek BS, Kumar AA, Arrington J, Mikolaitis RA, Utset TO, et al. Trait-stratified genome-wide association study identifies novel and diverse genetic associations with serologic and cytokine phenotypes in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R151. doi: 10.1186/ar3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pothlichet J, Niewold TB, Vitour D, Solhonne B, Crow MK, Si-Tahar M. A loss-of-function variant of the antiviral molecule MAVS is associated with a subset of systemic lupus patients. EMBO Mol Med. 2011;3:142–52. doi: 10.1002/emmm.201000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orozco G, Eyre S, Hinks A, Bowes J, Morgan AW, Wilson AG, et al. Study of the common genetic background for rheumatoid arthritis and systemic lupus erythematosus. Ann Rheum Dis. 2011;70:463–8. doi: 10.1136/ard.2010.137174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fransen K, Visschedijk MC, van Sommeren S, Fu JY, Franke L, Festen EA, et al. Analysis of SNPs with an effect on gene expression identifies UBE2L3 and BCL3 as potential new risk genes for Crohn's disease. Hum Mol Genet. 2010;19:3482–8. doi: 10.1093/hmg/ddq264. [DOI] [PubMed] [Google Scholar]
- 11.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet. 2009;41:1228–33. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissman AM. Themes and variations on ubiquitylation. Nat Rev Mol Cell Biol. 2001;2:169–78. doi: 10.1038/35056563. [DOI] [PubMed] [Google Scholar]
- 13.Nuber U, Schwarz S, Kaiser P, Schneider R, Scheffner M. Cloning of human ubiquitin-conjugating enzymes UbcH6 and UbcH7 (E2-F1) and characterization of their interaction with E6-AP and RSP5. J Biol Chem. 1996;271:2795–800. doi: 10.1074/jbc.271.5.2795. [DOI] [PubMed] [Google Scholar]
- 14.Chuang TH, Ulevitch RJ. Triad3A, an E3 ubiquitin-protein ligase regulating Toll-like receptors. Nat Immunol. 2004;5:495–502. doi: 10.1038/ni1066. [DOI] [PubMed] [Google Scholar]
- 15.Salloum R, Niewold TB. Interferon regulatory factors in human lupus pathogenesis. Transl Res. 2011;157:326–31. doi: 10.1016/j.trsl.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8:492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niewold TB, Swedler WI. Systemic lupus erythematosus arising during interferon-alpha therapy for cryoglobulinemic vasculitis associated with hepatitis C. Clin Rheumatol. 2005;24:178–81. doi: 10.1007/s10067-004-1024-2. [DOI] [PubMed] [Google Scholar]
- 18.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–5. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 19.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis Rheum. 2008;58:2481–7. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kariuki SN, Crow MK, Niewold TB. The PTPN22 C1858T polymorphism is associated with skewing of cytokine profiles toward high interferon-alpha activity and low tumor necrosis factor alpha levels in patients with lupus. Arthritis Rheum. 2008;58:2818–23. doi: 10.1002/art.23728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salloum R, Franek BS, Kariuki SN, Rhee L, Mikolaitis RA, Jolly M, et al. Genetic variation at the IRF7/PHRF1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis Rheum. 2010;62:553–61. doi: 10.1002/art.27182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson T, Kariuki SN, Franek BS, Kumabe M, Kumar AA, Badaracco M, et al. Autoimmune disease risk variant of IFIH1 is associated with increased sensitivity to IFN-a and serologic autoimmunity in lupus patients. J Immunol. 2011;187:1298–303. doi: 10.4049/jimmunol.1100857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kariuki SN, Niewold TB. Genetic regulation of serum cytokines in systemic lupus erythematosus. Transl Res. 2010;155:109–17. doi: 10.1016/j.trsl.2009.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weckerle CE, Franek BS, Kelly JA, Kumabe M, Mikolaitis RA, Green SL, et al. Network analysis of associations between serum interferon-alpha activity, autoantibodies, and clinical features in systemic lupus erythematosus. Arthritis Rheum. 2011;63:1044–53. doi: 10.1002/art.30187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–33. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 27.Niewold TB, Adler JE, Glenn SB, Lehman TJ, Harley JB, Crow MK. Age- and sex-related patterns of serum interferon-alpha activity in lupus families. Arthritis Rheum. 2008;58:2113–9. doi: 10.1002/art.23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niewold TB, Kariuki SN, Morgan GA, Shrestha S, Pachman LM. Elevated serum interferon-alpha activity in juvenile dermatomyositis: associations with disease activity at diagnosis and after thirty-six months of therapy. Arthritis Rheum. 2009;60:1815–24. doi: 10.1002/art.24555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niewold TB, Rivera TL, Buyon JP, Crow MK. Serum type I interferon activity is dependent on maternal diagnosis in anti-SSA/Ro-positive mothers of children with neonatal lupus. Arthritis Rheum. 2008;58:541–6. doi: 10.1002/art.23191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Budarf ML, Goyette P, Boucher G, Lian J, Graham RR, Claudio JO, et al. A targeted association study in systemic lupus erythematosus identifies multiple susceptibility alleles. Genes Immun. 2011;12:51–8. doi: 10.1038/gene.2010.47. [DOI] [PubMed] [Google Scholar]
- 32.Harley IT, Niewold TB, Stormont RM, Kaufman KM, Glenn SB, Franek BS, et al. The role of genetic variation near interferon-kappa in systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:706825. doi: 10.1155/2010/706825. Epub 2010 Jul 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lovgren T, Eloranta ML, Bave U, Alm GV, Ronnblom L. Induction of interferon-alpha production in plasmacytoid dendritic cells by immune complexes containing nucleic acid released by necrotic or late apoptotic cells and lupus IgG. Arthritis Rheum. 2004;50:1861–72. doi: 10.1002/art.20254. [DOI] [PubMed] [Google Scholar]
- 34.Kirou KA, Lee C, George S, Louca K, Peterson MG, Crow MK. Activation of the interferon-alpha pathway identifies a subgroup of systemic lupus erythematosus patients with distinct serologic features and active disease. Arthritis Rheum. 2005;52:1491–503. doi: 10.1002/art.21031. [DOI] [PubMed] [Google Scholar]
- 35.Landolt-Marticorena C, Bonventi G, Lubovich A, Ferguson C, Unnithan T, Su J, et al. Lack of association between the interferon-alpha signature and longitudinal changes in disease activity in systemic lupus erythematosus. Ann Rheum Dis. 2009;68:1440–6. doi: 10.1136/ard.2008.093146. [DOI] [PubMed] [Google Scholar]
- 36.Petri M, Singh S, Tesfasyone H, Dedrick R, Fry K, Lal P, et al. Longitudinal expression of type I interferon responsive genes in systemic lupus erythematosus. Lupus. 2009;18:980–9. doi: 10.1177/0961203309105529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weckerle CE, Niewold TB. The unexplained female predominance of systemic lupus erythematosus: Clues from genetic and cytokine studies. Clin Rev Allergy Immunol. 2011;40:42–9. doi: 10.1007/s12016-009-8192-4. [DOI] [PMC free article] [PubMed] [Google Scholar]