Abstract
The LRP (low-density lipoprotein receptor-related protein) can bind a wide range of structurally diverse ligands to regions composed of clusters of ~40 residue Ca2+-dependent, disulfide-rich, CRs (complement-like repeats). Whereas lysine residues from the ligands have been implicated in binding, there has been no quantification of the energetic contributions of such interactions and hence of their relative importance in overall affinity, or of the ability of arginine or histidine residues to bind. We have used four representative CR domains from the principal ligand-binding cluster of LRP to determine the energetics of interaction with well-defined small ligands that include methyl esters of lysine, arginine, histidine and aspartate, as well as N-terminally blocked lysine methyl ester. We found that not only lysine but also arginine and histidine bound well, and when present with an additional proximal positive charge, accounted for about half of the total binding energy of a protein ligand such as PAI-1 (plasminogen activator inhibitor-1). Two such sets of interactions, one to each of two CR domains could thus account for almost all of the necessary binding energy of a real ligand such as PAI-1. For the CR domains, a central aspartate residue in the sequence DxDxD tightens the Kd by ~20-fold, whereas DxDDD is no more effective. Together these findings establish the rules for determining the binding specificity of protein ligands to LRP and to other LDLR (low-density lipoprotein receptor) family members.
Keywords: affinity, complement-like repeat domain (CR domain), ligand specificity, lysine binding, low-density lipoprotein receptor, low-density lipoprotein receptor-related protein (LRP), model compound, thermodynamics
Abbreviations: α2M, α2-macroglobulin; 2-ME, 2-mercaptoethanol; CR, complement-like repeat; GST, glutathione transferase; IPTG, isopropyl β-D-thiogalactoside; LDLR, low-density lipoprotein receptor; LPL, lipoprotein lipase; LRP, low-density lipoprotein receptor-related protein; PAI-1, plasminogen activator inhibitor-1; RAP, receptor-associated protein; RP-HPLC, reverse-phase HPLC; TEV, tobacco etch virus; uPA, urokinase-type plasminogen activator; VLDLR, very-low-density lipoprotein receptor
INTRODUCTION
The LRP (low-density lipoprotein receptor-related protein) is a multidomain mosaic-like receptor present on many mammalian cells [1]. It is one of the largest members of the LDLR (low-density lipoprotein receptor) family of proteins, with a Mw of ~600 kDa for the two-chain processed form. It is a constitutively active cell surface receptor capable of binding and internalizing a large range of structurally and functionally different types of protein ligand, and has been shown to be essential for life [1]. Ligands include members of the serpin family and their proteinase complexes, proteinase complexes of the pan-proteinase inhibitor α2M (α2-macroglobulin), LPL (lipoprotein lipase), β-amyloid precursor protein and RAP (receptor-associated protein). A common feature of all of these proteins and complexes is their ability to bind to heparin, suggesting the presence of one or more highly positively charged regions within each. That this might also be of importance regarding the ability to bind to LRP is suggested by the identification of critical lysine residues for binding of a number of ligands to LRP or other LPL receptor family members [2–5].
Ligand binding occurs to regions of LRP composed exclusively of clusters of up to 11 CR (complement-like repeat) domains, which are flanked at each end by EGF (epidermal growth factor)-like domains and one or more β-propellor, or YWTD, domains. The CR domains are small, ~40 residue, domains that contain three disulfides and a Ca2+-binding site that is required for structural integrity [6] and hence for ligand binding [7,8]. Consistent with the implication of lysine residues being important for ligand binding, as well as with the requirement for Ca2+, an X-ray structure of a fragment of two CR domains from LDLR with one of the three domains of RAP showed direct engagement of each of the two CR domains with lysine residues from the RAP domain, with binding occurring to acidic residues that form part of the Ca2+-binding site or are immediately adjacent to it [9].
Whereas these findings directly implicate lysine engagement as being involved in ligand binding by LRP and other LDLR family members, they do not establish how great the energetic contribution of such interactions is to the overall affinity of a complex. It is therefore unresolved whether these interactions play a dominant role in binding affinity or are of lesser importance, with other contributions making up most of the binding energy, and being variable from domain to domain. Also unresolved are whether arginine or histidine residues can also interact as effectively as lysine and whether all CR domains are equivalent in their ability to bind lysine. This latter point is an important consideration, since ligand binding to LRP has been reported to occur preferentially to only two of the four clusters of CR domains [10], and even to show some preference within a given cluster for certain groupings of domains [4,11]. There is semi-quantitative evidence to suggest that such a preference is related to distinct patterns of acidic residues [12]. Of the three patterns, the most common involves a DxDxD motif for the pentapeptide that occurs between the fourth and fifth cysteine residues of the domain. A less common motif involves an additional aspartate or glutamate residue as the fourth residue in the pentapeptide, whereas another motif, which interestingly is more common in the cluster of CR domains that binds the fewest ligands (cluster III), is DxxxD [10,13]. How tightly does each of these motifs engage lysine residues?
With the goal of answering each of these three questions and thereby of shedding light on the nature and basis of ligand specificity to LDLR family members, we have employed a simple model system to systematically examine the thermodynamics of interaction of positively charged residues with representative CR domains from LRP. Using single CR domains that contain each of the three patterns of acidic residues described above, and simple amino acid ligands, we have dissected the contributions to binding affinity from positive charges on the ligand and acidic residues of the CR domain. Our findings establish a preponderant contribution to ligand binding from engagement of a CR domain that contains a DxDxD or DxDDD motif with a ligand that has a proximal pair of positive charges, though these charges may be contributed by lysine, arginine or even protonated histidine side chains. Although the aspartate residue at position 4 in the pentapeptide makes little difference to affinity, the loss of the middle aspartate residue reduces the energy of interaction by ~7 kJ/mol. These results have major consequences for the affinity of ligands and the specificity of an LDLR family member.
EXPERIMENTAL
Expression, purification and refolding of CR constructs
CR3, CR8 and CR8 D1085A were cloned into pGEX-2T and expressed as GST (glutathione transferase)-fusion proteins in Escherichia coli BL21 cells in 2YT medium [1.6% (w/v) tryptone, 1% (w/v) yeast extract and 0.05% NaCl]. Cells were grown to D600=0.6–1.0 before induction with 1 mM IPTG (isopropyl β-D-thiogalactoside), and the cells were harvested after 5–6 h at 37°C. The GST-fusion proteins were purified from the cleared cell lysate by GSH–Sepharose chromatography, and the GST tag was removed by thrombin cleavage [1:5000 dilution at room temperature (21°C) for 1 h] after overnight dialysis against 4 litres 20 mM Tris/HCl, pH 8.0, 50 mM a NaCl and 4 mM EDTA, containing 7 mM 2-ME (2-mercaptoethanol). GST and uncleaved GST–CR fusion proteins were removed by passage through the GSH column.
CR5 was cloned into pQE-30, modified to contain a TEV (tobacco etch virus) cleavage site. The His6-tagged fusion protein was expressed in E. coli SG13009 cells containing the plasmid pRARE. Cells were grown to a D600 of 0.6–1.0 before induction with 1 mM IPTG, and the cells were harvested after 5–6 h at 37°C. CR5 was purified from cell lysate by Ni2+ chromatography, and the His6-tag was removed by TEV cleavage (1:10000 dilution) during overnight dialysis against PBS containing 14 mM 2-ME.
All CR domains were purified and concentrated by Q-Sepharose HP chromatography, using a gradient of 0–1000 mM NaCl in 20 mM Tris/HCl, pH 8.0, 6 M urea and 0.1% 2-ME. The denatured domains were further purified by RP-HPLC (reverse-phase HPLC) on a Discovery wide-pore C18 column (21.2 mm×150 mm), using a linear gradient of 10–60% acetonitrile in 0.1% TFA (trifluoroacetic acid) over 60 min at 4 ml/min.
The reduced CR domains were diluted with 50 mM Tris/HCl, pH 8.5, 50 mM NaCl and 10 mM CaCl2, containing 14 mM 2-ME and 8 mM 2-hydroxyethyl disulfide (refolding buffer). Final CR concentration in the refolding mixture was 0.1 mg/ml. The CR species were refolded by dialysis against refolding buffer for 24 h at room temperature with N2 bubbling, followed by 24 h at 4°C without N2. Finally, the CR domains were dialysed against 4 litres of 20 mM Tris/HCl, pH 7.8, 50 mM NaCl and 5 mM CaCl2 and 4 litres of 50 mM sodium acetate, pH 5.0, 50 mM NaCl and 5 mM CaCl2, at 4°C to remove the redox buffer. Folded CR domains were purified by RP-HPLC on a Discovery wide-pore C18 column as described above. Concentrations of the CR stock solutions were determined spectrophotometrically, using the molar absorption coefficient (ϵ) based on the presence of one tryptophan residue and three disulfides in each domain and an additional tyrosine in the linker for CR5. This gave values of 5.86×103 M−1·cm−1 for CR3, CR8 and D1085A and of 6.01×103 M−1·cm−1 for CR5.
Fluorescence measurements
All fluorescence measurements were made on a PTI Quantamaster spectrofluorimeter, equipped with double monochromators on both the excitation and emission sides. λex was at 280 nm for both complete emission spectra and measurements of emission intensity at a single wavelength, used in most ligand titrations. For single-wavelength measurements, the λem was chosen as the wavelength of maximum change, on the basis of previously recorded spectra in the presence of saturating ligand and in its absence. This was usually ~347 nm, but in some cases was as low as 336 nm or as high as 371 nm, and depended on whether there was a large change in quantum yield as well as a shift in emission maximum. For emission spectra, slits of 1 nm for λex and 2 nm for λem were used. For single-wavelength measurements, slits of 1 nm for λex and 8–10 nm for λem were used.
Samples of 1.1 ml in 1-cm-pathlength cuvettes contained 1–3 μM CR domain, depending on the fluorescence intensity of the domain. Experiments at pH 5.5 were in buffer containing 20 mM Mes, 1 mM Ca2+ and 0.1% PEG [poly(ethylene glycol)] 8000, whereas those at pH 7.4 had 20 mM Tris/HCl instead of Mes. Additional NaCl was present according to the desired final ionic strength. Titrations with ligand were made from stock solutions of 0.05, 0.1, 1 or 2 M, depending on the anticipated affinity. Stock solutions were at the same pH as the protein solution.
Emission spectra were recorded from 310 to 450 nm in 2 nm steps, with 6 s dwell time. Single-wavelength measurements at a given ligand concentration were made over 120 s and averaged. Data in each case were corrected for contributions from a buffer blank. Correction to the fluorescence intensity was also made for any dilution of the protein resulting from addition of ligand. In most cases, such dilution was less than 2%. Temperature was maintained by a circulating water bath. All titrations were carried out at 298 K, except for those needed for the van't Hoff plot.
Kd values were determined by non-linear least-squares fitting of the binding data to a simple single site-binding isotherm. Fitting was carried out using Kaleidagraph (Synergy Software). Except in a few instances where the fluorescence change was small (~10%), all fits were excellent, with very small errors (see Tables 1–3). Between nine and thirteen different concentrations were used for each titration, which usually extended to ~5 times the Kd value, except for the very weakest-binding ligand, aspartate methyl ester, where this was impossible to achieve. For titrations of lysine and arginine, methyl esters at pH 7.4 and for histidine methyl ester at pH 5.5, an adjustment to the concentration was made to take into account the fraction of the ligand that had protonated α-amino group (lysine and arginine methyl esters) or protonated side chain (histidine methyl ester). pKa values of 7.2 for lysine and arginine methyl esters [14] and 5.33 for histidine methyl ester [15] were used.
Table 1. Affinity of Nα-acetyl-lysine methyl ester for different cluster II CR domains as a function of pH and ionic strength (I).
n.d., not determined.
| CR3 (DxDxD) | CR5 (DxDDD) | CR8 (DxDxD) | D1085A (DxxxD) | |||||
|---|---|---|---|---|---|---|---|---|
| Kd (mM) | ΔG° (kJ/mol) | Kd (mM) | ΔG° (kJ/mol) | Kd (mM) | ΔG° (kJ/mol) | Kd (mM) | ΔG° (kJ/mol) | |
| I=0.02 | ||||||||
| pH 5.5 | 2.9±0.12 | −14.5 | 1.8±0.16 | −15.7 | 3.1±0.44 | −14.3 | 12.8±2.0 | −10.8 |
| pH 7.4 | 1.9±0.09 | −15.5 | 1.6±0.15 | −15.9 | 1.8±0.19 | −15.7 | 12.7±2.9 | −10.8 |
| I=0.17 | ||||||||
| pH 5.5 | 2.9±0.28 | −14.5 | 2.3±0.49 | −15.0 | 2.6±0.28 | −14.7 | n.d. | n.d. |
| pH 7.4 | 2.4±0.12 | −14.9 | 2.5±0.32 | −14.8 | 3.1±0.25 | −14.3 | n.d. | n.d. |
RESULTS
Choice of CR domains
LRP contains 31 CR domains organized into four clusters. Counting from the N-terminal end, these clusters are numbered I through IV and contain 2, 8, 10 and 11 CR domains respectively [16]. Nearly all ligand binding has been reported to occur to clusters II and IV, with each cluster able to bind a similar repertoire of ligands [10]. We therefore focused the present study on CR domains from cluster II, as they are likely to be representative of ligand-binding domains. We further restricted our study to domains that correspond to the different patterns of aspartate residues present between the fourth and fifth cysteine residues of the CR domains from both clusters II and IV. Within cluster II the motif DxDxD is present in CR3, CR4, CR6, CR7, CR8 and CR9 domains. The motif DxDDD is present only in CR5, whereas the motif DxxxD is present only in CR10 (Figure 1). The significance of this pentapeptide sequence, and hence its relevance to the present study, is that the first and last aspartate residues co-ordinate the essential Ca2+ in all CR domain structures determined to date, whereas the backbone carbonyl of the central ‘x’ residue co-ordinates the Ca2+, thereby leaving the carboxy group in the side chain free to potentially interact with ligand. A similar preponderance of domains (eight out of eleven) with either the middle ‘x’ alone, or both the middle and fourth residues being aspartate (or glutamate) is found in cluster IV. In contrast, only 4 out of 10 domains from cluster III, which is reported not to bind most ligands, contain three or four acidic residues, suggesting an important relationship between three or more acidic residues in this pentapeptide and the ability to bind either a single ligand or a range of ligands. We therefore chose CR3, and for comparison CR8, as representative domains with the DxDxD motif and CR5 as representative of the DxDDD motif. Although CR10 would have been the appropriate choice for the DxxxD motif, the absence of a tryptophan residue from this domain, which we used as a non-perturbing endogenous reporter for ligand binding in the other domains, made this an unsuitable choice. Rather than making a drastic change to CR10 by mutating the corresponding lysine residue to tryptophan (see Figure 1), we created a variant of CR8 in which the central ‘x’ was mutated from aspartate to alanine. Thus our choice of CR10 surrogate was a CR8 variant, D1085A.
Figure 1. Pattern of residues between the third and fifth cysteine residues of the eight CR domains from cluster II of LRP.
The critical pentapeptide that provides two aspartate carboxy group and a backbone carbonyl group to co-ordinate Ca2+ lies between the fourth and fifth cysteine residues. The tryptophan (bold) used as a fluorescent reported and which also co-ordinates Ca2+ through its backbone carbonyl lies two residues N-terminal to the fourth cysteine. The three aspartate residues constituting the DxDxD motif are shown in bold against a grey background, whereas the additional aspartate residue that is present in CR5 is bold and underlined. The remaining Ca2+-co-ordinating residues are a conserved aspartate residue and adjacent glutamate residue that lie between the fifth and sixth cysteine residues (not shown).
Fluorescence approach to monitoring ligand binding
We have shown previously that binding of the protein ligands PAI-1 (plasminogen activator inhibitor-1) [17], α2M receptor binding domain [18] and domains from the RAP [19] to double and triple CR domain fragments of LRP results in a characteristic blue shift of the tryptophan fluorescence that is most likely to be associated with the LRP component rather than the protein ligand. Indeed, such a blue shift has also been seen for binding of an ApoE (apolipoprotein E) fragment, which contains no tryptophan residues, to CR17 of LRP [20]. This is understandable given the known co-ordination of the CR domain tryptophan carbonyl to the essential Ca2+, and the proximity of the indole ring to lysine residues from the ligand in the known X-ray structures of CR complexes with protein ligands. In addition, such fluorescence perturbations for binding of LRP fragments to protein ligands correlated with gel shifts on native PAGE and species that sedimented as well-defined 1:1 complexes in the analytical ultracentrifuge [19].
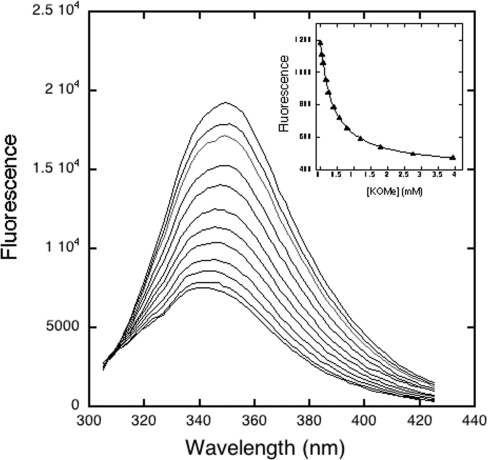
We found that binding of our simple amino acid ligands to the four CR domains gave saturable perturbation of CR fluorescence, as expected from a specific binding interaction. Binding almost always resulted in sufficient perturbation of the CR domain tryptophan fluorescence to permit accurate monitoring of ligand binding. Depending primarily on whether one or two positive charges were present in the ligand, the fluorescence perturbation consisted of a quench in addition to a blue shift. This can clearly be seen for the binding of lysine methyl ester to CR8, which gave a 66% quench at saturation and a blue shift of approximately 7 nm (Figure 2). These perturbations varied with ligand and domain, requiring appropriate adjustment of the optimal wavelength for observing the maximum perturbation caused by ligand association.
Figure 2. Fluorescence emission spectra of CR8 as a function of added lysine methyl ester, at pH 7.4, I=0.17 and 294 K.
The highest intensity spectrum is CR8 alone, whereas addition of ligand caused progressive reduction in intensity and blue shift. The final ligand concentration, corrected for the doubly protonated form, was 3.9 mM. Inset is a non-linear least-squares fit of these data, using the integrated area of each spectrum, to a single binding isotherm, with a resulting Kd of 0.37 mM and maximum quench of 66%.
Is there specificity of binding to different domains?
To examine whether there is preferential binding of ligand to the four different CR domains, we examined binding of the simplest ligand, Nα-acetyl-lysine methyl ester, to each domain. Since both the α-amino and carboxy groups are blocked, this should closely correspond to an accessible lysine side chain on a protein ligand. For the three domains that contain a central aspartate residue (CR3, CR5 and CR8) binding was very similar for a given CR domain for a given set of conditions of ionic strength and pH and showed relatively small change when compared at two ionic strengths and at pH 5.5 and 7.4 (Table 1). The whole range of Kd values was from 1.6 to 3.1 mM (Table 1), corresponding to ΔG° values of −15.9 to −14.3 kJ/mol. Only for the CR10 surrogate (D1085A), which lacked the central aspartate, was binding clearly weaker, with a Kd value 6–9-fold higher (Table 1), corresponding to a loss of binding energy of 3.5–5.1 kJ/mol. Of great interest, CR5, which contains a fourth aspartate residue, bound Nα-acetyl-lysine methyl ester under physiological conditions of pH and ionic strength with almost the same affinity as CR3 and CR8, each of which lacked this additional aspartate residue.
Does a second proximal positive charge modify affinity or specificity?
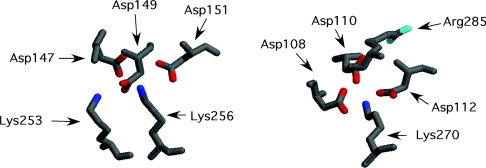
Although the X-ray structure of RAP D3 with the two domain CR construct LB3–LB4 from LDLR demonstrated the clear engagement of a lysine side chain by carboxy groups surrounding the Ca2+-binding site, it also showed that a second positively charged residue from the ligand was very close by and also interacted with these aspartate residues (Lys253 to LB4, and, somewhat further away, Arg285 to LB3) (Figure 3). By using lysine methyl ester, in which the α-amino group is no longer blocked, we were able to examine the effect on binding of such a second positive proximal positive charge. Binding was examined at both pH 5.5 and 7.4, though a correction needed to be made to the Kd values determined at pH 7.4 for the fraction of the α-amino group in the protonated state, since the reported pKa of lysine methyl ester is 7.2.
Figure 3. Engagement of positive charges on RAP D3 by LB3–LB4 from LDLR (from PDB code 2FCW [9]).
Only the interacting aspartate residues from the two CR domains (LB4, left, and LB3, right) and lysine and arginine residues from the RAP D3 ligand that engage them are shown. Each CR domain has three aspartate residues in a DxDxD motif, all of which are separately engaged in coordinating to lysine ϵ-amino groups or possibly the arginine guanidinium group. In this structure, the ϵ-amino of Lys253 engages one aspartate carboxy group (Asp149), whereas the ϵ-aminos of Lys256 and Lys270 interact with all three aspartate carboxy groups. The separations between carboxy oxygens and lysine nitrogens are as follows: Lys270–Asp108, 2.82 Å (1 Å=0.1 nm); Lys270–Asp110, 2.90 Å; Lys270–Asp112, 2.79 Å; Lys256–Asp147, 2.66 Å; Lys256–Asp149, 2.83 Å; Lys256–Asp151, 2.93 Å; Lys253–Asp149, 2.75 Å. In addition, the guanidine of Arg285 is within 3.06 Å of the carboxy group of Asp110. Side chain nitrogens are shown in blue (lysine) or cyan (arginine), carboxy oxygens are in red and other atoms in grey. As a measure of the distance between the binding sites on the two CR domains, the separation between the ϵ-amino of Lys256 (co-ordinated to domain LB4) and the ϵ-amino of Lys270 (co-ordinated to domain LB3) is 21 Å. The indole side chain of Trp144 from domain LB4 (results not shown for clarity) stacks with the methylene groups of the Lys256 side chain.
For each of the three CR domains that contain a middle aspartate residue, the affinity for the ligand was greatly enhanced, at both pH values, compared with Nα-acetyl-lysine methyl ester, with a 5–18-fold reduction in Kd at low ionic strength (I=0.02), corresponding to an increase in ΔG° of 4.3–7.2 kJ/mol (Table 2). Depending on the pH and ionic strength, there was also discrimination between binding of lysine methyl ester to the three domains, with the largest variation occurring at low pH and low ionic strength (~3-fold difference in Kd). However, this variation did not correlate with the number of aspartate residues in the pentapeptide, since CR5 with four and CR8 with three bound more tightly than CR3, which has three. At physiological pH and ionic strength, binding affinities were very similar for all three domains. Even D1085A showed enhanced binding for lysine methyl ester compared with Nα-acetyl-lysine methyl ester, though the Kd value was decreased less than 4-fold at pH 5.5 and I=0.02. This Kd of 3.7 mM represents a greatly weakened interaction compared with the value of 0.17 mM for the parent CR8 (Table 2).
Table 2. Affinity of lysine and arginine methyl esters to CR domains with different patterns of acidic residues.
| Lysine methyl ester | Arginine methyl ester | |||||
|---|---|---|---|---|---|---|
| Kd (mM) | ΔG° (kJ/mol) | ΔGu (kJ/mol) | Kd (mM) | ΔG° (kJ/mol) | ΔGu (kJ/mol) | |
| CR3 (DxDxD) | ||||||
| I=0.02 | ||||||
| pH 5.5 | 0.53±0.05 | −18.7 | −28.6 | 0.68±0.07 | −18.1 | −28.0 |
| pH 7.4 | 0.22±0.03 | −20.8 | −30.7 | 0.76±0.06 | −17.8 | −27.7 |
| I=0.17 | ||||||
| pH 5.5 | 1.19±0.11 | −16.7 | −26.6 | 2.09±0.16 | −15.3 | −25.2 |
| pH 7.4 | 0.37±0.04 | −19.6 | −29.5 | 0.78±0.04 | −17.7 | −27.6 |
| CR5 (DxDDD) | ||||||
| I=0.02 | ||||||
| pH 5.5 | 0.18±0.01 | −21.4 | −31.2 | 0.29±0.01 | −20.2 | −30.1 |
| pH 7.4 | 0.28±0.01 | −20.3 | −30.2 | 0.58±0.03 | −18.5 | −28.3 |
| I=0.17 | ||||||
| pH 5.5 | 0.60±0.04 | −18.4 | −28.3 | 0.84±0.05 | −17.5 | −27.4 |
| pH 7.4 | 0.40±0.02 | −19.4 | −29.3 | 0.76±0.08 | −17.8 | −27.7 |
| CR8 (DxDxD) | ||||||
| I=0.02 | ||||||
| pH 5.5 | 0.17±0.01 | −21.5 | −31.4 | 0.21±0.01 | −21.0 | −30.9 |
| pH 7.4 | 0.18±0.01 | −21.4 | −31.2 | 0.37±0.02 | −19.6 | −29.5 |
| I=0.17 | ||||||
| pH 5.5 | 0.57±0.02 | −18.5 | −28.4 | 0.78±0.01 | −17.7 | −27.6 |
| pH 7.4 | 0.35±0.02 | −19.7 | −29.6 | 0.56±0.02 | −18.5 | −28.4 |
| D1085A (DxxxD) | ||||||
| I=0.02 | ||||||
| pH 5.5 | 3.7±0.13 | −13.9 | −23.8 | 2.8±0.11 | −14.6 | −24.5 |
| pH 7.4 | 3.9±0.27 | −13.7 | −23.6 | 5.0±0.5 | −13.1 | −23.0 |
Do other side chains bind?
Two other amino acid side chains possess both the positive charge and hydrophobic moiety of lysine and so might be expected to bind to CR domains with good affinity. These are histidine at pHs where it is protonated and arginine. Arginine methyl ester bound to all four domains with mostly slightly weaker affinities than did lysine methyl ester, though by not more than 1.8 kJ/mol (Table 2). Although histidine methyl ester also bound, measurements were possible under fewer conditions, since the low side chain pKa of 5.33 meant that, even at pH 5.5, only ~40% of the imidazole is protonated, whereas at pH 7.4 it is largely unprotonated. At pH 5.5 and low ionic strength, and making correction for the concentration of protonated side chain, histidine methyl ester bound only 1.1–2.6-fold weaker than did lysine methyl ester to any of the four domains, corresponding to ΔΔG°s of 0.6–2 kJ/mol (Table 3).
Table 3. Affinity of other amino acid methyl esters for the four cluster II CR domains.
n.d., not determined.
| Histidine methyl ester | Aspartate methyl ester | Alanine methyl ester | ||||
|---|---|---|---|---|---|---|
| Kd (mM) | ΔG° (kJ/mol) | Kd (mM) | ΔG° (kJ/mol) | Kd (mM) | ΔG° (kJ/mol) | |
| CR3 | ||||||
| pH 5.5 | 1.2±0.07 | −16.7 | 83±17 | −6.2 | n.d. | |
| pH 7.4 | n.d. | 66±11 | −6.7 | n.d. | ||
| CR5 | ||||||
| pH 5.5 | 0.33±0.01 | −19.9 | 59±2 | −7.0 | 4.1±0.24 | −13.6 |
| pH 7.4 | n.d. | 63±5 | −6.8 | n.d. | ||
| CR8 | ||||||
| pH5.5 | 0.29±0.01 | −20.2 | 78±3 | −6.3 | 4.9±0.25 | −13.2 |
| pH 7.4 | n.d. | 119±17 | −5.3 | n.d. | ||
| D1085A | ||||||
| pH 5.5 | 4.6±0.05 | −13.3 | 235±15 | −3.6 | n.d. | |
| pH 7.4 | n.d. | 263±19 | −3.3 | n.d. | ||
As controls, binding of the methyl esters of alanine and aspartate residues was also examined under selected conditions. For alanine, the elimination of the positive charge and much of the hydrophobic portion of the side chain resulted in weakening of the Kd value 23- and 28-fold for CR5 and CR8 respectively (ΔΔG° of 7.8–8.3 kJ/mol) (Table 3). It was not possible to measure the Kd value for CR3 since the fluorescence perturbation was too small to permit reliable monitoring of binding. For aspartate the affinities were, not surprisingly, even weaker, with Kdvalues that were 65–450-fold weaker than for lysine methyl ester (ΔΔG° of 10.3–15.2 kJ/mol) (Table 3). Here, it is likely that the side chain orients such that it points away from the acidic cluster surrounding the Ca2+-binding site and so does not contribute at all to binding, leaving only the α-amino group to interact. That this extremely weak binding was nevertheless probably specific was shown by repeating the titrations in the presence of EDTA, to remove the Ca2+ from the CR domain and make it incapable of ligand binding. Under such conditions, no perturbation of the CR domain tryptophan fluorescence was observed with aspartate (results not shown).
Finally, we also examined how blocking the α-carboxy group influenced affinity. For CR3, CR5 and CR8, we compared the affinity of lysine methyl and ethyl esters and found negligible difference in binding affinity, irrespective of the ionic strength or pH used (results not shown). In contrast, unmodified lysine, with a free carboxy group, bound to CR8 with a 6-fold weaker Kd value than did lysine methyl ester (Kd of 1.05 mM compared with 0.17 mM at pH 5.5 and I=0.02). This might be due either to a zwitterionic interaction between α-amino and carboxy groups precluding other interactions with the CR domain, or else the favourable interaction of the α-amino with the CR domain being offset by a negative interaction from the α-carboxy group and aspartate residues on the CR domain.
How many charge–charge interactions are involved?
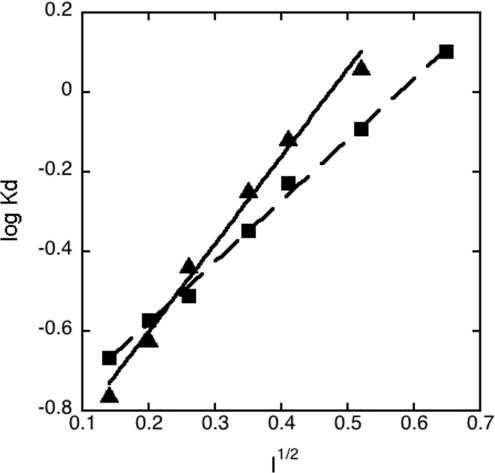
Given the higher affinity of ligands containing both a positively charged side chain and a positively charged α-amino group, we expected that both groups are involved in favourable charge–charge interactions with groups on the CR domain. To quantify this, we determined the ionic-strength-dependence of binding of lysine methyl ester to CR8, at both pH 5.5 and 7.4. This gave a linear Debye–Hückel plot at both pH values, with slopes (−zA×zB) of 2.2 at pH 5.5 and of 1.5 at pH 7.4, roughly consistent with interaction between two exposed pairs of singly positively and negatively charged species (Figure 4). The extrapolated affinities at I=0 were 91 μM at pH 5.5 and 130 μM at pH 7.4, when corrected for the concentration of protonated ligand.
Figure 4. Debye–Hückel plot for binding of lysine methyl ester to CR8 at pH 5.5 and 7.4.
Fits to the data at pH 5.5 (▲) and 7.4 (■) gave slopes of 2.20 and 1.53 respectively.
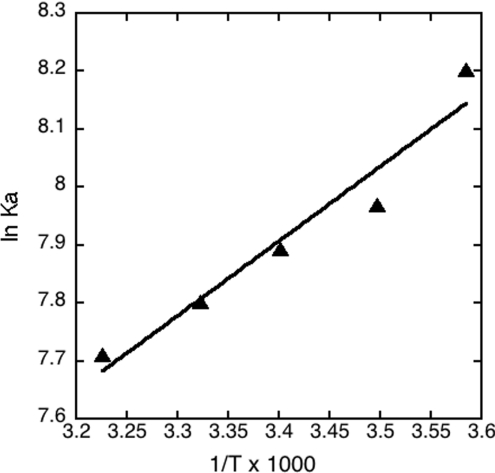
Is binding enthalpically driven?
To better understand the separate contributions of ΔH and ΔS to binding, we examined the temperature-dependence of binding of lysine methyl ester to CR8 at pH 7.4 and physiological ionic strength. A van't Hoff plot over the temperature range of 279–310 K was linear, with a positive slope corresponding to an exothermic binding reaction with ΔH° of −10.7 kJ/mol (Figure 5). Using the interpolated Kd value of 0.38 mM at 298 K, this gives ΔG° of −19.5 kJ/mol and ΔS° of 29 J·K−1·mol−1. Thus both enthalpy and entropy contribute about equally to binding, despite the unfavourable entropy contribution of bringing ligand and receptor together to form one species. It should be noted that the ΔG under physiological conditions of 310 K and I=0.17 is −19.1 kJ/mol.
Figure 5. van't Hoff plot for binding of lysine methyl ester to CR8 at pH 7.4 and I=0.17.
DISCUSSION
Using a model system of individual CR domains in solution and very simple defined ligands we have explored the specificity, thermodynamics and pH-dependence of ligand binding to representative domains from one of the two principal ligand binding regions of LRP. In so doing, we have discovered some common features that should allow general conclusions to be made with regard to the requirements for binding of protein ligands not only to LRP, but also to other LDL family receptors, or even other unrelated proteins that contain CR domains, such as members of the transmembrane serine proteinase family [21,22].
Dominant importance of two proximal positive charges
We found that each of the three CR domains that contain a central aspartate residue in the intra-cysteine pentapeptide sequence DxXxD binds a single lysine side chain with very similar affinity, corresponding to ΔG° of approximately −15 kJ/mol. An additional aspartate residue at position 4, had minimal beneficial effect, whereas loss of the central aspartate (D1085A) reduced affinity compared with its parent CR8 by ~4-fold, corresponding to a reduction in ΔG° of approximately 3 kJ/mol.
Although a single positive charge thus made a major contribution to binding energy, we found that a second, immediately adjacent, positive charge, here exemplified by an α-amino group, added significantly to overall affinity for each of the four CR domains, so that such pairs of positive charges are likely to be highly preferred binding loci on protein ligands compared with single lysine residues. The contribution of two pairs of charge–charge interactions to binding is supported by the ionic-strength-dependence measurements of CR8 to lysine methyl ester, which gave a Debye–Hückel plot at pH 5.5 with a slope of 2.2.
Comparison of the affinities of lysine methyl ester and Nα-acetyl-lysine methyl ester at pH 5.5 shows that the second amino group contributes between 3.1 and 7.1 kJ/mol, depending on the domain. Interestingly, the largest and smallest contributions are for CR8 and its D1085A variant respectively, suggesting that the central aspartate residue contributes not only ~3 kJ/mol to binding the ϵ-amino group (see above) but an additional ~4 kJ/mol to the second positive charge. This is also consistent with the ~3-fold weaker Kd of the D1085A variant than of CR8 for aspartate methyl ester, where only the ‘second’ positive charge is present. Furthermore, since the Kd value of D1085A for lysine methyl ester is ~3-fold higher than for Nα-acetyl-lysine methyl ester, this suggests that interaction of the second positive charge with acidic residues other than the central aspartate contribute ~3–4 kJ/mol. These conclusions highlight a fundamental difference between those CR domains that contain the central aspartate, abundant in ligand-binding clusters II and IV of LRP, and those that do not. Other things being equal, a CR domain possessing the central aspartate should engage a proximal pair of positive charges with ~7 kJ/mol more binding energy than one lacking the aspartate residue. These findings provide quantification of the earlier more qualitative observations of Andersen et al. [12].
An additional important finding is that, for those CR domains that do contain a central aspartate residue, the variation in affinity, at least at physiological ionic strength and pH, between different domains is quite small when engaging two positive charges, and represents a modulation of the overall ΔG° of less than 5% (Table 2). This is despite the large variation in sequence of these domains outside of the Ca2+-binding residues and residues of the inter-cysteine pentapeptide.
Arginine and histidine also interact well
Our results also show that both arginine and protonated histidine side chains can engage all CR domains, with affinities only slightly lower than those of lysine to the same domain. This suggests that appropriately accessible arginine and histidine side chains, which contain both positive charges and large hydrophobic moieties, could also serve as effective recognition elements, either in combination with a second lysine residue or even another arginine or histidine residue as the second proximal positive charge. This is supported by some studies of protein–receptor interactions. In addition to the interaction of Arg285 with Asp110 in the RAPLB3–LB4 structure mentioned above, the X-ray structure of the ectodomain of LDLR, in which the β-propellor, YWTD, domain binds intramolecularly to two of the seven CR domains, shows engagement not only of Lys560 of the YWTD domain by Asp147 and Asp149 of CR domain 4, but also of His586 of the YWTD by Asp149 and Asp151 [23]. In this case, the proximal positively charged Lys560–His586 pair seems to be the one responsible for binding to CR domain 4, with the expectation that this will be much weaker at higher pH when the histidine residue deprotonates. A previous study examined the effects of mutations of both selected arginine and lysine residues of PAI-1 on the ability of its complex with uPA (urokinase-type plasminogen activator) to bind to three different LDLR family receptors, VLDLR (very-low-density lipoprotein receptor), LRP and sorLA [4]. Mutation of either Arg78 or Arg120 separately reduced binding to all three receptors, with affinity reduced by over 90% to both VLDLR and LRP. Mutation of other arginine residues also reduced binding, though by lesser amounts. In contrast, however, with these clear demonstrations of the importance of arginine or histidine residues, a recent study on the binding of RAP D3 domain by LRP found that arginine could not substitute well in binding experiments for the three lysine residues [24] found to interact with CR domains 3 and 4 of LDLR [9,25], though strangely the substitution did not adversely affect uptake of intact RAP.
pH-dependence of binding
Although there is much evidence for a strong pH-dependence of ligand association with members of the LDLR family, we found mostly small differences in affinity for a given ligand–CR pair between pH 5.5 and 7.4 at physiological ionic strength. Binding is favoured at pH 7.4, but the effects are relatively modest, with differences in ΔG° ranging from 0.3 (arginine methyl ester to CR5) to 2.9 kJ/mol (lysine methyl ester to CR3). Consequently, protein ligands that use pairs of lysine or arginine residues to bind to LDLR family receptors should be able to bind with only a small variation in affinity between extracellular and endosomal pHs. This is indeed what we found with several protein ligands binding to two- and three-domain fragments of LRP's cluster II [18,19].
Thermodynamics of binding
Although charge–charge interactions have been shown above to be major contributors to binding, the large positive ΔS for lysine methyl ester binding to CR8 suggests that displacement of ordered water molecules, either from the hydrophobic portions of the lysine side chain of the ligand or the tryptophan residue of CR8, or from the polar side chains that interact in the complex, also contribute. Such major contributions of ΔS to binding have been seen in the somewhat analogous binding studies on lysine and related species to kringle domains from plasminogen. There it was found that ΔS made a major favourable contribution to binding, for binding to both kringles 1 and 4, with the largest contribution arising from the ligand with the longest hydrophobic moiety (7-aminoheptanoic acid) [26,27].
Consequences for ligand binding to LRP and other LDLR family members
The above findings have major implications for protein ligand binding to LRP and other LDLR family members. For the subsequent discussion of CR domain binding contributions unitary free energies, ΔGu, rather than ΔG° are used, since they eliminate the unfavourable entropic component of simply bringing two binding partners together to form one complex [28]. Consider two ligands reported to bind to LRP with high affinity, PAI-1 with a Kd of ~30 nM and RAP domain D3 with a Kd of ~3 nM. These correspond to ΔGu values of −52.8 and −58.5 kJ/mol respectively. For single CR domains that contain a central aspartate residue in the critical pentapeptide region, e.g. CR3, CR5 or CR8, the ΔGu values for binding a CR domain through engagement of a pair of positive charges range from −27.7 to −31.2 kJ/mol at pH 7.4 and I=0.17. This is already close to half of the required binding energy for the real protein ligands. It should also be noted that the fixed distance relationship of the positive charge on the side chain and the α-amino group in our model studies may result in underestimation of the binding interaction, compared with two side chains in a real protein ligand, which might have greater freedom of movement and thus of more optimal positioning. Consequently, a pair of linked CR domains, each of which contains central aspartate residues in the pentapeptide region, could readily generate binding energy of up to −62 kJ/mol and thus potentially form a high affinity binding site for PAI-1 or RAP such as is found experimentally. In addition, since the requirement for a DxDxD motif for high affinity binding is met by many adjacent CR domains within the principal ligand-binding clusters of receptors such as LRP and VLDLR, there may be a number of overlapping high affinity ligand-binding sites within the cluster, thus giving even tighter binding by this statistical factor within the intact receptor.
Significantly, this multiplicity of sites is exactly what we found previously when examining the binding of PAI-1, PAI-1–uPA complexes, PN-1 (proteinase nexin-1) and RAP domains, D1, D2 and D3 to pairs and triplets of CR domains [17,19]. For all ligands studied, there were multiple pairs of CR domains, all taken from cluster II of LRP, that could generate a binding site with affinity very close to that reported for the ligand to intact LRP. Furthermore, addition of a third CR domain only very modestly enhanced, at best, the overall affinity.
Can there be specific ligand binding to LDLR members?
The above discussion might be taken to imply that there is little specificity of ligand binding to LDLR members, in that optimal engagement by two proximal positive charges by any two CR domains that contain DxDxD or DxDDD motifs could be expected to generate a binding affinity in the low nanomolar range. However, there are several ways in which specificity could result. One is that there may be other residues on the ligand close to the pairs of positive charges that result in a repulsive interaction with surface residues of many, though not necessarily all, CR domains. Such repulsion could modify the ability of the positively charged pair to properly engage the CR domain, or simply reduce the overall affinity for interaction at that site. In a receptor where there are multiple potential ligand-binding sites, this could result in preferential binding only to sites where such repulsion is absent. In a receptor with only a single potential binding site, the presence of repulsion for certain ligands might preclude them binding with high affinity at all and thus give selectivity for only those ligands where repulsion is absent.
An example of this may be the interaction of RAP with the LDLR. Whereas RAP binds with low nanomdar affinity to many members of the LDLR family [29], it binds much less tightly to LDLR itself [30]. Consistent with this lower affinity for the intact receptor, the Kd value for the interaction of the two domain fragment of LDLR, LB3–LB4, with domain D3 of RAP is ~1 μM at I=0.15 [31]. The low binding affinity seems surprising given that LB3 and LB4 are the two LDLR domains that each contains the DxDxD motif and the D3 domain of RAP contains two critical lysine residues (Lys256 and Lys270) that have been implicated in binding to LRP [32], each of which occurs as a part of a proximal pair of lysine residues (Lys256 with Lys253 and Lys270 with Lys289). However, the X-ray structure of RAP domain D3 bound to LB3–LB4 from LDLR shows that, whereas LB4 engages both Lys253 and Lys256, LB3 engages only a single lysine, Lys270 [9]. On the basis of the above results of the effect of the second positive charge on binding (5–18-fold increase in binding affinity, depending on domain and ionic strength) the absence of an interaction with the second nearby lysine (Lys289) in the D3–LB3–LB4 complex might be expected to reduce the affinity of LB3–LB4 significantly. Whereas Arg285 is close by (3.06 Å from Asp110) it may not be close enough to act as the second positive charge. The reason that Lys289 of RAP does not appear to engage LB3 is not clear, but may be due to the presence of an arginine as the fourth residue of the DxDxD motif of LB3, which is juxtaposed to two arginine residues on the RAP domain (Arg282 and Arg285). This may give rise to an unfavourable interaction. Perhaps significantly, none of the CR domains from the principal ligand-binding regions of LRP or VLDLR that contains a DxDxD repeat has either an arginine or lysine residue in this location, whereas each of these regions can bind RAP tightly.
A second way in which specificity might result is where the ligand engages only one CR domain by the mechanism of two proximal positive charges and derives the remaining binding energy from a different specific interaction with another CR domain. An example of this may be the binding of α2M, whose receptor-binding domain binds to LRP with ΔGu of approximately −49 kJ/mol. Of this, −31.6 kJ/mol has been shown to result from interaction with a single CR domain [18,33], almost certainly the result of binding to the only two adjacent lysine residues that have been shown to be critical for binding [3,34]. This leaves only ~17 kJ/mol of binding energy to be accounted for. This is unlikely to come from another pair of lysine residues engaging a second CR domain since (i) it is only half of the expected energy of such an interaction, and (ii) there are no other pairs of lysine residues on the receptor-binding domain that have been shown to be critical for binding. An alternative explanation is that the remaining 17 kJ/mol come from a different specific interaction that might then be highly selective for individual CR domains. This could then explain both why some, but not all, LDLR family members bind α2M and why there appears to be preferential binding of α2M to the N-terminal portion of cluster II of LRP [18,35,36].
AUTHOR CONTRIBUTION
Klavs Dolmer prepared the proteins. Peter Gettins designed the experiments, carried out the fluorescence measurements and wrote the paper.
ACKNOWLEDGEMENT
We thank Dr Steven Olson for helpful comments on the paper.
FUNDING
This work was supported by the National Institutes of Health [grant number GM54414].
References
- 1.Herz J., Strickland D. K. LRP: a multifunctional scavenger and signaling receptor. J. Clin. Invest. 2001;108:779–784. doi: 10.1172/JCI13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lalazar A., Weisgraber K. H., Rall S.C.J., Giladi H., Innerarity T. L., Levanon A. Z., Boyles J. K., Amit B., Gorecki M., Mahley R. W. Site-specific mutagenesis of human apolipoprotein E. Receptor binding activity of variants with single amino acid substitutions. J. Biol. Chem. 1988;263:3542–3545. [PubMed] [Google Scholar]
- 3.Arandjelovic S., Hall B. D., Gonias S. L. Mutation of lysine 1370 in full-length human α2-macroglobulin blocks binding to the low density lipoprotein receptor-related protein. Arch. Biochem. Biophys. 2005;438:29–35. doi: 10.1016/j.abb.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Skeldal S., Larsen J. V., Pedersen K. E., Egelund R., Christensen A., Jensen J., Andreasen P. A. Binding areas of urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex for endocytosis receptors of the low density lipoprotein receptor family, determined by site-directed mutagenesis. FEBS J. 2006;273:5143–5159. doi: 10.1111/j.1742-4658.2006.05511.x. [DOI] [PubMed] [Google Scholar]
- 5.Horn I. R., Van den Berg B.M.M., Van der Meijden P. Z., Pannekoek H., van Zonneveld A. J. Molecular analysis of ligand binding to the second cluster of complement-type repeats of the low density lipoprotein receptor-related protein: evidence for an allosteric component in receptor-associated protein-mediated inhibition of ligand binding. J. Biol. Chem. 1997;272:13608–13613. doi: 10.1074/jbc.272.21.13608. [DOI] [PubMed] [Google Scholar]
- 6.Blacklow S. C., Kim P. S. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat. Struct. Biol. 1996;3:758–761. doi: 10.1038/nsb0996-758. [DOI] [PubMed] [Google Scholar]
- 7.Davis C. G., Goldstein J. C., Sudhof T. C., Anderson R.G.W., Russell D. W., Brown M. S. Acid-dependent ligand dissociation and recycling of LDL receptor mediated by growth factor homology region. Nature. 1987;326:760–765. doi: 10.1038/326760a0. [DOI] [PubMed] [Google Scholar]
- 8.Simonovic M., Dolmer K., Huang W., Strickland D. K., Volz K., Gettins P.G.W. Calcium coordination and pH dependence of the calcium affinity of ligand-binding repeat CR7 from the LRP. Comparison with related domains from the LRP and the LDL receptor. Biochemistry. 2001;40:15127–15134. doi: 10.1021/bi015688m. [DOI] [PubMed] [Google Scholar]
- 9.Fisher C., Beglova N., Blacklow S. C. Structure of an LDLR-RAP complex reveals a general mode for ligand recognition by lipoprotein receptors. Mol. Cell. 2006;22:277–283. doi: 10.1016/j.molcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 10.Neels J. G., Van den Berg B.M.M., Lookene A., Olivecrona G., Pannekoek H. P., van Zonneveld A.-J. The second and fourth cluster of class A cysteine-rich repeats of the low density lipoprotein receptor-related protein share ligand-binding properties. J. Biol. Chem. 1999;274:31305–31311. doi: 10.1074/jbc.274.44.31305. [DOI] [PubMed] [Google Scholar]
- 11.Andersen O. M., Petersen H. H., Jacobsen C., Moestrup S. K., Etzerodt M., Andreasen P. A., Thøgersen H. C. Analysis of a two-domain binding site for the urokinase-type plasminogen activator-plasminogen activator inhibitor-1 complex in low-density-lipoprotein-receptor-related protein. Biochem. J. 2001;357:289–296. doi: 10.1042/0264-6021:3570289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen O. M., Christensen L. L., Christensen P. A., Sorensen E. S., Jacobsen C., Moestrup S. K., Etzerodt M., Thøgersen H. C. Identification of the minimal functional unit in the low density lipoprotein receptor-related protein for binding the receptor-associated protein (RAP) J. Biol. Chem. 2000;275:21017–21024. doi: 10.1074/jbc.M000507200. [DOI] [PubMed] [Google Scholar]
- 13.Bu G., Rennke S. Receptor-associated protein is a folding chaperone for low density lipoprotein receptor-related protein. J. Biol. Chem. 1996;271:22218–22224. doi: 10.1074/jbc.271.36.22218. [DOI] [PubMed] [Google Scholar]
- 14.del Castillo L. M., Davila G., Dorantes L., Olivier C., Ibarra R., Castañeda-Agullo M. The influence of a free alpha-ammonium group in the substrate upon trypsin-catalyzed transesterification. Biochim. Biophys. Acta. 1969;191:354–361. doi: 10.1016/0005-2744(69)90254-x. [DOI] [PubMed] [Google Scholar]
- 15.Sachs D. H., Schechter A. N., Cohen J. S. Nuclear magnetic resonance titrations of histidine ring protons. I. Influence of neighboring charged groups. J. Biol. Chem. 1971;246:6576–6580. [PubMed] [Google Scholar]
- 16.Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., Stanley K. K. Surface location and high calcium affinity of a 500 kD liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen J. K., Dolmer K., Gettins P.G.W. Specificity of binding of the low density lipoprotein receptor-related protein (LRP) to different conformational states of the clade E serpins PAI-1 and PN-1. J. Biol. Chem. 2009;284:17989–17997. doi: 10.1074/jbc.M109.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolmer K., Gettins P.G. W. Three complement-like repeats compose the complete α2-macroglobulin binding site in the second ligand binding cluster of the low density lipoprotein receptor-related protein. J. Biol. Chem. 2006;281:34189–34196. doi: 10.1074/jbc.M604389200. [DOI] [PubMed] [Google Scholar]
- 19.Jensen J. K., Dolmer K., Schar C., Gettins P.G.W. Receptor associated protein (RAP) has two high affinity binding sites for the low density lipoprotein receptor related protein (LRP). Consequences for the chaperone function of RAP. Biochem. J. 2009;421:273–282. doi: 10.1042/BJ20090175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guttman M., Prieto J. H., Handel T. M., Domaille P. J., Komives E. A. Structure of the minimal interface between ApoE and LRP. J. Mol. Biol. 2010;398:306–319. doi: 10.1016/j.jmb.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hooper J. D., Clements J. A., Quigley J. P., Antalis T. M. Type II transmembrane serine proteases: insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 2001;276:857–860. doi: 10.1074/jbc.R000020200. [DOI] [PubMed] [Google Scholar]
- 22.Netzel-Arnett S., Hooper J. D., Szabo R., Madison E. L., Quigley J. P., Bugge T. H., Antalis T. M. Membrane anchored serine proteases: a rapidly expanding group of cell surface proteolytic enzymes with potential roles in cancer. Cancer Metastasis Rev. 2003;22:237–258. doi: 10.1023/a:1023003616848. [DOI] [PubMed] [Google Scholar]
- 23.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. Structure of the LDL receptor extracellular domain at endosomal pH. Science. 2002;298:2353–2358. doi: 10.1126/science.1078124. [DOI] [PubMed] [Google Scholar]
- 24.van den Biggelaar M., Sellink E., Klein Gebbink J.W.T.M., Mertens K., Meijer A. B. A single lysine of the two-lysine recognition motif of the D3 domain of receptor-associated protein is sufficient to mediate endocytosis by low-density lipoprotein receptor-associated protein. Int. J. Biochem. Cell Biol. 2011;43:431–440. doi: 10.1016/j.biocel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 25.Lee D., Walsh J. D., Mikhailenko I., Yu P., Migliorini M., Wu Y., Krueger S., Curtis J. E., Harris B., Lockett S., et al. RAP uses a histidine switch to regulate its interaction with LRP in the ER and Golgi. Mol. Cell. 2006;22:423–430. doi: 10.1016/j.molcel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Sehl L. C., Castellino F. J. Thermodynamic properties of the binding of α-,ω-amino acids to the isolated kringle 4 region of human plasminogen as determined by high sensitivity titration calorimetry. J. Biol. Chem. 1990;265:5482–5486. [PubMed] [Google Scholar]
- 27.Menhart N., Sehl L. C., Kelley R. F., Castellino F. J. Construction, expression and purification of recombinant kringle 1 of human plasminogen and analysis of its interaction with ω-amino acids. Biochemistry. 1991;30:1948–1957. doi: 10.1021/bi00221a031. [DOI] [PubMed] [Google Scholar]
- 28.Lewis S. D., Shields P. P., Shafer J. A. Characterization of the kinetic pathway for liberation of fibrinopeptides during assembly of fibrin. J. Biol. Chem. 1985;260:10192–10199. [PubMed] [Google Scholar]
- 29.Bu G., Marzolo M. P. Role of RAP in the biogenesis of lipoprotein receptors. Trends Cardiovasc. Med. 2000;10:148–155. doi: 10.1016/s1050-1738(00)00045-1. [DOI] [PubMed] [Google Scholar]
- 30.Medh J. D., Fry G. L., Bowen S. L., Pladet M. W., Strickland D. K., Chappell D. A. The 39-kDa receptor-associated protein modulates lipoprotein catabolism by binding to LDL receptors. J. Biol. Chem. 1995;270:536–540. doi: 10.1074/jbc.270.2.536. [DOI] [PubMed] [Google Scholar]
- 31.Estrada K., Fisher C., Blacklow S. C. Unfolding of the RAP-D3 helical bundle facilitates dissociation of RAP-receptor complexes. Biochemistry. 2008;47:1532–1539. doi: 10.1021/bi702076y. [DOI] [PubMed] [Google Scholar]
- 32.Migliorini M., Behre E. H., Brew S., Ingham K. C., Strickland D. K. Allosteric modulation of ligand binding to low density lipoprotein receptor-related protein by the receptor-associated protein requires critical lysine residues within its carboxyl domain. J. Biol. Chem. 2003;278:17986–17992. doi: 10.1074/jbc.M212592200. [DOI] [PubMed] [Google Scholar]
- 33.Dolmer K., Huang W., Gettins P.G.W. NMR solution structure of complement-like repeat CR3 from the low density lipoprotein receptor-related protein (LRP). Evidence for specific binding to the receptor binding domain of human α2-macroglobulin. J. Biol. Chem. 2000;275:3264–3271. doi: 10.1074/jbc.275.5.3264. [DOI] [PubMed] [Google Scholar]
- 34.Huang W., Dolmer K., Liao X., Gettins P. G. W. NMR solution structure of the receptor binding domain of human α2-macroglobulin. J. Biol. Chem. 2000;275:1089–1094. doi: 10.1074/jbc.275.2.1089. [DOI] [PubMed] [Google Scholar]
- 35.Mikhailenko I., Battey F. D., Migliorini M., Ruiz J. F., Argraves K., Moayeri M., Strickland D. K. Recognition of α2-macroglobulin by the low density lipoprotein receptor-related protein requires the cooperation of two ligand binding cluster regions. J. Biol. Chem. 2001;276:39484–39491. doi: 10.1074/jbc.M104382200. [DOI] [PubMed] [Google Scholar]
- 36.Andersen O. M., Christensen P. A., Christensen L. L., Jacobsen C., Moestrup S. K., Etzerodt M., Thøgersen H. C. Specific binding of α-macroglobulin to complement-type repeat CR4 of the low-density lipoprotein receptor-related protein. Biochemistry. 2000;39:10627–10633. doi: 10.1021/bi000498h. [DOI] [PubMed] [Google Scholar]