Abstract
The neocortex contains excitatory neurons and inhibitory interneurons. Clones of neocortical excitatory neurons originating from the same progenitor cell are spatially organized and contribute to the formation of functional microcircuits. In contrast, relatively little is known about the production and organization of neocortical inhibitory interneurons. We found that neocortical inhibitory interneurons were produced as spatially organized clonal units in the developing ventral telencephalon. Furthermore, clonally related interneurons did not randomly disperse but formed spatially isolated clusters in the neocortex. Individual clonal clusters consisting of interneurons expressing the same or distinct neurochemical markers exhibited clear vertical or horizontal organization. These results suggest that the lineage relationship plays a pivotal role in the organization of inhibitory interneurons in the neocortex.
The neocortex contains two major classes of neurons: glutamatergic excitatory neurons and γ-aminobutyric acid (GABA)–ergic inhibitory interneurons. Proper functioning of the neocortex critically depends on the production of a correct number of excitatory and inhibitory neurons, which largely occurs during the embryonic stages. Extensive studies over the past decade have provided a comprehensive view of excitatory neuron neurogenesis in the developing neocortex. In contrast, our knowledge of neocortical interneuron neurogenesis remains incomplete.
Most, if not all, neocortical interneurons are generated in the developing ventral (i.e., subcortical) telencephalon, including the ganglionic eminences (GEs) and the preoptic area (PoA), and migrate tangentially over long distances to reach their destination in the neocortex (1–3). Genetic and transplantation studies have demonstrated that different regions of the GEs generate distinct interneuron subgroups that differ in morphology, expression of neurochemical markers, biophysical properties, and synaptic connectivity (4–9). Distinct temporal origins of physiologically defined interneuron subgroups have also been reported (5, 10). However, little is known about the cellular processes that produce neocortical interneurons.
The neocortex is thought to be functionally organized into columns consisting of excitatory neurons and inhibitory interneurons (11). Clonal analysis of neocortical excitatory neuron production and migration has revealed that individual radial glial progenitor cells in the ventricular zone of the dorsal telencephalon undergo consecutive rounds of asymmetric cell division, producing a number of lineage-related sister excitatory neurons (12). Despite some lateral dispersion during migration (13–15), clonally related sister excitatory neurons are often spatially organized into vertical clusters and, to a lesser extent, horizontal arrays in the mature neocortex (16–19). Moreover, specific chemical synapses preferentially form between vertically aligned sister excitatory neurons, suggesting that spatially organized ontogenetic clones of excitatory neurons contribute to the formation of functional columnar microcircuits in the neocortex (18). Whether inhibitory interneurons are spatially organized with respect to the formation of repetitive functional columns in the neocortex remains an outstanding question.
Selective labeling of neocortical interneuron progenitor cells in the medial GE (MGE) and the PoA
Progenitor cells in the MGE and the PoA expressing the homeobox transcription factor Nkx2.1 are responsible for producing a majority of neocortical interneurons in the mouse neocortex (6, 7, 20, 21). It is therefore imperative to selectively label individual progenitor cells in the ventricular zone of the MGE and the PoA for clonal analysis of neocortical interneuron production and organization. We took advantage of the exquisite fidelity of the subgroup A avian sarcoma leukosis virus (ASLV)–receptor interaction (22) and combined it with mouse genetics. We generated a knockin mouse line, LSL-R26TVAiLacZ, to conditionally express the avian tumor virus receptor A (TVA) and β-galactosidase (gene LacZ) in a Cre recombinase-dependent manner (23). By crossing the LSL-R26TVAiLacZ mouse line with the Nkx2.1-Cre mouse line (24), we obtained Nkx2.1-Cre;LSL-R26TVAiLacZ mice (Fig. 1A) in which the TVA receptor was specifically expressed in the neocortical interneuron progenitor cells in the MGE and the PoA, as manifested by β-galactosidase (β-gal) staining (Fig. 1B).
Fig. 1.
Selective labeling of progenitor cells in the MGE and the PoA. (A) Schematic diagram of the strategy. (B) Image of a section of an E14 mouse brain stained for β-gal (blue). β-gal is expressed specifically in the MGE and the PoA. Ncx indicates Neocortex; E, eye. Scale bar indicates 400 µm. (C) Image of a section of an E16 mouse brain injected with EGFP-expressing RCAS retrovirus (green) at E12 and stained with the DNA dye 4´,6´-diamidino-2-phenylindole (DAPI) (blue). There is robust labeling of progenitor cells in the ventricular zone of the MGE (arrow) but not the LGE or the Ncx (arrowheads). Scale bar, 400 µm. (D) Images of a section of an E16 mouse brain injected with EGFP-expressing RCAS retrovirus (green) at E12 and stained with nestin (red) and DAPI (blue). (Right) High-magnification images of the MGE (area 1) and the Ncx (area 2). EGFP-expressing cells are present in the ventricular zone of the MGE but not the LGE or the Ncx, and they are nestin-positive (arrowheads, inset). Scale bars (from left to right): 400 µm, 20 µm, 100 µm, and 50 µm. (E) Images of a section of an E14 mouse brain injected with EGFP-expressing RCAS retrovirus (green) at E12 and stained with Nkx2.1 (red) and DAPI (blue). (Right) High-magnification images of the MGE (area 1), the LGE (area 2), and the Ncx (area 3). EGFP-expressing progenitor cells in the ventricular zone, including those in mitosis with condensed DNA (arrowheads and broken circles, middle), are restricted to the MGE (broken rectangle, area 1) and the PoA and express Nkx2.1. (Inset) An EGFP-expressing cell in the Ncx with the characteristic morphology of a migrating interneuron. Scale bars, 400 µm, 50 µm, 10 µm, 100 µm, and 10 µm.
To label dividing progenitor cells expressing the TVA receptor in the ventricular zone of the MGE and the PoA, we performed in utero intraventricular injections of RCAS (replication-competent ASLV long terminal repeat with a splice acceptor) retroviruses expressing enhanced green fluorescence protein (EGFP) into the mouse embryos at embryonic day 11 to 12 (E11–12), the beginning of the peak phase of interneuron neurogenesis in this region (10). These injections led to a robust and specific labeling of cells in the ventricular zone of the MGE and the PoA (arrow, Fig. 1C) but not the lateral GE (LGE) or the neocortex (arrowheads, Fig. 1C). Some scattered cells in the brain were also labeled (Fig. 1C). These scattered EGFP-expressing cells are likely to be the progeny of labeled progenitor cells in the MGE and the PoA, because retrovirus infection of the dividing progenitor cells results in integration of the viral genome into their daughter cell genome. No labeling was observed in the brains of animals that did not express TVA (fig. S1).
To confirm specificity of the labeling, we used the antibodies against nestin (Fig. 1D), a progenitor cell marker, and Nkx2.1 (Fig. 1E), which is distinctly expressed in the cells of the MGE and the PoA but not the LGE or the neocortex (6, 20). EGFP-expressing cells in the ventricular zone of the MGE and the PoA (arrowheads, Fig. 1D) were positive for nestin, indicating a progenitor cell fate. EGFP-expressing cells in the ventricular zone, including those in mitosis at the ventricular zone surface, also expressed Nkx2.1 (broken circles and arrowheads, Fig. 1E).
Nkx2.1 expression is selectively down-regulated in the progeny of the MGE and the PoA progenitor cells that adopt a neocortical interneuron fate (25), and this down-regulation is required for their migration to the neocortex (26). We found that many EGFP-expressing cells located away from the ventricular zone of the MGE and the PoA no longer expressed Nkx2.1 and gradually acquired the characteristic bipolar morphology of tangentially migrating interneurons 2 to 3 days after infection (arrow, Fig. 1E). To test whether the labeled cells do indeed reach the neocortex and become neocortical inhibitory interneurons, we examined the distribution and fate specification of EGFP-expressing cells in the brain at postnatal day 7 (P7), when neuronal migration is largely complete in the neocortex (27). Nearly all EGFP-expressing cells were in the neocortex, as well as the hippocampus (96.1 ± 0.2%, n = 3 brains; fig. S2A). Furthermore, all EGFP-expressing cells in the neocortex expressed GABA (fig. S2B), suggesting that they adopted the fate of neocortical GABAergic inhibitory interneurons.
Neocortical interneurons are remarkably diverse and can be subdivided into distinct subgroups (28). Individual neocortical interneurons originating from the MGE and the PoA express parvalbumin (PV), somatostatin (SOM), or neuropeptide tyrosine (NPY; some overlap exists with the SOM-expressing subgroup) (1, 2). Indeed, we found that EGFP-expressing cells in the neocortex expressed PV, SOM, or NPY at P21 to P30, suggesting that they develop into characteristic neocortical interneurons (fig. S2C).
Spatially organized ontogenetic clonal units in the MGE and the PoA
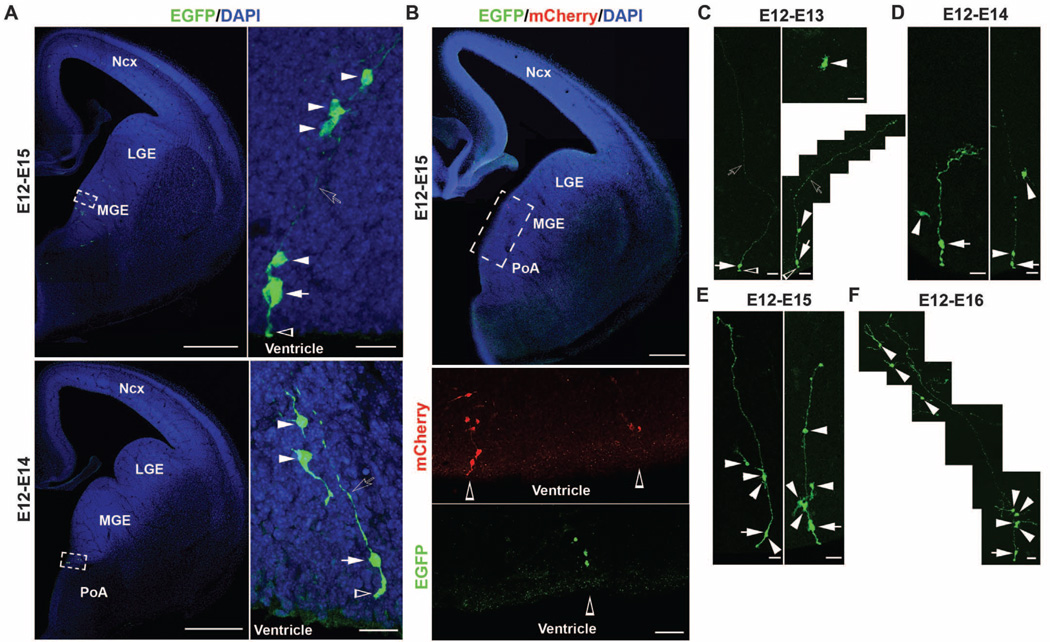
Having established a method for selectively labeling mitotic progenitor cells in the MGE and the PoA that produce neocortical interneuron progeny, we proceeded to perform clonal analysis of neocortical interneuron production and organization. We used in utero intraventricular injections of serially diluted RCAS retrovirus expressing EGFP at E11–12 to label dividing progenitor cells in the ventricular zone of the MGE and the PoA at clonal density. Two to 3 days after the injection of low-titer retrovirus, a few radially arrayed isolated clusters of EGFP-expressing cells were reliably observed in the MGE (Fig. 2A, top) and the PoA (Fig. 2A, bottom). Each cluster contained a single radial glia–like cell (Fig. 2A, arrow) with the defining morphological characteristics, including a nucleus in the ventricular zone, a short process reaching the ventricular surface with a large endfoot (Fig. 2A, open arrowhead), and a long fine radial process pointing toward the pial surface (Fig. 2A, open arrow), as well as several cells with short processes that were arrayed along the long radial process of the radial glia–like cell (Fig. 2A, arrowheads). The same radial clusters in the MGE and the PoA were also reliably labeled by using Moloney murine leukemia retrovirus expressing EGFP (Fig. 2, C to F). The repetitive organization of radial clusters of cells in the MGE and the PoA suggested that individual clusters were clonally related.
Fig. 2.
Spatially organized ontogenetic radial clonal units in the MGE and the PoA. (A) Images of sections of E14 mouse brains injected with low-titer EGFP-expressing RCAS retrovirus (green) at E12 and stained with DAPI (blue). Individual radial clusters of cells express EGFP in the MGE (top) and the PoA (bottom). (Right) High-magnification images of individual radial clusters (broken rectangles). Individual radial clusters contained a radial glia–like cell (white arrows) with a short ventricular endfoot (open arrowheads) and a long radial process pointing toward the pial surface (open arrows) and several cells with short processes arrayed along the radial process (white arrowheads). Scale bars, 500 µm (left) and 20 µm (right). (B) Images of a section of an E14 mouse brain injected with a mixture of low-titer EGFP- or mCherry-expressing RCAS retrovirus and stained with DAPI (blue). (Bottom) High-magnification images of the ventricular zone of the MGE and the PoA (broken rectangle). Individual radial clusters of cells express either mCherry (red) or EGFP (green) (open arrowheads) in the MGE and the PoA. Scale bars, 250 µm (top) and 50 µm (bottom). (C to F) Images of isolated radial clusters in the MGE at 12 to 24 (C), 48 (D), 72 (E), and 96 (F) hours after EGFP-expressing Moloney retrovirus injection at E12. White arrows indicate the radial glia–like cell; white arrowheads, non–radial glia short-process cells of individual clones; open arrows, the long radial process pointing toward the pial surface; and open arrowheads, the ventricular endfoot of the radial glia–like cells. Scale bars, 25 µm.
To demonstrate the clonal identity of individual radial clusters, we injected E11–12 mouse embryos with a mixture of low-titer RCAS retroviruses expressing either EGFP or mCherry, a red fluorescence protein (Fig. 2B). If individual radial clusters were clonal, cells within individual clusters should be labeled by the same retrovirus and thereby express the same fluorescent protein(s). Two to 3 days after infection, we observed that spatially segregated radial clusters contained only EGFP- or mCherry-expressing cells (Fig. 2B, open arrowheads) but not a mixture of cells expressing different fluorescent proteins in the same radial clusters (Fig. 2B).
We next examined the progressive development of ontogenetic radial clones in the MGE and the PoA during the peak phase of interneuron neurogenesis. At 12 to 24 hours after retrovirus injection at E11–12, labeled cells were mostly single and either resembled a radial glial cell (arrows) or a multipolar cell with short processes (Fig. 2C, arrowheads). Some clusters at 24 hours after injection were composed of two cells: one radial glia–like cell (Fig. 2C, arrow) and the other a multipolar short-process cell (Fig. 2C, arrowhead) closely associated with the radial process of the radial glia–like cell (1.5 ± 0.1 cells per cluster, n = 33; Fig. 2C). At 48 hours after injection, we observed radially arrayed clones comprising a radial glia–like cell (Fig. 2D, arrows) with one to three short-process cells (Fig. 2D, arrowheads) arrayed along the radial process (2.5 ± 0.2 cells per cluster, n = 55; Fig. 2D). At 72 hours after injection, typical clones consisted of a radial glia–like cell and two to five short-process cells distributed along the radial process (3.3 ± 0.2 cells per cluster, n = 42; Fig. 2E). Furthermore, at 48 to 96 hours after injection, we observed an increase in short-process cells situated away from the radial process of radial glia– like cells with the typical morphology of migrating interneurons (Fig. 2, D to F).
Radial clones contain mitotic progenitors and differentiating interneurons
To test whether the radial glia–like cells in individual MGE and PoA clonal radial clusters are indeed radial glial cells, we used antibodies against astrocyte-specific glutamate transporter (GLAST) and brain-lipid-binding protein (BLBP), two proteins specifically expressed in radial glial progenitor cells (29).We found that EGFP-labeled radial glia–like cells in individual clones were positive for both GLAST (fig. S3A) and BLBP (fig. S3B).
The progressive increase in the number of cells in individual MGE and PoA clonal clusters beside the radial glial cell suggests that radial glial cells are mitotically active progenitors responsible for producing these clones. Consistent with this, we found that EGFP-labeled radial glial cells in individual clones expressed the neural progenitor cell marker Nestin (fig. S3C).We pulse-labeled S-phase cells with 5-bromo-2′-deoxyuridine (BrdU) 24 hours after retrovirus injection and found that radial glial cells in the labeled clones were BrdU-positive (fig. S3D), indicating that they are indeed mitotically active.
Besides the radial glial progenitor cell and possibly the intermediate progenitor cell (Fig. 3B) (30), individual radial clones contained cells located at a substantial distance from the ventricular zone that adopt the typical morphology of migrating interneurons (Figs. 2 and 3), indicating the presence of differentiating interneurons in the clone. We performed immunohistochemistry experiments by using the antibody against TuJ1, a neuron-specific marker (fig. S4A). The bipolar cells located at a distance from the ventricular zone in individual radial clones were positive for TuJ1 (cell 5). Although the radial glial progenitor cell (cell 1) and the multipolar cell close to the ventricular zone (cell 2) did not express TuJ1, the multipolar cells away from the ventricular zone (cells 3 and 4) did (fig. S4, A and B). These results suggest a progressive differentiation of cells within individual radial clones toward a neuronal fate, which is further supported by their acquisition of Na+ conductance (fig. S4C). The bipolar cell and the multipolar cell located at a distance from the ventricular zone likely represent the early-born cells, whereas the multipolar cells located close to the ventricular zone likely represent the late-born cells.
Fig. 3.
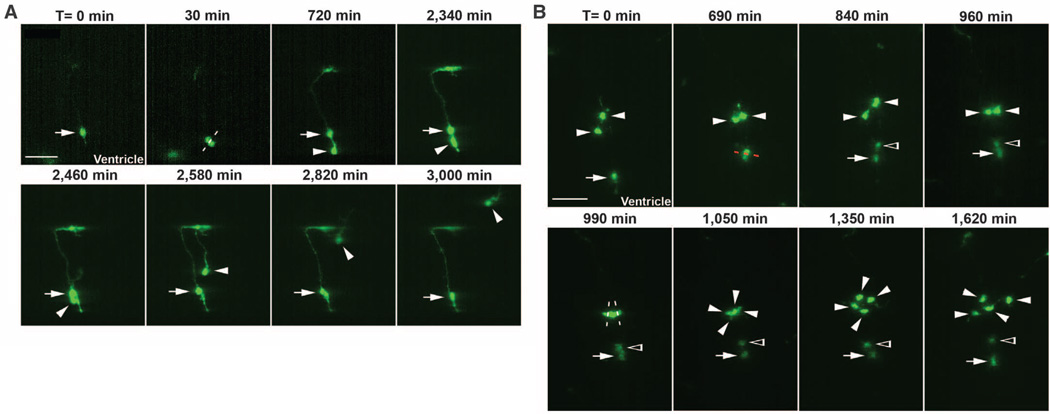
Asymmetric division of a radial glial progenitor cell and symmetric division of an intermediate progenitor cell. (A) Time-lapse images of an MGE radial glial progenitor cell dividing asymmetrically at the ventricular zone surface. Time is indicated at the top of the images. Arrows indicate the original and renewed radial glial cell, and arrowheads indicate the differentiating daughter cell that initially migrates along the radial glial cell, away from the ventricular zone surface, and later acquires bipolar morphology and migrates tangentially toward the neocortex. The white broken line indicates the orientation of the cleavage plane. Scale bar, 50 µm. (B) Time-lapse images of symmetric divisions of intermediate progenitor cells in the subventricular zone of the MGE. White arrows indicate the radial glial cell; white arrowheads, the intermediate progenitor cells and their daughter cells of the radial clone; and open arrowheads, the new multipolar daughter cell of the dividing radial glial progenitor cell. White and red broken lines indicate the orientation of the cleavage plane of dividing intermediate progenitor cells and radial glial progenitor cell, respectively. Scale bar, 50 µm.
Asymmetric division of radial glial progenitors and symmetric division of intermediate progenitors
Clonal clusters labeled in the MGE and the PoA contained a single radial glial progenitor cell and a few non–radial glial cells, including differentiating interneurons, suggesting that the radial glial progenitor undergoes asymmetric cell division to self-renew and, at the same time, to produce other cell types. We performed time-lapse imaging experiments to trace the mitotic behavior of individual EGFP-expressing MGE or PoA radial glial cells in organotypic slice cultures (Fig. 3A). Labeled radial glial cells in the MGE or the PoA were identified by using morphological criteria (Fig. 3A, arrow). Radial glial cells underwent interkinetic nuclear migration and divided at the ventricular zone surface. After mitosis, while one of the two daughter cells became a radial glial cell (Fig. 3A, arrows), the other daughter cell initially moved radially along the mother radial glial cell away from the ventricular zone surface and then migrated tangentially toward the neocortex (Fig. 3A, arrowheads).We observed a total of seven radial glial progenitor cell divisions at the ventricular zone surface, and each of them resulted in an asymmetric fate specification for the two daughter cells. These asymmetric divisions could be classified into two subtypes. In about half of the asymmetric radial glial cell divisions, the non–radial glia daughter cell gradually adopted a bipolar morphology, departed the mother radial glial cell, and migrated tangentially toward the neocortex (arrowheads, Fig. 3A). In the rest of the asymmetric divisions, the resulting non–radial glia daughter cell reentered the cell cycle and divided again in the subventricular zone (Fig. 3B).
The direct observation of cell divisions away from the ventricular zone surface further indicates the existence of intermediate progenitor cells in the MGE and the PoA. In time-lapse imaging experiments, we observed a total of 15 cell divisions in the subventricular zone of the MGE, and each of them appeared to be symmetric, producing two daughter cells with similar morphology and migratory behavior (Fig. 3B).
Clustering of interneuron clones in the neocortex
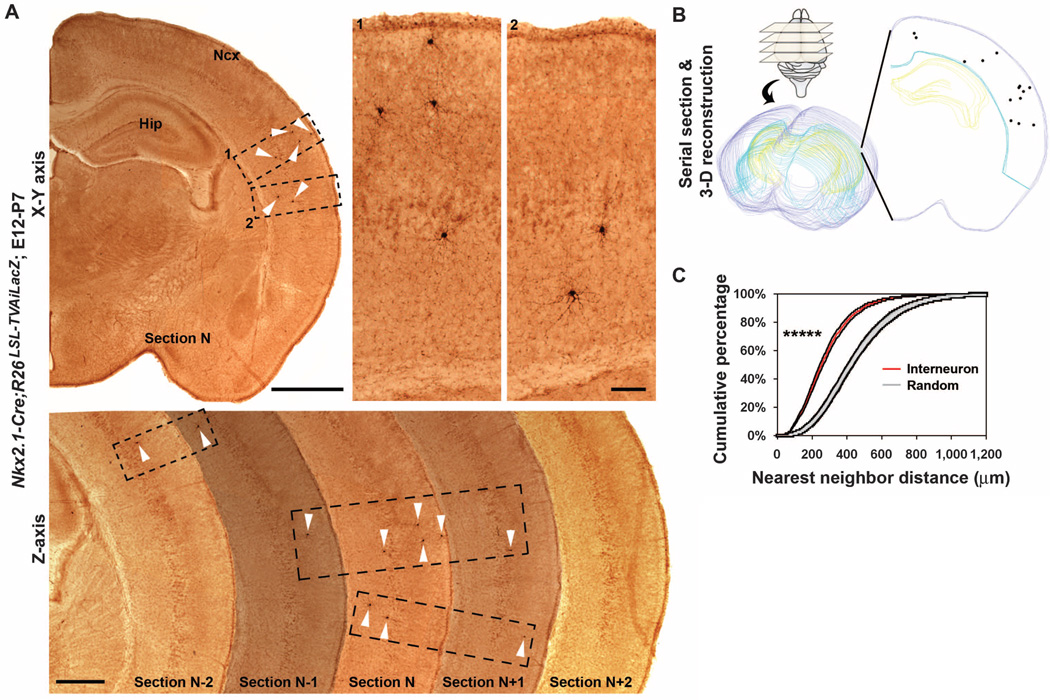
Individual ontogenetic clones of future neocortical interneurons in the MGE and the PoA are initially organized into radial arrays, similar to ontogenetic clones of excitatory neurons (12, 18). This raises the question of whether individual ontogenetic clones of interneurons remain spatially organized after they reach their final destination in the neocortex. We examined the distribution of clonally labeled interneurons in the neocortex at P7, when the migration of neocortical interneurons largely ends (27). Clonally labeled interneurons did not randomly disperse but formed spatially isolated clusters that were often more than 500 µm apart (Fig. 4A). To systematically examine the distribution of clonally labeled interneurons, we performed serial coronal sectioning and three-dimensional reconstruction of the neocortex (Fig. 4, A and B). We frequently observed clusters of clonally labeled interneurons, which were confined to three consecutive sections (70 µm each), and individual clusters contained three to eight interneurons often distributed across different neocortical laminar layers.
Fig. 4.
Clustering of clonally labeled neocortical interneurons. (A) Images of sections of a P7 mouse brain injected with low-titer EGFP-expressing RCAS retrovirus at E12. (Top left) Image of a single section showing clonally labeled interneurons (arrowheads) forming clusters in the neocortex. (Top right) High-magnification images of the interneuron clusters (broken rectangles 1 and 2). (Bottom) Images of five consecutive coronal sections showing clonally labeled interneurons in the neocortex (arrowheads) that form spatially isolated clusters confined to three consecutive sections (broken rectangles). Hip indicates Hippocampus. Scale bars, 1000 µm, 100 µm, and 500 µm. (B) Serial sectioning and three-dimensional reconstruction of a P7 mouse brain. Purple lines indicate the outer boundary of the section; blue lines, the inner boundary of the neocortex; yellow lines, the hippocampus; and individual dots, the cell bodies of labeled interneurons (~six times larger than their relative size). (C) NND analysis showing that clonally labeled interneurons do not randomly distribute but form clusters in the neocortex at P7 (*****P < 1 × 10−10; n = 23 brains).
To quantitatively analyze the distribution of clonally labeled interneurons, we applied nearest neighbor distance (NND) analysis to the three-dimensional reconstruction data sets, which reports the spacing pattern of the data points within the data set (fig. S5) (31). We compared three-dimensional neocortex data sets of clonally labeled interneurons with spatially random simulated data sets containing the same number of data points within the same volume repeated 100 times. The NND distribution of clonally labeled interneurons in every neocortex was outside the upper and lower boundaries of the random simulated data set NND distribution (n = 23 brains; Fig. 4C and fig. S6). Furthermore, the NND distribution of clonally labeled neocortical interneurons was shifted toward shorter distances compared with that of the random simulation data sets (P< 1 × 10−10, n = 23 brains; Fig. 4C), suggesting that clonally labeled interneurons form clusters when they reach their final destination in the neocortex. This clustering feature was not a transient phenomenon but persisted to P21–30, when neocortical interneurons are mature (interneuron versus randomsimulation: P < 1 × 10−10, n = 13 brains; fig. S7).
We then injected a mixture of low-titer RCAS retroviruses expressing EGFP or mCherry and examined the probability of observing spatially isolated clusters containing cells expressing the same fluorescent protein versus those containing cells expressing different fluorescent proteins in the neocortex. Individual clusters of clonally labeled interneurons in the neocortex were comprised of cells expressing either EGFP or mCherry (fig. S8). This segregation of cells expressing EGFP or mCherry into individual clusters was unlikely to have occurred randomly (interneuron versus random simulation: P < 5 × 10−5; n = 8 brains).
Neurochemical marker expression of neocortical interneuron clonal clusters
Nkx2.1-expressing progenitor cells in the MGE and the PoA produce neocortical interneurons that express the neurochemical marker PV, SOM, or NPY (1, 2, 7, 20, 21). To assess the subgroup composition of labeled interneuron clonal clusters in the neocortex, we used the antibodies against EGFP, PV, and SOM at P30, when neocortical interneurons are mature (Fig. 5). Although 19 out of 75 spatially isolated clusters of clonally labeled interneurons in the neocortex shared the same neurochemical marker (Fig. 5A and fig. S9), the remaining clusters contained interneurons expressing different neurochemical markers (Fig. 5B and fig. S9).
Fig. 5.
Neurochemical marker expression of neocortical interneuron clonal clusters. (A and B) (Left) Images of sections of P30 mouse brains injected with low-titer EGFP-expressing RCAS retrovirus at E12 and stained with the antibodies against EGFP (green), SOM (red), PV (white), and DAPI (blue). (Right) High-magnification images of individual cells (arrowheads and numbers 1 to 3) of the clonally labeled interneuron clusters. Although some clusters of clonally labeled neocortical interneurons share the same neurochemical marker (A), many clusters contain interneurons expressing distinct neurochemical markers (B). Scale bars, 200 µm and 20 µm.
Spatial organization of neocortical interneuron clonal clusters
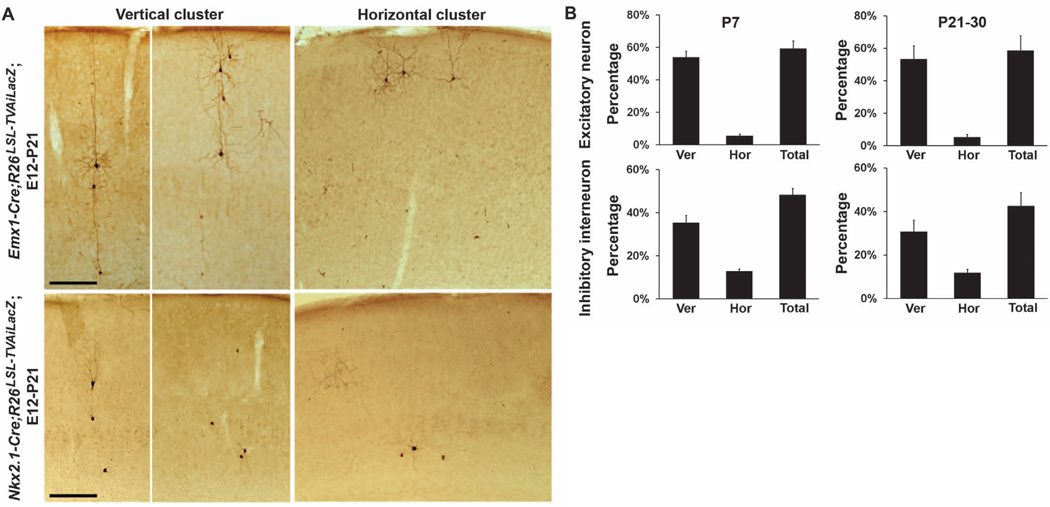
When we selectively labeled ontogenetic clones of excitatory neurons by using the RCAS-TVA approach in conjunction with the dorsal forebrain-specific Emx1-Cre knockin mouse line (32), we found that clonally related excitatory neurons formed spatially isolated clusters in the neocortex (Fig. 6A, top, and fig. S10). Sister excitatory neurons within individual clusters were often organized into vertical and, to a lesser extent, horizontal arrays relative to the pial surface (Fig. 6A, top) (16, 18, 19). Clonally labeled interneuron clusters displayed similar spatial organization, and sister interneurons within individual clusters were frequently arranged into vertical or horizontal arrays (Fig. 6A, bottom).
Fig. 6.
Spatial organization of clonally labeled neocortical interneurons. (A) Images of sections of P21mouse brains injected with low-titer EGFP expressing RCAS retrovirus at E12. Similar to clonally labeled excitatory neurons (top), clonally labeled inhibitory interneurons (bottom) form spatially isolated vertical and horizontal clusters. Scale bar, 250 µm. (B) Quantification of the percentages of clonally labeled excitatory neurons (P7, n = 10 brains; P21–30, n = 8 brains) and inhibitory interneurons (P7, n = 23 brains; P21–30, n = 13 brains) that form vertical and horizontal clusters in the neocortex. Error bars indicate SEM.
We established a method for detecting vertical and horizontal neuronal clusters in the three-dimensional neocortex reconstruction data sets (fig. S11).We first obtained the spatial dispersion parameters of representative vertical and horizontal clusters of clonally labeled excitatory neurons in isolation in the neocortex. By using these spatial parameters, we defined a vertical or horizontal cuboidal matrix to systematically scan through the three-dimensional reconstruction data sets and identify vertical or horizontal neuron clusters, respectively. After confirming that this method reliably detected vertical and horizontal clusters of clonally labeled excitatory neurons in the neocortex (percentage of all labeled neocortical excitatory neurons: P7, 59.1 ± 4.7%, n = 10 brains; P21–30, 58.6 ± 9.2%, n = 8 brains; Fig. 6B and fig. S12), we applied this analysis to the three-dimensional neocortex reconstruction data sets of clonally labeled interneurons by using the same spatial parameters. A similar percentage of all labeled neocortical interneurons were organized into spatially isolated vertical and horizontal clusters (P7, 48.4 ± 3.0%, n = 23 brains; excitatory neuron versus interneuron, P = 0.1; P21–30, 42.5 ± 6.2%, n = 13 brains; excitatory neuron versus interneuron, P = 0.2; Fig. 6B and figs. S12 and S13). We applied this analysis to random simulated data sets and found that the spatial organization of clonally labeled neocortical interneurons was unlikely to have occurred randomly (interneuron versus random simulation: P < 5 × 10−18; n = 23 brains).
Clonal analysis of excitatory neuron production and organization has provided crucial insights into the formation of a functional neocortex (12–16, 18, 19, 33); however, no systematic clonal analysis of inhibitory interneuron production and organization has been reported. We adapted a method for selectively labeling individual mitotic progenitor cells in the MGE and the PoA that produce a majority of neocortical interneurons and performed clonal analysis of the production and organization of neocortical interneurons (fig. S14). The progenitor cells in the ventricular zone of the MGE and the PoA were radial glial cells in nature. They underwent interkinetic nuclear migration and divided asymmetrically at the ventricular zone surface to self-renew and to simultaneously produce differentiating interneurons or intermediate progenitor cells that migrate away from the ventricular zone. Intermediate progenitor cells further divided symmetrically within the subventricular zone to produce postmitotic interneurons. The progeny of the same radial glial progenitor cell closely associated with the radial process of the mother radial glial cell, forming a radial clone. During development, the early-born cells progressively moved away from the ventricular zone, acquired the characteristic morphology and biochemical and biophysical properties of differentiating interneurons, and migrated tangentially toward the neocortex. These findings suggest that, similar to excitatory neurons, neocortical inhibitory interneurons originating from the MGE and the PoA are generated in spatially organized ontogenetic clonal units. The progenitor cell behavior and division pattern responsible for producing neocortical interneurons in the MGE are also similar to those producing neocortical excitatory neurons, despite the distinct transcriptional networks present in the progenitor cells that specify inhibitory interneurons versus excitatory neurons (1–3).
However, neocortical interneurons are remarkably diverse and often classified by distinctive morphology, expression of neurochemical markers, firing pattern, and synaptic connectivity (28, 34, 35). Proper production of a correct number of each of these different subgroups of neocortical interneurons is essential for constructing a functional neocortex. The diversity and precision in generating neocortical interneurons most certainly requires intricate regulation of progenitor cell division pattern and dynamics.
Radial migration of daughter cells along the mother radial glial cell has been extensively characterized in the dorsal telencephalon during excitatory neuron neurogenesis and migration. Our results suggest that, before tangential migration, differentiating daughter cells in the MGE and the PoA, which include neocortical interneurons, migrate radially along the mother radial glial progenitor cell. The physiological importance of this initial radial migration is unclear. It may allow proper neuronal differentiation of the daughter cells before they begin migrating tangentially to reach the neocortex. It has been suggested that direct contact with radial glia promotes GABAergic interneuron differentiation (36). Consistent with this, we found that cells within individual clones with the most-pronounced neuronal characteristics are located furthest away from the ventricular zone. They possess the typical bipolar morphology of a tangentially migrating interneuron, indicating that they are poised for tangential migration. Cells with less-pronounced neuronal characteristics are located close to the ventricular zone.
Neocortical interneurons generated in the MGE and the PoA undertake complex migration routes to reach their final destination in the neocortex (37, 38). They migrate tangentially over long distances to enter the neocortex through the marginal zone or the intermediate and subventricular zones before turning radially to reach their ultimate location in the neocortex. Similar to excitatory neurons, inhibitory interneurons generated in the MGE and the PoA display birth date–dependent laminar distribution in the neocortex (1, 39, 40), thereby arguing for a regulated process of interneuron migration. However, the long-distance tangential migration of interneurons has been considered to be mostly random (39, 41). It is unclear how random tangential migration of individual interneurons could lead to an organized distribution pattern in the neocortex for constructing functional circuits, for example, repetitive columnar microcircuits. We found that clonally related interneurons do not randomly disperse but form spatially organized vertical and horizontal clusters in the neocortex (fig. S14).
Nearly all neocortical neuronal circuits are composed of excitatory neurons and inhibitory interneurons. Similar to excitatory neurons, inhibitory interneurons in the neocortex develop highly specific synaptic connections for the assembly assembly of functional circuits (42–44) [but see (45)]. The synaptic connections from local inhibitory interneurons to excitatory neurons exhibit stereotypic spatial pattern (46), suggesting a high degree of spatial and functional organization of neocortical interneurons. The predictable spatial organization of clonally related sister inhibitory interneurons raises the possibility of a lineage-dependent functional organization of inhibitory interneurons in the mammalian neocortex.
Supplementary Material
Acknowledgments
We thank A. L. Joyner and M. E. Ross for comments on the manuscript; the members of the Shi laboratory for valuable discussion and input; E. C. Holland for RCAS-EGFP plasmid and advice on the production of RCAS retroviruses; F. H. Gage for providing 293 gp NIT-EGFP retrovirus packaging cell line; J. A. Kaltschmidt for R26LSL-TVAilacZ mice; and S. Li for technical assistance.
This work was supported by grants from the NIH (R01DA024681 and R21NS072483 to S.-H.S., R21MH083624 to S.-H.S. and K.H., R01MH066912 and K02MH070031 to S.A.A.), the McKnight Foundation, and the March of Dimes Foundation (to S.-H.S.).
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/334/6055/480/DC1
Materials and Methods
Figs. S1 to S14
References
References and Notes
- 1.Batista-Brito R, Fishell G. Curr. Top. Dev. Biol. 2009;87:81. doi: 10.1016/S0070-2153(09)01203-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wonders CP, Anderson SA. Nat. Rev. Neurosci. 2006;7:687. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 3.Gelman DM, Marin O. Eur. J. Neurosci. 2010;31:2136. doi: 10.1111/j.1460-9568.2010.07267.x. [DOI] [PubMed] [Google Scholar]
- 4.Wonders CP, et al. Dev. Biol. 2008;314:127. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butt SJ, et al. Neuron. 2005;48:591. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 6.Flames N, et al. J. Neurosci. 2007;27:9682. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fogarty M, et al. J. Neurosci. 2007;27:10935. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi G, et al. J. Neurosci. 2010;30:1582. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Q, et al. Neuron. 2010;65:328. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyoshi G, Butt SJ, Takebayashi H, Fishell G. J. Neurosci. 2007;27:7786. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mountcastle VB. Brain. 1997;120:701. doi: 10.1093/brain/120.4.701. [DOI] [PubMed] [Google Scholar]
- 12.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Nature. 2001;409:714. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 13.Walsh C, Cepko CL. Nature. 1993;362:632. doi: 10.1038/362632a0. [DOI] [PubMed] [Google Scholar]
- 14.Reid CB, Liang I, Walsh C. Neuron. 1995;15:299. doi: 10.1016/0896-6273(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 15.O’Rourke NA, Sullivan DP, Kaznowski CE, Jacobs AA, McConnell SK. Development. 1995;121:2165. doi: 10.1242/dev.121.7.2165. [DOI] [PubMed] [Google Scholar]
- 16.Kornack DR, Rakic P. Neuron. 1995;15:311. doi: 10.1016/0896-6273(95)90036-5. [DOI] [PubMed] [Google Scholar]
- 17.Rakic P. Science. 1988;241:170. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 18.Yu YC, Bultje RS, Wang X, Shi SH. Nature. 2009;458:501. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price J, Thurlow L. Development. 1988;104:473. doi: 10.1242/dev.104.3.473. [DOI] [PubMed] [Google Scholar]
- 20.Gelman DM, et al. J. Neurosci. 2009;29:9380. doi: 10.1523/JNEUROSCI.0604-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butt SJ, et al. Neuron. 2008;59:722. doi: 10.1016/j.neuron.2008.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young JA, Bates P, Varmus HE. J. Virol. 1993;67:1811. doi: 10.1128/jvi.67.4.1811-1816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seidler B, et al. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10137. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q, Tam M, Anderson SA. J. Comp. Neurol. 2008;506:16. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 25.Sussel L, Marin O, Kimura S, Rubenstein JL. Development. 1999;126:3359. doi: 10.1242/dev.126.15.3359. [DOI] [PubMed] [Google Scholar]
- 26.Nóbrega-Pereira S, et al. Neuron. 2008;59:733. doi: 10.1016/j.neuron.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bortone D, Polleux F. Neuron. 2009;62:53. doi: 10.1016/j.neuron.2009.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ascoli GA, et al. Petilla Interneuron Nomenclature Group. Nat. Rev. Neurosci. 2008;9:557. doi: 10.1038/nrn2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anthony TE, Klein C, Fishell G, Heintz N. Neuron. 2004;41:881. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- 30.Sheth AN, Bhide PG. J. Comp. Neurol. 1997;383:220. [PubMed] [Google Scholar]
- 31.Diggle PJ. Statistical Analysis of Spatial Point Patterns. New York: Oxford Univ. Press; 2003. [Google Scholar]
- 32.Gorski JA, et al. J. Neurosci. 2002;22:6309. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavdas AA, Mione MC, Parnavelas JG. Cereb. Cortex. 1996;6:490. doi: 10.1093/cercor/6.3.490. [DOI] [PubMed] [Google Scholar]
- 34.Markram H, et al. Nat. Rev. Neurosci. 2004;5:793. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 35.Huang ZJ, Di Cristo G, Ango F. Nat. Rev. Neurosci. 2007;8:673. doi: 10.1038/nrn2188. [DOI] [PubMed] [Google Scholar]
- 36.Li H, et al. Dev. Neurobiol. 2008;68:1549. doi: 10.1002/dneu.20679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marín O, Rubenstein JL. Annu. Rev. Neurosci. 2003;26:441. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 38.Corbin JG, Nery S, Fishell G. Nat. Neurosci. 2001;4 suppl.:1177. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- 39.Ang ES, Jr, Haydar TF, Gluncic V, Rakic P. J. Neurosci. 2003;23:5805. doi: 10.1523/JNEUROSCI.23-13-05805.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyoshi G, Fishell G. Cereb. Cortex. 2011;21:845. doi: 10.1093/cercor/bhq155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka DH, et al. J. Neurosci. 2009;29:1300. doi: 10.1523/JNEUROSCI.5446-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta A, Wang Y, Markram H. Science. 2000;287:273. doi: 10.1126/science.287.5451.273. [DOI] [PubMed] [Google Scholar]
- 43.Yoshimura Y, Callaway EM. Nat. Neurosci. 2005;8:1552. doi: 10.1038/nn1565. [DOI] [PubMed] [Google Scholar]
- 44.Thomson AM, Lamy C. Front. Neurosci. 2007;1:19. doi: 10.3389/neuro.01.1.1.002.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fino E, Yuste R. Neuron. 2011;69:1188. doi: 10.1016/j.neuron.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kätzel D, Zemelman BV, Buetfering C, Wölfel M, Miesenböck G. Nat. Neurosci. 2011;14:100. doi: 10.1038/nn.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.