How did the study come about?
According to World Health Organization (WHO) estimates, 33.4 million people were infected with the human immunodeficiency virus (HIV) type 1 globally at the end of 2008.1 Sub-Saharan Africa and Asia are the two regions that have the highest HIV prevalence, with 22.4 and 4.7 million people infected, respectively.1 During the 5 years prior, access to combination antiretroviral therapy (ART) in low- and middle-income countries increased 10-fold to reach 4 million people, providing coverage to 28% of those in need.2 Several studies have reported significant reductions in HIV-related morbidity and mortality for individuals with access to treatment in these regions.3–5 In resource-limited settings, to facilitate the rapid expansion of access to ART, WHO recommends a standardized, public-health approach.6 This is in contrast to the individualized patient-management strategies in developed countries, based on routinely available diagnostic monitoring.7 Standardized first-line ART regimens consist of a non-nucleoside reverse transcriptase inhibitor (NNRTI) and a dual nucleoside/nucleotide reverse transcriptase inhibitor (NRTI) backbone, available in some countries as generic fixed-dose combinations.6 Recommended second-line regimens combine a ritonavir-boosted protease inhibitor (PI) with two previously unused and/or recycled NRTIs.6
Routine HIV viral-load monitoring is not generally available in resource-limited countries and treatment failure is frequently identified based on immunological definitions and/or the occurrence of clinical events.6 Virological breakthrough may be detected late while the failing regimen is continued, thus facilitating the acquisition and accumulation of drug resistance-associated mutations.8 Drug-resistant HIV variants may compromise the effectiveness of subsequent lines of treatment and their transmission to newly infected individuals has severe public health consequences.9,10 To date, ART programmes have been implemented without accompanying HIV drug resistance (HIVDR) monitoring. Monitoring studies are hampered by the lack of a molecular laboratory infrastructure required for genotypic resistance testing, logistical challenges related to maintaining specimen integrity in remote settings and the high costs of testing.11 Challenges to scaling up ART in resource-limited countries, such as absence of routine virological monitoring and limited choices of drug regimen, advocate for the development of a global public-health framework to monitor and prevent the emergence of HIVDR and thus maximize long-term ART effectiveness.12
HIV-1 subtype B is the predominant viral subtype in North America, Western Europe and Australia, and antiretroviral (ARV) drugs have been developed on this subtype. However, in sub-Saharan Africa and Asia, the genetic diversity in HIV subtypes and circulating recombinant forms (CRFs), resulting from recombination between subtypes within a dually infected person, is extensive.13 Although current evidence is limited, some reports have suggested that the propensity to develop HIVDR and the spectrum of mutations that emerge during drug selective pressure, may differ across subtypes and CRFs.14–17 Viral heterogeneity may, therefore, have implications for rates of disease progression and patient response to ART, warranting further study of inter-subtype differences in mutational pathways to resistance.
To help assess the extent of HIVDR in sub-Saharan Africa and Asia, a collaborative bi-regional programme was established, called LAASER [Linking African and Asian Societies for an Enhanced Response (LAASER) to HIV/AIDS; http://www.laaser-hivaids.org] with the primary aim of increasing regional capacities for the monitoring of HIVDR. PharmAccess Foundation, a non-profit organization dedicated to the strengthening of health systems and improving access to quality basic health care in sub-Saharan Africa, has developed the PharmAccess African Studies to Evaluate Resistance (PASER). TREAT Asia (Therapeutics, Research, Education and AIDS Training in Asia) is a network of clinics, hospitals and research institutions working to ensure safe and effective delivery of HIV/AIDS treatment throughout the Asia-Pacific and has developed the TREAT Asia Studies to Evaluate Resistance (TASER). Both PASER and TASER programmes incorporate a monitoring and evaluation (M) and a surveillance (S) protocol. Laboratories providing genotyping results for PASER and TASER are required to participate in the TREAT Asia Quality Assurance Scheme (TAQAS), which is an ongoing assessment programme to build genotyping laboratory capacity, described elsewhere.18 The focus of this cohort profile is the monitoring and evaluation protocols, PASER-M and TASER-M.
How are PASER and TASER set up and how are they funded?
Through the LAASER programme, PASER and TASER receive financial support from the Dutch Ministry of Foreign Affairs through a partnership with Stichting Aids Fonds, PharmAccess Foundation, TREAT Asia (a programme of amfAR, The Foundation for AIDS research) and International Civil Society Support. PASER-M is coordinated by PharmAccess Foundation, in collaboration with the Amsterdam Institute for Global Health and Development (AIGHD) and the Virology Department at the University Medical Center Utrecht, The Netherlands. TASER-M is coordinated by TREAT Asia and its statistical and data management centre is the National Centre in HIV Epidemiology and Clinical Research (NCHECR), The University of New South Wales in Sydney, Australia.
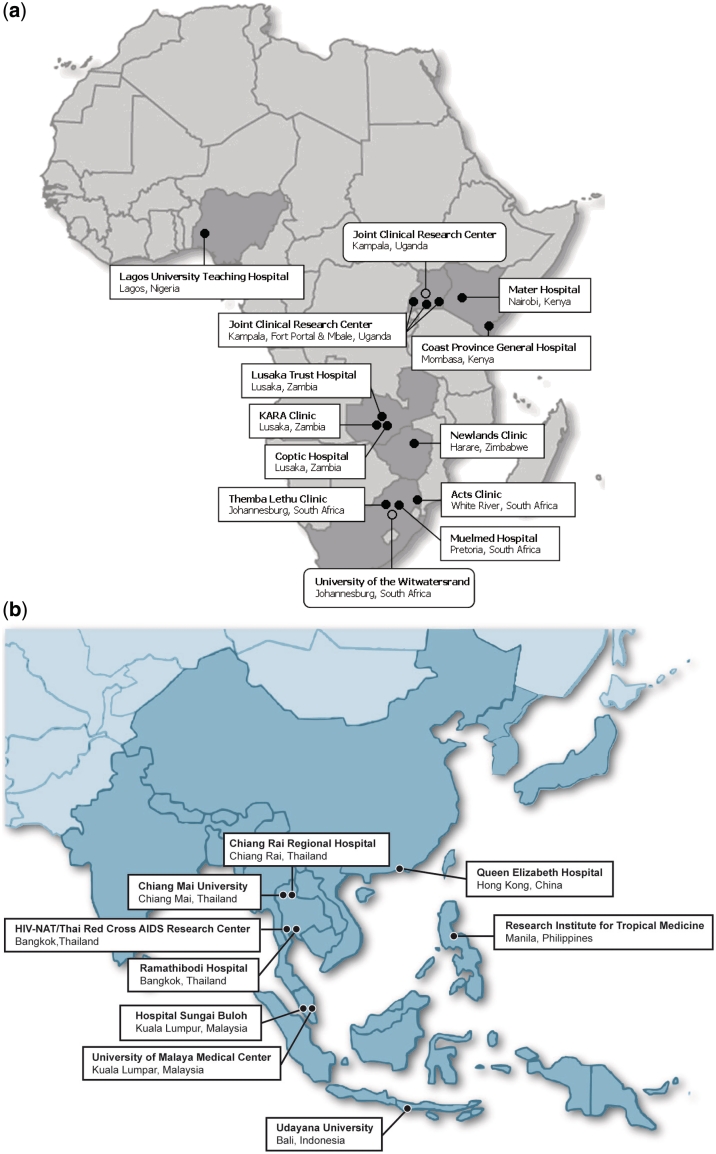
PASER constitutes a newly established collaboration between HIV treatment clinics, laboratories with the capacity to perform genotypic sequencing and research centres. Thirteen clinical sites and two reference laboratories in six African countries (Kenya, Nigeria, South Africa, Uganda, Zambia and Zimbabwe) are collaborating on PASER-M (Figure 1a). Details of the PASER-M collaborating clinical sites are summarized in Supplementary Table 1 available as Supplementary data at IJE online. Ethics approvals were obtained from the Academic Medical Center Institutional Review Board (IRB) and local IRBs. Sites are government, non-government, faith-based or private clinics and hospitals, situated in major cities or urban areas. ART was introduced at the sites at various time points between 1992 and 2006 (median: 2004). Of the 13 sites, 11 provided drugs, consultations and routine laboratory testing free of charge. HIV viral load testing is available at 8 of the 13 sites.
Figure 1.
Geographical location of (a) PASER-M collaborating sites and (b) TASER-M collaborating sites
TASER collaborating sites are selected from within the existing TREAT Asia network, based on their laboratory capacity to perform genotypic sequencing, as described elsewhere.18,19 Sites that do not have internal laboratory genotyping capacity can participate through collaboration with a TAQAS-certified laboratory. Eleven clinical and laboratory sites in six Asian countries (China, Indonesia, Malaysia, Philippines, South Korea and Thailand) are collaborating on TASER-M (Figure 1b). Ethics approvals were obtained from local IRBs having Federal wide Assurances (FWAs) in place from the United States Office for Human Research Protections. FWAs are required for TASER sites as they participate in the International Epidemiologic Databases to Evaluate AIDS (IeDEA) initiative, described elsewhere.20 Sites are generally government- or university-based clinics and hospitals or private clinics, situated in major cities and other urban areas. Those with ethics approvals prior to June 2010 are shown in Figure 1b. ART has been available in Asia for more than 10 years, even in less-resourced countries in the region, and all TASER-M clinical sites have on-site viral load testing.
Clinical sites follow their national guidelines to assess eligibility for ART initiation in accordance with the WHO recommendations.6 Genotypic resistance testing on PASER and TASER clinical specimens are performed in TAQAS-certified genotyping laboratories.18 Laboratories are encouraged to become accredited members of the WHO/HIVResNet HIV Drug Resistance Laboratory network.21 Population-based nucleotide sequencing of the HIV protease (PR) and partial reverse transcriptase (RT) gene regions is performed on plasma specimens, which have HIV RNA of more than 1000 copies/ml. Plasma is obtained from blood collected in EDTA tubes which is locally stored at −80°C and, if required, batch-shipped on dry ice to a genotyping laboratory.
PASER-M genotypic testing is concentrated in two central reference laboratories and thus depends on a robust cold-chain and web-based specimen tracking system for managing specimen shipments. Approximately half of TASER-M clinical sites have an on-site or local genotyping laboratory. Most genotyping laboratories amplify viral sequences using in-house methods, based on assembled commercially available assay components and laboratory-specific sequencing and amplification primers. One TASER laboratory uses the commercial kit TruGene (Bayer HealthCare, Tarrytown, NY, USA). The online Stanford interpretation system is used by most laboratories to identify drug resistance-associated mutations.22 Resistance genotyping is generally performed retrospectively (i.e. not real-time) for all participants. Details of the genotyping laboratories are summarized in Supplementary Table 2 available as Supplementary data at IJE online. PASER and TASER sequences are submitted to the ViroScore database (Advanced Biological Laboratories SA, France) for data storage.
What do PASER-M and TASER-M cover and who is included in the sample?
The monitoring studies are multi-centre prospective cohort designs with sequential patient enrolment. Patient eligibility criteria are listed in Table 1. The main study objectives are to assess prevalence and incidence of HIVDR, mutational patterns and factors associated with HIVDR in persons initiating first-line ART or switching to a second-line regimen due to treatment failure under routine circumstances. Participants are required to sign informed consent prior to study enrolment and must initiate or switch ART within 30 days (PASER-M) or 181 days (TASER-M) following baseline specimen collection. Regimen switch due to treatment failure may be determined clinically, as assessed by disease progression, immunologically, by CD4 cell count or virologically, by HIV viral load. A single drug substitution, due to toxicity or intolerance, is not considered a regimen switch. Each site aims to recruit a total of 240 participants. Second-line participants are recruited among first-line participants failing therapy and the clinical site patient population. The recommended maximum site-specific enrolment period is 18 months.
Table 1.
Patient eligibility criteria for PASER-M and TASER-M
| Inclusion criteria |
| Confirmed HIV-1 infection |
| ≥18 years of age |
| Eligiblea for initiation of a first-line ART regimen or switch from a first-line ART regimen (containing at least three antiretroviral drugs and taken for ≥6 months) to a second-line ART regimen due to virological, immunological and/or clinical failure |
| Signed informed consent for study participation prior to enrolment |
| Exclusion criteria |
| Currently taking ART (minimum of three-drug regimen), if initiating a first-line ART regimenb |
| Pregnancy at enrolmentc |
| HIV-1/2 dual infection (in endemic countries only)c |
aEligibility for ART initiation defined in accordance with national ART guidelines (i.e. advanced immunodeficiency as defined by CD4 cell count less than 200 or less than 350 cells/µl, or advanced clinical disease according to WHO clinical stage/CDC classification).
bSpecified PASER-M definition: re-initiation of a first-line ART regimen <30 days after stopping previous first-line ART (previous use of antiretroviral prophylaxis or mono/dual therapy is not an exclusion criterion).
cExclusion criteria applicable to PASER-M only.
How often are participants followed-up? What data are being collected?
Participants are followed-up as per local standard of care guidelines. The frequency of follow-up visits for patients varies by site (range: every 1–6 months). The studies make use of clinical data collected during routine visits and recorded in medical records. HIV viral load measurement and, if the HIV RNA value is more than 1000 copies/ml, genotypic resistance testing is performed on plasma specimens taken at baseline, prior to regimen switch due to treatment failure and at annual follow-up. For patients failing a first-line regimen, the treatment failure data collection becomes the new baseline for the second-line regimen. Annual follow-up is then calculated from this point. For patients failing a second-line ART, the treatment failure data collection is the final assessment prior to the patient going off study.
PASER-M clinical data are recorded on standardized hard-copy data forms, which are completed at 3-monthly intervals and entered in a web-based clinical data system, called the HAART Monitoring System. PharmAccess performs quality assurance measures, which include (i) source data verification during 3- to 6-monthly site audits, (ii) checks to identify data entry inconsistencies or suspect data values and (iii) specimen tracking. TASER-M site personnel extract clinical data from site databases and medical records collected as part of usual care. From March 2008 to March 2009, TASER-M data were submitted electronically to NCHECR on a quarterly basis, as part of study start up, then at 6-monthly intervals. At each transfer, NCHECR performs quality assurance measures, which include (i) checks to identify data entry inconsistencies or suspect data values, (ii) specimen tracking and (iii) ARV history completeness. Annually, a random 10% of TASER-M patients are selected for internal site audit where submitted data are compared with patient medical records.
The studies capture standardized virological and genotypic data at protocol determined intervals. Genotyping data consist of HIV subtype and HIVDR mutations, including insertions and deletions. TASER-M also records discordant subtypes, i.e. when the PR and RT region subtypes differ. Laboratory specimen tracking information is recorded during specimen processing, allowing assessment of pre-analytical and assay validity. Genotyping laboratories complete an annual laboratory survey that includes the dynamic range of the virological assay used, the regions of PR and RT genome routinely sequenced and the interpretation algorithm used. Observational patient data includes demographic parameters, physical measures, Centres for Disease Control and Prevention (CDC) class (TASER-M) or WHO clinical stage (PASER-M), serology of hepatitis and syphilis (TASER-M), opportunistic infections, current ART regimen, ARV history, concomitant medications, routine laboratory parameters (including CD4 counts) and assessment of drug adherence. Main analyses will include age, sex, ethnicity, HIV exposure category, WHO clinical stage (PASER-M) or CDC class (TASER-M), viral hepatitis co-infection status, CD4 count, HIV viral load, HIV subtype, drug adherence, ARV history and ART regimen as covariates. Predictors of drug resistance will be assessed using logistic regression models. Incidence of drug resistance will be summarized using person-years methods and Kaplan–Meier plots. Cox proportional hazards models will be used to assess risk factors associated with developing drug resistance.
What is the anticipated attrition?
The actual attrition in PASER-M and TASER-M cannot currently be accurately estimated because the duration of follow-up in the databases is still limited. In sub-Saharan Africa patient retention in routine ART programmes has been estimated at 61.623 to 66.8%2 at 24 months on ART, attrition being mainly due to loss of follow-up and early death.23 Therefore, in PASER-M, the original site-specific sample size was calculated accounting for 20% loss to follow-up and 25% mortality after 24 months. Attrition is expected to vary between sites as a result of differences in patient populations, care provided and provisions for tracing lost to follow-up. TASER-M sites are generally sourced from the ongoing TREAT Asia HIV Observational Database (TAHOD).19 Loss to follow-up for TAHOD was 6.9/100 person-years for the 12-month period from September 2007 to September 2008. Since TASER-M monitors specified outcomes, we speculate that TASER-M follow-up will be similar to TAHOD or better.
What has been found?
PASER-M
Patient recruitment commenced in March 2007 and was completed in September 2009. Of the 13 sites, 12 reached the site-specific target of 240 participants, enrolling a total of 3005 participants. Excluding patients with protocol violations (n = 16) and key data missing (n = 4), 2985 patients were included in the analysis. Of these, 2736 (91.6%) were eligible for a first-line ART regimen and 249 (8.3%) were eligible for second-line ART due to treatment failure. Patient characteristics are summarized in Table 2. For first-line patients, the median age was 36.8 years [inter-quartile range (IQR) 31.3–42.6] and 58% were women. HIV exposure was predominantly reported as heterosexual contact. More than 60% had advanced disease (classified as WHO stages III or IV) and 37% had pre-therapy CD4 counts of less than 100 cells/µl. Across all 13 sites, median baseline CD4 counts of first-line patients were less than 200 cells/µl (site median 135 cells/μl, range 93–191). Median baseline HIV viral load was 4.9 log10 copies/ml (IQR 4.2–5.5). The most frequently prescribed first-line regimens were based on NNRTIs (99.7%), i.e. efavirenz (EFV) and nevirapine (NVP) at 60 and 40%, respectively. First-line dual NRTI backbones were predominantly lamivudine (3TC)/zidovudine (AZT) (37%), emtricitabine (FTC)/tenofovir (TDF) (34%) and 3TC/stavudine (d4T) (26%). Overall, 67% of patients started a 3TC-containing first-line regimen. Among patients initiating first-line ART, 95% (n = 2 598) reported to be ARV-naive and 5% (n = 138) had previous ARV experience, which included ART (n = 60), mono/dual therapy (n = 6), single-dose NVP for prevention of mother-to-child transmission of HIV (PMTCT) (n = 35), combination therapy for PMTCT (n = 19), and unspecified (n = 22). Compared with ARV-naive first-line patients, ARV-experienced first-line patients had higher median CD4 counts (177 vs 133 cells/μl, P < 0.0001), were younger (median 34.7 vs 37.0 years, P < 0.0001) and were more likely to be female (76.1 vs 57.5%; P < 0.001). Other baseline characteristics did not differ between ARV-naive and ARV-experienced patients.
Table 2.
Baseline patient characteristics, by region and line of ART
| PASER-M (Africa) |
TASER-M (Asia) |
||||||
|---|---|---|---|---|---|---|---|
| Initiation of first-line ARTa |
Switch to second-line ARTb | Initiation of first-line ARTa |
Switch to second-line ARTb | ||||
| Total | ARV-naive | ARV-experiencedc | ARV-naive | ARV-experiencedc | |||
| Patients, n (%) | 3713 | 2598 (87.0) | 138 (4.6) | 249 (8.3) | 693 (95.2) | 10 (1.4) | 25 (3.4) |
| Sex | |||||||
| Female, n (%) | 1988 (53.5) | 1494 (57.5) | 105 (76.1) | 124 (49.8) | 239 (34.5) | 10 (100.0) | 16 (64.0) |
| Male, n (%) | 1725 (46.5) | 1104 (42.5) | 33 (23.9) | 125 (50.2) | 454 (65.5) | 0 (0.0) | 9 (36.0) |
| Age (years), median (IQR) | 36.9 (31.7–43.3) | 37.0 (31.2–42.8) | 34.7 (29–2–40.2) | 38.6 (32.9–44.2) | 36.5 (31.1–43.2) | 33.1 (27.4–38.4) | 36.5 (32.4–41.9) |
| 18–29 | 707 (19.0) | 490 (18.9) | 39 (28.3) | 28 (11.2) | 143 (20.6) | 3 (30.0) | 4 (16.0) |
| 30–39 | 1668 (44.9) | 1176 (45.3) | 65 (47.1) | 112 (45.0) | 298 (43.0) | 5 (50.0) | 12 (48.0) |
| ≥40 | 1338 (36.0) | 932 (35.9) | 34 (24.6) | 109 (43.8) | 252 (36.4) | 2 (20.0) | 9 (36.0) |
| HIV exposure, n (%) | |||||||
| Heterosexual contact | 2583 (69.6) | 1731 (66.6) | 108 (78.3) | 191 (76.7) | 520 (75.0) | 10 (100.0) | 23 (92.0) |
| Homosexual contact | 134 (3.6) | 4 (0.2) | 0 (0.0) | 0 (0.0) | 128 (18.5) | 0 (0.0) | 2 (8.0) |
| Otherd | 996 (26.8) | 863 (33.2) | 30 (21.7) | 58 (23.3) | 45 (6.5) | 0 (0.0) | 0 (0.0) |
| WHO clinical stages, n (%) | |||||||
| I | 493 (16.5) | 393 (15.1) | 25 (18.1) | 75 (30.1) | na | na | na |
| II | 711 (23.8) | 623 (24.0) | 33 (23.9) | 55 (22.1) | na | na | na |
| III | 1281 (42.9) | 1145 (44.1) | 59 (42.8) | 77 (30.9) | na | na | na |
| IV | 500 (16.7) | 437 (16.8) | 21 (15.2) | 42 (16.9) | na | na | na |
| CDC classification, n (%) | |||||||
| A | 302 (41.5) | na | na | na | 296 (42.7) | 0 (0.0) | 6 (24.0) |
| B | 166 (22.8) | na | na | na | 152 (21.9) | 10 (100.0) | 4 (16.0) |
| C | 260 (35.7) | na | na | na | 245 (35.4) | 0 (0.0) | 15 (60.0) |
| Ever pulmonary tuberculosis, n (%) | 741 (20.0) | 569 (21.9) | 25 (18.1) | 74 (29.7) | 69 (10.0) | na | 4 (16.0) |
| Hepatitis Be, n (%) | 36 (4.9) | na | na | na | 35 (5.1) | 0 (0.0) | 1 (4.0) |
| Hepatitis Cf, n (%) | 56 (7.7) | na | na | na | 55 (7.9) | 0 (0.0) | 1 (4.0) |
| History of ARV drug use, n (%) | 422 (11.4) | na | 138 (100.0) | 249 (100.0) | na | 10 (100.0) | 25 (100.0) |
| ART | 334 (9.0) | na | 60 (43.5) | 249 (100.0) | na | 0 (0.0) | 25 (100.0) |
| Mono or dual therapy | 10 (0.3) | na | 6 (4.3) | 4 (1.6) | na | 0 (0.0) | 0 (0.0) |
| Single-dose NVP for PMTCT | 36 (1.0) | na | 35 (25.4) | 1 (0.4) | na | 0 (0.0) | 0 (0.0) |
| Combination therapy for PMTCT | 31 (0.8) | na | 19 (13.8) | 2 (0.8) | na | 10 (100.0) | 0 (0.0) |
| Unspecified | 22 (0.6) | na | 22 (15.9) | 0 (0.0) | na | 0 (0.0) | 0 (0.0) |
| CD4 cell count (cells/μl), median (IQR) | 129 (56–205) | 133 (62–204) | 177 (92–262) | 125 (46–-196) | 99 (33.5–201) | 169 (151–222) | 197 (109–299) |
| <100 | 1456 (39.2) | 975 (37.5) | 38 (27.5) | 102 (41.0) | 337 (48.6) | 1 (10.0) | 3 (12.0) |
| 100–199 | 1215 (32.7) | 914 (35.2) | 42 (30.4) | 81 (32.5) | 164 (23.7) | 6 (60.0) | 8 (32.0) |
| ≥200 | 1007 (27.1) | 702 (27.0) | 57 (41.3) | 64 (25.7) | 171 (24.7) | 3 (30.0) | 10 (40.0) |
| Unknown | 25 (0.7) | 7 (0.3) | 1 (0.7) | 2 (0.8) | 21 (3.0) | 0 (0.0) | 4 (16.0) |
| HIV-1 RNA (log10 copies/ml), median (IQR) | 4.9 (4.3–5.5) | 4.9 (4.3–5.6) | 4.8 (4.2–5.5) | 4.1 (3.2–5.0) | 5.0 (5.4–6.8) | 4.8 (4.5–5.0) | 4.0 (3.6–4.5) |
| ART regimen | |||||||
| NNRTI-based triple regimen | 3330 (89.7) | 2590 (99.7) | 135 (97.8) | 2 (0.8) | 593 (85.6) | 10 (100.0) | 0 (0.0) |
| AZT-containing | 1302 (35.1) | 964 (37.2) | 51 (37.8) | 1 (0.4) | 170 (28.7) | 10 (100.0) | 0 (0.0) |
| TDF-containing | 1088 (29.3) | 868 (33.5) | 49 (36.3) | 1 (0.4) | 110 (18.5) | 0 (0.0) | 0 (0.0) |
| d4T-containing | 833 (22.4) | 690 (26.6) | 33 (24.4) | 0 (0.0) | 276 (46.5) | 0 (0.0) | 0 (0.0) |
| ABC-containing | 106 (2.9) | 68 (2.6) | 2 (1.5) | 0 (0.0) | 37 (6.2) | 0 (0.0) | 0 (0.0) |
| 3TC-containing | 2387 (71.7) | 1746 (67.4) | 89 (65.9) | 0 (0.0) | 542 (91.4) | 10 (100.0) | 0 (0.0) |
| FTC-containing | 942 (28.3) | 843 (32.5) | 48 (35.6) | 0 (0.0) | 49 (8.3) | 0 (0.0) | 0 (0.0) |
| PI-based triple regimen | 351 (9.5) | 6 (0.2) | 0 (0.0) | 247 (99.2) | 73 (10.5) | 0 (0.0) | 25 (100.0) |
| Triple NRTI regimen | 29 (0.8) | 2 (0.1) | 3 (2.2) | 0 (0.0) | 24 (3.5) | 0 (0.0) | 0 (0.0) |
| NNRTI+PI-based triple regimen | 3 (0.1) | 0 (0.0) | 0 (0.0 | 0 (0.0) | 3 (0.4) | 0 (0.0) | 0 (0.0) |
Data are n (%) of patients, unless otherwise indicated. na, not available; ART, combination antiretroviral therapy; ARV, antiretroviral; CDC, US Center for Disease Control and Prevention; WHO, World Health Organization; PMTCT, prevention of mother-to-child transmission of HIV-1; TB, tuberculosis; IQR, interquartile range; NVP, nevirapine; d4T, stavudine; AZT, zidovudine; TDF, tenofovir; ABC, abacavir; 3TC, lamivudine; FTC, emtricitabine; NRTI, nucleoside reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, protease inhibitor.
aEligibility for ART initiation in accordance with national ART guidelines (i.e. advanced immunodeficiency as defined by CD4 cell count less than 200 or less than 350 cells/µl, or advanced clinical disease according to WHO clinical stages or CDC classification).
bRegimen switch due to treatment failure, defined by local standard of care guidelines as determined clinically, immunologically or virologically.
cARV-experienced is defined as any previous use of ARVs, i.e. (first-line) ART, mono/dual therapy and/or PMTCT.
dIncludes recipients of blood products, injecting drug users, perinatal transmission and unknown exposures.
eHepatitis B positive status was defined as being HBsAg positive.
fHepatitis C positive status was defined as being HCV antibody positive.
For the 249 (8.3%) patients switching to second-line ART, the median age was 38.6 years (IQR 32.9–44.2) and sex was equally distributed. HIV exposure was predominantly heterosexual contact and 48% of patients had advanced disease (classified as WHO stages III or IV). Median CD4 count was 125 cells/μl (IQR 46–196). Median pre-switch HIV viral load was 4.1 log10 copies/ml (IQR 3.2–5.0). Ritonavir-boosted lopinavir (LPV) was the PI used almost exclusively (98%).
As shown in Table 3, analysis of the first available 1795 viral sequences demonstrated that the most common HIV subtypes in the cohort were C (1216, 68.7%), A (338, 18.7%) and D (179, 10.0%). The first PASER report published in 2008 reviewed the available data on HIVDR in sub-Saharan Africa.11 Baseline HIVDR data from Lusaka, Zambia, has recently been published.24 International presentations have summarized preliminary baseline HIVDR mutations and subtype distributions.25,26
Table 3.
HIV subtypes and circulating recombinant formsa by region and country
| PASER-M (Africa) |
TASER-M (Asia)b |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total | South Africa | Zambia | Uganda | Kenya | Nigeria | Zimbabwe | Thailand | Hong Kong | Malaysia | |
| n (%) | 2523 (100.0) | 624 (24.7) | 583 (23.1) | 410 (16.3) | 140 (5.5) | 21 (0.8) | 17 (0.7) | 542 (21.5) | 111 (4.4) | 75 (3.0) |
| A | 340 (13.6) | 2 (0.3) | 4 (0.7) | 235 (57.3) | 97 (69.1) | 1 (4.8) | 1 (0.2) | |||
| B | 114 (4.5) | 3 (0.5) | 37 (7.0) | 55 (49.5) | 19 (25.3) | |||||
| C | 1236 (49.2) | 617 (98.9) | 571 (97.9) | 9 (2.2) | 19 (13.6) | 17 (100.0) | 2 (1.8) | 1 (1.3) | ||
| D | 179 (7.1) | 1 (0.2) | 1 (0.2) | 160 (39.0) | 17 (12.1) | |||||
| G | 11 (0.4) | 2 (0.3) | 1 (0.2) | 2 (1.4) | 6 (28.6) | |||||
| CRF01_AE | 585 (23.2) | 485 (89.5) | 52 (46.8) | 48 (64.0) | ||||||
| CRF02_AG | 20 (0.8) | 1 (0.2) | 3 (0.5) | 1 (0.2) | 14 (66.7) | 1 (0.2) | ||||
| Other recombinants or discordant subtypes | 32 (1.3) | 2 (0.3) | 1 (0.2) | 2 (1.4) | 18 (3.3) | 2 (1.8) | 7 (9.3) | |||
| Unclassified | 6 (0.2) | 3 (0.7) | 3 (2.1) | |||||||
Data are n (%) of subtypes/CRFs, unless otherwise indicated. CRF, circulating recombinant form.
aHIV subtypes were determined from the pol sequences, using the REGA HIV-1 subtyping algorithm version 2.0 (http://dbpartners.stanford.edu/RegaSubtyping) and/or Stanford HIV drug resistance database (http://hivdb.stanford.edu).
bThe collaborating sites in Philippines, South Korea and Indonesia had not yet provided sequence data at time of the current analysis.
TASER-M
Patient recruitment commenced in March 2007 and for the March 2009 transfer, seven sites from Thailand, Hong Kong and Malaysia provided data. Of 773 patients, 755 (97.7%) commenced ART within 181 days of baseline specimen collection and 728 (96.4%) participants had genotypic data available. Of these, 693 (95.2%) ARV-naive patients and 10 (1.4%) ARV-experienced patients, following prior PMTCT, were eligible for first-line regimens. A further 25 (3.5%) patients were eligible for second-line ART following first-line treatment failure. Patient characteristics are summarized in Table 2. For ARV-naive first-line patients, the median age was 36.5 years. Almost two-thirds of patients were male and HIV exposure was predominantly heterosexual contact. More than one-third of patients were classified as CDC class C and almost half of the patients had pre-therapy CD4 counts less than 100 cells/µl. Median baseline HIV viral load was 5.0 log10 copies/ml (IQR 5.4–6.8) and the most common first-line regimens were based on NNRTIs (85.6%). Excluding 14 (2.4%) patients on a randomized clinical trial with a blinded NNRTI component, NVP was more commonly prescribed than EFV at 56 and 42%, respectively. First-line dual NRTI backbones were predominantly 3TC/d4T (47%), 3TC/AZT (29%) and 3TC/TDF (10%). For first-line PI regimens, the favoured NRTI backbone was FTC/TDF (33%) compared with 3TC/d4T (30%) or 3TC/AZT (18%). For the ritonavir-boosted PI component, atazanavir (ATZ) was only slightly favoured over LPV at 43 vs 41%, respectively. Overall, 542 (91.4%) of ARV-naive patients started a 3TC-containing first-line regimen. The 10 PMTCT patients received perinatal prophylaxis of AZT/3TC/NVP (n = 7), AZT (n = 2) or AZT/NVP (n = 1) for between 14 and 102 days and all were prescribed AZT/3TC/NVP as first-line regimens.
For the 25 (3.4%) second-line patients, 22 (88%) were of Thai ethnicity and the median age was 36.5 years (IQR 32.4–41.9). Females were in the majority (64%), HIV exposure was predominantly heterosexual (92%) and 60% of patients had experienced at least one CDC class C event. Of 21 patients with CD4 counts available within 6 months of starting a second-line therapy, the median CD4 count was 197 (IQR 109–299). Median pre-switch HIV viral load was 4.0 log10 copies per/ml (IQR 3.6–4.5). All patients were on PI-based regimens, following failure on first-line NNRTI-based regimens (median duration 30.3 months). The most commonly prescribed PI component was ritonavir-boosted LPV (88%).
As shown in Table 3, from analysis of the 728 available viral sequences, the most common subtypes were CRF01_AE (584, 80.2%) and subtype B (111, 15.2%). Non-CRF01_AE recombinants were identified in eight (1.1%) patient specimens. For 21 (2.9%) specimens, the subtype differed between PR and RT regions, suggesting dual infection or recombination. International presentations have summarized 2009 baseline HIVDR mutations and subtype distributions.27,28
Complete baseline and prospective outcome data for PASER-M and TASER-M are anticipated to become available in 2010–13.
What are the main strengths and weaknesses?
Programmes that monitor national and regional levels of primary and secondary HIVDR contribute to evidence-based recommendations to inform treatment guidelines and provide feedback on the success of HIV treatment and prevention programmes. PASER and TASER, with TAQAS, are developing capacity in sub-Saharan Africa and the Asia-Pacific for coordinated HIVDR monitoring and genotypic laboratory testing. The study protocols are harmonized with the WHO HIV Drug Resistance Prevention Survey protocol.29 An important strength is the large number of patients and sites participating, representing a diverse spectrum of patient populations, clinic types, ART regimens and HIV subtypes. Opportunities exist to investigate the impact of drug resistance on HIV natural history, rates of disease progression and response to treatment in non-B subtypes. Data from genotypic resistance testing will also provide insight into the population genetics and dynamics of transmitted HIVDR in the region.
PASER-M and TASER-M have several limitations. First, patient samples at each site are not necessarily representative of the site, country or region. Second, data quality depends on the completeness of clinical information captured through routine patient care. In PASER-M, data may have been collected under varying conditions, since some sites had no or limited research experience at study initiation. Third, at some sites, study initiation was delayed by several months due to the time required for contract negotiation, IRB study approval and, in TASER-M, procurement of FWAs. After study initiation, recruiting the required number of patients within the recommended 18-month period proved difficult for some sites, due to asymptomatic patients not seeking care or treatment, cost of medication or low-prevalence in their setting. Fourth, HIVDR monitoring activities in resource-limited countries in sub-Saharan Africa are limited by high costs of laboratory testing. To address this challenge, PASER has initiated a public–private consortium, called Affordable Resistance Test for Africa (ART-A), which aims to develop affordable test algorithms for the detection and interpretation of HIVDR for use in laboratories and clinics (http://www.arta-africa.org).
How can I collaborate? Where can I find out more?
Ownership of individual site data remains with the contributing site. Sites are represented by their principal investigators on the respective PASER and TASER Steering Committees. Research is to be the subject of peer-reviewed publications and analysis priorities are driven by a concept sheet process. Both studies accept concept proposals from external researchers for review, if submitted in collaboration with one or more of the site principal investigators. The PASER and TASER protocols contribute data under the LAASER partnership (http://www.laaserhivaids.org) and TASER also contributes data to IeDEA.20 Collaborating sites are also encouraged to make an appropriate subset of their data available to Ministry of Health in their respective countries in order to contribute to local efforts in monitoring HIVDR. Questions regarding participation, research concepts or requests for data should be sent to Tobias Rinke de Wit, email: t.rinkedewit@pharmaccess.org (PASER), or Thida Singtoroj, email: thida.singtoroj@treatasia.org (TASER).
Supplementary Data
Supplementary Data are available at IJE online.
Funding
The PharmAccess African Studies to Evaluate Resistance is an initiative of PharmAccess Foundation, supported by the Ministry of Foreign Affairs of The Netherlands through a partnership with Stichting Aids Fonds (grant no 12454). The TREAT Asia Studies to Evaluate Resistance is an initiative of TREAT Asia, a programme of amfAR, The Foundation for AIDS Research, with major support provided by the Ministry of Foreign Affairs of The Netherlands through a partnership with Stichting Aids Fonds (grant no 12454), and with additional support from amfAR and the National Institute of Allergy and Infectious Diseases (NIAID) of the U.S. National Institutes of Health (NIH) and the National Cancer Institute (NCI) as part of the International Epidemiologic Databases to Evaluate AIDS (IeDEA) (grant no U01AI069907). Queen Elizabeth Hospital and the Integrated Treatment Centre are supported by the Hong Kong Council for AIDS Trust Fund. The National Centre in HIV Epidemiology and Clinical Research is funded by the Australian Government Department of Health and Ageing and is affiliated with the Faculty of Medicine, The University of New South Wales. The funders had no role in the study design, data collection, data analysis, data interpretation, decision to publish or writing of the report. The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of any of the institutions mentioned above.
Supplementary Material
Acknowledgements
The authors wish to thank the participants, site staff and support staff at PharmAccess Foundation, Contract Laboratory Services, TREAT Asia and NCHECR. Special thanks to Donald Sutherland and Diane Bennett (WHO) for encouraging the development of PASER and TASER.
Conflict of interest: None declared.
Appendix 1
The PharmAccess African Studies to Evaluate Resistance
M Siwale* and C Njovu, Lusaka Trust Hospital, Lusaka, Zambia;
M Labib, Coptic Hospital, Lusaka, Zambia;
J Menke, KARA Clinic and Laboratory, Lusaka, Zambia;
ME Botes*, Muelmed Hospital, Pretoria, South Africa;
F Conradie* and I Sanne, Themba Lethu Clinic, Clinical HIV Research Unit, University of the Witwatersrand, Johannesburg, South Africa;
CL Wallis and WS Stevens‡, Department of Molecular Medicine and Haematology, University of the Witwatersrand, and National Health Laboratory Services, Johannesburg, South Africa;
M Hardman, Acts Clinic, White River, South Africa;
N Ngorima*, M Wellington and R Luthy, Newlands Clinic, Harare, Zimbabwe;
K Mandaliya*, S Abdallah and I Jao, Coast Province General Hospital, International Center for Reproductive Health Kenya, Mombasa, Kenya;
M Dolan, Mater Misericordiae Hospital, Nairobi, Kenya;
C Kityo*, G Namayanja, L Nakatudde, M Kiconco, M Abwola and P Mugyenyi, Joint Clinical Research Center, Fort Portal, Mbale and Kampala, Uganda;
N Ndembi‡ and P Kaleebu*, Medical Research Council, Uganda Virus Research Institute, Entebbe, Uganda;
A Osibogun* and S Akanmu, Lagos University Teaching Hospital, Lagos, Nigeria;
AM Wensing, I Derdelinckx and R Schuurman‡, Department of Virology, University Medical Center, Utrecht, The Netherlands;
RL Hamers*, K Sigaloff, E Straatsma, M van Vugt*, TF Rinke de Wit*†, PharmAccess Foundation, Amsterdam Institute for Global Health and Development, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands
*PASER Steering Committee member
†Principal Investigator
‡PASER Scientific Advisor
The TREAT Asia Studies to Evaluate Resistance
PCK Li*¶ and MP Lee, Queen Elizabeth Hospital, and KH Wong, Integrated Treatment Centre, Hong Kong, China;
N Kumarasamy*§ and S Saghayam, YRG Centre for AIDS Research and Education, Chennai, India;
S Pujari* and K Joshi, Institute of Infectious Diseases, Pune, India;
TP Merati* and F Yuliana, Faculty of Medicine, Udayana University & Sanglah Hospital, Bali, Indonesia;
CKC Lee* and LL Low, Hospital Sungai Buloh, Kuala Lumpur, Malaysia;
A Kamarulzaman*¶ and LY Ong, University of Malaya, Kuala Lumpur, Malaysia;
M Mustafa and N Nordin, Hospital Raja Perempuan Zainab II, Kota Bharu, Malaysia;
R Ditangco† and RO Bantique, Research Institute for Tropical Medicine, Manila, Philippines;
YMA Chen*§, WW Wong and YW Yang, Taipei Veterans General Hospital and AIDS Prevention and Research Centre, National Yang-Ming University, Taipei, Taiwan;
P Phanuphak* and S Sirivichayakul, HIV-NAT/Thai Red Cross AIDS Research Centre, Bangkok, Thailand;
S Sungkanuparph* and B Piyavong, Faculty of Medicine, Ramathibodi Hospital, Mahidol University, Bangkok, Thailand;
T Sirisanthana* and J Praparattanapan, Research Institute for Health Sciences, Chiang Mai University, Chiang Mai, Thailand;
P Kantipong‡ and P Kambua, Chiang Rai Regional Hospital, Chiang Rai, Thailand;
JY Choi* and SH Han, Division of Infectious Diseases, Department of Internal Medicine, Yonsei University College of Medicine, Seoul, South Korea;
W Ratanasuwan and R Sriondee, Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand;
R Kantor, Brown University, RI, USA;
AH Sohn, L Messerschmidt* and T Singtoroj, TREAT Asia, amfAR – The Foundation for AIDS Research, Bangkok, Thailand;
DA Cooper, MG Law*, A Jiamsakul, and J. Zhou, National Centre in HIV Epidemiology and Clinical Research, The University of New South Wales, Sydney, Australia.
*TASER Steering Committee member
†Steering Committee Chair, ‡Co-Chair
§Protocol Chair
¶Protocol Co-Chair
References
- 1.Joint United Nations Programme on HIV/AIDS and World Health Organization. AIDS Epidemic Update 2009. Geneva, Switzerland: http://data.unaids.org/pub/Report/2009/JC1700_Epi_Update_2009_en.pdf (1 July 2010, date last accessed) [Google Scholar]
- 2.World Health Organization. Towards Universal Access: Scaling Up Priority HIV/AIDS Interventions in the Health Sector: Progress Report 2009. http://www.who.int/hiv/pub/tuapr_2009_en.pdf (1 July 2010, date last accessed) [Google Scholar]
- 3.Braitstein P, Brinkhof MW, Dabis F, et al. Mortality of HIV-1-infected patients in the first year of antiretroviral therapy: comparison between low-income and high-income countries. Lancet. 2006;367:817–24. doi: 10.1016/S0140-6736(06)68337-2. [DOI] [PubMed] [Google Scholar]
- 4.Ivers LC, Kendrick D, Doucette K. Efficacy of antiretroviral therapy programs in resource-poor settings: a meta-analysis of the published literature. Clin Infect Dis. 2005;41:217–24. doi: 10.1086/431199. [DOI] [PubMed] [Google Scholar]
- 5.Jahn A, Floyd S, Crampin AC, et al. Population-level effect of HIV on adult mortality and early evidence of reversal after introduction of antiretroviral therapy in Malawi. Lancet. 2008;371:1603–11. doi: 10.1016/S0140-6736(08)60693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. Antiretroviral Therapy for HIV Infection in Adults and Adolescents: Recommendations for a Public Health Approach, 2006 Revision. Geneva, 2006. http://www.who.int/hiv/pub/guidelines/artadultguidelines.pdf (1 October 2009, date last accessed) [Google Scholar]
- 7.Keiser O, Orrell C, Egger M, et al. Public-health and individual approaches to antiretroviral therapy: township South Africa and Switzerland compared. PLoS Med. 2008;5:e148. doi: 10.1371/journal.pmed.0050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RK, Hill A, Sawyer AW, et al. Virological monitoring and resistance to first-line highly active antiretroviral therapy in adults infected with HIV-1 treated under WHO guidelines: a systematic review and meta-analysis. The Lancet Infect dis. 2009;9:409–17. doi: 10.1016/S1473-3099(09)70136-7. [DOI] [PubMed] [Google Scholar]
- 9.Rahim S, Fredrick LM, da Silva BA, Bernstein B, King MS. Geographic and temporal trends of transmitted HIV-1 drug resistance among antiretroviral-naive subjects screening for two clinical trials in North America and Western Europe. HIV Clin Trials. 2009;10:94–103. doi: 10.1310/hct1002-94. [DOI] [PubMed] [Google Scholar]
- 10.Wensing AM, van de Vijver DA, Angarano G, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis. 2005;192:958–66. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar]
- 11.Hamers RL, Derdelinkx I, Van Vugt M, Stevens W, Rinke de Wit TF, Schuurman R. The status of HIV-1 resistance to antiretroviral drugs in sub-Saharan Africa. Antivir Ther. 2008;13:625–39. [PubMed] [Google Scholar]
- 12.Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl. 2):1–13. [PubMed] [Google Scholar]
- 13.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 14.Dumans AT, Soares MA, Machado ES, et al. Synonymous genetic polymorphisms within Brazilian human immunodeficiency virus Type 1 subtypes may influence mutational routes to drug resistance. J Infect Dis. 2004;189:1232–38. doi: 10.1086/382483. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez LM, Brindeiro RM, Aguiar RS, et al. Impact of nelfinavir resistance mutations on in vitro phenotype, fitness, and replication capacity of human immunodeficiency virus type 1 with subtype B and C proteases. Antimicrob Agents Chemother. 2004;48:3552–55. doi: 10.1128/AAC.48.9.3552-3555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kantor R, Katzenstein DA, Efron B, et al. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: results of a global collaboration. PLoS Med. 2005;2:e112. doi: 10.1371/journal.pmed.0020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Cajas JL, Pant-Pai N, Klein MB, Wainberg MA. Role of genetic diversity amongst HIV-1 non-B subtypes in drug resistance: a systematic review of virologic and biochemical evidence. AIDS Rev. 2008;10:212–23. [PubMed] [Google Scholar]
- 18.Land S, Cunningham P, Zhou J, et al. TREAT Asia Quality Assessment Scheme (TAQAS) to standardize the outcome of HIV genotypic resistance testing in a group of Asian laboratories. J Virol Methods. 2009;159:185–93. doi: 10.1016/j.jviromet.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J, Kumarasamy N, Ditangco R, et al. The TREAT Asia HIV Observational Database: baseline and retrospective data. J Acquir Immune Defic Syndr. 2005;38:174–79. doi: 10.1097/01.qai.0000145351.96815.d5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGowan CC, Cahn P, Gotuzzo E, et al. Cohort Profile: Caribbean, Central and South America Network for HIV research (CCASAnet) collaboration within the International Epidemiologic Databases to Evaluate AIDS (IeDEA) programme. Int J Epidemiol. 2007;36:969–76. doi: 10.1093/ije/dym073. [DOI] [PubMed] [Google Scholar]
- 21.Bertagnolio S, Derdelinckx I, Parker M, et al. World Health Organization/HIVResNet Drug Resistance Laboratory Strategy. Antivir Ther. 2008;13(Suppl. 2):49–57. [PubMed] [Google Scholar]
- 22.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosen S, Fox MP, Gill CJ. Patient retention in antiretroviral therapy programs in sub-Saharan Africa: a systematic review. PLoS Med. 2007;4:e298. doi: 10.1371/journal.pmed.0040298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamers RL, Siwale M, Wallis C, et al. HIV-1 drug resistance mutations are present in 6 percent of persons initiating antiretroviral therapy in Lusaka, Zambia. J Acquir Immune Defic Syndr. 2010;55:95–101. doi: 10.1097/QAI.0b013e3181e544e0. [DOI] [PubMed] [Google Scholar]
- 25.Hamers RL, Kityo C, Siwale M, et al. Cohort Profile: The PharmAccess African Studies to Evaluate Resistance Monitoring Study (PASER-M) – HIV-1 Drug Resistance in sub-Saharan Africa. XVIII International AIDS Conference, July 18–23 2010, Vienna, Austria. Abstract no THPE0236. 2010. [Google Scholar]
- 26.Hamers RL, Wensing AM, Siwale M, et al. Prevalence of drug-resistant HIV-1 variants at initiation of standard first-line HAART in Africa. XVIII International HIV Drug Resistance Workshop. Fort Myers, Fl, USA. June 9–13, 2009. Abstract 155. p. 155. [Google Scholar]
- 27.Sungkanuparph S, Oyomopito R, Sirivichayakul S, et al. HIV-1 drug resistance among antiretroviral-naive HIV-1-infected patients in Asia: results from the TREAT Asia Studies to Evaluate Resistance-Monitoring Study (TASER-M) European AIDS Conference, 11–14 November 2009. Cologne, Germany. [Google Scholar]
- 28.Oyomopito R, Phanuphak P, Sungkanuparph S, et al. Cohort profile: the TREAT Asia studies to evaluate resistance monitoring study (TASER-M) – HIV drug resistance in the Asia-Pacific. XVIII International AIDS Conference, July 18–23, 2010, Vienna, Austria. Abstract no THPE0443. 2010. [Google Scholar]
- 29.Jordan MR, Bennett DE, Bertagnolio S, Gilks CF, Sutherland D. World Health Organization surveys to monitor HIV drug resistance prevention and associated factors in sentinel antiretroviral treatment sites. Antivir Ther. 2008;13(Suppl. 2):15–23. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.