Abstract
Over the last two decades, the mutation of mitochondrial DNA (mtDNA) has emerged as a major cause of inherited human disease. The disorders present clinically in at least 1 in 10 000 adults, but pathogenic mutations are found in approximately 1 in 200 of the background population. Mitochondrial DNA is maternally inherited and there can be marked phenotypic variability within the same family. Heteroplasmy is a significant factor and environmental toxins also appear to modulate the phenotype. Although genetic and biochemical studies have provided part of the explanation, a comprehensive understanding of the incomplete penetrance of these diseases is lacking—both at the population and family levels. Here, we review the potential role of epigenetic factors in the pathogenesis of mtDNA diseases and the contribution that epidemiological approaches can make to improve our understanding in this area. Despite being previously dismissed, there is an emerging evidence that mitochondria contain the machinery required to epigenetically modify mtDNA expression. In addition, the increased production of reactive oxygen species seen in several mtDNA diseases could lead to the epigenetic modification of the nuclear genome, including chromatin remodelling and alterations to DNA methylation and microRNA expression, thus contributing to the diverse pathophysiology observed in this group of diseases. These observations open the door to future studies investigating the role of mtDNA methylation in human disease.
Keywords: DNA methylation, epigenomics, mitochondrial diseases, mtDNA, mitochondrial
Introduction
Mitochondrial biogenesis
Mitochondria are the primary source of intracellular energy in the form of adenosine triphosphate (ATP). ATP is synthesized by more than 100 proteins organized into five respiratory chain complexes on the inner mitochondrial membrane. Reduced cofactors generated from the intermediary metabolism of carbohydrates, proteins and fats donate electrons to complexes I and II. Electron flux through the respiratory chain is linked to the expulsion of protons from within the mitochondrial matrix into the inter-membrane space. This generates an electrochemical gradient that is harnessed by complex V (ATP synthase) to synthesize ATP. ATP is required for all active cellular processes, from the maintenance of ionic gradients across cell membranes to muscle contraction, so a deficiency of ATP synthesis can have catastrophic effects for the cell, organ and individual.1
The mitochondrial respiratory chain has a dual genetic origin. Thirteen key proteins are synthesized from multiple copies of circular double-stranded DNA present within each mitochondrion: the mitochondrial genome or mtDNA. mtDNA also codes for 24 RNA genes that are required for intra-mitochondrial protein synthesis.2 However, the vast majority of respiratory chain components and proteins required for the synthesis, expression and regulation of mitochondrial genes are encoded by the cell nucleus.3 Efficient mitochondrial function is thus critically dependent on the concerted action of two genomes, and mitochondrial diseases can be due to mutations of either nuclear DNA or mtDNA (Figure 1).4 This review will focus on mtDNA and disease, although some of the downstream consequences could equally apply to mitochondrial disorders due to primary nuclear gene defects, especially if these are mediated through secondary mtDNA damage.
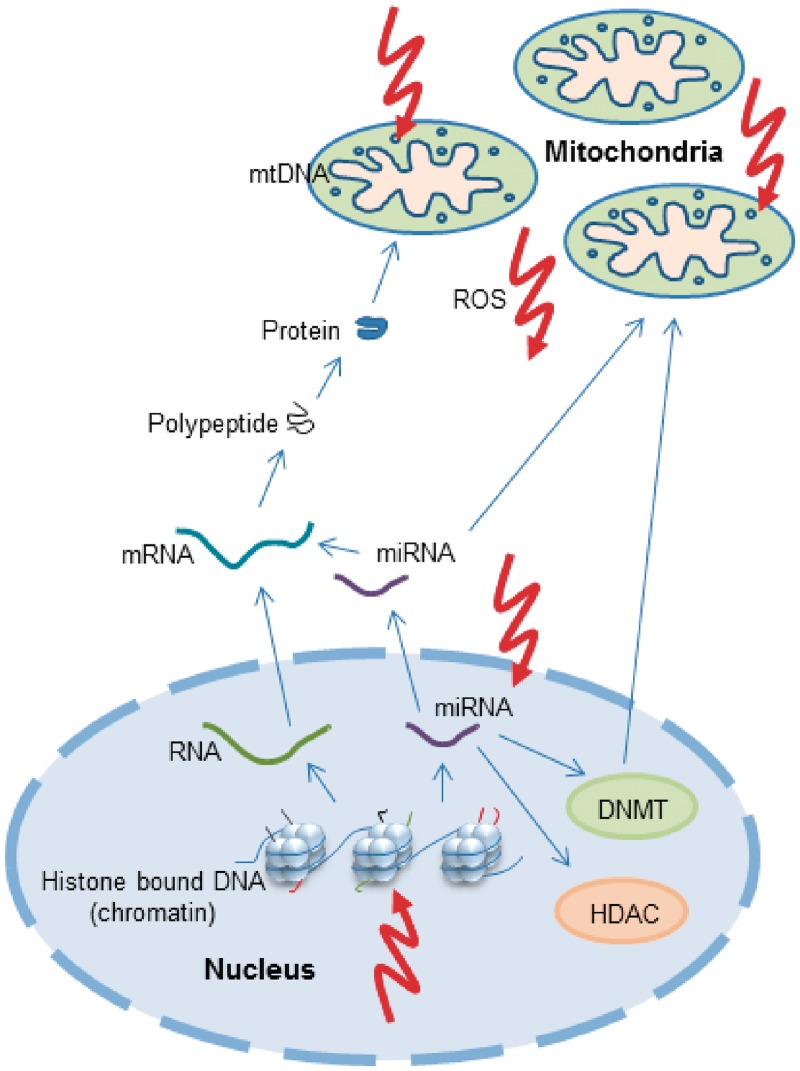
Figure 1.
A schematic representation of mitochondrial and nuclear genomes and their inter-relation with epigenetic factors. The nuclear genome is coiled around histone octamers to form nucleosomes. The tails of histone proteins are decorated with a variety of modifications that influence the regulation of gene expression. Permissive histone markings allow transcription from DNA to RNA, post-transcription processing to mRNA and translation to polypeptides and thus proteins. Nuclear-encoded mitochondrial proteins are then translocated into the mitochondrion. mtDNA also encodes genes essential for intra-mitochondrial protein synthesis, but this genome is not histone bound. In addition to mRNA, miRNA are also transcribed from nuclear DNA and can interfere with mRNA to induce degradation or suppress translation. miRNAs can influence mitochondrial metabolism and some miRNAs are known to directly activate the generation of ROS. Furthermore, miRNAs influence the expression of DNMT and HDAC enzymes. ROS produced by mitochondria or other endogenous sources can damage the mtDNA genome directly as well as influence epigenetic machinery at several levels, either through damage to miRNA or through the alteration of histone modifications. DNMTs translocate to the mitochondria and bind to mtDNA, although evidence that this is to effect epigenetic regulation remains elusive. HDAC, histone deacetylase; mRNA, messenger RNA; ROS, reactive oxygen species
MtDNA and human diseases
Human mtDNA was first sequenced in 1981,2 and the first pathogenic mutations were identified <10 years later.5,6 More than 200 different molecular defects have subsequently been described in patients with mitochondrial diseases.7 Point mutations can affect the various structural subunits of the respiratory chain, or compromise protein synthesis through the RNA genes. Alternatively, large-scale deletions of mtDNA remove one or more essential genes. Duplications of mtDNA have been described in association with mtDNA deletions, but it remains unclear whether they cause a biochemical defect.8
There are three major differences between mtDNA and nuclear DNA, which are relevant for our understanding of human disease. First, mammalian cells contain many copies of mtDNA, ranging from a few hundred to hundreds of thousands, depending on the cell type. Although the amount of mtDNA appears to be tightly regulated in a tissue-specific manner, this can change over time through poorly understood regulatory mechanisms.9 Secondly, pathogenic mutations can affect a varying proportion of the many mtDNA molecules—from 1% to 100%, and every possibility in between. This situation is known as heteroplasmy. The percentage level of the mutation can vary from person to person and even between adjacent cells within the same individual.10 Cells that contain a high percentage level of mutant mtDNA express a biochemical defect. This is often associated with a reduced amount of wild-type mtDNA.11 Tissue and organs that contain many affected cells lead to the clinical features of disease. Thirdly, for all practical purposes, mtDNA is exclusively inherited down the maternal line.12 This means that men with mtDNA disease cannot pass on the disorder to their offspring. Women harbouring heteroplasmic mtDNA mutations pass on very different levels of mutation to each of their children. This occurs because only a small group of maternal mtDNA is transmitted to the offspring (the mtDNA genetic bottleneck),13,14 which leads to a statistical sampling effect and rapid shifts in the heteroplasmy level within a single generation.
A further consequence of strict maternal transmission is that the mtDNA undergoes negligible inter-molecular recombination at the population level.15 As a result, mtDNA in the human population has acquired polymorphic variants that subdivide the population into geographical clusters.16 The major clusters are called mtDNA haplogroups. There is emerging evidence that these haplogroups have subtle biochemical consequences,17 providing a potential explanation for the genetic association of common polymorphic variants of mtDNA with complex common human diseases such as ischaemic stroke, diabetes and Parkinsons disease (reviewed in ref. 17).17 Although contentious, these polymorphic variants may also have shaped human evolution in response to the environment.18
Unanswered questions
Although phenotypic heterogeneity of primary mtDNA diseases can be partly explained by differences in mtDNA heteroplasmy, several major questions remain unanswered. For some common mtDNA diseases, all maternal relatives carry only the mutated mtDNA—a situation known as ‘homoplasmic mutant’. Leber hereditary optic neuropathy (LHON) is a common cause of inherited visual failure.19 In Europeans, LHON is primarily due to one of three pathogenic mutations of the mtDNA: m.11778G>A; m.3460G>A; m.14484T>C, which are found in >90% of the affected individuals.20 The disease is maternally transmitted and typically presents in young adult life with sequential painless visual loss, but only ~40% of the men and ~10% of the women become affected, despite all usually being homoplasmic for the causative mtDNA mutation.21 Several additional factors have been implicated in the pathophysiology, including the background mtDNA haplogroup,22 which appears to involve epistatic effects through common polymorphisms in the mtDNA cytochrome b gene.23,24 Cigarette smoking and alcohol intake are the major risk factors for the visual loss in LHON families,25 and nuclear genetic mapping studies implicate interacting nuclear genes.26,27 Finally, the homoplasmic mutations that cause LHON are found in approximately 1 in 300 of the background population,28 but only cause blindness in approximately 1 in 20 000.29 LHON can therefore be considered as a complex disease trait, with several genetic and environmental factors interacting to cause the disease.
Other examples include the m.1555A>G mutation that causes isolated (non-syndromic) deafness both spontaneously and in response to environmental exposure to aminoglycoside antibiotics.30 Again, the m.1555A>G mutation is found in approximately 1 in 300 of the population.28 These, and other examples, highlight a recurring theme: that homoplasmic mtDNA mutations are present in control subjects, only rarely cause disease and most strikingly cause tissue-specific phenotypes that differ from mutation to mutation. The reasons for the variable penetrance and tissue specificity are not known, and could well be due to the epigenetic modification of the mitochondrial genome, which influences intra-mitochondrial transcription or nuclear genes known to influence or modulate mitochondrial function.
Could epigenetic mechanisms involving mtDNA provide the answer?
Evidence for mtDNA associated changes in DNA methylation
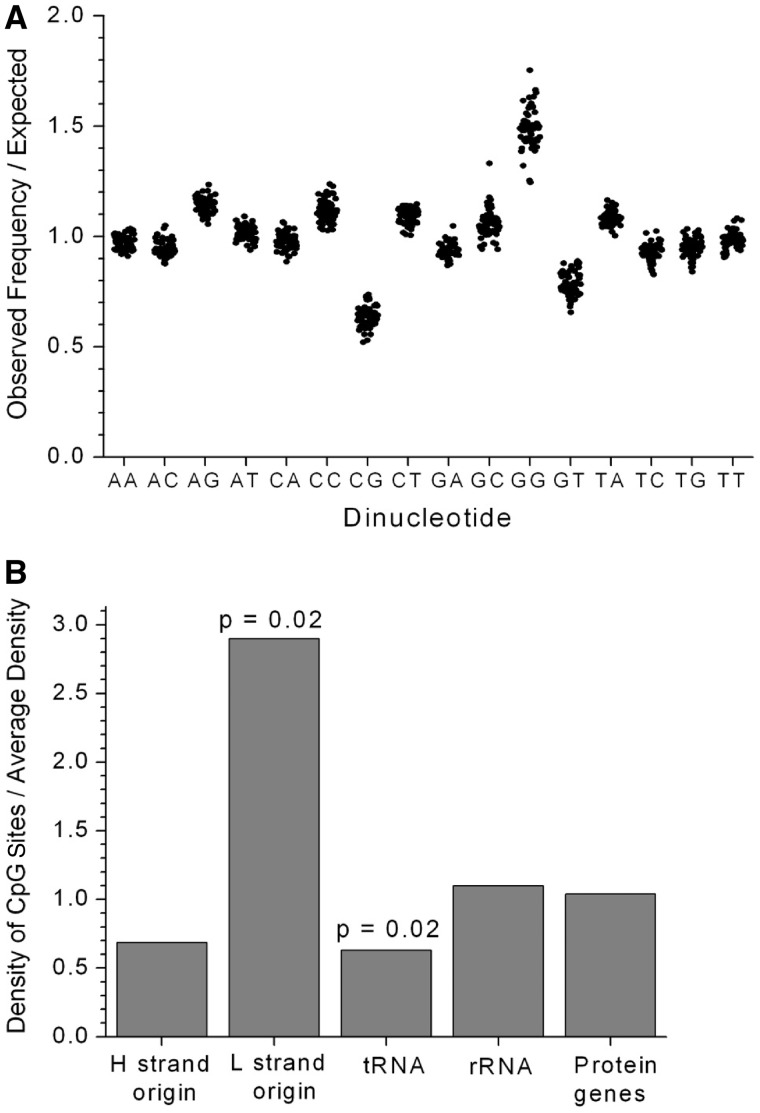
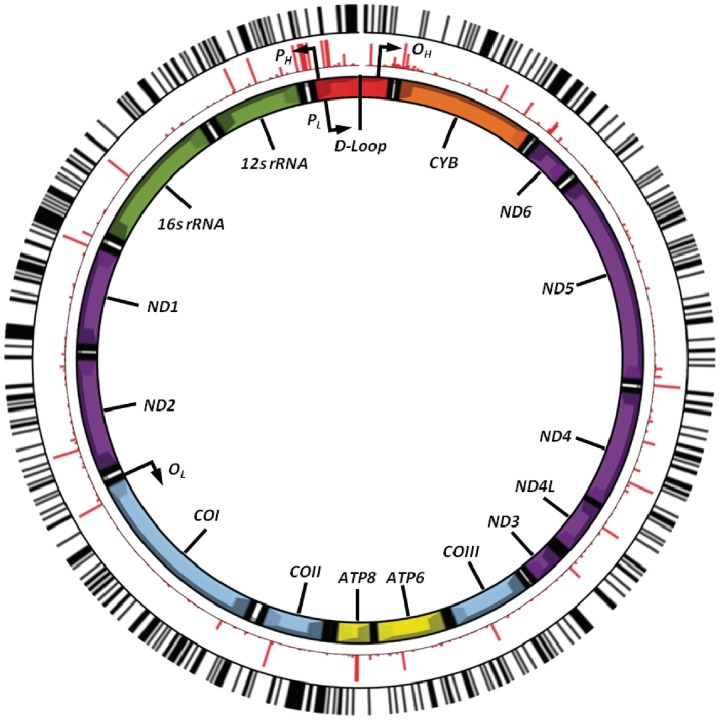
MtDNA molecules are arranged in clusters, called nucleoids, which are tethered to the mitochondrial membrane and devoid of histones.9,31 Based on early experimental evidence in frogs and HeLa cells,32 the prevailing view has been that mitochondria lack the machinery required for DNA methylation. However, several strands of evidence suggest that this is not the case. Bioinformatic analysis across several species shows a lower frequency of CG dinucleotides than would be expected by chance (Figure 2a), implying a selective force acting against CpG sites in mtDNA. For comparison, the CpG fraction in the human nuclear genome is only 25% of the expected frequency, but this rises to 45% in exons.33 Secondly, the distribution of CpG sites is not even across the human mtDNA sequence (Figure 2b), with a low frequency in tRNA genes and a high frequency in OL, the origin of mtDNA light strand replication. This raises the possibility that the methylation of OL could influence mtDNA replication. The location of the 435 predicted CpG sites present in the mitochondrial genome is shown in Figure 3. It can be seen that the CpG sites appear to cluster, and when considered in relation to the 3358 polymorphic variants in the mitochondrial genome, more than half of the CpG sites in the mitochondrial genome co-locate with polymorphic variants. These observations clearly require further investigation.
Figure 2.
The distribution of CpG sites in mtDNA. (a) The observed frequencies of the 16 dinucleotides divided by the expected frequencies for a random sequence based on the individual nucleotide frequencies. Values are plotted for the human mtDNA sequence plus 60 other mammalian species. (b) The density of the CpG sites in different sections of the human mtDNA sequence (rCRS), normalized by the average density of 0.026 per site. P-values are calculated by chi-square tests with Yates correction. OL and OH locations were taken from the mtDNA function locations list at MITOMAP (www.MitoMap.org) without alteration. OL was defined as 5721–5798 and OH was defined as 110–441
Figure 3.
The location of CpG sites in the mitochondrial genome relative to mitochondrial haplogroup defining polymorphisms. The image shows the mitochondrial genome (centre), showing the position and relative size of each of the 13 major mitochondrial genes, the origins of heavy-strand and light-strand replication (OH and OL, respectively) as well as both the light-strand and heavy-strand promoter sites (PL and PH, respectively). Shown in the middle (in red) are the relative frequencies and positions of 3358 mtDNA variants (MAF = 0.01–49.6% of 2147 full European mtDNA sequences). The outer ring (in black) shows the relative position of each of the 435 predicted CpG sites
Patterns of mtDNA methylation were first described in the mouse in the early 1980s,34 but at very low levels (<5%). These data comprised a ‘global’ measure of DNA methylation but without single base pair resolution, which is now far more tractable following recent technical developments. More recently, several key regions involved in the regulation of mtDNA gene expression were shown to be relatively protected from methylation in vitro within cultured human cell lines.35 Intriguingly, these regions included OL, despite the high CpG content. Inducing mtDNA transcription appeared to decrease in vivo methylation, whereas inducing mtDNA replication led to increased methylation. The relatively low level of mtDNA methylation under standard cell-culture conditions could be related to the structural arrangement of mtDNA within nucleoids and associated nucleoid proteins.35
Finally, DNA methyltransferase 1 (DNMT1) has recently been shown to translocate into mitochondria through a mitochondrial targeting pre-peptide sequence found upstream of the mature peptide, enabling direct access to mtDNA.36 DNMT1 binds to the mtDNA within the mitochondrial matrix, and the level of intra-mitochondrial DNMT1 is up-regulated by genes known to regulate mitochondrial biogenesis, including NRF1 and PGC1α.36 There, therefore, appears to be a mechanism in place to enable the regulation of intra-mitochondrial gene expression through the methylation of mtDNA cytosine residues.
A key observation is the modulation of mtDNA methylation in response to oxidative stress. The induction of reactive oxygen species (ROS) with sublethal doses of H2O2 has been shown to decrease mtDNA methylation (although it is also possible that nucleoids remodel into a more compact state during oxidative stress to provide a more insulated environment for mtDNA).35 Although controversial, there is evidence implicating increased ROS production in mtDNA diseases,1 raising the possibility that the suppression of mtDNA methylation acts as a compensatory response to mtDNA damage, thus increasing gene expression from residual intact mtDNA molecules. Furthermore, the influence of endogenous exposures, including ROS, has also been implicated in mitochondrial disease due to nuclear gene as well as mtDNA defects.37,38 The altered DNA methylation of the nuclear genome could therefore mediate the downstream consequences of pathogenic mtDNA mutations through hitherto unknown pathways, perhaps not directly related to ATP synthesis, nor indeed even directly related to mitochondrial function. If correct, then the tissue-specific expression of nuclear genes could contribute to the tissue selectivity in mtDNA diseases, and a variable epigenetic signature could explain the phenotypic variability of mitochondrial disorders.
In keeping with these hypotheses, recent evidence using an agnostic global assessment of DNA methylation showed that cells deplete in mtDNA showed altered DNA methylation of the nuclear genome, which was rescued upon the repletion of mtDNA—implicating the mitochondrion as an important determinant of epigenetic status.36 This raises the possibility that the increased ROS production in mitochondrial diseases downregulates nuclear–mitochondrial genes, further compromising respiratory chain activity. This would establish a vicious cycle, generating further ROS and ultimately leading to bio-energetic failure.
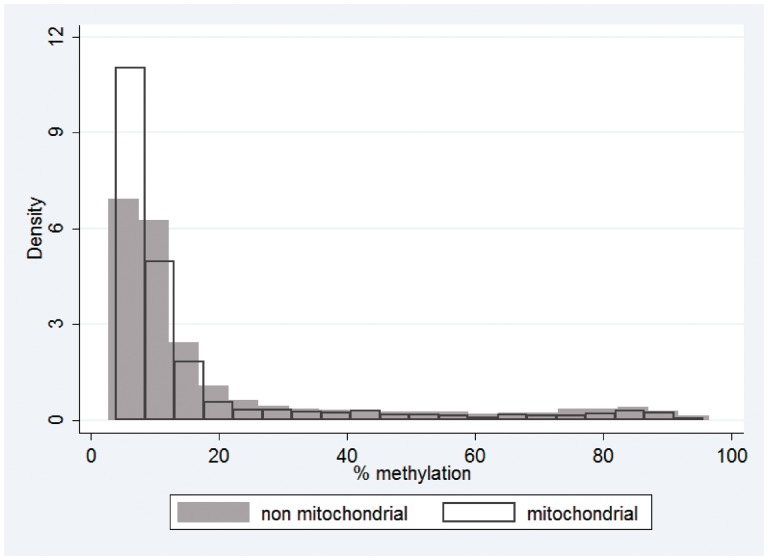
Figure 4 provides a preliminary appraisal of the level of variation in DNA methylation in CpG island probes in nuclear-encoded mitochondrial genes in a group of 24 normal individuals free of mitochondrial disease. Array probes were assigned to ‘non-mitochondrial’ or ‘mitochondrial’ groups based on the CpG location. Mitochondrial gene IDs were extracted from the MitoProteome resource (www.mitoproteome.org) and matched to gene IDs on the methylation array. More than half (85%) of the gene IDs extracted from mitoproteome were present on the methylation array. It should also be noted that the non-mitochondrial group is likely to include a number of probes targeting proteins of unknown function and cellular location that target the mitochondria. A total of 1239 mitochondrial probes and 26 339 non-mitochondrial probes were analysed. One thousand and forty-two mitochondrial probes and 18 964 non-mitochondrial probes were located in CpG islands. Mitochondrial probes were preferentially located within CpG islands compared with non-mitochondrial probes (Fisher's exact P = 1.058e−22). Outside CpG islands, there was no difference in methylation level between mitochondrial and non-mitochondrial genes. However, within CpG islands, mitochondrial probes had lower levels of methylation than non-mitochondrial probes. Mitochondrial median (IQR) was 8.4 (6.5–13.7) and non-mitochondrial median (IQR) was 9.7 (6.7–18.5), with P = 2.967e−09. These observations relate to DNA methylation patterns in peripheral blood cells and may conceivably differ in other tissues, perhaps reflecting metabolic activity.
Figure 4.
DNA methylation levels in nuclear-encoded mitochondrial genes in a control population. Twenty-four healthy female adult samples were analysed for genome-wide methylation using Illumina® HumanMethylation27 arrays. Probes were assigned to ‘non-mitochondrial’ or ‘mitochondrial’ groups as described in the main text. Histogram shows the DNA methylation distribution of CpG island probes assigned to non-mitochondrial or mitochondrial groups
Since there is a general trend between the methylation and level of transcription, this observation suggests that nuclear mitochondrial genes might be more actively expressed than their non-mitochondrial gene counterparts. Further analysis of functional groupings of genes could provide evidence of coordinated regulation mediated through epigenetic mechanisms. Direct measurement of methylation in different regions of the mtDNA would also give further insight; this is the focus of future work. Knowledge of mtDNA methylation and DNA methylation of nuclear mitochondrial genes may play an important role in the future epidemiological understanding of mitochondrial disorders.
A link between mitochondrial function and nuclear DNA histone modifications
Histones are nuclear proteins around which the nuclear DNA is wound. Nucleosomes are formed from assembled groups of histones, and these nucleosomes are able to fold and wind into a higher order structure. Histones and DNA that have assembled into a condensed higher order structure form chromatin. Histones can be modified by a number of post-transcriptional modifications including acetylation, phosphorylation, ubiquitination and methylation. These modifications change their ability to regulate or repair cellular processes. In general, DNA that is less bound to histones is more transcriptionally active than DNA that is bound.39
Although the mtDNA is known for its lack of protective histones (one of the reasons why mtDNA mutations are relatively common), there is evidence that some types of histones localize to the mitochondrial membrane.40 However, these levels are recorded at much lower levels than in the nucleus. In the nuclear DNA, histone modifications are important in transcriptional control. They can become altered in diseases affecting nuclear-encoded mitochondrial proteins, of which Friedreich ataxia is an example. This disorder is caused by transcriptional silencing of the FXN gene, which encodes a mitochondrial protein involved in biosynthesis of iron–sulphur clusters. It is characterized by a tri-nucleotide expansion, which leads to the formation of densely packed heterochromatin. The possibility of using histone deacetylase inhibitors in the treatment of this disorder is being investigated.41 The role of histone modifications in the transcriptional regulation of other nuclear mitochondrial genes warrants further attention.
MicroRNA and the mitochondrion
MicroRNAs (miRNAs) are highly conserved, ~21-mer, non-coding RNAs and are important regulators of gene expression in human cells. Post-transcriptional regulation by miRNAs has been reported in numerous diseases, particularly in cancer.42 An explosion of interest has led to more than 1000 mammalian miRNAs being described. Following transcription and a number of processing steps, mature miRNAs regulate gene expression by binding to target mRNAs, usually in the 3'-untranslated regions. miRNAs may elicit their effect through a number of routes, most commonly via the targeting of mRNA for degradation or by suppression of mRNA translation. The mechanisms of maturation and action of miRNAs have been reviewed elsewhere.43 miRNAs are important regulators of cellular function, and since their effect is not part of the transcriptional machinery, they are able to quickly alter the cell's ability to translate mRNA in response to the surrounding cellular environment. In the case of an mtDNA disorder, the immediate cellular environment may be altered, hypothetically causing an miRNA-mediated response.
Although there is no evidence, to date, to demonstrate that mitochondrial disorders directly affect miRNA expression, an increasing number of miRNAs has been shown to be involved in the regulation of mitochondrial metabolism. For example, miR-210 is postulated to target both units of the electron transport chain and tri-carboxylic acid cycle, reducing the rate of mitochondrial metabolism.44 Evidence suggests that another miRNA, miR-30, regulates mitochondrial fission.44,45 In rats, it has been postulated that a small pool of miRNAs is associated with the mitochondria; these miRNAs appear to modulate the expression of genes associated with apoptosis, cell proliferation and differentiation.46 Many of these pathways are associated with response to environmental stress such as hypoxia and may lead to downstream ROS production. As described earlier, this can affect other epigenetic marks including methylation. Furthermore, a number of miRNAs (including miR-210 and miR-128a) are known to activate the generation of ROS directly.44,47
To highlight the complexity and inter-related nature of epigenetic regulation, recent studies have demonstrated that DNA methylation and histone modification not only regulate the expression of miRNA genes but these marks are themselves regulated by some miRNAs.48 A subset of miRNAs influences the expression of DNA methyltransferases and histone deacetylases.49,50 Thus, it could be envisaged that, for example, ROS-induced alteration of DNA methylation patterns in the nuclear genome might alter miRNA expression and this, in turn, could impact upon other aspects of epigenetic regulation and ultimately, possibly pleiotropically, upon disease phenotype. Such complex interaction of epigenetic regulation, however, is not unique to mitochondrial disease.
Epidemiological approaches applicable to investigating epigenetic variation in the context of mitochondrial diseases
Conventional epidemiological study designs can be applied to investigate the role of epigenetic variation in relation to a variety of intermediate phenotypes or diseases,51 including mitochondrial disease. Epigenetic variation can be considered as a continuous variable or phenotype and analysed using standard epidemiological methods, although with some caveats relating to the largely non-normal distribution of epigenetic data. Selecting the most appropriate study design requires consideration not only of epigenetic phenomena but also of the idiosyncrasies of mitochondrial genetics and the inheritance patterns of mitochondrial diseases.
A case–control study design would allow the comparison of epigenetic patterns in subjects with mitochondrial disease and those without clinical signs of disease and ideally without disease-causing mitochondrial mutations. Family-based studies, including trios of mother, father and child, probably offer the most powerful design to interrogate the relationship between epigenetic variation and mitochondrial disease, this design having been widely used in the study of mitochondrial diseases, given their maternal inheritance patterns.52 The use of paternal controls has been advocated in conventional epidemiological investigations when assessing maternal in utero effects.53 For example, assessing the relationship of maternal smoking and offspring birthweight and then comparing this with paternal smoking and offspring birthweight will give some indication of likely confounding. Similar risk estimates from both paternal and maternal analyses would be suggestive of confounding, as maternal smoking, if a causal biological birthweight-reducing mechanism, should have a much larger influence than paternal smoking.
Epigenetic modifications have inherent features that must be considered in designing epidemiological studies. Epigenetic patterns change over time with a high degree of plasticity in early life and stochastic, age-related loss of fidelity in older age.54 Therefore, any cross-sectional appraisal, for example, of DNA methylation could only be confidently associated with any specific outcome at the time of sampling. This, however, should not detract from the potential utility of epigenetic marks as diagnostic or prognostic biomarkers. Serial sampling or longitudinal study designs can overcome some of these limitations. Unlike genetic epidemiology, which is robust to the vagaries of confounding and reverse causation that plague conventional observational epidemiology, epigenetic epidemiology is not. Potential confounders such as age, smoking and diet should be routinely considered in epigenetic studies for this reason.
Numerous environmental factors are known to impact upon the epigenome;55 indeed, this has fuelled interest in epigenetics as a biological mechanism mediating environmental influences on complex disease risk,56 including mitochondrial diseases. As mentioned earlier in the context of LHON, environmental factors of particular relevance to the aetiology of mitochondrial diseases include smoking and alcohol intake.
For the reasons outlined in the introduction, further complexity arises when studying the potential role of the epigenetic modification of mtDNA in mitochondrial diseases. Different levels of heteroplasmy7—both within and between individuals—and the influence of background polymorphic variation (haplogroups)24 confound the relationship between mtDNA genotype and clinical phenotype for common pathogenic mtDNA mutations. The factors must be accounted for in any epidemiological approach.
Methods of measuring epigenetic variation relevant to investigating mitochondrial diseases
Technologies available for the analysis of epigenetic variation have undergone rapid development in recent years, harnessing the methodological advances from the broader field of genomics. The highly detailed annotation of nuclear genome-wide DNA methylation is now a realistic proposition,57,58 and less detailed, high-throughput appraisal of genome-wide DNA methylation is a cost-effective approach to highlight areas of differential nuclear DNA methylation.59 A wide repertoire of techniques is available to quantify DNA methylation, assess histone modifications or measure miRNA expression levels in the context of nuclear DNA (reviewed in detail elsewhere),60 and could be applied to investigate the influence of mitochondrial dysfunction on the nuclear epigenome.
DNA from peripheral blood samples is frequently used in population-based studies, primarily due to its ease of collection and storage. The suitability of using DNA from peripheral blood for analysing DNA methylation has been questioned, especially since the DNA methylation pattern may not be representative of the disease-relevant target tissue. Measuring DNA methylation in blood has recently been reviewed.61 Additionally, DNA methylation is known to vary between blood cell types, further complicating analysis.62,63 However, DNA methylation variation in peripheral blood has been proposed as a marker for a number of cancers63,64 and complex diseases including type 2 diabetes.65 More recent studies have begun to account for blood-cell type differences.63,65The tissue-specific nature of epigenetic patterns may hold additional relevance for mitochondrial studies, where the heteroplasmic load of mtDNA mutations varies between tissues59 and peripheral blood DNA might only be appropriate to analyse if heteroplasmy levels reflect those of the relevant target tissue.
Some of the approaches that can be readily applied to the direct analysis of mtDNA are summarized below. These are not intended to be exhaustive, merely illustrative.
Bisulphite sequencing
The bisulphite modification of DNA forms the basis of a number of methods to detect and quantify the level of methylation in a given sample, including bisulphite sequencing. The bisulphite modification of DNA provides a way of differentiating between cytosine bases in CpG sites that are methylated and those which are not. Methylated cytosine bases have a methyl group bound to the carbon-5 position of their pyrimidine ring. During bisulphite treatment, 5-methylcytosine (5-mC) residues are protected, but cytosine residues that are unmethylated are converted to uracil. Although widely used, bisulphite treatment denatures a high proportion of the template DNA, thus reducing the number of longer length templates for downstream applications. The availability of longer fragments has been studied in depth.66 Despite this length limitation, a number of downstream methods can be employed to measure how much of the original template was methylated. The simplest of these methods is sequencing of the bisulphite-modified DNA. Bisulphite sequencing is a well-established method, is able to assess single CpG sites and is relatively quick to perform, and is a tractable undertaking in the context of the 16 kb mitochondrial genome. However, it does not allow the percentage methylation at a given site to be quantified. During sequencing, methylated and unmethylated bases are differentiated on the basis of detecting a cytosine or thymidine base at a given CpG site. More recently, second-generation sequencing has been used to quantify methylation state at high resolution;57,67 this approach could also be utilized in the quantification of methylation in the mitochondrial genome.
It is worth noting that bisulphite modification does not differentiate between 5-mC and 5-hydroxymethylcytosine (5-hmC), which occurs when a methyl group followed by a hydroxyl group is added to a cytosine residue. The existence of 5-hmC to date has been reported in the brain and ES cells, but it is unknown whether this form of covalent epigenetic modification occurs in mtDNA. The relative biological roles of 5-mC and 5-hmC have been described.68 5-hmC has also been shown to be involved in mammalian transcriptional regulation.69 To measure the contribution from each of these methylation species, another method should be used, for example, methylated DNA immunoprecipitation (MeDIP) using antibodies to 5-mC or 5-hmC. Platforms are now coming online that have the capability to undertake single molecular, real-time sequencing that can distinguish between 5-mC and 5-hmC. For example, the Pacific Biosciences PACBIO RS platform and Helicos BioSciences Corporation's Genetic Analysis platforms currently lead the way in this area.
Pyrosequencing®
Pyrosequencing® is a sequence by synthesis method to detect the methylation of CpG sites over a short region of DNA, typically between 50 and 100 bp in length.
It uses a bisulphite-modified DNA template, from which amplicons are generated in a standard polymerase chain reaction (PCR) using a biotin-labelled primer. A sequencing primer is annealed to single-stranded PCR amplicons and incubated with a cocktail of enzymes and substrates. During this sequencing by synthesis method, deoxyribonucleotide triphosphates (dNTPs) are added sequentially to the template, and light produced in the luciferase-catalysed reaction is recorded. The light detected is proportional to the number of nucleotides incorporated. By recording the signal from the incorporation of C or T dinucleotides at a CpG site, the site-specific percentage of methylated DNA template can be quantified. This method has been reported to have a detection limit of 2–5%.70 Pyrosequencing® has been widely used as a method for detecting methylation. Drawbacks include the low to medium throughput (96-well plates) and the small number of CpG sites that can be investigated in one read, thus requiring multiple amplicons to cover a sizeable region.
Sequenom® EpiTYPER®
The EpiTYPER® platform, as with Pyrosequencing®, relies on the initial generation of bisulphite-modified DNA and measures DNA methylation. Modified DNA is amplified by PCR using primers designed to target the region of interest, then is subjected to in vitro transcription. This step generates a single-stranded RNA molecule that is cleaved in a base-specific manner. The resulting fragments are analysed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). This method is rapid and high throughput. Small amounts of input DNA are required (10–20 ng) and CpG methylation in up to 400 bases can be detected. The detection limit of the method is ~5%,71 although some reports suggest it is slightly lower at ~3%.72 One of the main drawbacks of this method is that CpG sites in close proximity to each other are often not resolved following RNA cleavage. This results in CpG methylation values that are averages of a pair or group of CpG sites. Overlapping and duplicated signals arising from fragments of the same mass also complicate analysis. However, this method has been used widely to measure methylation in genomic DNA and could be used in future to assess mtDNA methylation.
Whole-genome methylation arrays
Whole-genome DNA methylation arrays have been developed, which allow high throughput analysis of DNA methylation, the most recent of which is the Illumina® HumanMethylation450 BeadChip.73 This array utilizes beads with target-specific probes to measure methylation at individual CpG sites. Although none of these probes anneal to mtDNA, a number of them bind nuclear targets that are important for mitochondrial function (as discussed earlier).
Chromatin modifications
As described earlier, mtDNA lacks protective histones and so cannot condense to form nucleosomes. Instead, mtDNA is packaged with proteins to form nucleoids. Protein–DNA interactions are commonly analysed by chromatin immunoprecipitation (ChIP) followed by a further downstream analysis of the extracted DNA (and protein).74 mtDNA-packaging proteins such as Abf2 have also been successfully characterized using ChIP-like procedures.75
miRNA expression
Expression of characterized miRNAs can be measured using array-based methods, which are currently able to measure in the region of 1100 human miRNAs.76,77 Quantitative PCR methods are also commonly used, either in profiling miRNAs using one of a number of commercially available panels or in analysis of single miRNAs.77 RNA is typically extracted from tissue or cells with enrichment for small RNA molecules.78 Following the identification of miRNAs, which have altered expression, the downstream evaluation of target gene expression in putative miRNA targets can be employed.
Transcriptome sequencing
Relative transcript abundance (between individual tRNAs, mRNAs and rRNAs), RNA variation and post-translational modification can be investigated by using next-generation sequencing (NGS) of purified mitochondria RNA. By marrying the depth of NGS, by resequencing a relatively small genome, with parallel analysis of RNA ends (PARE), the global schema of polycistron cleavage can be investigated.79
Summary and future direction
In summary, there is a strong motivation to explore the role of epigenetic mechanisms in mtDNA disease, as epigenetic factors may serve to explain the observed phenotypic heterogeneity, variable penetrance and pronounced environmental triggers in this group of disorders.
Epidemiological approaches can contribute to knowledge and understanding in this nascent field, as they have done in defining the prevalence of pathogenic mtDNA mutations in the general population.28 Well-characterized patient populations, including the detailed information of polymorphic variation in the mitochondrial genome and haplogroup categories, will greatly assist in this endeavour, as epigenetic variation can not only be investigated in relation to the presence or absence of clinically diagnosed disease status but can also probe the relationship between the mitochondrial genome, epigenetic signatures and a multitude of phenotypic traits. Furthermore, the evolutionary role of CpG sites in the mitochondrion remains unexplored and the epidemiological investigation of their distribution at a population level may well provide insight into the processes of mitochondrial genetic selection.
Technologies are now available to directly assess epigenetic variation in both the mitochondrial and nuclear genome in relation to mitochondrial disease. The challenge remains, should correlative observations be made, to discern whether these are causal in the pathogenesis of mitochondrial disease itself, in a broader array of common complex diseases in which mitochondrial dysfunction has been implicated, or whether such associations are non-causal epiphenomena.
Funding
P.F.C. is a Wellcome Trust Senior Fellow in Clinical Science and a UK National Institute for Health Research (NIHR) Senior Investigator who also receives funding from the Medical Research Council (UK), the Association Française contre les Myopathies and the UK NIHR Biomedical Research Centre for Ageing and Age-related Disease award to the Newcastle upon Tyne Foundation Hospitals NHS Trust. H.R.E. is a UK Medical Research Council-funded postdoctoral fellow and G.H. is a Wellcome Trust-funded postdoctoral fellow. D.C.S. is supported by a National Institute of Health/National Institute of General Medical Sciences grant R01GM073744. C.L.R. receives funding from the Medical Research Council (MRC), Biotechnology and Biological Sciences Research Council (BBSRC), Wellcome Trust and numerous charitable sources.
Acknowledgements
Professor Debbie Lawlor is gratefully acknowledged for use of Illumina® HumanMethylation27 BeadChip data generated as part of a Wellcome Trust-funded project she leads.
KEY MESSAGES.
Mitochondrial DNA mutations cause disease that is often influenced by environmental factors and that is characterized by variable penetrance.
Epigenetic mechanisms may play a role at multiple levels: within the mitochondrial genome itself, through the regulation of expression of mitochondrially targeted genes or through changes induced more widely in the nuclear genome as a consequence of mitochondrial dysfunction.
Technologies are now available to investigate the relationship between epigenetic patterns and mitochondrial and nuclear genomes. Epidemiological approaches can contribute to advancing knowledge in this field.
References
- 1.Wallace DC. Colloquium paper: bioenergetics, the origins of complexity, and the ascent of man. Proc Natl Acad Sci U S A. 2011;107(Suppl 2):8947–53. doi: 10.1073/pnas.0914635107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–65. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 3.Smeitink J, van den Heuvel L, DiMauro S. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet. 2001;2:342–52. doi: 10.1038/35072063. [DOI] [PubMed] [Google Scholar]
- 4.Zeviani M, Di Donato S. Mitochondrial disorders. Brain. 2004;127:2153–72. doi: 10.1093/brain/awh259. [DOI] [PubMed] [Google Scholar]
- 5.Holt I, Harding AE, Morgan-Hughes JA. Deletion of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–19. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 6.Wallace DC, Singh G, Lott MT, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1988;242:1427–30. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 7.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schon E, Hirano M, DiMauro S. Mitochondrial encephalomyopathies: clinical and molecular analysis. J Bioenerg Biomemb. 1994;26:291–99. doi: 10.1007/BF00763100. [DOI] [PubMed] [Google Scholar]
- 9.Holt IJ. Zen and the art of mitochondrial DNA maintenance. Trends Genet. 2010;26:103–9. doi: 10.1016/j.tig.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet. 1997;13:450–55. doi: 10.1016/s0168-9525(97)01266-3. [DOI] [PubMed] [Google Scholar]
- 11.Durham SE, Samuels DC, Cree LM, Chinnery PF. Normal levels of wild-type mitochondrial DNA maintain cytochrome c oxidase activity for two pathogenic mitochondrial DNA mutations but not for m.3243A–>G. Am J Hum Genet. 2007;81:189–95. doi: 10.1086/518901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poulton J, Bredenoord AL. 174th ENMC international workshop: Applying pre-implantation genetic diagnosis to mtDNA diseases: implications of scientific advances 19–21 March 2010, Naarden, The Netherlands. Neuromuscul Disord. 2010;20:559–63. doi: 10.1016/j.nmd.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Cree LM, Samuels DC, de Sousa Lopes SC, et al. A reduction of mitochondrial DNA molecules during embryogenesis explains the rapid segregation of genotypes. Nat Genet. 2008;40:249–54. doi: 10.1038/ng.2007.63. [DOI] [PubMed] [Google Scholar]
- 14.Wai T, Teoli D, Shoubridge EA. The mitochondrial DNA genetic bottleneck results from replication of a subpopulation of genomes. Nat Genet. 2008;40:1484–88. doi: 10.1038/ng.258. [DOI] [PubMed] [Google Scholar]
- 15.Elson JL, Andrews RM, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Analysis of European mtDNAs for recombination. Am J Hum Genet. 2001;68:145–53. doi: 10.1086/316938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torroni A, Achilli A, Macaulay V, Richards M, Bandelt HJ. Harvesting the fruit of the human mtDNA tree. Trends Genet. 2006;22:339–45. doi: 10.1016/j.tig.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Duran A, Pacheu-Grau D, Lopez-Gallardo E, et al. Unmasking the causes of multifactorial disorders: OXPHOS differences between mitochondrial haplogroups. Hum Mol Genet. 2010;19:3343–53. doi: 10.1093/hmg/ddq246. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–26. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 19.Sadun AA, Morgia CL, Carelli V. Leber's hereditary optic neuropathy. Curr Treat Options Neurol. 2011;13:109–17. doi: 10.1007/s11940-010-0100-y. [DOI] [PubMed] [Google Scholar]
- 20.Harding AE, Sweeney MG, Govan GG, Riordan-Eva P. Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am J Hum Genet. 1995;57:77–86. [PMC free article] [PubMed] [Google Scholar]
- 21.Riordan-Eva P, Sanders MD, Govan GG, Sweeney MG, Da Costa J, Harding AE. The clinical features of Leber's hereditary optic neuropathy defined by the presence of a pathogenic mitochondrial DNA mutation. Brain. 1995;118:319–37. doi: 10.1093/brain/118.2.319. [DOI] [PubMed] [Google Scholar]
- 22.Brown MD, Sun F, Wallace DC. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am J Hum Genet. 1997;60:381–87. [PMC free article] [PubMed] [Google Scholar]
- 23.Carelli V, Achilli A, Valentino ML, et al. Haplogroup effects and recombination of mitochondrial DNA: novel clues from the analysis of Leber hereditary optic neuropathy pedigrees. Am J Hum Genet. 2006;78:564–74. doi: 10.1086/501236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudson G, Carelli V, Spruijt L, et al. Clinical expression of Leber hereditary optic neuropathy is affected by the mitochondrial DNA-haplogroup background. Am J Hum Genet. 2007;81:228–33. doi: 10.1086/519394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkman MA, Yu-Wai-Man P, Korsten A, et al. Gene–environment interactions in Leber hereditary optic neuropathy. Brain. 2009;132:2317–26. doi: 10.1093/brain/awp158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hudson G, Keers S, Man PY, et al. Identification of an x-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am J Hum Genet. 2005;77:1086–91. doi: 10.1086/498176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shankar SP, Fingert JH, Carelli V, et al. Evidence for a novel x-linked modifier locus for Leber hereditary optic neuropathy. Ophthalmic Genet. 2008;29:17–24. doi: 10.1080/13816810701867607. [DOI] [PubMed] [Google Scholar]
- 28.Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008;83:254–60. doi: 10.1016/j.ajhg.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Man PY, Griffiths PG, Brown DT, Howell N, Turnbull DM, Chinnery PF. The epidemiology of Leber hereditary optic neuropathy in the north east of England. Am J Hum Genet. 2003;72:333–39. doi: 10.1086/346066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan MX, Enriquez JA, Fischel-Ghodsian N, et al. The deafness-associated mitochondrial DNA mutation at position 7445, which affects tRNASer(UCN) precursor processing, has long-range effects on NADH dehydrogenase subunit ND6 gene expression. Mol Cell Biol. 1998;18:5868–79. doi: 10.1128/mcb.18.10.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satoh M, Kuroiwa T. Organisation of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp Cell Res. 1991;196:137–40. doi: 10.1016/0014-4827(91)90467-9. [DOI] [PubMed] [Google Scholar]
- 32.Dawid IB. 5-methylcytidylic acid: absence from mitochondrial DNA of frogs and HeLa cells. Science. 1974;184:80–81. doi: 10.1126/science.184.4132.80. [DOI] [PubMed] [Google Scholar]
- 33.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–17. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pollack Y, Kasir J, Shemer R, Metzger S, Szyf M. Methylation pattern of mouse mitochondrial DNA. Nucleic Acids Res. 1984;12:4811–24. doi: 10.1093/nar/12.12.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rebelo AP, Williams SL, Moraes CT. In vivo methylation of mtDNA reveals the dynamics of protein-mtDNA interactions. Nucleic Acids Res. 2009;37:6701–15. doi: 10.1093/nar/gkp727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shock LS, Thakkar PV, Peterson EJ, Moran RG, Taylor SM. DNA methyltransferase 1, cytosine methylation, and cytosine hydroxymethylation in mammalian mitochondria. Proc Natl Acad Sci U S A. 2011;108:3630–35. doi: 10.1073/pnas.1012311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carelli V, Ghelli A, Bucchi L, et al. Biochemical features of mtDNA 14484 (ND6/M64V) point mutation associated with Leber's hereditary optic neuropathy. Ann Neurol. 1999;45:320–28. [PubMed] [Google Scholar]
- 38.Carelli V, Ross-Cisneros FN, Sadun AA. Mitochondrial dysfunction as a cause of optic neuropathies. Prog Retin Eye Res. 2004;23:53–89. doi: 10.1016/j.preteyeres.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 39.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 40.Choi YS, Hoon Jeong J, Min HK, et al. Shot-gun proteomic analysis of mitochondrial D-loop DNA binding proteins: identification of mitochondrial histones. Mol Biosyst. 2011;7:1523–36. doi: 10.1039/c0mb00277a. [DOI] [PubMed] [Google Scholar]
- 41.Rai M, Soragni E, Chou CJ, et al. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich's ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farazi TA, Spitzer JI, Morozov P, Tuschl T. miRNAs in human cancer. J Pathol. 2011;223:102–15. doi: 10.1002/path.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–68. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kren BT, Wong PY, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting Bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5:e10748. doi: 10.1371/journal.pone.0010748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato F, Tsuchiya S, Meltzer SJ, Shimizu K. MicroRNAs and epigenetics. FEBS J. 2011;278:1598–609. doi: 10.1111/j.1742-4658.2011.08089.x. [DOI] [PubMed] [Google Scholar]
- 49.Yan H, Choi AJ, Lee BH, Ting AH. Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS One. 2011;6:e20628. doi: 10.1371/journal.pone.0020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H, Wu J, Meng X, et al. MicroRNA-342 inhibits colorectal cancer cell proliferation and invasion by directly targeting DNA methyltransferase 1. Carcinogenesis. 2011;32:1033–42. doi: 10.1093/carcin/bgr081. [DOI] [PubMed] [Google Scholar]
- 51.Michels KB. The promises and challenges of epigenetic epidemiology. Exp Gerontol. 2010;45:297–301. doi: 10.1016/j.exger.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 52.Chinnery PF, Thorburn DR, Samuels DC, et al. The inheritance of mitochondrial DNA heteroplasmy: random drift, selection or both? Trends Genet. 2000;16:500–5. doi: 10.1016/s0168-9525(00)02120-x. [DOI] [PubMed] [Google Scholar]
- 53.Macdonald-Wallis C, Tobias JH, Davey Smith G, Lawlor DA. Parental smoking during pregnancy and offspring bone mass at age 10 years: findings from a prospective birth cohort. Osteoporos Int. 2011;22:1809–19. doi: 10.1007/s00198-010-1415-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–88. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 55.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 56.Bell CG, Beck S. The epigenomic interface between genome and environment in common complex diseases. Brief Funct Genomics. 2010;9:477–85. doi: 10.1093/bfgp/elq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li Y, Zhu J, Tian G, et al. The DNA methylome of human peripheral blood mononuclear cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandoval J, Heyn HA, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 60.Beck S. Taking the measure of the methylome. Nat Biotechnol. 2010;28:1026–28. doi: 10.1038/nbt1010-1026. [DOI] [PubMed] [Google Scholar]
- 61.Terry MB, Delgado-Cruzata L, Vin-Raviv N, Wu HC, Santella RM. DNA methylation in white blood cells: association with risk factors in epidemiologic studies. Epigenetics. 2011;6:828–37. doi: 10.4161/epi.6.7.16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu HC, Delgado-Cruzata L, Flom JD, et al. Global methylation profiles in DNA from different blood cell types. Epigenetics. 2011;6:76–85. doi: 10.4161/epi.6.1.13391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teschendorff AE, Menon U, Gentry-Maharaj A, et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One. 2009;4:e8274. doi: 10.1371/journal.pone.0008274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore LE, Pfeiffer RM, Poscablo C, et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. Lancet Oncol. 2008;9:359–66. doi: 10.1016/S1470-2045(08)70038-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toperoff G, Aran D, Kark JD, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–83. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ehrich M, Nelson MR, Stanssens P, et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc Natl Acad Sci U S A. 2005;102:15785–90. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bock C, Tomazou EM, Brinkman AB, et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–14. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H, D'Alessio AC, Ito S, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–84. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lillycrop KA, Phillips ES, Torrens C, Hanson MA, Jackson AA, Burdge GC. Feeding pregnant rats a protein-restricted diet persistently alters the methylation of specific cytosines in the hepatic PPAR alpha promoter of the offspring. Br J Nutr. 2008;100:278–82. doi: 10.1017/S0007114507894438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ehrich M, Field JK, Liloglou T, et al. Cytosine methylation profiles as a molecular marker in non-small cell lung cancer. Cancer Res. 2006;66:10911–18. doi: 10.1158/0008-5472.CAN-06-0400. [DOI] [PubMed] [Google Scholar]
- 72.Wong HL, Byun HM, Kwan JM, et al. Rapid and quantitative method of allele-specific DNA methylation analysis. Biotechniques. 2006;41:734–39. doi: 10.2144/000112305. [DOI] [PubMed] [Google Scholar]
- 73.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–95. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–72. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kucej M, Kucejova B, Subramanian R, Chen XJ, Butow RA. Mitochondrial nucleoids undergo remodeling in response to metabolic cues. J Cell Sci. 2008;121:1861–68. doi: 10.1242/jcs.028605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–64. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 77.Camarillo C, Swerdel M, Hart RP. Comparison of microarray and quantitative real-time PCR methods for measuring MicroRNA levels in MSC cultures. Methods Mol Biol. 2011;698:419–29. doi: 10.1007/978-1-60761-999-4_30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schmittgen TD, Lee EJ, Jiang J, et al. Real-time PCR quantification of precursor and mature microRNA. Methods. 2008;44:31–38. doi: 10.1016/j.ymeth.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mercer TR, Neph S, Dinger ME, et al. The human mitochondrial transcriptome. Cell. 2011;146:645–58. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]