Abstract
Background Gestational age at birth strongly predicts neonatal, adolescent and adult morbidity and mortality through mostly unknown mechanisms. Identification of specific genes that are undergoing regulatory change prior to birth, such as through changes in DNA methylation, would increase our understanding of developmental changes occurring during the third trimester and consequences of pre-term birth (PTB).
Methods We performed a genome-wide analysis of DNA methylation (using microarrays, specifically CHARM 2.0) in 141 newborns collected in Baltimore, MD, using novel statistical methodology to identify genomic regions associated with gestational age at birth. Bisulphite pyrosequencing was used to validate significant differentially methylated regions (DMRs), and real-time PCR was performed to assess functional significance of differential methylation in a subset of newborns.
Results We identified three DMRs at genome-wide significance levels adjacent to the NFIX, RAPGEF2 and MSRB3 genes. All three regions were validated by pyrosequencing, and RAGPEF2 also showed an inverse correlation between DNA methylation levels and gene expression levels. Although the three DMRs appear very dynamic with gestational age in our newborn sample, adult DNA methylation levels at these regions are stable and of equal or greater magnitude than the oldest neonate, directionally consistent with the gestational age results.
Conclusions We have identified three differentially methylated regions associated with gestational age at birth. All three nearby genes play important roles in the development of several organs, including skeletal muscle, brain and haematopoietic system. Therefore, they may provide initial insight into the basis of PTB's negative health outcomes. The genome-wide custom DNA methylation array technology and novel statistical methods employed in this study could constitute a model for epidemiologic studies of epigenetic variation.
Keywords: Epigenetic epidemiology, differentially methylated regions, pre-term birth, gestational age, genome-wide DNA methylation
Introduction
Gestational age is the most important indicator of perinatal mortality in developed countries,1 and also contributes to childhood and adult morbidity and mortality.2–4 In 2005, approximately 13% of infants in the USA were born pre-term (<37 weeks), a rise from <10% in 1990.5 The mechanism by which pre-term birth (PTB) increases morbidity and mortality is largely unknown. Recognition of specific genes that are still undergoing regulatory change prior to birth would not only increase our understanding of the developmental changes that are occurring during late pregnancy, but also it would aid in identifying genetic, epigenetic and environmental factors that could lead to PTB. The risks of negative public health consequences of PTB are many, including mortality, learning disabilities and respiratory illnesses.6 Identification of environmental and epigenetic factors has the potential to prevent or ameliorate these adverse impacts.
From the point of view of a developmental change that is associated with health risk and environmental mediators, epigenetic changes in the fetus are potentially important, since epigenetic information affects gene expression, and its function varies within an individual across developmental stages. A significant challenge in understanding the role of epigenetic changes in epidemiology is integrating novel molecular, epidemiological and biostatistical tools at a genome-scale level. Unlike classical genome sequence analyses, the methods and study designs for whole-genome epigenetic epidemiology are not yet well established. The approach we have taken here is to design a genome-scale epidemiological analysis a priori from this joint conceptual perspective. We focused on DNA methylation because it is a key primary epigenetic process, with a well-established mechanism for propagating non-sequence-based information during cell division. The DNA methylation analysis presented here can serve as a paradigm for other epidemiological studies intending to characterize epigenetic profiles in specimen repositories, in which DNA methylation but not other epigenetic marks (e.g. histone modifications) are preserved. We have applied a significant technological extension of our previously described comprehensive high-throughput array-based relative methylation (CHARM) approach7 that can now detect 5.2 million cytosine–guanine dinucleotide (CpG) sites which can be subject to DNA methylation. We also formally define an epigenetic variable, termed differentially methylated region (DMR), which we have used previously, but now have advanced its genome-wide detection to include novel statistical strategies to improve signal to noise detection, as well as the concept of regional methylation detection (Jaffe et al., companion paper8).
While one would expect large-scale epigenetic changes to occur between early embryogenesis and the end of gestation, at present nothing is known about epigenetic changes in the fetus that occur relatively late in pregnancy, covering intervals relevant to the variation in gestational ages at birth that represent dramatic changes in health outcomes. Epigenetic changes in placental samples across gestation have been observed, implying the importance of such modifications for support of a growing fetus,9 but genome-scale site-specific methylation data on the fetus itself, and with respect to the late gestational ages associated with most births, have not yet been reported to our knowledge. For these reasons, we performed a genome-scale comprehensive analysis of DNA methylation on 141 newborns to identify regions of the genome with DNA methylation levels correlated to gestational age at birth. We then validated these microarray results via bisulphite sequencing and further characterized the relationship between developmental age and DNA methylation at the DMRs by comparing these newborn results to the same regions among adult DNA samples.
Methods
Study sample
Cord blood samples were obtained from the Baltimore Tracking Health Related to Environmental Exposures [THREE] Study.10 THREE is a cross-sectional sample of newborns born at the Johns Hopkins Hospital in Baltimore, MD, between November 2004 and March 2005. Of the 603 children delivered during that time window, 300 were eligible (24 twin births removed, 291 did not have any or ample cord blood available). Of these, 187 contributed a cord blood clot from which DNA could be isolated for this epigenetic project. Clots were saved during the second half of the data collection period. Those with available cord blood clots are similar to the rest of the study population with respect to gestational age, birthweight and maternal age, race, body mass index and smoking status (data not shown). Study personnel abstracted data from maternal and infant medical records and study clinicians reviewed a 10% random sample for accuracy; gestational age was taken as the best obstetrical estimate. Information on potential confounders was based on clinical records. Women who reported smoking during pregnancy or had an umbilical cord serum cotinine measurement >10 ng/ml were considered active smokers; the remainder were considered passive smokers or non-smokers (not reporting smoking and cotinine <1 ng/ml).11 Copper (previously found to be associated with gestational age in this population) was measured in umbilical cord serum using inductively coupled plasma dynamic reaction cell mass spectrometry (ICP–DRC–MS)12 at Centers for Disease Control and Prevention (CDC) laboratories, with 4 mg/dl as the limit of detection. The THREE study was reviewed and approved by the Johns Hopkins School of Medicine Institutional Review Board.
For comparison of newborn methylation results with adult samples, CHARM 2.0 data were available on 156 adult samples obtained as unrelated controls for a schizophrenia case–control epigenetics consortium.13–15 This sample was 40% male and had a broad age range of between 16 and 89 years (interquartile range 31–55 years). DNA was obtained from the Rutgers University Cell & DNA Repository (RUCDR). DNA had been isolated from whole blood using Qiagen Autopure LS and pellets were hydrated in 1× Tris-EDTA (TE) buffer. Sample concentration and integrity were verified locally using NanoDrop and gel electrophoresis. DNA methylation was measured using the CHARM 2.0 assay.
Laboratory analyses
CHARM DNA methylation
DNA was isolated from cord blood clot samples using the DNeasy® Blood & Tissue kit (Qiagen), following the manufacturer's instructions. From the 187 fetal cord blood clot samples available, 167 (89.3%) yielded enough DNA for methylation array analysis. DNA methylation was measured via the CHARM 2.0 assay, a customized microarray method extended from our previous CHARM procedure,7 a genome-scale microarray technique for DNA methylation that identifies differential DNA methylation without assumptions regarding where such changes would be, interrogating all CpG islands, as well as CpG island ‘shores’.16 CHARM 2.0 now includes 2.1 million probes, which cover 5.2 million CpGs arranged into probe groups (where consecutive probes are within 300 bp of each other) that tile regions of at least moderate CpG density. It includes all annotated and non-annotated promoters and microRNA sites on top of the features that are present in the original CHARM method. The design specifications are freely available on our website (rafalab.jhu.edu). For the CHARM 2.0 assay, 5 μg of purified genomic DNA was sheared, digested, purified, amplified, labeled as described,17 but hybridized onto our new CHARM 2.0 array. We dropped 26 arrays with <80% of their probes above background intensities, resulting in 141 samples for DNA methylation analysis. We then filtered probes where signal was below background in <25% of arrays (542 055) and removed sex chromosomes (39 454) to improve the batch correction methods, leaving 1 569 888 autosomal probes covering 4 254 946 CpGs spread across 114 984 probe groups. Subsequent pre-processing, normalization and correction for batch effects are described in the Statistical Methods subsection. CHARM hybridization and processing for these samples were performed across 5 separate days, with the following numbers of samples per day: 40, 36, 38, 21, 6, reflecting a potential source of batch effects that was addressed through the surrogate variable analysis (SVA) described in the Statistical Methods subsection.
Bisulphite pyrosequencing
Individual CpGs inside the DMRs meeting our significance threshold were chosen for validation based on MethPrimer software.18 Of the 141 samples for which CHARM data were generated, 139 had ample DNA for subsequent pyrosequencing. Genomic DNA (200 ng) from each sample was bisulphite treated using an EZ DNA Methylation-Gold™ Kit (Zymo research) according to the manufacturer's instructions. Bisulphite-treated genomic DNA was PCR amplified using unbiased nested primers, and DNA methylation was subsequently assessed quantitatively by pyrosequencing using a PSQ HS96 (Biotage). Quantitative measurements (percentage methylation at each CpG) from the pyrosequencing results were determined using the Q-CpG methylation software (Biotage). Control titration standards of 0, 25, 50, 75 and 100% methylated samples were generated using appropriate mixtures of Whole Genome Amplified (WGA) Human Genomic DNA: Male (Promega) using a REPLI-g Mini Kit (Qiagen) and SSsI-treated WGA DNA. Primer sequences used for the bisulphite pyrosequencing reactions can be found in Supplementary Table S2 available as Supplementary Data at IJE online.
Quantitative real-time PCR
To examine the correlation between DNA methylation and gene expression in cord blood clots for each of the top three DMRs, we performed real-time PCR assays. Primers were designed to determine the mRNA expression of the gene closest to each DMR. Since this analysis required isolation of mRNA from cord blood clots, we were only able to perform these expression analyses on a subset of newborns with cord blood clots available. This included 10 babies with gestational age at birth <35 weeks, 15 with gestational age at 40 weeks and 17 with gestational ages ≥41 weeks. For isolation of RNA, fetal cord blood clot samples were treated with TRIzol (Invitrogen) and RNA was purified using a PureLink™ RNA Mini Kit (Invitrogen) according to the manufacturer's instructions. cDNA was synthesized using a QuantiTect Reverse Transcription Kit (Qiagen) and random hexamers. Real-time PCR amplification was performed by using a Fast SYBR® Green Master Mix (Applied Biosystems), and transcript levels were quantified using an ABI 7900 Sequence Detection Systems (Applied Biosystems). Relative expression level for each gene was calculated based on the standard curve and normalized by the relative expression of β-actin. Primer sequences used for the real-time PCR reactions are in Supplementary Table S3 available as Supplementary Data at IJE online.
Statistical analyses
Descriptive statistics (median or percentage) for gestational age at birth and potential confounders were calculated and compared using chi-squared tests for categorical variables and Mann–Whitney U-tests for continuous variables.
The CHARM microarray data were pre-processed and normalized as previously described.19,20 We employed a novel statistical approach (see companion paper, Jaffe et al.8) for identifying regions of the epigenome associated with gestational age in days. Briefly, we fit a linear model predicting methylation at each probe as a function of gestational age at birth, adjusted for surrogate variables, estimated via SVA21,22 to account for unmeasured potential confounding often due to batch effects. SVA identifies combinations of probes in the data associated with heterogeneity of DNA methylation, conditioned on the covariate of interest, in this case, gestational age, and then constructs a ‘surrogate variable’ for each set. A value for each individual based on each surrogate variable can then be used for adjustment in subsequent regression. Measured variables in our data set most associated with these surrogate variables (assessed through pruned regression trees of all possible variables) were array quality control score and hybridization date/batch. We did not adjust for sex, but did remove sex chromosome probes from the initial genome-wide screen. The estimated regression coefficients from these linear models for gestational age at each probe were then smoothed within the CHARM array's pre-defined probe groups. Consecutive smoothed slopes above a fixed cut-off of the 99.5th percentile of all smoothed slopes were summed into a region-level statistic reflecting the area of the DMR (see companion paper, Jaffe et al.8). We then ranked DMRs by their areas and calculated two measures of statistical uncertainty, a P-value and q-value, for each DMR by permutation that accounts for genome-wide testing. Gestational ages were permuted 1000 times, and each time, the above regression, smoothing, and thresholding procedure was repeated exactly as on the observed data to get 1000 sets of declared DMRs that occurred solely by chance. Empirical P-values, defined as the fraction of the maximum areas from each permutation greater than the observed area, were calculated (‘Pmax’) to compare with a specified family-wise error rate control of 10%. False discovery rate (FDR) q-values were obtained by pooling all areas across all permutations, calculating the proportion of these ‘null’ areas greater than the observed area, then converting this to a q-value for comparison to an FDR control of 5%.23 DMRs with an empirical Pmax < 0.10 or an FDR q-value < 0.05 were examined visually via plots of the methylation curve within the DMR. Average methylation for each newborn across all probes within a DMR was plotted against gestational age at birth with slopes and P-values estimated via linear regression and Wald statistics.
Univariate relationships between potential confounders and methylation at DMRs were also estimated via linear regression. Although some potential confounding due to these variables may already be addressed via the SVA adjustment, we also explicitly estimated relationships between average DNA methylation for each DMR and each confounder through linear models adjusted for the same surrogate variables used in our discovery. To do this, we applied SVA analysis to the methylation data first, then took SVA-adjusted methylation as the methylation metric for linear regression with the covariate, to ensure the same SVA adjustment was applied in each analysis. Also, as a sensitivity analysis to assess the influence of sex on our list of identified DMRs, we repeated the original DMR identification procedure adjusting for sex. We further performed the original discovery procedure after omitting samples with mothers who had pregnancy-induced hypertension (PIH), intrapartum fever or diabetes, separately, to assess influence of these variables on our results.
For analysis of DNA methylation data from pyrosequencing, we fit a linear model at every CpG predicting DNA methylation as a function of gestational age. We assessed the functional implications of differential methylation at each gene by fitting linear models at each CpG assessing the linear association between DNA methylation and gene expression. Heavily skewed gene expression values were transformed to log2 scale.
Results
To identify epigenetic changes that occur throughout later stages of gestation in an unselected population of newborns, we performed the CHARM 2.0 assay, which now includes approximately one-third of all single-copy CpG sites including all islands and shores, as well as all annotated promoters and microRNAs. Bisulphite pyrosequencing and real-time PCR were performed to validate DNA methylation levels and functional significance of the DMRs associated with gestational age at birth. Of the 141 newborns with CHARM data, there were 18 PTBs (<37 weeks) and the range of gestational ages in days was 208–292 (see Supplementary Figure S1 available as Supplementary Data at IJE online for full distribution). The pre-term newborns did not differ in the distributions of sex or maternal age, race or diabetes status compared with newborns born after 37 weeks (Table 1). Birthweight differed strongly between the two groups, as did smoking and serum copper levels, which had been previously reported for the full study sample of 300 newborns.24
Table 1.
Characteristics of THREE study newborns included in this epigenetics project
| Pre-term (<37 weeks), N = 18 | Term/post-term (≥37 weeks), N = 123 | P-value* | |
|---|---|---|---|
| Male sex (%) | 56 | 52 | 0.98 |
| Maternal age, median (IQR) (years) | 28 (22–30) | 24 (20–29) | 0.14 |
| Maternal race (%) | 0.32 | ||
| Caucasian | 33 | 21 | |
| AA | 67 | 73 | |
| Asian | 0 | 6 | |
| Maternal smoking (%) | <0.01 | ||
| Non-smoker | 56 | 75 | |
| Passive smoker | 0 | 11 | |
| Active smoker | 44 | 14 | |
| Birthweight, median (IQR) (g) | 2422 (2102–2689) | 3279 (2906–3648) | <0.01 |
| Pregnancy-induced hypertension (%) | 22 | 6 | 0.04 |
| Intrapartum fever (%) | 6 | 8 | 1.00 |
| Serum copper, median (IQR) (μg/dl) | 26 (22–34) | 41 (30–55) | <0.01 |
| Elected delivery (%)a | 44 | 37 | 0.59 |
aCaesarean section or induced delivery.
*P-values based on chi-squared tests for categorical variables and Mann–Whitney tests for quantitative variables.
IQR = interquartile range;
AA = African American.
Previous research indicates that increasing gestational ages at birth through 39–41 weeks is advantageous for neurodevelopment25,26 and confers a lower risk of respiratory morbidity,27 suggesting the need to study gestational age on a continuum. Thus, treating gestational age as a continuous variable in linear regression, compared with pre-term and term birth categories can be useful. Using this approach, we identified 8611 candidate DMRs associated with gestational age at birth (Supplementary Table S1 available as Supplementary Data at IJE online), of which the top three DMRs met our genome-wide threshold of protecting family-wise error rates <10% and false discovery rates <5% (Table 2). The first of these DMRs was found to be positioned in the first intron of the nuclear factor I/X (NFIX) gene, encoding a transcription factor known to be responsible for fetal-specific transcription regulation during skeletal muscle development.28 Another was positioned in the first intron of the alternative transcript of the Rap guanine nucleotide exchange factor (RAPGEF2) gene, which encodes one of the RAS protein family activators that maintains the GTP-bound state of RAS. Although this DMR was not located at the promoter of the canonical gene, the DMR contains strong DNase I hypersensitive sites and a number of strong transcription factor-binding sites including Gata-2 and PU.1, which are the critical transcription factors in haematopoiesis.29,30 The third DMR was located next to the promoter region of the methionine-S-sulphoxide reductase 3 (MSRB3) gene, which encodes the enzyme involved in the methionine cycle and is responsible for antioxidant repairing by converting methionine sulphoxide to methionine.31 Two of the three DMRs are located at the CpG island shore, suggesting that these DMRs may be associated with alternative transcription or splicing.16
Table 2.
Differentially methylated regions for gestational age identified via CHARM 2.0
| Chr | Nearest gene | DMR area | P-value | q-value | DMR start position | DMR end position | Location relative to gene |
|---|---|---|---|---|---|---|---|
| 19 | NFIX | 0.343 | 0.001 | 0.012 | 13 130 686 | 13 133 039 | Inside intron |
| 4 | RAPGEF2 | 0.223 | 0.047 | 0.029 | 160 026 138 | 160 028 079 | Upstream, intron of alt. transcript |
| 12 | MSRB3 | 0.197 | 0.098 | 0.041 | 65 671 230 | 65 672 140 | Promoter |
P-values and q-values based on comparison of observed DMR area ranks to ranks among 1000 permutations of gestational age values. All coordinates are based on hg19/build 37. Chr, Chromosome; Alt., Alternative.
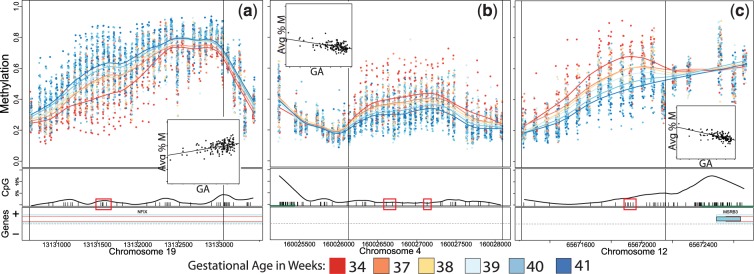
The methylation values at each probe for each of these DMRs are shown in Figure 1 according to gestational age in weeks (calculated from days). Smoothed lines indicate the average methylation curve for each week of gestational age at birth, and show a dose–response trend between gestational age and methylation levels across all weeks for each DMR. To further illustrate this point, Figure 1 also shows the relationship between the average methylation across all probes in the DMR and gestational age, and the linear fit of this relationship (see insets in each panel). For the DMR near NFIX, DNA methylation levels of each probe are greater in high gestational age neonates when compared with low gestational age neonates (Figure 1a), and the average DNA methylation level of each sample in the DMR exhibits a linear correlation with gestational age, with an estimated increase of 1.57% DNA methylation per week of gestation [95% confidence interval (CI) 1.02–2.12], or an increase of 7.85 between Weeks 35 and 40, roughly corresponding to late pre-term vs term births (P = 8.6 × 10−8 for Wald statistic; see Figure 1a, inset). In contrast, the DMRs at RAPGEF2 and MSRB3 show lower DNA methylation levels of each probe in higher gestational age neonates when compared with lower gestational age neonates (Figure 1b and c), and the average DNA methylation levels of each sample in these DMRs exhibit inverse linear correlation with gestational age. For RAPGEF2, there is a 1.33 decrease in %DNA methylation (95% CI −1.76 to −0.9) per week of gestation or a decrease of 6.65 between Weeks 35 and 40; (Wald P = 9.9 × 10−9) and for MSRB3, a 2.08 decrease (95% CI −2.51 to −1.64) per week or 10.4 between Weeks 35 and 40 (Wald P = 1.3 × 10−16; see Figure 1b and c insets). Also note the progressive change in DNA methylation within each gestational age bin, a dose–response relationship consistent with a functional relationship between methylation and gestational age.
Figure 1.
Methylation plots for three identified DMRs for gestational age at birth. (a) NFIX, (b) RAPGEF2, (c) MSRB3. Top half of panels show individual methylation levels at each probe by genomic position, with coloured lines reflecting the average methylation curve for samples binned by gestational age—gestational ages in weeks were split into equal sized bins, and the average age for each bin is shown in the legend. Bottom half of panels show location of CpG dinucleotides (black tick marks) and CpGs validated by bisulphite pyrosequencing (black tick marks contained in red box) as well as the CpG density by position (black curve) and the location of refseq annotated genes (bar, + and − represent the direction of the gene, green bar indicates CpG island). Vertical lines represent boundaries of the DMR. Inset box: linear regression plot of average methylation across the DMR (Avg %M) per sample by gestational age (GA)
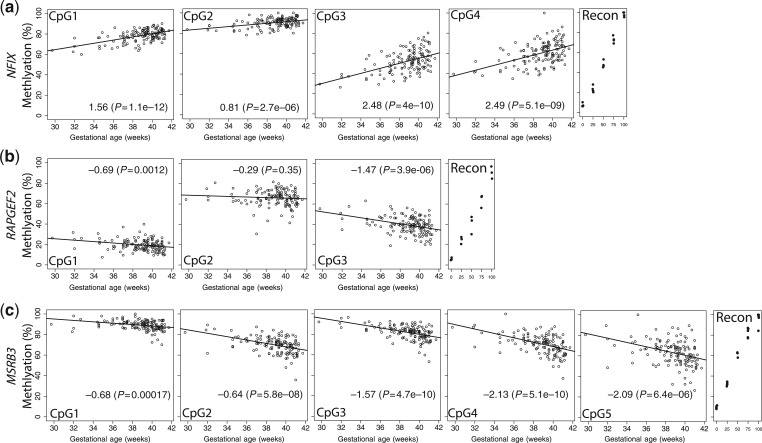
To validate these findings on a separate platform, we designed bisulphite pyrosequencing assays for CpGs within each DMR (indicated as red blocks in Figure 1). The individual CpG results within each DMR were correlated (average pair-wise correlation for neighbouring CpG methylation was 0.85 for NFIX, 0.68 for RAPGEF2 and 0.82 for MSRB3) and confirmed the CHARM differences in methylation by gestational age. For NFIX, four CpGs were assayed (see Figure 1 for locations), each showing an incremental increase in methylation with increase in gestational age at birth, consistent with the pattern detected in CHARM (Figure 2a). All three of the CpGs assayed in RAPGEF2 (see Figure 1 for locations) showed greater methylation with early gestational age at birth, consistent with the CHARM results (Figure 2b). For MSRB3, all five CpGs assayed showed greater methylation in earlier gestational age samples as seen in CHARM (Figure 2c). Thus, these DNA methylation analyses on an independent measurement platform confirmed the differential methylation by gestational age for each of the three genes identified via CHARM.
Figure 2.
Bisulphite pyrosequencing results for each DMR. (a) NFIX, (b) RAPGEF2, (c) MSRB3. Circles represent methylation values (y-axis) at individual CpGs for their corresponding gestational age in weeks (x-axis). Lines represent predicted values from linear regression. Reconstitution controls (represented as black dots) with explicitly designed % methylation (x-axis) are located at the right of each panel (Recon). The numbers on the bottom of each figure represent effect size/slope estimate from the regression of methylation on gestational age and P-value for a Wald test of this slope
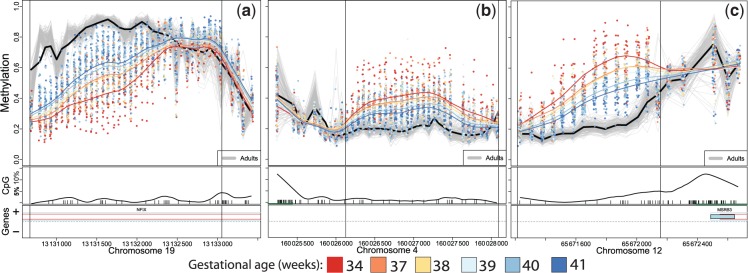
Since the three DMRs we identified reflect variability in methylation corresponding to late-stage development in utero, we considered whether adult DNA methylation at these same sites would show any variability and whether adult levels would be similar to those of full-term births. We compared CHARM 2.0 data for each DMR among healthy adult blood DNA samples with our newborn sample results. Although the three DMRs appear very dynamic and progressive with gestational age in the newborn sample, these exact same regions have little variability in the adult population. Given the span of adult ages represented, this suggests that these sites are stable in adulthood. The magnitude of adult DNA methylation levels is similar to or more extreme than those of the latest gestational ages in a direction consistent with the newborn sample correlations to gestational age (Figure 3). These results provide compelling support for maturation-related changes in DNA methylation at these loci, and also indicate that the process continues beyond birth, but reaches a maximum at some time at or before adult life.
Figure 3.
Methylation plots for three identified DMRs for gestational age at birth with adult methylation results included. Individual adult methylation levels are represented as grey lines, and the black line represents mean adult methylation level. (a) NFIX, (b) RAPGEF2 and (c) MSRB3
To address potential confounding by sex, maternal age, race, maternal smoking, presence of PIH, intrapartum fever, maternal smoking and serum copper levels, we estimated the linear relationships between each of these variables and gestational age at birth. Consistent with the general characteristics comparing pre-term babies to the rest of the newborns, maternal smoking, PIH and serum copper were associated with gestational age (Table 3). To further address whether these potential confounders were associated with methylation at the identified DMRs, we estimated the linear relationship between these variables and the average methylation value per DMR as well. PIH and serum copper were also associated with methylation at each of these DMRs (Table 3), suggesting the potential for confounding. However, the strong association between methylation and gestational age remained even after adjusting for PIH and copper in both CHARM and pyrosequencing data. For example, in the CHARM data, the coefficient for gestational age at birth in linear models predicting average methylation at each DMR with and without adjustment for copper (which had a stronger effect than PIH) changed from 1.57 to 1.37 for NFIX, −1.33 to −1.17 for RAPGEF2 and −2.08 to −1.87 for MSRB3, and all remained statistically significant. We in fact examined the potential influence of each potential confounder on the detected associations with these three DMRs and saw no substantial change in effect sizes after adjustment for any of these covariates (Supplementary Table S4 available as Supplementary Data at IJE online).
Table 3.
Co-efficient (95% CIs) of linear relationship between potential confounders and gestational age at birth or average methylation at each of the identified differentially methylated regions
| GA (in weeks) | NFIXa | RAPGEF2a | MSRB3a | |
|---|---|---|---|---|
| Male sex | −0.12 (−0.84 to 0.60) | −5.72 (−8.16 to −3.27) | 1.85 (−0.21 to 3.91) | 1.07 (−1.32 to 3.46) |
| Maternal age | −0.06 (−0.13 to −0.00) | −0.17 (−0.40 to 0.05) | 0.17 (−0.01 to 0.35) | 0.24 (0.04 to 0.44) |
| Caucasian race (vs African American) | −0.38 (−1.26 to 0.49) | −1.09 (−4.27 to 2.10) | 0.53 (−2.01 to 3.07) | 0.14 (−2.76 to 3.04) |
| Maternal smokingb | −0.62 (−1.06 to −0.18) | 0.86 (−0.83 to 2.54) | 0.39 (−0.95 to 1.73) | −0.21 (−1.75 to 1.33) |
| PIH | −2.25 (−3.54 to −0.96) | −8.41 (−13.09 to −3.73) | 5.01 (1.23 to 8.79) | 6.77 (2.46 to 11.09) |
| Intrapartum fever | 0.60 (−0.7 to 1.91) | −1.93 (−6.76 to 2.9) | −0.24 (−4.1 to 3.62) | 0.13 (−4.28 to 4.55) |
| Serum copper (µg/dl) | 0.04 (0.02 to 0.06) | 0.13 (0.06 to 0.20) | −0.10 (−0.16 to −0.05) | −0.15 (−0.21 to −0.08) |
| Birthweight (kg) | 2.07 (1.63 to 2.51) | 2.86 (0.87 to 4.84) | −2.89 (−4.43 to −1.34) | −3.97 (−5.71 to −2.22) |
| Elected delivery | −0.12 (−0.84 to 0.61) | −1.84 (−4.50 to 0.82) | 1.10 (−1.03 to 3.22) | −0.56 (−3.00 to 1.88) |
Bold indicates statistical significance at P < 0.05.
aAverage residual DNA methylation across DMR adjusted for the same surrogate variables from SVA used in the primary analysis.
bOrdinal variable: 0 = non-smoker, 1 = passive smoker, 2 = active smoker.
Birthweight was also correlated with both gestational age and with methylation at each of the three DMRs. This is expected given the strong relationship between gestational age and birthweight. Gestational age is the best indicator of maturation of the newborn including growth parameters. Since birthweight is largely a consequence of gestational age, removing birthweight variability would almost completely restrict variability for gestational age in our analyses, so we did not condition on birthweight for these analyses. When we considered birthweight for gestational age as a separate phenotype, we saw no relationship to methylation at the three DMRs (Supplementary Table S5 available as Supplementary Data at IJE online).
To explore the functional significance of the differential methylation, we measured the expression of the NFIX, RAPGEF2 and MSRB3 using real-time PCR. RAPGEF2 showed an inverse linear correlation between expression and DNA methylation levels in two of the three CpGs at this DMR (CpG1: P = 0.37; CpG2: P = 0.013, CpG3: P = 0.014, Supplementary Figure S2 available as Supplementary Data at IJE online).
Discussion
Using a genome-wide custom DNA methylation array technology and novel statistical methods, we have identified three differentially methylated regions associated with gestational age at birth. Array-based methylation results for all three regions were validated via bisulphite pyrosequencing. These regions target areas of the genome likely to be under developmental regulation in late gestation, which may have implications for understanding the reasons for immediate as well as long-term health effects of gestational age at birth. The observed incremental progression between methylation and gestational age at birth is further supported by the observation that adults are not variable at these DMRs, but rather appear to be stable at levels similar to or more extreme than newborns with the latest gestational ages at birth.
The genes nearest the identified DMRs may play important roles in late-stage fetal development. NFIX is known to be responsible for regulating skeletal muscle,28 brain and bone development,32–34 which show substantial growth during late gestation. This finding offers face validity that our approach can identify epigenomic regions relevant to late gestational development. RAPGEF2 plays a critical role in embryonic haematopoiesis 35 and brain development (i.e. commissures).36 Although this DMR was not located at the promoter of the canonical gene, the DMR contains strong DNase I hypersensitive sites and a number of strong transcription factor-binding sites including Gata-2 and PU.1, which are the critical transcription factors in haematopoiesis.29,30 In utero, a fetus has a higher haematocrit (given lower available oxygen in utero), low B-cell function (given ready transplacental passage of maternal antibodies) as well as lower platelet counts than seen in babies (born at term). This methylation change with gestational age could be involved in the ontogeny of the haematopoietic system and the switch from production of erythrocytes to increased production of B-lymphocytes and megakaryocytes in preparation for birth and, respectively, secretion of antibodies in response to antigenic assaults as well as production of platelets to prepare for possible birth trauma. Furthermore, anaemia of prematurity is known to cause morbidity in pre-term infants; disruption of regulation of this system may contribute to anaemia of prematurity, due to higher haematocrits and restricted erythropoiesis at birth. The differential methylation detected in our newborn sample did correlate with expression of RAPGEF2 in cord blood cells, lending support for involvement in development of the haematopoietic system. Finally, MSRB3 encodes a methionine sulphate reductase enzyme involved in antioxidant repair, converting methionine sulphoxide to methionine. This specific reductase has been found to be present in many tissues including the human lens and the cochlea and has been suspected to be involved in cataracts caused by oxidative damage to lens cells.31 Most congenital cataracts are idiopathic; however, PTB and the administration of certain drugs in utero have been identified as risk factors,37 pointing to a possible role for oxidative stress for cataract formation in infants as well as adults. Generally, a number of morbid conditions associated with term birth have been tied to oxidative stress, from administration of oxygen, including retinopathy of prematurity, bronchopulmonary dysplasia, necrotizing enterocolitis and intraventricular haemorrhage.38 MSRB3 and other Methionine Sulfoxide Reductases (MSRs) may play a role in this sensitivity to oxidative stress. Mutations in MSRB3 also cause hereditary deafness39 and variants in this gene have been associated with primary tooth development during infancy in a recent genome-wide association study.40
These results do not appear to be sensitive to confounding by measured variables. Furthermore, it is possible that methylation may be part of the mechanism relating factors to gestational age at birth. In this case, one would not want to adjust for such factors in analysis. Thus, we were conservative in our approach to adjustment. Nonetheless, inclusion of potential confounders in our models did not attenuate the relationship between methylation and gestational age at these DMRs. Our use of SVA to reduce the impact of measurement issues, such as batch effects, may also have adjusted for potential residual confounding not captured by a measured variable. It is worth noting that serum copper levels have previously been shown in this sample to be related to gestational age at birth and, therefore, a potential confounder. Nonetheless, the relationship between DMR methylation and gestational age did not attenuate after adjustment for copper. We did, however, observe a relationship between copper levels and methylation in these adjusted models, suggesting an independent effect of copper on methylation, consistent with the growing interest in environmental impacts on the epigenome and their implications for human health.41,42 Also, although we saw a relationship between birthweight and these DMRs, this appeared to be a function of the relationship between gestational age and birthweight, rather than specific to birthweight itself. Although a recent report did see a relationship between global DNA methylation and birthweight for gestational age,43 we did not see an association with these particular DMRs when considering birthweight adjusted for gestational age (Supplementary Table S5 available as Supplementary Data at IJE online).
An important caveat in this study is that we measured DNA methylation from a surrogate tissue, blood, for which methylation changes may not reflect those of tissues undergoing developmental epigenetic changes. Despite this, one of these genes, RAPGEF2, showed the expected inverse relationship of DNA methylation and gene expression. Consistent with this idea, RAPGEF2 regulates embryonic haematopoiesis,35 whereas NFIX and MSRB3 play in the development of organs such as brain, tooth, skeletal muscle and bone.32,33,40,44 Thus, differential expression by methylation patterns of the latter two genes may not be detectable in cord blood, or these DMRs may regulate the enhancer function of distal genes or focally modify the high-order chromatin structure and thus not manifest a change in cord blood expression of NFIX or MSRB3. These results are quite encouraging for epigenetic epidemiology in general, since they indicate that DNA methylation differences may be widespread, and methylation profiles in blood may be a useful indicator of developmental change even in tissues that do not utilize the differentially methylated genes in normal developmental processes.
Overall, the results obtained here by genome-wide DNA methylation analysis are encouraging for the field of epigenetic epidemiology, since they indicate that DNA methylation differences are detectable with this strategy. Specifically, this work identifies epigenetic changes associated with gestational age at birth. The underlying reason for this correlation cannot be determined in this cross-sectional study, but there are at least two implications of these findings for the epidemiology of PTB. First, regions of the genome that are still undergoing DNA methylation variation late in gestation may be functionally related to the health consequences of PTB, and our findings can inform new epidemiologic research and biological mechanisms towards understanding the reasons for negative outcomes in premature babies and lessening these negative infant, childhood or even adult health consequences related to gestational age at birth. Secondly, it is possible that these results reflect involvement of DNA methylation in the aetiology of PTB. There are a number of mechanisms (including infections leading to inflammation,45 preeclampsia46 and stress47) and risk factors (African American race, bacterial vaginosis, cigarette smoking and low maternal pregnancy body mass index48,49) associated with PTB, which could be associated with epigenetic changes themselves, although this explanation is less consistent with the function of the particular genes identified in our study. In addition, the use of assisted reproductive technology and nutritional deficiencies have been identified as possible risk factors for PTB50,51 and also have the potential to alter the epigenome.52,53 Identification of epigenetic changes associated with PTB potentially could be useful for identifying, among the many factors associated with PTB, which are most likely to be causal factors, although our design did not contain a large number of spontaneous PTBs and thus the relationship between methylation and causes of PTB may be best suited for subsequent studies in different samples.
Further work is required to determine whether the detection of DNA methylation in non-primary proxy tissues (in this instance, blood) indeed is a useful indicator of developmental change in the primary tissue for expression of affected genes. However, the work presented here shows that DNA methylation changes progressively during late fetal development, and thus opens the door to studies of the epigenetic epidemiology of PTB.
Supplementary Data
Supplementary Data are available at IJE online.
Funding
National Institutes of Health (R01ES017646 to M.D.F. and A.F., 5R37CA054358-19 to A.F.); Johns Hopkins Bloomberg School of Public Health, The THREE study (to L.R.G.); the Maryland Cigarette Restitution Program Research Grant (to R.U.H.); National Institute of Environmental Health Sciences (grant 1R01ES015445 to R.U.H.); Heinz Family Foundation (to L.R.G.).
Supplementary Material
Acknowledgements
The authors thank the participants in the THREE study, Hopkins Labor and Delivery staff, Benjamin Apelberg, PhD, Ellen Wells, PhD, and, from the US Centers for Disease Control and Prevention, Robert Jones, PhD and Kathleen Caldwell, PhD (copper analysis), John Bernert, PhD (cotinine analysis), and Rolf Halden, PhD (THREE study). The authors thank Walter Kaufmann MD for comments, and Eirikur Briem and Unner Unnsteinsdottir for assisting in the CHARM 2.0 data generation. We also thank the principle investigators on the contributing studies of the schizophrenia epigenetics consortium from which adult comparison sample data were available: David Braff, MD (UCSD), Rodney Go, PhD (UAB), Vishwajit L. Nimgaonkar, MD, PhD (University of Pittsburgh), Raquel Gur, MD, PhD (University of Pennsylvania). H.L. performed DNA extraction and generated gene-specific DNA methylation expression data. A.E.J performed the genome-wide and site-specific data analyses with assistance from R.A.I. J.I.F, and R.T performed CHARM 2.0 assays, with the array designed by M.J.A. S.B. was a study coordinator. C.M. provided unpublished region-specific methylation data. J.H., F.R.W. and L.R.G. were involved with aspects of conception and design of the THREE study and collection of THREE data. A.P.F. and M.D.F. designed and oversaw the study, and wrote the paper with H.L. and A.E.J.
Conflict of interest: None declared.
KEY MESSAGES.
There is a need for established statistical methodology for performing genome-wide DNA methylation studies in epidemiologic samples.
Using a novel statistical approach for DNA methylation micro-array data, we identify three regions of the genomic containing methylation levels that are associated with gestational age at birth in a sample of 141 newborns.
Differential methylation by gestational age at NFIX is consistent with the role of this gene in skeletal and brain development; at RAPGEF2 may implicate the haematopoietic system in ways relevant to anaemia of prematurity; and at MSRB3 may relate to the role of this gene in protection from oxidative damage, which may have implications for several health consequences of PTB.
References
- 1.Goldenberg RL, Culhane JF, Iams JD, Romero R. Epidemiology and causes of preterm birth. Lancet. 2008;371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crump C, Sundquist K, Sundquist J, Winkleby MA. Gestational age at birth and mortality in young adulthood. JAMA. 2011;306:1233–40. doi: 10.1001/jama.2011.1331. [DOI] [PubMed] [Google Scholar]
- 3.Saigal S, Doyle LW. An overview of mortality and sequelae of preterm birth from infancy to adulthood. Lancet. 2008;371:261–9. doi: 10.1016/S0140-6736(08)60136-1. [DOI] [PubMed] [Google Scholar]
- 4.Swamy GK, Ostbye T, Skjaerven R. Association of preterm birth with long-term survival, reproduction, and next-generation preterm birth. JAMA. 2008;299:1429–36. doi: 10.1001/jama.299.12.1429. [DOI] [PubMed] [Google Scholar]
- 5.Martin JA, Hamilton BE, Sutton PD, et al. Births: final data for 2007. Natl Vital Stat Rep. 2010;58:1–85. [PubMed] [Google Scholar]
- 6.Beck S, Wojdyla D, Say L, et al. The worldwide incidence of preterm birth: a systematic review of maternal mortality and morbidity. Bull World Health Organ. 2010;88:31–8. doi: 10.2471/BLT.08.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–90. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaffe AE, Murakami P, Lee H et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 2012;41:200–09. [DOI] [PMC free article] [PubMed]
- 9.Novakovic B, Yuen RK, Gordon L, et al. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Apelberg BJ, Goldman LR, Calafat AM, et al. Determinants of fetal exposure to polyfluoroalkyl compounds in Baltimore, Maryland. Environ Sci Technol. 2007;41:3891–97. doi: 10.1021/es0700911. [DOI] [PubMed] [Google Scholar]
- 11.Bernert JT, Jr, Turner WE, Pirkle JL, et al. Development and validation of sensitive method for determination of serum cotinine in smokers and nonsmokers by liquid chromatography/atmospheric pressure ionization tandem mass spectrometry. Clin Chem. 1997;43:2281–91. [PubMed] [Google Scholar]
- 12. CDC. CfDCaP. Serum multi-element DLS method ITS005A. Division of Laboratory Science CfDCaP, editor. Atlanta, GA, 2008. pp. 1–63.
- 13.Calkins ME, Dobie DJ, Cadenhead KS, et al. The Consortium on the genetics of endophenotypes in schizophrenia: model recruitment, assessment, and endophenotyping methods for a multisite collaboration. Schizophr Bull. 2007;33:33–48. doi: 10.1093/schbul/sbl044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gur RE, Nimgaonkar VL, Almasy L, et al. Neurocognitive endophenotypes in a multiplex multigenerational family study of schizophrenia. Am J Psychiatry. 2007;164:813–19. doi: 10.1176/ajp.2007.164.5.813. [DOI] [PubMed] [Google Scholar]
- 15.Aliyu MH, Calkins ME, Swanson CL, Jr, et al. Project among African-Americans to explore risks for schizophrenia (PAARTNERS): recruitment and assessment methods. Schizophr Res. 2006;87:32–44. doi: 10.1016/j.schres.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ladd-Acosta C, Aryee MJ, Ordway JM, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Curr Protoc Hum Gene. 2010 doi: 10.1002/0471142905.hg2001s65. Chapter 20:Unit 20 1 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 19.Aryee MJ, Wu Z, Ladd-Acosta C, et al. Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostatistics. 2010;12:197–210. doi: 10.1093/biostatistics/kxq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffe AE, Feinberg AP, Irizarry RA, Leek JT. Significance analysis and statistical dissection of variably methylated regions. Biostatistics. 2012;13:166–78. doi: 10.1093/biostatistics/kxr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leek JT, Storey JD. A general framework for multiple testing dependence. Proc Natl Acad Sci USA. 2008;105:18718–23. doi: 10.1073/pnas.0808709105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–45. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wells EM, Jarrett JM, Lin YH, et al. Body burdens of mercury, lead, selenium and copper among Baltimore newborns. Environ Res. 2011;111:411–17. doi: 10.1016/j.envres.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang S, Platt RW, Kramer MS. Variation in child cognitive ability by week of gestation among healthy term births. Am J Epidemiol. 2010;171:399–406. doi: 10.1093/aje/kwp413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis EP, Buss C, Muftuler LT, et al. Children's brain development benefits from longer gestation. Front Psychol. 2011;2:1. doi: 10.3389/fpsyg.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen AK, Wisborg K, Uldbjerg N, Henriksen TB. Risk of respiratory morbidity in term infants delivered by elective caesarean section: cohort study. BMJ. 2008;336:85–87. doi: 10.1136/bmj.39405.539282.BE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Messina G, Biressi S, Monteverde S, et al. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell. 2010;140:554–66. doi: 10.1016/j.cell.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 29.Vicente C, Conchillo A, Garcia-Sanchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. 2011 doi: 10.1016/j.critrevonc.2011.04.007. doi:10.1016/j.critrevonc.2011.04.007 [Epub 24 May 2011] [DOI] [PubMed] [Google Scholar]
- 30.Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. 2010;22:177–84. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchetti MA, Pizarro GO, Sagher D, et al. Methionine sulfoxide reductases B1, B2, and B3 are present in the human lens and confer oxidative stress resistance to lens cells. Invest Ophthalmol Vis Sci. 2005;46:2107–12. doi: 10.1167/iovs.05-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campbell CE, Piper M, Plachez C, et al. The transcription factor Nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- 34.Driller K, Pagenstecher A, Uhl M, et al. Nuclear factor I X deficiency causes brain malformation and severe skeletal defects. Mol Cell Biol. 2007;27:3855–67. doi: 10.1128/MCB.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satyanarayana A, Gudmundsson KO, Chen X, et al. RapGEF2 is essential for embryonic hematopoiesis but dispensable for adult hematopoiesis. Blood. 2010;116:2921–31. doi: 10.1182/blood-2010-01-262964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilasy SE, Satoh T, Terashima T, Kataoka T. RA-GEF-1 (Rapgef2) is essential for proper development of the midline commissures. Neurosci Res. 2011;71:200–9. doi: 10.1016/j.neures.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Rahi JS, Dezateux C. Congenital and infantile cataract in the United Kingdom: underlying or associated factors. British Congenital Cataract Interest Group. Invest Ophthalmol Vis Sci. 2000;41:2108–14. [PubMed] [Google Scholar]
- 38.Walsh BK, Brooks TM, Grenier BM. Oxygen therapy in the neonatal care environment. Respir Care. 2009;54:1193–202. [PubMed] [Google Scholar]
- 39.Ahmed ZM, Yousaf R, Lee BC, et al. Functional null mutations of MSRB3 encoding methionine sulfoxide reductase are associated with human deafness DFNB74. Am J Hum Genet. 2011;88:19–29. doi: 10.1016/j.ajhg.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pillas D, Hoggart CJ, Evans DM, et al. Genome-wide association study reveals multiple loci associated with primary tooth development during infancy. PLoS Genet. 2010;6:e1000856. doi: 10.1371/journal.pgen.1000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolinoy DC, Jirtle RL. Environmental epigenomics in human health and disease. Environ Mol Mutagen. 2008;49:4–8. doi: 10.1002/em.20366. [DOI] [PubMed] [Google Scholar]
- 42.Sutherland JE, Costa M. Epigenetics and the environment. Ann NY Acad Sci. 2003;983:151–60. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- 43.Michels KB, Harris HR, Barault L. Birthweight, maternal weight trajectories and global DNA methylation of LINE-1 repetitive elements. PLoS One. 2011;6:e25254. doi: 10.1371/journal.pone.0025254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messina G, Biressi S, Monteverde S, et al. Nfix regulates fetal-specific transcription in developing skeletal muscle. Cell. 140:554–66. doi: 10.1016/j.cell.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 45.Leitich H, Kiss H. Asymptomatic bacterial vaginosis and intermediate flora as risk factors for adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol. 2007;21:375–90. doi: 10.1016/j.bpobgyn.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Moldenhauer JS, Stanek J, Warshak C, Khoury J, Sibai B. The frequency and severity of placental findings in women with preeclampsia are gestational age dependent. Am J Obstet Gynecol. 2003;189:1173–77. doi: 10.1067/s0002-9378(03)00576-3. [DOI] [PubMed] [Google Scholar]
- 47.Smith R. Parturition. N Engl J Med. 2007;356:271–83. doi: 10.1056/NEJMra061360. [DOI] [PubMed] [Google Scholar]
- 48.Behrman RE, Butler AS. Preterm birth: causes, consequences, and prevention. Natl Academy Pr. 2007 [Epub ahead of print] [PubMed] [Google Scholar]
- 49.Kramer MR, Hogue CJ, Dunlop AL, Menon R. Preconceptional stress and racial disparities in preterm birth: an overview. Acta Obstet Gynecol Scand. 2011;90:1307–16. doi: 10.1111/j.1600-0412.2011.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henderson JJ, McWilliam OA, Newnham JP, Pennell CE. Preterm birth aetiology 2004-2008. Maternal factors associated with three phenotypes: spontaneous preterm labour, preterm pre-labour rupture of membranes and medically indicated preterm birth. J Matern Fetal Neonatal Med. 2011 doi: 10.3109/14767058.2011.597899. [Epub 10 August 2011] [DOI] [PubMed] [Google Scholar]
- 51.Dunlop AL, Kramer M, Hogue CJ, Menon R, Ramakrishan U. Racial disparities in preterm birth: an overview of the potential role of nutrient deficiencies. Acta Obstet Gynecol Scand. 2011;90:1332–41. doi: 10.1111/j.1600-0412.2011.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–60. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chmurzynska A. Fetal programming: link between early nutrition, DNA methylation, and complex diseases. Nutr Rev. 2010;68:87–98. doi: 10.1111/j.1753-4887.2009.00265.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.