The revellers depicted in the painting by the Dutch artist Judith Leyster (1609–60) (Figure 1) will not have given epigenetics a passing thought. Little were they to know that indulgences, such as drinking alcohol and smoking, would be contributing to their ‘exposome’1 and marking their epigenome to potentially compromise their future health. The skeleton proffering an hourglass is perhaps a portent of the perils of such indulgence. Epigenetic alterations have been linked—sometimes tentatively—to a wide array of exposures and health outcomes, from smoking2 and alcohol3,4 to lung cancer5 and psychoses,6 and the field will surely witness a glut of further literature in the near future.
Figure 1.

‘The Last Drop’. Judith Leyster, c.1630–1631. Philadelphia Museum of Art, reproduced by kind permission of Philadelphia Museum of Art
Epigenetics has undoubtedly recently taken the world of medical research by storm,7 offering the promise of prediction, prevention and treatment of a wide spectrum of common complex diseases.8 The current special issue brings together a collection of reviews and articles with epigenetics as a common theme to consider the contribution that epidemiology can make in defining the role of epigenetics in common complex disease, or conversely, to ask the question—is epidemiology ready for epigenetics?
The emergence of epigenetics
The emergence of the modern usage of epigenetics is attributed to the landmark article by Conrad Waddington, reprinted in this issue of the IJE.9 In this article, Waddington asserts that
We certainly need to remember that between genotype and phenotype, and connecting them to each other, there lies a whole complex of developmental processes.
Waddington termed this complex the epigenotype.9 In this article, Waddington recognizes that genes are the fundamental regulators of the developmental process, dictating every organism’s developmental trajectory. As Gilbert notes, this was a paradigm-changing idea.10 Perturbation of this ‘genetic regulation’ thus alters development, with critical periods when even minor events can have disproportionate importance and ‘far-reaching consequences’—a concept that resonates with the contemporary Developmental Origins of Health and Disease hypothesis. As highlighted by Jablonka and Lamm11 in their commentary on Waddington’s article, he had already, prior to this publication, articulated the concept of not only the interaction of genotype and epigenotype to shape development, but also the reaction of these elements with the external environment. This still forms the basis of much of the scientific enquiry in the field of epigenetics today in our attempts to establish the role of epigenetic mechanisms in health and development. These interactions so insightfully recognized by Waddington are now, in modern parlance, articulated as ‘network-oriented’ approaches and invoke ‘systems biology’ methods in an attempt to understand these complex dynamic systems.12
The epigenome as an integrator of environmental and germ-line genetic perturbation
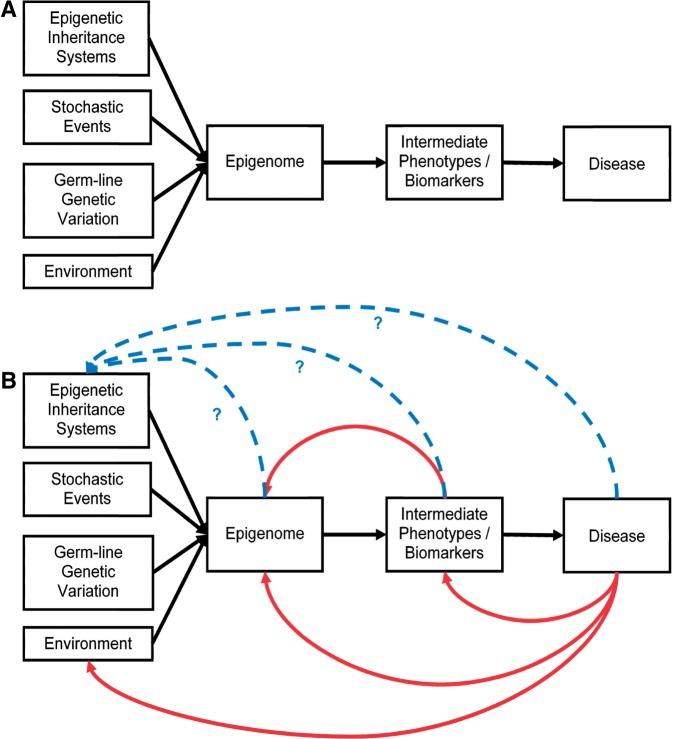
The notion of the epigenome as an ‘integrator’ of multiple signals—environmental exposures, germ-line genetic variation, stochastic events and possibly inherited non-germ-line phenomena—is presented in Figure 2. This schema is wholly consistent with Waddington’s concept of the epigenotype being the product of many contributory factors and can be used to highlight some of the complexities facing epidemiological investigation of this subject area.
Figure 2.
The epigenome as a biosensor of exposure and/or outcome. (A) Multiple factors have been proposed to influence epigenetic patterns including epigenetic inheritance systems, stochastic events, germ-line genetic variation and the environment. The epigenome is commonly postulated to mediate the influence of these factors upon intermediate phenotypes and subsequently disease. This linear unidirectional relationship is simplistic and does not account for the complexities of various feedback loops, as shown in (B). The red arrows highlight how intermediate phenotypes and/or disease states might impact directly upon the epigenome (reverse causation). Additionally, disease might alter the environmental exposure and thus its observed association with the epigenome. The blue dashed arrows highlight the potential for feedback from all stages of the pathway to influence epigenetic inheritance systems. The question marks indicate that these links have, as yet, no robust evidence base in humans
Delineating the relative influences of the multiple factors shown in Figure 2a poses many challenges. The concept of gene–environment equivalence13 could be extended to incorporate stochastic and epigenetic inheritance systems in this framework—each factor potentially able to elicit an epigenetic ‘phenocopy’. It is unclear at this stage whether different factors have unique or generic influences on the epigenome; there are simply insufficient data to confidently distinguish exposure-specific ‘epigenetic fingerprints’. A wide range of environmental factors have been associated with epigenetic alterations,4,14 but far fewer studies have described the relationship between germ-line genetic variation and epigenetic patterns.15 The contribution of stochastic events in determining epigenetic patterns is recognized16–20 and may be one mechanism through which chance events influence disease risk;20 however, it is difficult to quantify in population-based studies. The fourth component in this framework that might plausibly impact upon the epigenome is entitled ‘epigenetic inheritance systems’21 and represents the possibility that environmentally induced epigenetic perturbations persist across generations. Often, the seamless transition (or profound leap) is made from the discussion of within-generation epigenetic factors that might impact upon the regulation of gene expression and phenotype to the evolutionary impact of epigenetic variation via the transduction of environmental exposures into molecular events capable of multi-generational transmission. The evidence base for the latter is largely based upon animal models and plant biology, with some emerging but difficult to interpret human literature.22
The enthusiasm focused upon establishing the mediating role of epigenetic variation on the pathway from environmentally modifiable exposures to disease must be tempered in the knowledge that epigenetic aberrations can arise as a consequence of disease23 or, indeed, as a consequence of exposure to intermediate phenotypes such as elevated blood glucose.24 These feedback loops are depicted in Figure 2b, with red arrows highlighting the potential routes of reverse causation and blue dashed arrows representing the more speculative links between components of the exposure–disease pathway that might feed back to influence epigenetic inheritance systems. Considerations from the classic studies of ‘pure lines’ – i.e. essentially genetically identical organisms selected by phenotypic extreme – suggest that in most circumstances such effects cannot be quantitatively large, and certainly are minor compared to the influence of germ-line genetic variation on intergenerational similarity in phenotype.25
The complexity of the interplay of the factors illustrated in Figure 2 is not uncommon to scenarios encountered in conventional observational epidemiology, where confounding and reverse causation are routinely considered as components of any association. In conventional genetic epidemiology such problems are largely circumvented, because of the lack of confounding of germ-line genetic variants with other exposures26 and the absence of potential reverse causation in such studies. Epigenetic epidemiology is susceptible to all the problems of conventional observational epidemiology, and the suspension of epidemiological principles in the design of some successful genetic epidemiology studies cannot be maintained in the context of epigenetic epidemiology.
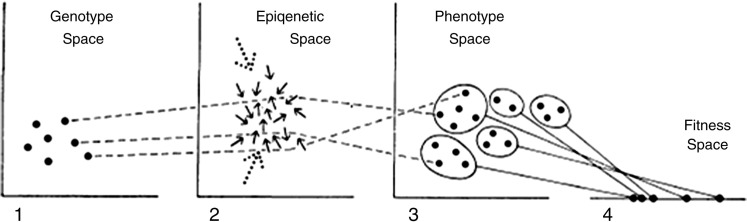
With the plethora of potential bidirectional influences on the epigenome, it will be challenging to decipher real signals among the variations that exist in any population. This concept is wonderfully depicted in the representation of Waddington’s ‘Epigenetic Space’ (Figure 3),27 with the random arrows influencing the genetically determined trajectory from genotype to phenotype in what would appear to be a disordered, perhaps unpredictable, way. In this space, environmental and stochastic influences may be exerting their effect. Given this level of disorder, it is likely that characterization of individuals will make little epidemiological sense, but group-level comparisons will yield robust and replicable findings. Since epidemiology is essentially a group-level discipline,20 it provides the appropriate methodologies for identifying the major determinants of epigenetic variation and their putative contribution to health and disease.
Figure 3.
Waddington's Epigenetic Space (reproduced from Scarr and McCartney27), where genotype, environment and chance coalesce to produce phenotype
What can epidemiology offer?
A fundamental aspect of the field of epigenetic epidemiology centres on understanding exactly what it is that we are measuring when we refer to ‘epigenetic variation’. This is illustrated by the numerous articles in the current special issue that include empirical epigenetic data that not only focus upon DNA methylation but also report the measurement of multiple different indices of methylation from global to genome-wide to the level of the single gene. As highlighted by Heijmans and Mill28 in their commentary, we are still grappling with the issue of where to look and what to look for when considering epigenetic variation. Approaches that help in defining where to look are emerging.29,30 Furthermore, with respect to genetic and environmental influences on the epigenome, effect sizes, where known, are relatively small and much more information is needed in this area to define the biological impact of such modest shifts in epigenetic patterns on health outcomes. Epidemiological approaches can clearly assist in addressing this.
Conventional epidemiological approaches have begun to be applied in epigenetic studies with examples of twin studies, family-based studies and cross-sectional studies all apparent in the literature. One approach that will undoubtedly contribute to our understanding of the role of epigenetic mechanisms in the evolution of complex disease is the ability to utilize longitudinal cohort studies. Resources that are particularly valuable in this regard are studies that have collected and stored biological samples prospectively, and thus allow for the analysis of epigenetic profiles well before the onset of disease, which is highly likely to confound any cross-sectional study. The serial sampling of biological samples, at multiple time points across the life course, will provide further value, allowing insights into the temporal variation in epigenetic signatures over time. Epigenome-wide association studies (EWAS) are also beginning to emerge, applying the fruitful methods of the GWAS era.31
However as mentioned above it must be remembered that epigenetic association studies can be expected to generate a large number of associations due to confounding and reverse causation, with the epigenomic profiles behaving like phenotypes in conventional epidemiological designs rather than genotypes.26 In this regard the special–case study design issues which allowed genetic epidemiological studies to utilise control groups that were not truly representative of the source population of the cases (e.g. the 1958 birth cohort and blood donor controls in the Wellcome Trust Case Control Consortium) will not apply, and utilisation of samples from such studies in epigenetic investigations is likely to generate a plethora of associations generated by bias, confounding and reverse causation.
Inherent in the above developments is a requirement for statistical modelling of epigenetic variables that demonstrate inter-individual variation and change over time. Statistical methods that have been developed and applied in longitudinal epidemiological studies will be invaluable in this regard.
Among the battery of epidemiological tools are those developed to strengthen causal inference, and it is these that find a particularly pertinent application in the context of epigenetics. The Mendelian randomization approach32 that has now been used extensively to establish causal relationships between environmentally modifiable exposures and common diseases33 has been developed to include the mediating role of DNA methylation in causal pathways to disease.34 This approach has begun to be applied to resolve issues of confounding and reverse causation35 and promises to be a useful tool in instances such as deciphering the direction of causality in the observed association of blood lipid biomarkers and DNA methylation patterns.36,37 As epigenetic profiles are essentially phenotypic, other methods for strengthening causal inference in observational epidemiological studies can be applied to the investigation of epigenetic influences on health and disease.38,39
Population-based studies of epigenetic variation inevitably face limitations,28 including a reliance on the use of easily accessible sources of DNA, such as saliva, buccal scrapes or peripheral blood DNA. These sources may not accurately reflect epigenetic perturbations in the disease-specific target tissue of interest, even though they may be suitable sentinels of environmentally induced epigenetic changes. Nonetheless, the field of epidemiology has much to offer the field of epigenetics by providing a framework for population-based studies with well-established methods for circumventing fundamental issues such as confounding and reverse causation as well as more recent developments of approaches to strengthen causal inference. After the brief detour into studies focused solely on germ-line genetic variation, in which epidemiological niceties could be ignored, the rapidly advancing field of epigenetics provides considerable opportunities for the application (and enrichment) of epidemiological methods.
Funding
CLR and GDS received funding from a number of sources including the Medical Research Council, Biotechnology and Biological Research Council, Wellcome Trust, European Union FP7 (IRSES GEoCoDE) and various medical charities. This work does not represent that funded from any single source.
References
- 1.Wild CP. The exposome: from concept to utility. Int J Epidemiol. 2012;41:24–32. doi: 10.1093/ije/dyr236. [DOI] [PubMed] [Google Scholar]
- 2.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–57. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shukla SD, Velazquez J, French SW, Lu SC, Ticku MK, Zakhari S. Emerging role of epigenetics in the actions of alcohol. Alcohol Clin Exp Res. 2008;31:1525–34. doi: 10.1111/j.1530-0277.2008.00729.x. [DOI] [PubMed] [Google Scholar]
- 4.Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41:126–39. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Herceg Z, Vaissiere T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics. 2011;6:804–19. doi: 10.4161/epi.6.7.16262. [DOI] [PubMed] [Google Scholar]
- 6.Dempster EL, Pidsley R, Schalkwyk LC, et al. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Hum Mol Genet. 2011;20:4786–96. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haig D. Commentary: The epidemiology of epigenetics. Int J Epidemiol. 2012;41:13–16. doi: 10.1093/ije/dyr183. [DOI] [PubMed] [Google Scholar]
- 8.Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention and treatment. PLoS Med. 2010;7:e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waddington CH. The epigenotype. Endeavour. 1942;1:18–20. Reprinted in Int J Epidemiol 2012;41:10–13. [Google Scholar]
- 10.Gilbert SF Commentary: ‘The Epigenotype’ by C.H. Waddington. Int J Epidemiol. 2012;41:20–23. doi: 10.1093/ije/dyr186. [DOI] [PubMed] [Google Scholar]
- 11.Jablonka E, Lamm E. Commentary: The epigenotype: a dynamic network view of development. Int J Epidemiol. 2012;41:16–20. doi: 10.1093/ije/dyr185. [DOI] [PubMed] [Google Scholar]
- 12.Huang S. The molecular and mathematical basis of Waddington’s epigenetic landscape: a framework for post-Darwinian biology? Bioessays. 2012;34:149–57. doi: 10.1002/bies.201100031. [DOI] [PubMed] [Google Scholar]
- 13.Zuckerkandl E, Villet R. Concentration–affinity equivalence in gene regulation: convergence of genetic and environmental effects. Proc Natl Acad Sci U S A. 1988;85:4784–88. doi: 10.1073/pnas.85.13.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou L, Zhang X, Wang D, Baccarelli A. Environmental chemical exposures and human epigenetics. Int J Epidemiol. 2012;41:79–105. doi: 10.1093/ije/dyr154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell JT, Pai AA, Pickrell JK, et al. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12:R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–88. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- 17.Martin GM. Epigenetic gambling and epigenetic drift as an antagonistic pleiotropic mechanism of aging. Aging Cell. 2009;8:761–64. doi: 10.1111/j.1474-9726.2009.00515.x. [DOI] [PubMed] [Google Scholar]
- 18.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler Colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation and disease. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1757–64. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novakovic B, Yuen RK, Gordon L, et al. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey Smith G. Epidemiology, epigenetics and the ‘Gloomy Prospect’: embracing randomness in population health research and practice. Int J Epidemiol. 2011;40:537–62. doi: 10.1093/ije/dyr117. [DOI] [PubMed] [Google Scholar]
- 21.Maynard SJ. Models of a dual inheritance system. J Theoret Biol. 1990;143:41–53. doi: 10.1016/s0022-5193(05)80287-5. [DOI] [PubMed] [Google Scholar]
- 22.Burdge GC, Hoile SP, Uller T, et al. Progressive, transgenerational changes in offspring phenotype and epigenotype following nutritional transition. PLoS One. 2011;6:e28282. doi: 10.1371/journal.pone.0028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin DI, Cropley JE, Suter CM. Epigenetics in disease: leader or follower? Epigenetics. 2011;6:843–48. doi: 10.4161/epi.6.7.16498. [DOI] [PubMed] [Google Scholar]
- 24.Pirola L, Balcerczyk A, Tothill RW, et al. Genome wide analysis distinguishes hyperglycaemia regulated epigenetic signatures of primary vascular cells. Genome Res. 2011;21:1601–15. doi: 10.1101/gr.116095.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey Smith G. Epigenesis for Epidemiologists: does evo-devo have implications for population health research and practice? Int J Epidemiol. 2012;41:236–47. doi: 10.1093/ije/dys016. [DOI] [PubMed] [Google Scholar]
- 26.Davey Smith G, Lawlor DA, Harbord R, Timpson NJ, Day I, Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scarr S, McCartney K. How people make their own environments: a theory of genotype → environment effects. Child Dev. 1983;54:424–35. doi: 10.1111/j.1467-8624.1983.tb03884.x. [DOI] [PubMed] [Google Scholar]
- 28.Heijmans BT, Mill J. The seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41:74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–09. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee H, Jaffe AE, Feinberg JI, et al. DNA methylation shows genome-wide association of NFIX, RAPGEF2 and MSRB3 with gestational age at birth. Int J Epidemiol. 2012;41:188–99. doi: 10.1093/ije/dyr237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davey Smith G. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 33.Timpson NJ, Wade KH, Davey Smith G. Mendelian randomization: application to cardiovascular disease. Curr Hypertens Rep. 2012;14:29–37. doi: 10.1007/s11906-011-0242-7. [DOI] [PubMed] [Google Scholar]
- 34.Relton CL, Davey Smith G. Two-step epigenetic Mendelian randomization: a strategy for establishing the causal role of epigenetic processes in pathways to disease. Int J Epidemiol. 2012;41:161–76. doi: 10.1093/ije/dyr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groom A, Potter C, Swan DC, et al. Postnatal growth and DNA methylation are associated with differential gene expression of the TACSTD2 gene and childhood fat mass. Diabetes. 2012;61:391–400. doi: 10.2337/db11-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGuiness D, McGlynn L, Johnson PCD, et al. Socio-economic status is associated with epigenetic differences in the pSoBid cohort. Int J Epidemiol. 2012;41:151–60. doi: 10.1093/ije/dyr215. [DOI] [PubMed] [Google Scholar]
- 37.Pearce MS, McConnell JC, Potter C, et al. Global LINE-1 DNA methylation is associated with blood glycaemic and lipid profiles. Int J Epidemiol. 2012;41:210–17. doi: 10.1093/ije/dys020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davey Smith G. Assessing intrauterine influences on offspring health outcomes: can epidemiological studies yield robust findings? Basic Clin Pharmacol Toxicol. 2008;102:245–56. doi: 10.1111/j.1742-7843.2007.00191.x. [DOI] [PubMed] [Google Scholar]
- 39.Brion MJ, Lawlor DA, Matijasevich A, et al. What are the causal effects of breastfeeding on IQ, obesity and blood pressure? Evidence from comparing high-income and middle-income cohorts. Int J Epidemiol. 2011;40:670–680. doi: 10.1093/ije/dyr020. [DOI] [PMC free article] [PubMed] [Google Scholar]